The crystal structure of the Fab region of a neutralizing fully human antibody against granulocyte-macrophage colony-stimulating factor is reported.
Keywords: monoclonal antibodies, Fab region, granulocyte-macrophage colony-stimulating factor, anti-GM-CSF, EV1007
Abstract
An increased level of granulocyte-macrophage colony-stimulating factor has a potential role in the development of autoimmune diseases, and the neutralization of its activity by monoclonal antibodies is a promising therapy for some diseases. Here, the crystal structure of the Fab region of EV1007, a fully human antibody expressed in Chinese hamster ovary cells that was developed from human peripheral blood mononuclear cells, is described. The structure closely resembles that of MB007, which is the Fab region of the same antibody expressed in Escherichia coli [Blech et al. (2012 ▸), Biochem. J. 447, 205–215], except at the hinge regions between the immunoglobulin domains and the H3 loop region. This paper presents evidence for the flexibility of the hinge and H3 loop regions of the antibody based on the comparison of two independently solved crystal structures.
1. Introduction
Granulocyte-macrophage colony-stimulating factor (GM-CSF) regulates the differentiation of granulocytes and macrophages from their precursor cells in the bone marrow. It is a well known agent for the supportive care of cancer patients that has been used in clinical settings. However, recent studies have suggested its potential role in the development of autoimmune diseases (Codarri et al., 2011 ▸; El-Behi et al., 2011 ▸), as well as in tumor progression in certain types of cancer (Hong, 2016 ▸). Neutralization of GM-CSF using antibodies has been shown to suppress a number of diseases, and there are a number of clinical trials targeting GM-CSF or its receptor using neutralizing monoclonal antibodies (Hamilton, 2015 ▸). A recent report has also suggested that antibody-mediated neutralization of GM-CSF is effective in preventing cytokine-release syndrome (CRS) in chimeric antigen receptor T-cell (CAR T-cell) therapy, suggesting its potential use as an anti-CRS treatment (Sachdeva et al., 2019 ▸).
EV1007 is a fully human antibody, derived from an autoimmune disease patient using Epstein–Barr virus technology, with a neutralizing effect on GM-CSF activity (Takada et al., 2014 ▸). Its Fab fragment produced in Escherichia coli is known as MB007, and its crystal structure and binding mode to GM-CSF have been determined using a combination of X-ray crystallography and computational and biophysical methods (Blech et al., 2012 ▸). Here, we describe a crystal structure of the Fab fragment of EV1007 (EV1007-Fab) produced from intact immunoglobulin G (IgG) that was expressed in a mammalian cell system. Compared with MB007, EV1007-Fab is a better representation of the potential active pharmaceutical ingredient that is currently being developed.
When compared with the MB007 structure, there is an approximately 70° rotation at the hinge between the constant and variable regions in the EV1007-Fab structure. In addition, a local structural difference of the H3 loop between the two structures was observed. The comparison adds information on the flexibility of antibody structures in the hinge and H3 loop regions.
2. Materials and methods
2.1. Macromolecule production
The genes encoding the heavy (H) and light (L) regions of human IgG against GM-CSF were ligated into a mammalian expression vector, which was used to transfect Chinese hamster ovary K1 (CHO-K1) cells. The transfected cells were grown in a serum-free medium in plastic culture dishes for one week for antibody production. The culture supernatant was collected and purified using Protein G column chromatography (GE Healthcare). The Fab fragment was prepared from the IgG antibody using the Pierce Fab Preparation Kit (Thermo Fisher Scientific) and was further purified by gel filtration on a HiLoad 16/60 Superdex 200 gel-filtration column (GE Healthcare) pre-equilibrated with 5 mM Tris–HCl pH 8.0 containing 100 mM NaCl. The purity of the Fab fragment was assessed by SDS–PAGE. The purified Fab fragment was concentrated to 10 mg ml−1 and used directly for crystallization screening or optimization. Macromolecule-production information is summarized in Table 1 ▸.
