Abstract
Lysozyme is a well-characterized protein in terms of its structure, dynamics, and functions. It has thus emerged as a potential target to understand protein–drug interactions. The aim of our study is to gain a biophysical outlook on the interaction of lysozyme (Lyz), a well-known model protein, with Noscapine, a potent tubulin-binding anticancer drug. Noscapine (Nos) is effective against a wide range of cancer and shows low toxicity and few side effects. We report the underlying mechanism of complex formation between Nos and Lyz using spectroscopic and advanced computational avenues. The spectroscopic techniques, that is, absorption and steady-state and time-resolved fluorescence, proved that Lyz–Nos forms a complex, and the quenching mechanism was of the static type. The binding constant was in the order of 103 indicative of moderate binding, while the stoichiometry of the protein–drug complex was 1:1 at 298 K. The secondary structural analysis using CD and UV thermal denaturation further confirmed the conformational changes in the protein upon binding with Nos. Molecular dynamics simulation studies confirmed the stable binding with minimum deviations in RMSD. The above conclusions are significant to the development of the pharmacokinetics and pharmacodynamic properties of Nos, and its successful interaction with a versatile protein like Lyz will help in overcoming its previous limitations.
Introduction
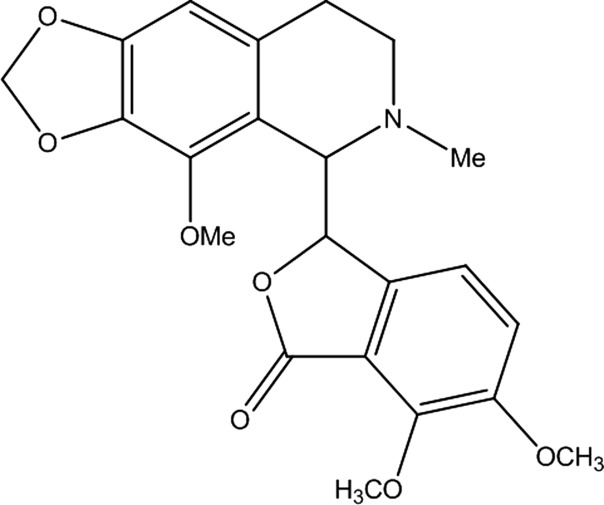
Noscapine (Nos) is a phthalideisoquinoline alkaloid found in the plant family of opium poppy. This non-narcotic drug has been significantly researched due to its potency against various types of cancer.1 The similarity in structure of Nos (Figure 1) to colchicine and podophyllotoxin led to the exploration of its binding with tubulin proteins. It was successfully demonstrated that the interaction of Nos with tubulins leads to the alteration of the microtubule assembly, causing mitotic arrest.2 Recently, Nos and its halogenated analogs have been biosynthesized using microbial fermentation, and it is a major step toward industrial production.3 The drug is emerging as a viable anticancer agent already undergoing clinical trial in phase I/II.4 Although it has a low-toxicity profile, the high-dosage requirement and low bioavailability have impeded its development as a commercially available drug. Several alternative methods for Nos drug delivery, including nanoparticles,5,6 soluble complexes,7 etc., have been reported, and the investigation is still underway to find the most suitable pathway to overcome the pharmacokinetic limitations. In order to aid the potency of the compound for cancer treatment, drug–protein binding studies are vital to assess its delivery and homing to specific target points.8,9 Previously, Nos and its analogues have been explored for their interaction with blood proteins (BSA and HSA) and pharmacological properties using biophysical approaches to aid reports on the potency of the compound for cancer treatment.10−13
Figure 1.
Chemical structure of Noscapine.
Biophysical studies provide deep insight into the drug interaction, in vivo half-life, solubility, efficacy, toxicity, heme metabolism, clearance, and elimination from the body.14−18 Therefore, it is necessary to study the biophysical parameters of the drug. Considering that fact and a literature survey, in this work, we have performed the binding mechanistic interaction studies of Nos with Lyz abundantly present in the body secretion. Hen egg-white lysozyme, an easily available and low-cost protein, is used as a model system due to its structural similarity (∼60% homology) to human lysozyme. Lyz is a small globular monomeric protein (129 amino acid residues, mol wt = 14.3 kDa) with six tryptophan and three tyrosine residues with four disulfide linkages providing structural stability.19 It has innumerable biologically and pharmaceutically important functions, including anti-inflammatory,20 antimicrobial,21 and antiviral functions.22 One of the most important functions of Lyz is its ability to transport drugs. In view of this property, the binding of Nos with Lyz was deciphered using different spectroscopic and in silico techniques. UV–visible and steady-state and time-resolved fluorescence spectroscopy elucidated the mechanism of binding, while circular dichroism spectroscopy offered an insight into the secondary structural changes upon interaction. Molecular docking has been performed to calculate binding free energy and interaction sites; molecular dynamic simulations have been used to assess the stability of the binding as many reports have employed to give a mechanistic interaction of regulatory networks.23 Molecular docking simulation analysis provides detailed information of the interaction at the atomic level of proteins by analyzing the protein conformations and root-mean-square deviations. Drug dynamics and stability parameters are potential factors to determine the biological functionality of the drug and specific binding of the drug to the binding grove of oncotargets.24,25 Since Lyz is extracted from the hen egg white, a boiled egg assay was also performed, an in silico technique to evaluate the gastrointestinal absorption and blood–brain barrier penetration potential. Overall, our current study offers a detailed outlook into the binding of Nos with Lyz, which will be useful in moving closer to realizing its full anticancer potential.
Results and Discussion
Absorption Spectroscopy
UV–visible spectroscopy is one of the most important techniques used to analyze drug–protein interactions. The absorption spectra of Lyz shows a characteristic peak at 281 nm, which is due to the π–π* transition of the three aromatic amino acids, tryptophan, tyrosine, and phenylalanine.26,27 The spectra of Lyz were recorded in the absence and presence of Nos to understand the mechanism of interaction (Figure 2). It can be seen that the absorbance of Lyz increases with the addition of Nos without any significant peak shift. Thus, it can be deduced that Lyz interacts with Nos. UV titration experiments are also employed to gain insight about the mode of interaction between the protein and ligands/drugs, that is, whether the drug is interacting with macromolecules via electrostatic interaction or intercalation. It is done to see if any wavelength shift occurs in the experiment along with any hypochromicity/hyperchromicity. But in our experiment, only hyperchromicity is observed without any shift in wavelength. So, it might be possible that the drug is exposing the tryptophan moiety by interacting as well as forming a complex with the protein.
Figure 2.
