Abstract
Background:
Multidrug-resistant Acinetobacter baumannii can cause complications in antibiotic therapy and increase the rate of morbidity and mortality in hospitalized patients. Patients with ventilator and burns are two specific groups at high risk for A. baumannii infections. This study aimed to determine antibiotic susceptibility patterns associated with biofilm production in A. baumannii and to assess its molecular epidemiology by random amplified polymorphic DNA polymerase chain reaction (RAPD PCR) in A. baumannii isolated from ventilator-associated pneumonia and burn wound colonization.
Materials and Methods:
In this study, 79 isolates of A. baumannii (32 ventilator-associated pneumonia [VAP] 47 burns) were collected in two teaching hospitals in Tehran, Iran, in 2018. Conventional biochemical and microbiological methods were used to identify bacteria. Antibiotic susceptibility was detected by disc diffusion methods according to the Clinical and Laboratory Standards Institute 2018. Tube test was examined for the detection of the biofilm formation rate in collected strains. The most prevalent carbapenemase genes were detected by PCR and molecular typing by RAPD PCR.
Results:
All of bacteria were extensively drug-resistant (XDR) except for two isolates. The results of tube test indicated that only 36% of XDR strains were in weak rate of biofilm formation group. Two major clonal genetic groups were found in VAP and burn strains. Oxa-23 was the most prevalent carbapenemase in collected A. baumannii.
Conclusion:
The presence of XDR strains of A. baumannii is considerable significant problem in hospitals. Further, similar genetic clonal identified in them indicated the nosocomial infection origin. Hence, these results are very important for control of nosocomial infection committee in health-care systems.
Keywords: Acinetobacter baumannii, extensively drug resistant, Oxa-23, random amplified polymorphic DNA polymerase chain reaction
Introduction
Acinetobacter baumannii is an opportunistic and predominant hospital microorganism that is responsible for the majority of nosocomial infections such as ventilator-associated infection (VAP) and burn wound infections.[1,2,3] Burn patients using ventilator and suffering an immune suppression are prone to get infected. A. baumannii can be associated with a wide range of antibiotic resistances such as third and fourth generations of cephalosporin, carbapenem, and even aminoglycosides.[4,5] Biofilm formation is one of the antibiotic resistance mechanisms in this bacterium which can lead to appearance of multidrug-resistant (MDR) and extensively drug-resistant (XDR) strains. Biofilms are complex mixtures of microbes that are attached to hard surfaces such as wounds of burn patients and ventilator.[6,7] Biofilms can reduce the permeability of antibiotics to bacterium, and as the result, the patients can face MDR and/or XDR bacterium.[6,7] On the other hand, the production of carbapenemase is the other antibiotic resistant that helps bacterium to resistance against all beta-lactam family except for aztreonam. Metallo-bata-lactamase (VIM and IMP), beta-lactamase group A (KPC and GES), and OXA types enzymes are carbapenemase, which can be carbapenem resistant in A. baumannii. Hence, infected patients with MDR and/or XDR strains of A. baumannii in VAP and burn patients can increase mortality and morbidity.[1,2,3,8,9] The detection of MDR and/or XDR strains of A. baumannii is a crucial step for nosocomial infection committees to control infectious spread of such organism. Molecular epidemiology of MDR and XDR A. baumannii with different methods such as molecular subtyping by random amplified polymorphic DNA polymerase chain reaction (RAPD PCR) can be helpful for the determination of genetic relationship of these strains and their possible nosocomial infection origin.[10,11]
The aim of this study was to determine antibiotic resistance patterns of A. baumannii that were isolated from VAP and wound burn infections and also to evaluate biofilms’ formation. Further, we assessed their molecular epidemiology by RAPD PCR in isolated strains to confirm nosocomial infection origin.
Materials and Methods
Sampling area
In this study, 79 samples of A. baumannii (32 VAP and 47 burns) were collected from patients in two of the teaching hospitals in Tehran, Iran, in 2018. Conventional biochemical and microbiological tests, including triple sugar iron, oxidase, and growth on 42°C, were used for phenotypic identification.