Table 1. Macromolecule-production information.
| Expression vector | pcDNA3.1(+) |
| Expression host | Chinese hamster ovary (CHO-K1) cells |
| Complete amino-acid sequence of EV1007 IgG | |
| Light chain | MAGFPLLLTLLTHCAGSWAQSVLTQPPSASGTPGQSVNISCSGSSSNIGNSYVYWYQQLPGTAPKLLIYRNNRRPSGVPDRFSGSKSDTSASLAISGLRSEDEADYYCATWDDSLSGRLFGGGTKLTVLGQPKAAPSVTLFPPSSEELQANKATLVCLISDFYPGAVTVAWKADGSPVKAGVETTTPSKQSNNKYAASSYLSLTPEQWKSHRSYSCQVTHEGSTVEMTVAPTECS |
| Heavy chain† | MDWTWRILFLVAAATGAPSQVHLVQSGSELKKPGASVKVSCKASGYSFSRYGIKWVRQAPGQGLEWMGWINTRSGVPAYAQGFTGRFVFSLDTSVDTAFLEISSLKTEDTGIYYCATRPPRFYDKTEYWEDGFDVWGRGTLVWSSASTKGPSVFPIIAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSWTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVWDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRWSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEGLHNHYTQKSLSLSPGK |
The H3 loop of the heavy chain is underlined.
2.2. Crystallization
Initial crystallization screening was performed using Crystal Screen and Crystal Screen 2 (Hampton Research) and Wizard Classic 1 & 2 (Rigaku Reagents). Crystals were obtained after 1–2 weeks of incubation at 293 K using the sitting-drop vapor-diffusion method. The drops consisted of 1 µl protein solution and 1 µl reservoir solution and were equilibrated against 70 µl reservoir solution. The best crystals were obtained from Crystal Screen 2 solution No. 14 (0.2 M sodium/potassium tartrate tetrahydrate, 0.1 M sodium citrate tribasic dihydrate, 2.0 M ammonium sulfate pH 5.6) and were used without further optimization. Crystallization information is summarized in Table 2 ▸.
Table 2. Crystallization conditions.
| Method | Sitting-drop vapor diffusion |
| Plate type | 48-well |
| Temperature (K) | 293 |
| Protein concentration (mg ml−1) | 10 |
| Buffer composition of protein solution | 5 mM Tris–HCl pH 8.0, 100 mM NaCl |
| Composition of reservoir solution | 0.2 M sodium potassium tartrate tetrahydrate, 0.1 M sodium citrate tribasic dihydrate, 2.0 M ammonium sulfate pH 5.6 |
| Volume and ratio of drop | 2 µl; 1:1 ratio |
| Volume of reservoir (µl) | 70 |
2.3. Data collection and processing
X-ray diffraction data were collected on the BL38B1 beamline at SPring-8 at a wavelength of 1.00 Å. Prior to data collection, the crystals were cryoprotected in mother-liquor solution containing 18% glycerol. X-ray diffraction data were obtained at 100 K. The crystals diffracted to 1.8–2.5 Å resolution and the crystal reported here was that with the highest resolution. Data reduction and scaling were performed using iMOSFLM (Battye et al., 2011 ▸) and POINTLESS (Evans, 2011 ▸), respectively. Data-collection and processing statistics are shown in Table 3 ▸.
Table 3. Data-collection and processing statistics.
Values in parentheses are for the highest resolution shell.