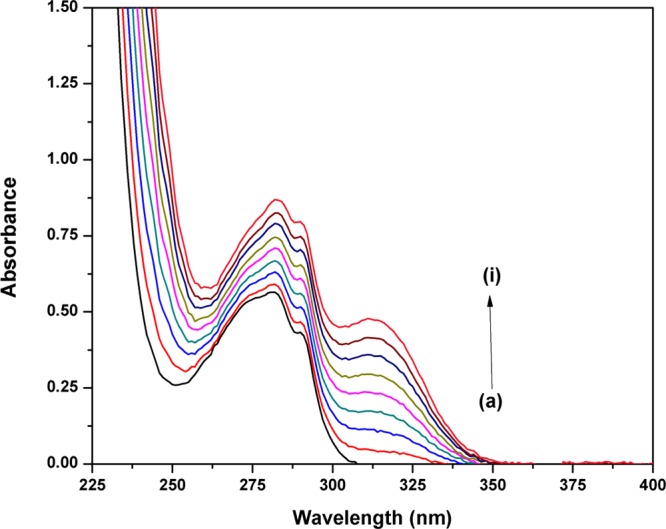
UV–visible absorption spectra of Lyz (15 μM) in the presence of Nos at different concentrations: (a) 0, (b) 15, (c) 30, (d) 45, (e) 60, (f) 75, (g)90, (h) 105, and (i) 120 μM.
Fluorescence Spectroscopy
Proteins have the unique property of showing fluorescence,28 which has greatly simplified the understanding of their structural and conformational changes during complex formation.29 The amino acids responsible for their fluorescent properties are tryptophan, tyrosine, and phenylalanine.30 To elucidate the mechanism of interaction between Lyz and Nos, the fluorescence spectra of Lyz in the absence and presence of the drug was carefully studied. Lyz shows an emission peak at 340 nm (Figure 3) at an excitation wavelength of 280 nm, and this fluorescence was quenched by the successive addition of Nos. Now, it is possible that the decrease in fluorescence can proceed via a static or dynamic mechanism. The two modes responsible can be differentiated on the basis of the value of the quenching constant. In general, static quenching is responsible when there is complex formation, while dynamic quenching comes into play due to collisional encounters. The binding mechanism was assessed by evaluating the fluorescence spectra using the Stern–Volmer equation31 (eq 1).
| 1 |
Figure 3.
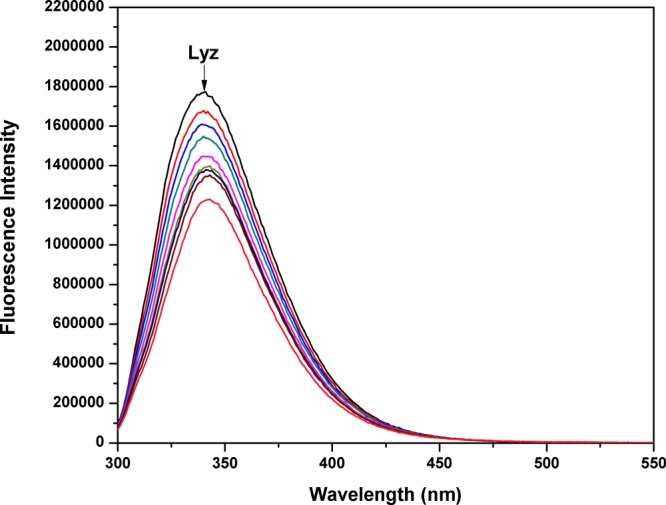
Fluorescence quenching spectra of Lyz (15 μM) in the presence of Nos (0 to 120 μM) at 298 K.
The symbol Fo denotes the fluorescence intensity of pure Lyz, while F is the intensity in the presence of Nos. [Q] is the concentration of Nos in M, τo is the average lifetime of a biomolecule, and Ksv and Kq are the Stern–Volmer constant and quenching constant, respectively. Kq is calculated simply by assuming the value of τo to be 10–8 s.32 The value of Kq (Table 1) was found to be higher than the maximum diffusion rate constant for biomolecules,33 in our case, proteins (2 × 1010 L mol–1 s–1). It can be concluded that the mechanism of quenching was due to the complex formation between Lyz and Nos.
Table 1. Stern–Volmer Quenching Constants and Binding Parameters of the Lyz–Nos System at Room Temperature.
| temperature (K) | 298 |
| Ksv (L mol–1) | 3.18 × 103 |
| Kq (L mol–1 s–1) | 3.18 × 1011 |
| n | 0.90 |
| Kb (L mol–1) | 1.41 × 103 |
The fluorescence data was further evaluated to determine the binding constant and the number of binding sites by following the modified Stern–Volmer equation34 (eq 2).
| 2 |
Here, Kb is the binding constant and n represents the number of binding sites.
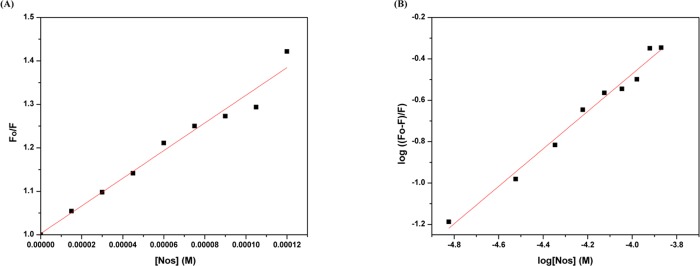
When eq 2 was used to plot the fluorescence data of the Lyz–Nos interaction, it was seen that the plot was linear and the slope came out to be 0.9, and the intercept gave the value for Kb. The Stern–Volmer and modified Stern–Volmer plots have been depicted in Figure 4. The binding constant was in the order of 103 (L mol–1) showing a moderate and reasonable binding, while the near-unity value of n (n = 0.9) shows the 1:1 stoichiometry of the Lyz–Nos complex. The UV–visible results were also in coherence with the steady-state fluorescence analysis because if the quenching was dynamic, the absorbance spectra would be unaltered.
Figure 4.
(A) Stern–Volmer plot of the Lyz–Nos
complex system
at 298 K. (B) Plot of log (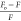 ) versus [Nos] for the Lyz–Nos system
at 298 K.
) versus [Nos] for the Lyz–Nos system
at 298 K.
Time-Resolved Fluorescence
The mechanism of quenching was concluded to be static based on steady-state fluorescence. Time-resolved fluorescence spectra can also be used to differentiate between static or dynamic quenching for any protein–drug interaction. When the quenching is static, the lifetime of the fluorophore remains unchanged in the presence of the ligand, while for dynamic quenching, a change is observed.35,36 On recording the spectra (Figure 5), it was seen that the lifetime of Lyz (1.8 ns) remained the same even in the presence of different concentrations of Nos (Table 2). Thus, the reason for the quenching of Lyz in the presence of the drug can be attributed to a static type.