Antibiotic susceptibility testing
Antibiotic susceptibility testing was carried out using Kirby–Bauer disc diffusion testing according to the 2015 Clinical and Laboratory Standards Institute guidelines[12] against piperacillin (100 μg), ampicillin/sulbactam (10-10 μg), cefepime (30 μg), ceftazidime (30 μg), cefotaxime (30 μg), imipenem (10 μg), piperacillin–tazobactam (100/10 μg), aztreonam (30 μg), ciprofloxacin (5 μg), gentamicin (10 μg), tobramycin (10 μg), amikacin (30 μg), tetracycline (30 μg), and trimethoprim/sulfamethoxazole (1.25/23.75 μg). The antibiotic disks used in this study were purchased from MAST Company (Mast Diagnostics, UK). Pseudomonas aeruginosa ATCC 27853 was used as control strain in the antibiotic susceptibility testing.
Phenotypic biofilm formation detection
Microtube method
Microtube method was used to determine biofilm formation and described previously.[6,13] This is a qualitative method for biofilm detection. A loop full of test organisms was inoculated in 1 mL of trypticase soy broth with 1% glucose in test tubes. The microtubes were incubated at 37°C for 24 h. After incubation, microtubes were decanted and washed with phosphate-buffered saline (pH 7.3) and dried. Microtubes were then stained with crystal violet (0.1%). Excess stain was washed with deionized water. Microtubes were dried in inverted position. The scoring for tube method was done according to the results of the control strains. Biofilm formation was considered in three types: (i) strong/high biofilm formation, (ii) moderate, and (iii) weak according to mass of visible film lined the wall of microtubes.[6,13]
Polymerase chain reaction amplification for carbapenemase production genes
Bacterial DNA extraction was performed by boiling. The boiling method was described by Higgins et al.[14] PCR for Vim, imp, oxa-23, oxa-48, NDM-1, and SPM-1 genes was performed. The lists of primers used are shown in Table 1.
Table 1.
Primers sequences of oprI, oprL
| Primers | Primer sequence | RCR product size (bp) | References |
|---|---|---|---|
| VIM-F VIM-F |
5’ TTGACACTCCATTTACDG -3’ 5’- GATYGAGAATTAAGCCACYCT -3’ |
390 | [15] |
| IMP-F IMP-R |
5’- GATGGTGTTTGGTCGCATA -3’ 5’- CGAATGCGCAGCACCAG -3’ |
139 | [15] |
| Oxa-23-F Oxa-23-R |
5’- GATGTGTCATAGTATTCGTCGT -3’ 5’- TCACAACAACTAAAAGCACTGT -3’ |
1050 | [8] |
| Oxa-48-F Oxa-48-R |
5’- CCAAGCATTTTTACCCGCATCKACC 5’- GYTTGACCATACGCTGRCTGCG -3’ |
389 | [8] |
| NDM-1-F NDM-1-R |
5’- CCCGGCCACACCAGTGACA -3’ 5’- GTAGTGCTCAGTGTCGGCAT -3’ |
129 | [8] |
| SPM-1-F SPM-1-R |
5’- GGGTGGCTAAGACTATGAAGCC -3’ | 447 | [8] |
| 5’- GCCGCCGAGCTGAATCGG -3’ |
PCR: Polymerase Chain Reaction
Then, the first set of PCR (Vim, imp genes) was performed in the following condition:
DNA thermal cycler was programmed as follows: the first denaturation at 94°C for 10 min and 30 cycles of 94°C for 40 s, annealing at 60°C for 40 s, extension at 72°C for 60 s, and at last, the final extension at 72°C for 7 min.
The second set of PCR (oxa-23, oxa-48 genes) was performed in the following condition:
DNA thermal cycler was programmed as follows: the first denaturation at 94°C for 60 s and 30 cycles of 94°C for 30 s, annealing at 55°C for 40s in, extension at 72°C for 60 s for extension, and the final extension at 72°C for 7 min.