| Temperature (K) | 100 |
| Space group | P212121 |
| a, b, c (Å) | 59.12, 66.69, 129.04 |
| α, β, γ (°) | 90, 90, 90 |
| Mosaicity (°) | 0.45–2.80 |
| Resolution range (Å) | 46.37–1.81 |
| Total No. of reflections | 662248 |
| No. of unique reflections | 43756 |
| Completeness (%) | 98.1 |
| Multiplicity | 13.8 (13.0) |
| 〈I/σ(I)〉 | 27.2 (11.5) |
| R merge | 0.068 (0.171) |
| R p.i.m. | 0.027 (0.069) |
| Overall B factor from Wilson plot (Å2) | 13.8 |
2.4. Structure solution, refinement and analysis
Phasing was performed with Phaser (McCoy et al., 2007 ▸) using the crystal structure of MB007 (PDB entry 4eow; Blech et al., 2012 ▸) as a model. Refinement and model improvement were performed using REFMAC5 (Murshudov et al., 2011 ▸) and Coot (Emsley et al., 2010 ▸). MolProbity (Chen et al., 2010 ▸) was used for Ramachandran analysis. Refinement statistics are shown in Table 4 ▸. DynDom (Poornam et al., 2009 ▸) was used to analyze hinge rotation. SUPERPOSE (Krissinel & Henrick, 2004 ▸) was used to analyze Cα atomic displacement and PyMOL (v.2.0; Schrödinger) was used to prepare the figures. The DALI server (Holm & Laakso, 2016 ▸) was used to compare the EV1007-Fab and MB007 structures against all structures in the PDB. Refinement statistics are summarized in Table 4 ▸.
Table 4. Refinement statistics.
| Resolution range (Å) | 46.37–1.81 (1.86–1.81) |
| Completeness (%) | 97.7 |
| σ Cutoff | None |
| No. of reflections, working set | 43716 (3133) |
| No. of reflections, test set | 2270 (154) |
| Final R cryst † | 0.161 (0.167) |
| Final R free† | 0.191 (0.200) |
| No. of non-H atoms | |
| Protein | 3316 |
| Ion | 34 |
| Solvent | 480 |
| Total | 3830 |
| R.m.s. deviations | |
| Bonds (Å) | 0.005 |
| Angles (°) | 0.796 |
| Average B factors (Å2) | |
| Protein | 19.3 |
| Ion | 38.0 |
| Water | 27.1 |
| Ramachandran plot | |
| Favored regions (%) | 96.8 |
| Outliers (%) | 0 |
R
cryst = 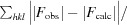
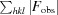 ; R
free is the R factor for a selected subset of the reflections (5%) that were not included in refinement calculations.
; R