Figure 5.
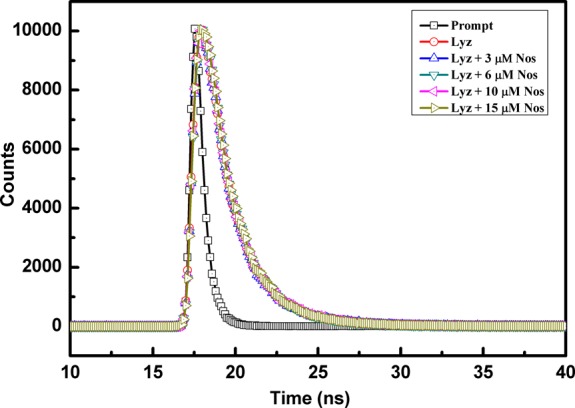
Fluorescence decay profile of Lyz in the absence and presence of Nos at different concentrations.
Table 2. Fluorescence Decay Profile of the Lyz–Nos System at Different Concentrations of Nos.
| [Nos] (μM) | τ1 (ns) | τ2 (ns) | a1 | a2 | τav (ns) |
|---|---|---|---|---|---|
| 0 | 1.05 | 2.59 | 51.28 | 48.72 | 1.80 |
| 3 | 1.1 | 2.63 | 55.23 | 44.77 | 1.785 |
| 6 | 1.09 | 2.56 | 52.46 | 47.54 | 1.788 |
| 10 | 1.1 | 2.6 | 54.18 | 45.82 | 1.787 |
| 15 | 1.05 | 2.49 | 48.28 | 51.72 | 1.795 |
UV-Thermal Denaturation
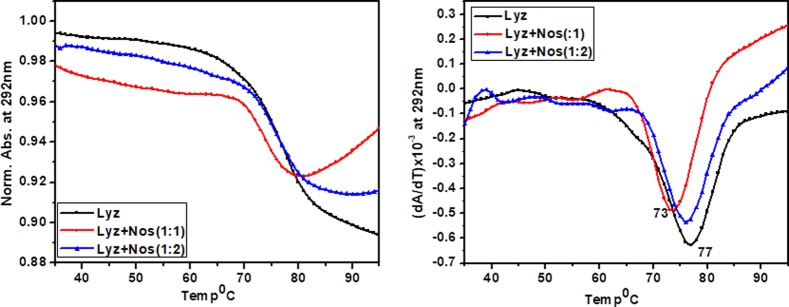
UV-Tm experiments have very important implications in interaction studies. It plays a significant role in interpreting the stabilizing/destabilizing effect of the ligand on biomolecules (protein, DNA, etc.). In the present study, the UV Tm of Lyz is monitored in the absence as well as in the presence of Nos. Figure 6 displays an inverted melting profile for Lyz in the absence as well as in the presence of a varied concentration of Nos. The thermal melting profile of Lyz is manifested with a melting temperature of 77 °C. On addition of Nos in the varied ratio (i.e., 1:1 and 1:2), a decrease in the melting temperature was observed, that is, 73 °C, which reflects destabilization of the protein. It might be possible that Nos interacts with the protein via intercalation to the hydrophobic pocket of the protein where tryptophan is located, thus distorting the structure of the protein.
Figure 6.
Thermal denaturation of Lyz with and without Nos monitored by UV spectroscopy at 292 nm. The overall melting rate was 0.5 °C min–1. The concentration of Lyz was 15 μM, and concentrations of Nos used were 15 and 30 μM.
CD Spectroscopy
Circular dichroism (CD) spectroscopy has been employed as an informative tool to investigate the folding of proteins as well as conformational changes in the protein upon interaction with ligands. Various structural elements (α helix, β sheets, peptide bonds, etc.) manifested characteristic CD signatures. CD spectroscopy of Lyz is carried out in phosphate buffer saline (pH 7.4) at room temperature (298 K). Hen egg protein Lyz has components of α + β with a larger α domain confining four α helices along with a comparatively smaller β domain. CD spectra recorded in the near-UV region (250–300 nm) are very significant in order to explain the folding of the proteins along with the presence of aromatic moieties, disulfide bonds, and prosthetic groups in the protein. When the protein is correctly folded, a prominent signal in the near-UV region is observed. Near-UV CD spectra significantly give information about the tertiary structure and environment of protein, whereas far-UV CD spectra reveal the important characteristic feature of the secondary structure in proteins.
Figure 7 demonstrates CD spectra of Lyz in the absence as well as in the presence of Nos. The far-ultraviolet CD spectrum of Lyz is manifested with two negative minima centered at 208 and 222 nm, reflecting the characteristic CD signature of an α-helical structure. It is in good agreement with a previous report.37 Minima at 208 nm correspond to the π–π* transition of the α helix, whereas minima at 222 nm belong to the n–π* transitions for both the α helix and the random coil. On subsequent addition of Nos, the far-UV CD spectrum of Lyz decreased in intensity with a shift of a 222 nm peak toward 208 nm, indicating secondary-structure conformational changes in the protein.38
Figure 7.
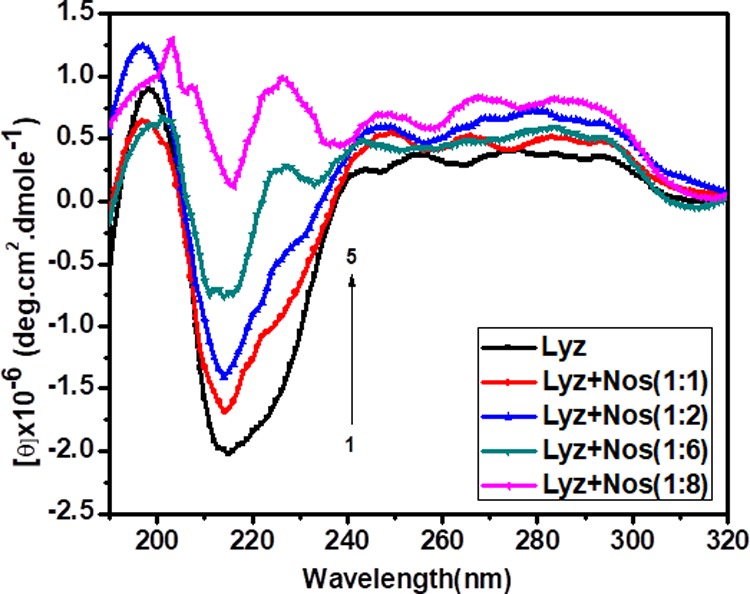
Circular dichroism spectra of Lyz (0.7 μM) with various concentrations of Nos. The curves (1–5) denote 0, 0.7, 1.4, 4.2, and 5.6 μM Nos in phosphate buffer saline (pH 7.4) at 298 K.
To gain more insight into secondary-structure conformational changes, the secondary structure is determined by using Yang software and tabulated in Table 3. The table clearly shows that native Lyz contains 30.5% α helices, 37% turns, and 32.5% random coils. On successive addition of Nos, the content of α helices decreases along with an increase in the content of β sheets for Lyz. Increasing the concentration of Nos at 1:1, 1:2, 1:6, and 1:8 manifested a decrease in the α-helix percentage, that is, 29.2, 24.4, 16, and 3%, respectively, along with an increment in the β-sheet percentage such as 24.1, 40.6, and 91.6% was observed. Herein, it is a clear indication of conformational changes occurring on the interaction of Lyz with Nos. It might be possible that Nos goes inside the hydrophobic pocket of the protein, binds there, and by doing so, induces conformation alteration/disruption of the protein’s native conformation.39 The CD experiment is in good correlation with the UV-thermal denaturation experiment.
Table 3. Percentage of Secondary Structures of the Lyz Protein in the Presence of Nos.
| protein | concentration | α helix | β sheet | β turn | random coil |
|---|---|---|---|---|---|
| lysozyme | lysozyme | 30.5% | 0.0% | 37.0% | 32.5% |
| lysozyme + Nos (1:1) | 29.2% | 0.0% | 36.0% | 34.9% | |
| lysozyme + Nos (1:2) | 24.4% | 24.1% | 24.3% | 27.2% | |
| lysozyme + Nos (1:6) | 16.0% | 40.6% | 20.9% | 22.5% | |
| lysozyme + Nos (1:8) | 3.0% | 91.6% | 0.0% | 5.4% |
Boiled-Egg Permeation Assay of Nos
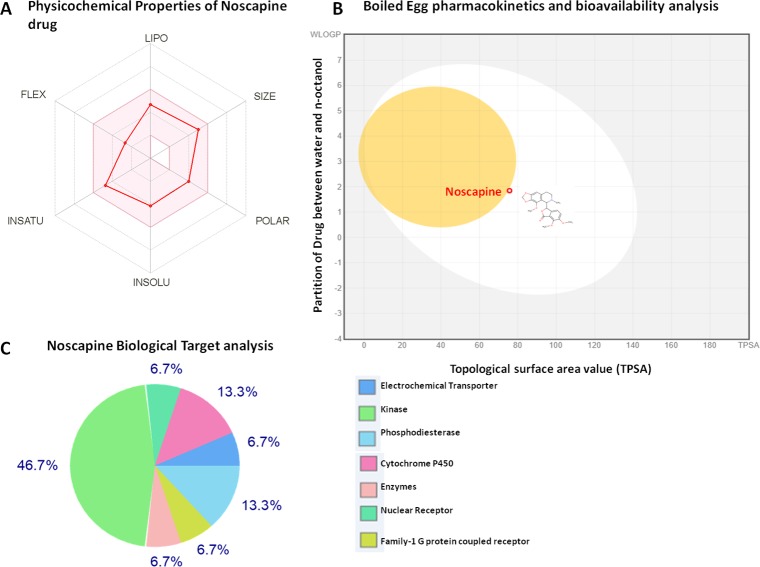
Pharmacokinetics and bioavailability gastrointestinal absorption for high efficacy of the drug were studied using the boiled-egg permeation assay. A boiled-egg model-based intuitive graphical analysis was performed for passive absorption of Nos through intestinal and brain penetration. The result showed that the Nos drug is found to fall inside the white ellipse, which indicated the high intestinal absorption. Also, Nos is found to fall very close and partially inside the yellow ellipse (yolk), which showed that the Nos has high values for permeation in the blood–brain barrier (BBB) (Figure 8).
Figure 8.
Pharmacokinetics and bioavailability property analysis of Nos. (A) Depiction of physicochemical properties of Nos. It is shown that Nos lies in an optimal range for various properties required for the potential drug. (B) Boiled-egg permeation assay of Nos. It lies inside the white ellipse and yellow ellipse (yolk). (C) Depiction of Nos biological targets.
Retrieval of the 3D structure of Lyz and Preparation for Molecular Docking
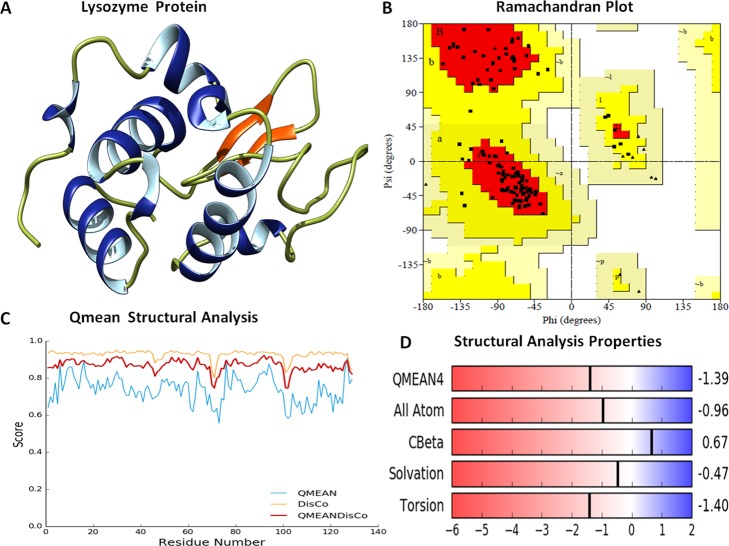
Prior to the molecular docking assay, the 3D structure of Lyz was downloaded from the Protein Data Bank (PDB ID: 2LYZ). Stereochemical properties were analyzed by the Ramachandran plot using the Saves server. The Ramachandran plot showed that ∼85% of residues lie in the most favored region, 15% of residues lie in the additionally allowed region, and no residue lies in the outlier region. Furthermore, physicochemical properties of the Lyz structure were evaluated by the Swiss server, and it showed the QMEAN score to be −1.39 for the structure, representing the overall absolute parameters, including the torsion angle energy, solvation energy, solvent accessibility, and atom pairwise energy. These results indicated the stability of the structure lying in a region close to the white region of the QMEAN plot. The precision of the structural assessment was determined by QMEANDisCo scores. It showed the consistent interatomic distances of the structure, and the local quality plot showed a high-quality structure by scoring values above 0.6, which was expected for the high-quality structure according to the QMEANDisCo algorithms (Figure 9).