free is the R factor for a selected subset of the reflections (5%) that were not included in refinement calculations.
3. Results and discussion
EV1007-Fab crystals were obtained using a crystallization solution that consisted of 0.2 M sodium/potassium tartrate tetrahydrate, 0.1 M sodium citrate tribasic dihydrate, 2.0 M ammonium sulfate pH 5.6. This crystallization condition is somewhat different from that for MB007, which was 19% PEG 4000, 0.1 M ammonium sulfate, 0.1 M sodium acetate pH 3.4 (Blech et al., 2012 ▸). Although the crystals used to determine both structures belonged to the same space group (P212121), the unit-cell parameters are slightly different. The differences in crystallization conditions and space-group properties may account for several notable differences, as described below.
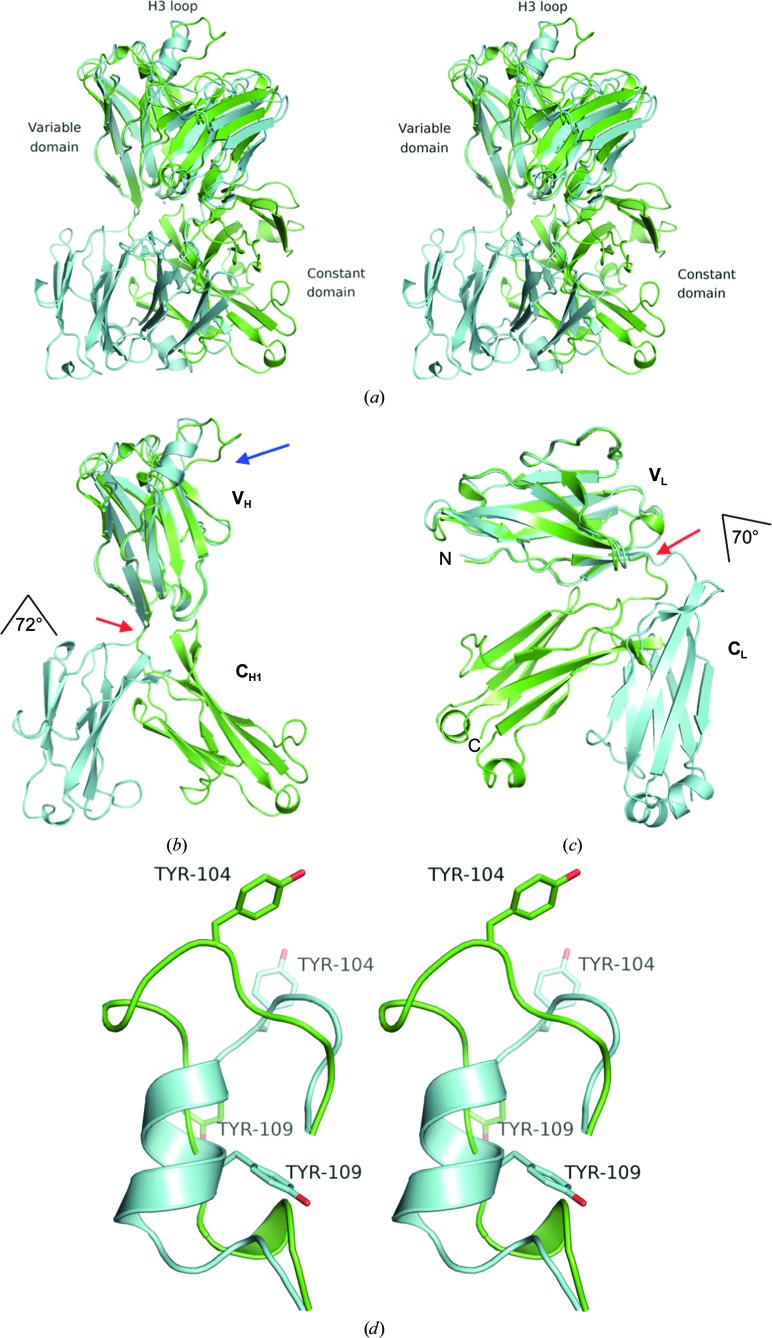
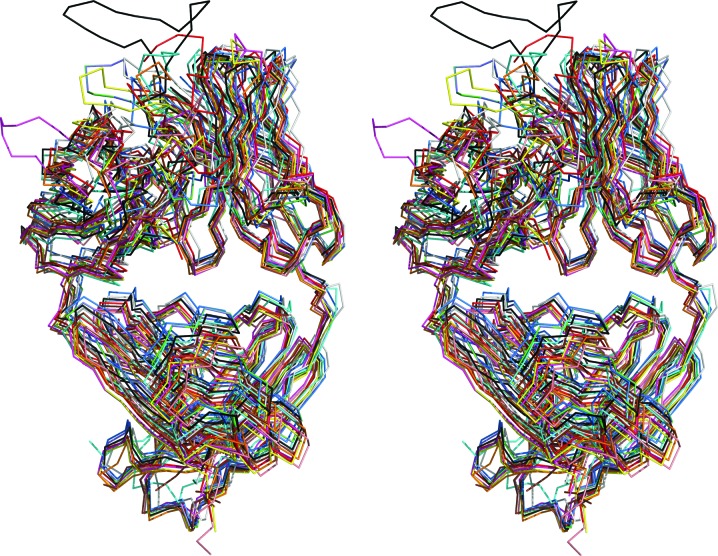
Firstly, there is an approximately 72° rotation at the hinge between the VH (variable/heavy) and CH1 (constant/heavy 1) regions of the H chain and a 70° rotation at the hinge between the VL (variable/light) and CL (constant/light) regions of the L chain of EV1007-Fab when compared with those of MB007 (Fig. 1 ▸). Such hinge-mediated domain movement is known to occur in immunoglobulin structures (Lesk & Chothia, 1988 ▸), as well as in many different proteins, including virus capsids (Gerstein et al., 1994 ▸). Heuristic comparison of the EV1007-Fab and MB007 structures against all structures in the PDB using the DALI server resulted more than 600 and 1500 structures with a Z-factor above 20.0, respectively, indicating that the hinge conformations of EV1007-Fab and MB007 are common. A superposition of the top ten nonredundant structures with the highest Z-factors with EV1007-Fab is shown in Fig. 2 ▸. Furthermore, some of those top ten nonredundant structures have antigen molecules or neighboring symmetry-related or pseudo-symmetry-related molecules bound to their CDR regions (not shown), indicating that the difference in the hinge structures of EV1007-Fab and MB007 is not caused by the binding of antigen or other molecules to the CDR regions.