Figure 9.
(A) Depiction of the three-dimensional structure of the Lyz protein (PDB 2LYZ), showing the secondary structures in different colors. (B) Ramachandran plot of Lyz with major residues (∼85%) lying in favorable regions. (C) QMEAN structural analysis of Lyz; all three QMEAN scores are above 0.6, indicating good physicochemical properties. (D) Structural proportion analysis of Lyz for various parameters (solvation energy and interatomic energy torsion energy scores).
Molecular Docking Assay
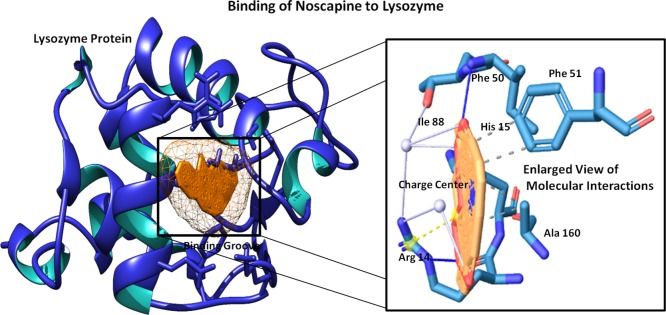
Molecular docking was performed to assess the binding of Nos to the Lyz protein. The Lyz protein was prepared by removing the water molecule to achieve the dry trajectory, and hydrogen atoms were added to stabilize the protein structure. HEX 8.0 parameters were set for the shape and conformations to analyze the binding in the 3D FFT mode with a whole-protein-range angle sampling method. The grid dimension of 3D Lyz was set to 0.6 Å, and the protein twist range of 360° and translation steps of 0.8 and 25 outputted the interacting complex system by the steric scan. The resulting top-score docked complex showed a strong interaction with a binding score of −215.32 kJ mol–1 for Nos, and it was found to possesses a strong interaction into the binding groove of the Lyz protein with two hydrogen bonds, one at Arg14 of a bond length of 2.85 Å with an angle of 147.48° and a second hydrogen bond at Ile88 of a bond length of 2.97 Å with an angle of 174.14° as well as strong hydrophobic interactions at Phe50, Phe51, and Ala160. Arginine14 was also found to form three water bridges with three different atoms of Nos with a distance of 3.94, 2.71, and 3.85 Å (Figure 10). Notably, Arg14 and His15 residues of Lyz were found to form the salt bridges with a distance of 4.74 and 5.06 Å. These molecular interaction dynamics and high binding scores suggested the potential binding of Nos to the Lyz protein.
Figure 10.
Molecular docking view of Nos to the binding groove of the Lyz protein (on the left) and depiction of an enlarged view of involved molecular interactions (on the right).
Molecular Dynamics Simulation Studies of the Nos–Lyz Complex
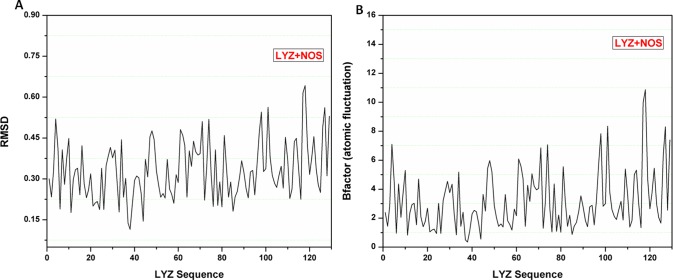
Molecular dynamics analysis of Nos interactions with the Lyz protein was done for 100 ns-long simulations that were run using the MDWeb modeling suite. Multiple frames for the complex system were retrieved for different structural conformations. Resulting trajectory files of the interacting complex system were taken and analyzed for RMSD, and they showed a minimal deviation in the Lyz protein in the range of 0.06 to 0.63 Å. Atomic fluctuation analysis showed the low atomic fluctuations in the range of 0.3 to 11 Å (Figure 11). These outcomes suggested the stable interaction of Nos with the Lyz protein with minimal deviations.
Figure 11.
Molecular dynamics simulation plots. (A) RMSD plot of the Nos–Lyz complex system, showing the minimal deviations. (B) Atomic fluctuation plot for the Nos–Lyz complex, showing the much fewer atomic fluctuation deviations and high stability of the interacting complex.
Conclusions
The mechanistic interaction of Nos and Lyz and their corresponding conformational changes have been investigated. The emergence of Nos as a versatile and non-narcotic anticancer drug has encouraged the studies to decipher its interactions with various model proteins. Here, in the current report, UV–vis absorption and steady-state and time-decay fluorescence confirmed the complex formation between Lyz and Nos, while CD spectra showed the decrease in alpha-helicity upon binding. Molecular docking and simulations provided a deeper insight into the phenomenon of binding, while the in silico boiled egg assay showed good gastrointestinal absorption of the drug. All these results are complimentary to each other and support the moderate binding of Nos with Lyz. This study is crucial in developing Nos for its oncological purposes and helps in understanding its pharmacokinetic properties.
Materials
Lysozyme (3× crystal, CAS no. 12650-88-3) was purchased from Sisco Research Laboratories (SRL), while Nos was purchased from Sigma-Aldrich, U.S.A. The chemicals used for all experiments were of analytical grade, and no further purification was done.
Methods
Sample Preparation
The stock solution of Lyz (25 μM) was prepared in phosphate buffer saline (PBS, 10 mM, pH 7.4) and diluted as per the requirement of the experiment. It was stored in a refrigerator and maintained at a temperature of 2–4 °C. The 10 mM stock solution of Nos was prepared in DMSO. The volume/volume ratio of DMSO/PBS was less than 1% in all experiments as it has been reported that, at this concentration, DMSO does not affect the structure of Lyz.40 All the experiments were performed at 298 K.
UV–Visible Spectroscopy
The UV–visible spectra were recorded using a Thermo Scientific Evolution 300 UV–vis spectrophotometer. The concentration of Lyz was kept constant at 15 μM, while the concentration of Nos was increased from 0 to 120 μM in intervals of 15 μM. The spectra were recorded in the UV range (200–400 nm).
Steady-State Fluorescence
The fluorescence emission spectra of Lyz in the absence and presence of Nos was recorded using Horiba PTI QM-8450-11-C. The excitation wavelength was set at 280 nm, and the spectra were recorded between 300 and 550 nm. The excitation and emission bandwidths were 3 nm.
Inner Filter Effect
The reason for the quenched fluorescence of the protein can also be attributed to the absorbance of the exciting light such that a less intense flux is encountered by the sample solution.41 The given equation can be used to correct the spectra for the inner filter effect.
Here, Fcorr is the corrected fluorescence intensity of the protein, while Fobs is the originally observed intensity. Aex and Aem are the absorbances of the drug or ligand at the excitation and emission wavelengths of the protein, respectively.
Time-Resolved Fluorescence Spectroscopy
The lifetime decay experiment was performed using a Horiba Yvon lifetime spectrometer. Time-correlated single photon counting (TCSPC) is the technique employed by the instrument to measure the decay in lifetime.42 An excitation pulse of 1.2 nm at a 1 MHz pulse repetition rate was sourced from NanoLED. The spectra were recorded for pure Lyz and in the presence of varying concentrations of the drug till a count of 10,000.
UV-Thermal Denaturation
The transition midpoint (Tm) of Lyz was monitored by the absorbance at 292 nm as a function of temperature with a Varian CARY 300 Conc UV spectrophotometer equipped with a peltier using the software Cary WinUV, thermal.43 A quartz cuvette of 10 mm path length is used to record the melting of samples. The concentration of Lyz was kept constant at 15 μM, while Nos is used at concentrations of 15 and 30 μM. The analysis of the melting curves for the measurement of TM was carried out using the first derivatives.
CD Experiment
CD spectroscopic analysis was carried out using a J-815 spectrophotometer (JASCO, Japan) at room temperature using a quartz cuvette of 10 mm path length. Camphorsulfonic acid was used for CD calibration. Samples were pre-equilibrated at the desired temperature for 15 min, and the scan speed is fixed at 100 nm min–1 with a response time of 1 s and a 1 nm bandwidth. To avoid water condensation in the cuvette chamber, it was flushed with constant purging of a N2 gas. The CD spectra were collected from 320 to 200 nm at wavelength steps of 1 nm, and the final reported spectra correspond to the average of three scans. The scans of the buffer alone were subtracted from the average scans for each sample for baseline correction. Data were collected in units of millidegrees and were normalized to total protein concentrations. Samples were prepared in 10 mM phosphate buffer saline of pH 7.4. To study the conformational changes of Lyz, a constant concentration of Lyz (0.7 μM) is titrated with increasing concentrations of Nos (0.7–5.6 μM).
The molar ellipticity [θ] is calculated from the observed ellipticity θ as
[θ]
= 100(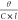 )
)
These experiments were performed for the 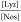 molar ratio in 1:1, 1:2, 1:6, and 1:8.
Secondary structure estimation is done by JASCO software Yang.jwr.
molar ratio in 1:1, 1:2, 1:6, and 1:8.
Secondary structure estimation is done by JASCO software Yang.jwr.
Boiled-Egg Permeation Assay of Nos
For potential drug development, the drug is required to possess the high values for pharmacokinetics and bioavailability gastrointestinal absorption for high efficacy of the drug at different stages. To assess gastrointestinal absorption and brain penetration, we have performed the intestinal estimated permeation method (boiled egg) using accurate predictive models. This method is based on determining the lipophilicity (W log P) and polar nature of the drug (TPSA) from a large dataset of drugs with reported 93% of accuracy.44,45
Molecular Docking Assay
Molecular docking assay was performed using the HEX 8.0 molecular docking suite. Hex uses the fast Fourier transformation algorithm (FTT). Hex outputs the multiple interacting ligand–protein complexes with the lowest energy scores; out of these results, a complex system with the lowest energy value was considered and further studied.46
For docking preparations, the three dimensional (3D) crystal structure of Lyz was retrieved from the Protein Data Bank (http://www.rcsb.org). Lyz was prepared and evaluated to perform molecular dynamics. Stereochemical parameters of Lyz were assessed through the Ramachandran plot and other in silico tools.47 The Nos compound was prepared for docking by drawing using ChemDraw and saving in the required format. After that, molecular docking parameters were set and docking was performed, and the resulting complex system (Nos–Lyz) was analyzed for involved molecular interactions (hydrogen bonds and hydrophobic bonds), and interacting conformations were analyzed using Chimera molecular modeling software.
Molecular Dynamics Simulation Studies of the Nos–Lyz Complex System
The complex of Nos–Lyz was evaluated for its stability through the molecular dynamics simulation analysis using the MDWeb program. The MDWeb suite works by performing the simulations through the NAMD full molecular dynamics set up to provide the molecular interaction trajectories.48 Simulation steps involve the complex-system cleaning, side-chain fixing, solvent addition, and energy minimization of system. After that, equilibration of the complex system was done by heating the solvent at 300 K, and the resulting dry trajectory was evaluated for RMSD and atomic fluctuations to assess the stability and flexibility of Nos binding to Lyz.
Acknowledgments
We are grateful to the Council of Scientific and Industrial Research (CSIR) for providing financial assistance and necessary funds to R.C. We all are grateful to CSIR for providing financial support and Delhi University USIC for research facilities. D.S. would like to thank UGC-SRF for their financial assistance and support. N.K. and V.T. are grateful to CSIR-RA for providing fellowship.
Author Contributions
# D.S. and N.K. contributed equally.
Author Contributions
D.S., N.K, R.C., and A.S. designed and performed the spectroscopic studies. N.K., V.T., and D.S. carried out the in silico experiments. The manuscript was written by D.S. and N.K., and A.S., S.K.D., and R.C. helped in preparing the figures and tables.
The authors declare no competing financial interest.