Figure 1.
(a) Superposition of the EV1007-Fab (green) and MB007 (cyan) structures. The structures were superposed at the variable domain of the heavy chain (VH). (b, c) Rotations at the hinges (indicated by red arrows) between VH and CH1 superposed at the VH region (b) or VL and CL superposed at the VL region (c) of EV1007-Fab (green) and MB007 (cyan). As indicated in the figure, the angle of rotation is 72° and 70° for the heavy and light chain, respectively. The H3 loop region is shown by a blue arrow. (d) Stereo image of the superposed H3 loops of MB007 and EV1007-Fab. The side chains of Tyr104 and Tyr109 are shown as stick models. For clarity, the rest of the protein is not shown. The top part of the figure (the direction of the side chain of Tyr104 in both structures) is the part where the protein is exposed to the solvent, and the bottom part of the figure (the direction of the side chain of Tyr109 in the MB007 structure) is where the rest of the protein is located.
Figure 2.
The superposition of the ten structures with the highest Z-scores with the EV1007-Fab structure (green). The following PDB entries were included: 5cck (cyan; Lee et al., 2015 ▸), 5d7s (magenta; Eylenstein et al., 2016 ▸), 5ghw (yellow; Rujas et al., 2016 ▸), 5nhw (salmon; Morvan et al., 2017 ▸), 5otj (gray; Mitropoulou et al., 2018 ▸), 6apc (blue; Goodwin et al., 2018 ▸), 6axk (red; Oyen et al., 2017 ▸), 6b0w (orange; Scally et al., 2018 ▸), 6mej (brown; Flyak et al., 2018 ▸) and 3mug (black; Pejchal et al., 2010 ▸).
MB007 has no visible ions and EV1007-Fab contains six sulfate ions. Three sulfate ions are bound to a single residue each [Gly162 (L chain), Arg67 (H chain) and Ser172 (L chain)], one is bound to the main-chain atoms of two consecutive residues (Lys160 and Ala161 in the L chain) and one is bound to two residues (Arg55 and Gly64 in the L chain) located in the variable domain of EV1007 that superimposes well with that of MB007. The last of the sulfate ions is bound to Tyr144 and Tyr176, which are located at the hinge of the light chain of EV1007. However, an examination of the other Fab structures that have highly similar overall conformations to that of EV1007 (Tyr140 and Tyr176 in PDB entry 5cck, Tyr138 and Tyr170 in PDB entry 5d7s, Tyr145 and Tyr177 in PDB entry 5otj, Tyr141 and Tyr173 in PDB entry 6apc and others) revealed that these conserved tyrosine residues assume the same conformation as those in EV1007-Fab without any ions present, thus eliminating the possibility that the sulfate ion contributes to the difference between the EV1007-Fab and MB007 hinge conformations. There is no observable electron density for sugar residues in the EV1007-Fab or MB007 structures, and the amino-acid sequences of both structures are identical. Sotriffer et al. (2000 ▸) also showed a high flexibility of the hinge area using a molecular-dynamics (MD) simulation and suggested that the lack of anchoring of this region to the other areas of the Fab is the main reason for its stability. Taken together, the difference in the hinge conformations of EV1007-Fab and MB007 is owing to the intrinsic flexibility of the hinge region. It is worth noting that the intrinsic flexibility of the hinge region has been mentioned as a problem in using Fab fragments as chaperones for crystallization (Bailey et al., 2017 ▸).