References
- Mahmoudian M.; Rahimi-Moghaddam P. The anti-cancer activity of noscapine: a review. Recent Pat. Anti-Cancer Drug Discovery 2009, 4, 92–97. 10.2174/157489209787002524. [DOI] [PubMed] [Google Scholar]
- Zhou J.; Panda D.; Landen J. W.; Wilson L.; Joshi H. C. Minor alteration of microtubule dynamics causes loss of tension across kinetochore pairs and activates the spindle checkpoint. J. Biol. Chem. 2002, 277, 17200–17208. 10.1074/jbc.M110369200. [DOI] [PubMed] [Google Scholar]
- Li Y.; Li S.; Thodey K.; Trenchard I.; Cravens A.; Smolke C. D. Complete biosynthesis of Noscapine and halogenated alkaloids in yeast. Proc. Natl. Acad. Sci. 2018, 115, E3922–E3931. 10.1073/pnas.1721469115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton R. A.; Gernert K. M.; Nettles J. H.; Aneja R. Drugs that target dynamic microtubules: a new molecular perspective. Med. Res. Rev. 2011, 31, 443–481. 10.1002/med.20242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebak S.; Mirzaei M.; Malhotra M.; Kulamarva A.; Prakash S. Human serum albumin nanoparticles as an efficient Noscapine drug delivery system for potential use in breast cancer: preparation and in vitro analysis. Int. J. Nanomed. 2010, 5, 525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdalla M. O.; Aneja R.; Dean D.; Rangari V.; Russell A.; Jaynes J.; Yates C.; Turner T. Synthesis and characterization of Noscapine loaded magnetic polymeric nanoparticles. J. Magn. Magn. Mater. 2010, 322, 190–196. 10.1016/j.jmmm.2009.07.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan J.; Dhiman N.; Parmar V. K.; Sardana S.; Bharatam P. V.; Aneja R.; Chandra R.; Katyal A. Inclusion complexes of Noscapine in β-cyclodextrin offer better solubility and improved pharmacokinetics. Cancer Chemother. Pharmacol. 2010, 65, 537. 10.1007/s00280-009-1060-3. [DOI] [PubMed] [Google Scholar]
- Tesseromatis C.; Alevizou A. The role of the protein-binding on the mode of drug action as well the interactions with other drugs. Eur. J. Drug Metab. Pharmacokinet. 2008, 33, 225–230. 10.1007/BF03190876. [DOI] [PubMed] [Google Scholar]
- Barreca D.; Laganà G.; Toscano G.; Calandra P.; Kiselev M. A.; Lombardo D.; Bellocco E. The interaction and binding of flavonoids to human serum albumin modify its conformation, stability and resistance against aggregation and oxidative injuries. Biochim. Biophys. Acta 2017, 1861, 3531–3539. 10.1016/j.bbagen.2016.03.014. [DOI] [PubMed] [Google Scholar]
- Sood D.; Kumar N.; Rathee G.; Singh A.; Tomar V.; Chandra R. Mechanistic Interaction Study of Bromo-Noscapine with Bovine Serum Albumin employing Spectroscopic and Chemoinformatics Approaches. Sci. Rep. 2018, 8, 16964. 10.1038/s41598-018-35384-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N.; Tomar R.; Pandey A.; Tomar V.; Singh V. K.; Chandra R. Preclinical evaluation and molecular docking of 1,3-benzodioxole propargyl ether derivatives as novel inhibitor for combating the histone deacetylase enzyme in cancer. Artif. Cells, Nanomed., Biotechnol. 2017, 46, 1288–1299. 10.1080/21691401.2017.1369423. [DOI] [PubMed] [Google Scholar]
- Chugh H.; Kumar P.; Tomar V.; Kaur N.; Sood D.; Chandra R. Interaction of Noscapine with human serum albumin (HSA): A spectroscopic and molecular modelling approach. J. Photochem. Photobiol., A. 2019, 372, 168–176. 10.1016/j.jphotochem.2018.12.001. [DOI] [Google Scholar]
- Maurya N.; Maurya J. K.; Singh U. K.; Dohare R.; Zafaryab M.; Moshahid Alam Rizvi M.; Kumari M.; Patel R. In vitro cytotoxicity and interaction of noscapine with human serum albumin: Effect on structure and esterase activity of HSA. Mol. Pharmaceutics 2019, 16, 952–966. 10.1021/acs.molpharmaceut.8b00864. [DOI] [PubMed] [Google Scholar]
- Kumar N.; Chugh H.; Sood D.; Singh S.; Singh A.; Awasthi A.D.; Tomar R.; Tomar V.; Chandra R. Biology of Heme: Drug Interactions and Adverse Drug Reactions with CYP450. Curr. Top. Med. Chem. 2019, 18, 2042–2055. 10.2174/1568026619666181129124638. [DOI] [PubMed] [Google Scholar]
- Patel R.; Maurya N.; Parray M. U. D.; Farooq N.; Siddique A.; Verma K. L.; Dohare N. Esterase activity and conformational changes of bovine serum albumin toward interaction with mephedrone: Spectroscopic and computational studies. J. Mol. Recognit. 2018, 31, e2734 10.1002/jmr.2734. [DOI] [PubMed] [Google Scholar]
- Maurya N.; Maurya J. K.; Kumari M.; Khan A. B.; Dohare R.; Patel R. Hydrogen bonding-assisted interaction between amitriptyline hydrochloride and hemoglobin: spectroscopic and molecular dynamics studies. J. Biomol. Struct. Dyn. 2017, 35, 1367–1380. 10.1080/07391102.2016.1184184. [DOI] [PubMed] [Google Scholar]
- Maurya N.; Ud Din Parray M.; Maurya J. K.; Kumar A.; Patel R. Interaction of promethazine and adiphenine to human hemoglobin: A comparative spectroscopic and computational analysis. Spectrochim. Acta A. 2018, 199, 32–42. 10.1016/j.saa.2018.03.023. [DOI] [PubMed] [Google Scholar]
- Rabbani G.; Lee E. J.; Ahmad K.; Baig M. H.; Choi I. Binding of tolperisone hydrochloride with human serum albumin: effects on the conformation, thermodynamics, and activity of HSA. Mol. Pharmaceutics 2018, 15, 1445–1456. 10.1021/acs.molpharmaceut.7b00976. [DOI] [PubMed] [Google Scholar]
- Eyles S. J.; Radford S. E.; Robinson C. V.; Dobson C. M. Kinetic consequences of the removal of a disulfide bridge on the folding of hen Lysozyme. Biochemistry 1994, 33, 13038–13048. 10.1021/bi00248a013. [DOI] [PubMed] [Google Scholar]
- Ogundele M.O. A novel anti-inflammatory activity of Lysozyme: modulation of serum complement activation. Mediators Inflammation 1998, 7, 363–365. 10.1080/09629359890893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragland S.A.; Criss A.K. From bacterial killing to immune modulation: Recent insights into the functions of Lysozyme. PLoS Pathog. 2017, 13, e1006512 10.1371/journal.ppat.1006512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari R.; Callerio C.; Podio G. Antiviral activity of Lysozyme. Nature 1959, 183, 548. 10.1038/183548a0. [DOI] [PubMed] [Google Scholar]