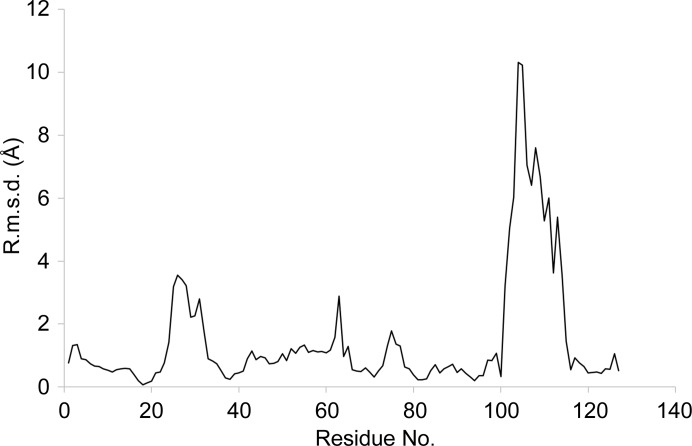
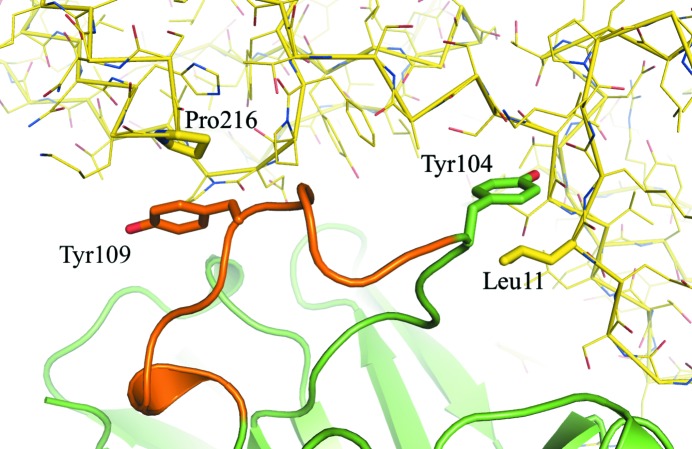
The second notable difference is the local structure of the H3 loop in the complementarity-determining region (CDR), as shown in Fig. 1 ▸(d) and by the Cα atomic displacement of the VH region in Fig. 3 ▸. The H3 loop in MB007 forms a short α-helical structure, whereas that in EV1007-Fab forms a random-coil structure (Fig. 1 ▸, blue arrow). Calculation using CONTACT from the CCP4 suite showed that there are 241 and zero water-free symmetry-related contacts at ≤5 Å distance in the H3 loop (residues 105–114 of the heavy chain, as identified by ABNUM; Abhinandan & Martin, 2008 ▸) in the EV1007-Fab and MB007 structures, respectively. Upon a more detailed inspection, it seems that the main contributors to symmetry-related H3 loop stabilization in EV1007-Fab are the hydrophobic interactions between Tyr109 of the H chain and Pro216 of the H chain of the symmetry-related molecule and between Tyr104 of the H chain and Leu11 of the H chain of the symmetry-related molecule (Fig. 4 ▸). In contrast, Tyr109 and Tyr104 of the helical structure formed by the H3 loop of MB007 are tucked inside the protein and exposed, respectively, and do not form any significant interactions with the surrounding residues [Fig. 1 ▸(d)]. It is worth noting that although Tyr109 of MB007 is in a tucked-in conformation, the packing is relatively loose. The H3 loop is known to be the key region that determines the ability of an antibody to bind a specific epitope and has the highest structural diversity among diverse antibodies (Zemlin et al., 2003 ▸).
Figure 3.
Cα atomic positional displacement of the VH region of EV1007-Fab compared with that of MB007 following least-squares superposition.
Figure 4.
Symmetry-assisted stabilization of the H3 loop structure of EV1007-Fab. The H3 loop identified by ABNUM (see text) is colored orange, in contrast to the rest of the heavy chain, which is colored green. The light chain is not shown for clarity. The symmetry-generated molecule is colored yellow.
Based on NMR epitope mapping and in silico (docking and MD simulation) data, Blech et al. (2012 ▸) showed that although the H3 loop of MB007 experiences a significant shift upon binding to its antigen, its helical conformation was preserved. Here, we show that the H3 loop structure of this antibody could change more drastically upon a nonspecific contact with a neighboring molecule in the crystal packing. This finding suggests that the H3 loop structure of this antibody is more flexible than previously thought and provides additional biological insights into the H3 loop flexibility of two identical antibody sequences, which can be used as a reference for further in silico docking of antibodies to their antigens.
The flexibility of the hinge region of Fab structures is well accepted and has been described in several papers (Lesk & Chothia, 1988 ▸; Sotriffer et al., 2000 ▸; Bailey et al., 2017 ▸). However, these publications compared the structures of different Fab fragments or are based on in silico studies. This paper offers the first clear-cut evidence that the same Fab fragment can assume highly varied hinge orientations under different crystallization conditions. The difference is not caused by the interaction between the protein and ions or other molecules in the crystallization mixture, indicating that the hinge region possesses intrinsic flexibility.
Supplementary Material
PDB reference: Fab region of EV1007, 5zmj
References
- Abhinandan, K. R. & Martin, A. C. R. (2008). Mol. Immunol. 45, 3832–3839. [DOI] [PubMed]
- Bailey, L. J., Sheehy, K. M., Dominik, P. W., Liang, W. G., Rui, H., Clark, M., Jaskolowski, M., Kim, Y., Deneka, D., Tang, W.-J. & Kossiakoff, A. A. (2017). J. Mol. Biol. 430, 337–347. [DOI] [PMC free article] [PubMed]
- Battye, T. G. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. W. (2011). Acta Cryst. D67, 271–281. [DOI] [PMC free article] [PubMed]
- Blech, M., Seeliger, D., Kistler, B., Bauer, M. T., Hafner, M., Hörer, S., Zeeb, M., Nar, H. & Park, J. E. (2012). Biochem. J. 447, 205–215. [DOI] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Codarri, L., Gyülvészi, G., Tosevski, V., Hesske, L., Fontana, A., Magnenat, L., Suter, T. & Becher, B. (2011). Nature Immunol. 12, 560–567. [DOI] [PubMed]
- El-Behi, M., Ciric, B., Dai, H., Yan, Y., Cullimore, M., Safavi, F., Zhang, G.-X., Dittel, B. N. & Rostami, A. (2011). Nature Immunol. 12, 568–575. [DOI] [PMC free article] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Evans, P. R. (2011). Acta Cryst. D67, 282–292. [DOI] [PMC free article] [PubMed]
- Eylenstein, R., Weinfurtner, D., Hartle, S., Strohner, R., Bottcher, J., Augustin, M., Ostendorp, R. & Steidl, S. (2016). MAbs, 8, 176–186. [DOI] [PMC free article] [PubMed]
- Flyak, A. I., Ruiz, S., Colbert, M. D., Luong, T., Crowe, J. E. Jr, Bailey, J. R. & Bjorkman, P. J. (2018). Cell Host Microbe, 24, 703–716. [DOI] [PMC free article] [PubMed]
- Gerstein, M., Lesk, A. M. & Chothia, C. (1994). Biochemistry, 33, 6739–6749. [DOI] [PubMed]
- Goodwin, E., Gilman, M. S. A., Wrapp, D., Chen, M., Ngwuta, J. O., Moin, S. M., Bai, P., Sivasubramanian, A., Connor, R. I., Wright, P. F., Graham, B. S., McLellan, J. S. & Walker, L. M. (2018). Immunity, 48, 339–349. [DOI] [PMC free article] [PubMed]