- Singh V.; Kumar N.; Chandra R. Structural Insights of Induced Pluripotent Stem Cell Regulatory Factors Oct4 and Its Interaction with Sox2 and Fgf4 Gene. Adv. Biochem. Eng./Biotechnol. 2017, J119, 1–9. [Google Scholar]
- Sood D.; Kumar N.; Singh A.; Sakharkar M. K.; Tomar V.; Chandra R. Antibacterial and Pharmacological Evaluation of Fluoroquinolones: A Chemoinformatics Approach. Genomics Inf. 2018, 16, 44–51. 10.5808/GI.2018.16.3.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A.; Kumar N.; Sood D.; Singh S.; Awasthi A.; Tomar V.; Chandra R. Designing of a Novel Indoline scaffold based Antibacterial Compound and Pharmacological Evaluation using Chemoinformatics approach. Curr. Top. Med. Chem. 2019, 18, 2056–2065. 10.2174/1568026619666181129125524. [DOI] [PubMed] [Google Scholar]
- Rudra S.; Jana A.; Sepay N.; Patel B. K.; Mahapatra A. Characterization of the binding of strychnine with bovine β-lactoglobulin and human Lysozyme using spectroscopic, kinetic and molecular docking analysis. New J. Chem. 2018, 42, 8615–8628. 10.1039/C8NJ00810H. [DOI] [Google Scholar]
- Sheng C. J.; Dian H. D.. Lysozyme; Shandong Science and Technology Press, 1982. p.50. [Google Scholar]
- Lakowicz J.R.Protein fluorescence. In Principles of fluorescence spectroscopy, Springer, Boston, MA, 1983, (pp. 341–381) [Google Scholar]
- Pasban Ziyarat F.; Asoodeh A.; Sharif Barfeh Z.; Pirouzi M.; Chamani J. Probing the interaction of Lysozyme with ciprofloxacin in the presence of different-sized Ag nano-particles by multispectroscopic techniques and isothermal titration calorimetry. J. Biomol. Struct. Dyn. 2014, 32, 613–629. 10.1080/07391102.2013.785919. [DOI] [PubMed] [Google Scholar]
- Mai W.; Hu C. cDNA cloning, expression and antibacterial activity of Lysozyme C in the blue shrimp (Litopenaeus stylirostris). Prog. Nat. Sci. 2009, 19, 837–844. 10.1016/j.pnsc.2008.10.008. [DOI] [Google Scholar]
- Stern O.; Volmer M. The extinction period of fluorescence. Phys. Z. 1919, 20, 183–188. [Google Scholar]
- Lakowicz J. R.; Weber G. Quenching of fluorescence by oxygen. Probe for structural fluctuations in macromolecules. Biochemistry 1973, 12, 4161–4170. 10.1021/bi00745a020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat I. A.; Bhat W. F.; Akram M.; Kabir-ud-Din Interaction of a novel twin-tailed oxy-diester functionalized surfactant with lysozyme: Spectroscopic and computational perspective. Int. J. Biol. Macromol. 2018, 109, 1006–1011. 10.1016/j.ijbiomac.2017.11.085. [DOI] [PubMed] [Google Scholar]
- Saha S.; Chowdhury J. Binding Interaction of Juglone with Lysozyme: Spectroscopic Studies Aided by In Silico Calculations. J. Photochem. Photobiol., B 2019, 193, 89–99. 10.1016/j.jphotobiol.2019.02.006. [DOI] [PubMed] [Google Scholar]
- Shanmugaraj K.; Anandakumar S.; Ilanchelian M. Probing the binding interaction of thionine with Lysozyme: a spectroscopic and molecular docking investigation. Dyes Pigm. 2015, 112, 210–219. 10.1016/j.dyepig.2014.07.003. [DOI] [Google Scholar]
- Millan S.; Satish L.; Bera K.; Konar M.; Sahoo H. Exploring the effect of 5-fluorouracil on conformation, stability and activity of Lysozyme by combined approach of spectroscopic and theoretical studies. J. Photochem. Photobiol., B 2018, 179, 23–31. 10.1016/j.jphotobiol.2017.12.019. [DOI] [PubMed] [Google Scholar]
- Jash C.; Kumar G. S. Binding of alkaloids berberine, palmatine and coralyne to lysozyme: a combined structural and thermodynamic study. RSC Adv. 2014, 4, 12514–12525. 10.1039/C3RA46053C. [DOI] [Google Scholar]
- Greenfield N. J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2876–2890. 10.1038/nprot.2006.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghili Z.; Taheri S.; Zeinabad H. A.; Pishkar L.; Saboury A. A.; Rahimi A.; Falahati M. Investigating the Interaction of Fe Nanoparticles with Lysozyme by Biophysical and Molecular Docking Studies. PLoS One 2016, 11, e0164878 10.1371/journal.pone.0164878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjya S.; Balaram P. Effects of organic solvents on protein structures: Observation of a structured helical core in hen egg-white Lysozyme in aqueous dimethylsulfoxide. Proteins: Struct., Funct., Genet. 1997, 29, 492–507. . [DOI] [PubMed] [Google Scholar]
- Fonin A. V.; Sulatskaya A. I.; Kuznetsova I. M.; Turoverov K. K. Fluorescence of Dyes in Solutions with High Absorbance. Inner Filter Effect Correction. PLoS One 2014, 9, e103878 10.1371/journal.pone.0103878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L.; Yan W.; Ho D. Recent advances in fluorescence lifetime analytical microsystems: Contact optics and CMOS time-resolved electronics. Sensors 2017, 17, 2800. 10.3390/s17122800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih P.; Kirsch J. F.; Holland D. R. Thermal stability determinants of chicken egg-white lysozyme core mutants: Hydrophobicity, packing volume, and conserved buried water molecules. Protein Sci. 1995, 4, 2050–2062. 10.1002/pro.5560041010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daina A.; Zoete V. A boiled-egg to predict gastrointestinal absorption and brain penetration of small molecules. ChemMedChem. 2016, 11, 1117–1121. 10.1002/cmdc.201600182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daina A.; Michielin O.; Zoete V. iLOGP: a simple, robust, and efficient description of n-octanol/water partition coefficient for drug design using the GB/SA approach. J. Chem. Inf. Model. 2014, 54, 3284–3301. 10.1021/ci500467k. [DOI] [PubMed] [Google Scholar]
- Macindoe G.; Mavridis L.; Venkatraman V.; Devignes M. D.; Ritchie D. W. HexServer: an FFT-based protein docking server powered by graphics processors. Nucleic Acids Res. 2010, 38, W445–W449. 10.1093/nar/gkq311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkert P.; Tosatto S. C. E.; Schomburg D. QMEAN: a comprehensive scoring function for model quality assessment. Proteins 2008, 71, 261–277. 10.1002/prot.21715. [DOI] [PubMed] [Google Scholar]
- Hospital A.; Andrio P.; Fenollosa C.; Cicin-Sain D.; Orozco M.; Gelpí J. L. MDWeb and MDMoby: an integrated web-based platform for molecular dynamics simulations. Bioinformatics 2012, 28, 1278–1279. 10.1093/bioinformatics/bts139. [DOI] [PubMed] [Google Scholar]