- Hamilton, J. A. (2015). Exp. Rev. Clin. Immunol. 11, 457–465. [DOI] [PubMed]
- Holm, L. & Laakso, L. M. (2016). Nucleic Acids Res. 44, W351–W355. [DOI] [PMC free article] [PubMed]
- Hong, I.-S. (2016). Exp. Mol. Med. 48, e242. [DOI] [PMC free article] [PubMed]
- Krissinel, E. & Henrick, K. (2004). Acta Cryst. D60, 2256–2268. [DOI] [PubMed]
- Lee, J. H., Leaman, D. P., Kim, A. S., Torrents de la Pena, A., Sliepen, K., Yasmeen, A., Derking, R., Ramos, A., de Taeye, S. W., Ozorowski, G., Klein, F., Burton, D. R., Nussenzweig, M. C., Poignard, P., Moore, J. P., Klasse, P. J., Sanders, R. W., Zwick, M. B., Wilson, I. A. & Ward, A. B. (2015). Nature Commun. 6, 8167. [DOI] [PMC free article] [PubMed]
- Lesk, A. M. & Chothia, C. (1988). Nature (London), 335, 188–190. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Mitropoulou, A. N., Bowen, H., Dodev, T. S., Davies, A. M., Bax, H. J., Beavil, R. L., Beavil, A. J., Gould, H. J., James, L. K. & Sutton, B. J. (2018). Proc. Natl Acad. Sci. USA, 115, E8707–E8716. [DOI] [PMC free article] [PubMed]
- Morvan, F., Rondeau, J.-M., Zou, C., Minetti, G., Scheufler, C., Scharenberg, M., Jacobi, C., Brebbia, P., Ritter, V., Toussaint, G., Koelbing, C., Leber, X., Schilb, A., Witte, F., Lehmann, S., Koch, E., Geisse, S., Glass, D. J. & Lach-Trifilieff, E. (2017). Proc. Natl Acad. Sci. USA, 114, 12448–12453. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Oyen, D., Torres, J. L., Wille-Reece, U., Ockenhouse, C. F., Emerling, D., Glanville, J., Volkmuth, W., Flores-Garcia, Y., Zavala, F., Ward, A. B., King, C. R. & Wilson, I. A. (2017). Proc. Natl Acad. Sci. USA, 114, E10438–E10445. [DOI] [PMC free article] [PubMed]
- Pejchal, R., Walker, L. M., Stanfield, R. L., Phogat, S. K., Koff, W. C., Poignard, P., Burton, D. R. & Wilson, I. A. (2010). Proc. Natl Acad. Sci. USA, 107, 11483–11488. [DOI] [PMC free article] [PubMed]
- Poornam, G., Matsumoto, A., Ishida, H. & Hayward, S. (2009). Proteins, 76, 201–212. [DOI] [PubMed]
- Rujas, E., Caaveiro, J. M., Partida-Hanon, A., Gulzar, N., Morante, K., Apellaniz, B., Garcia-Porras, M., Bruix, M., Tsumoto, K., Scott, J. K., Jimenez, M. A. & Nieva, J. L. (2016). Sci. Rep. 6, 38177. [DOI] [PMC free article] [PubMed]
- Sachdeva, M., Duchateau, P., Depil, S., Poirot, L. & Valton, J. (2019). J. Biol. Chem. 294, 5430–5437. [DOI] [PMC free article] [PubMed]
- Scally, S. W., Murugan, R., Bosch, A., Triller, G., Costa, G., Mordmuller, B., Kremsner, P. G., Sim, B. K. L., Hoffman, S. L., Levashina, E. A., Wardemann, H. & Julien, J.-P. (2018). J. Exp. Med. 215, 63–75. [DOI] [PMC free article] [PubMed]
- Sotriffer, C. A., Rode, B. M., Varga, J. M. & Liedl, K. R. (2000). Biophys. J. 79, 614–628. [DOI] [PMC free article] [PubMed]
- Takada, K., Nakajima, K., Kistler, B. & Park, J. (2014). US Patent 8679502.
- Zemlin, M., Klinger, M., Link, J., Zemlin, C., Bauer, K., Engler, J. A., Schroeder, H. W. & Kirkham, P. M. (2003). J. Mol. Biol. 334, 733–749. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: Fab region of EV1007, 5zmj