The third set of PCR (NDM-1 and SPM-1) was performed in the following condition:
DNA thermal cycler was programmed as follows: the first denaturation at 94°C for 60 s and 30 cycles of 94°C for 30 s, annealing at 60°C for 40s in, extension at 72°C for 60 s for extension, and the final extension at 72°C for 3 min.
PCR products were analyzed by electrophoresis on agarose 1.5% with SYBR Safe staining.
Molecular typing by random amplified polymorphic DNA polymerase chain reaction
Molecular typing for detection of molecular genetics relationships in Vancomycin-resistant Enterococcus (VRE) strains has been prepared by RAPD PCR with 5′ GCTTGTGAAC 3′.[16] The PCR condition and primers (Macrogen, Seoul, Korea) that were used in this PCR-based molecular typing method have been explained previously.[16] Amplification for random sequences by selected primers was done as follows: initial denaturation step at 94°C for 3 min followed by 34 cycles consisting of denaturation (92°C for 30 s), annealing (40°C for 1 min), extension (72°C for 3 min), and final extension step at 72°C for 10 min. Gel electrophoresis and visualized band patterns have been used for commentary of the results.
Results
Collected isolates were confirmed by microbiology and biochemistry methods. The results of antibiotic susceptibility testing showed that all of the tested strains were XDR except for two strains (one from VAP and one from burns). The rate of resistance to each antibiotic is shown in Tables 2 and 3.
Table 2.
Rate of antibiotic resistance in ventilator-associated pneumonia isolates
| Antimicrobial agent (disk content) | Percentage of resistant isolates (number of 32) | Percentage of intermediate isolates (number of 32) | Percentage of sensitive isolates (number of 32) |
|---|---|---|---|
| Imipenem (10 µg) | 75% (24) | 21.8% (7) | 3.1% (1) |
| Piperacillin (100 µg) | 96.8% (31) | 0% | 3.1% (1) |
| Aztreonam (30 µg) | 68.7% (22) | 15.6% (5) | 15.6% (5) |
| Trimethoprim/sulfamethoxazole (1.25/23.75 µg) | 81.2% (26) | 0% | 18.7% (6) |
| Ampicillin/sulbactam (10-10 µg) | 15.6% (5) | 6.2% (2) | 78.1% (25) |
| Tobramycin (10 µg) | 93.7% (30) | 0% | 6.2% (2) |
| Cefotaxime (30 µg) | 96.8% (31) | 0% | 3.1% (1) |
| Amoxicillin/clavulanic acid (20-10 µg) | 100% (32) | 0% | 0% |
| Tetracycline (30 µg) | 90.6% (29) | 0% | 9.3% (3) |
| Piperacillin/tazobactam (100-10 µg) | 90.6% (29) | 6.2% (2) | 3.1% (1) |
| Amikacin (30 µg) | 65.6% (21) | 21.8% (7) | 12.5% (4) |
| Ciprofloxacin (5 µg) | 96.8% (31) | 0% | 3.1% (1) |
| Ceftazidime (30 µg) | 96.8% (31) | 0% | 3.1% (1) |
| Gentamicin (10 µg) | 87.5% (28) | 6.2% (2) | 6.2% (2) |
| Cefepime (30 µg) | 78.1% (25) | 12.5% (4) | 9.3% (3) |
Table 3.
Rate of antibiotic resistance in burn isolates
| Antimicrobial agent (disk content) | Percentage of resistant isolates (number of 47) | Percentage of intermediate isolates (number of 47) | Percentage of sensitive isolates (number of 47) |
|---|---|---|---|
| Imipenem (10 µg) | 82.9% (39) | 12.7% (6) | 4.2% (2) |
| Piperacillin (100 µg) | 97.8% (46) | 0% | 2.1% (1) |
| Aztreonam (30 µg) | 93.6% (44) | 2.1% (1) | 4.2% (2) |
| Trimethoprim/sulfamethoxazole (1.25/23.75 µg) | 95.7% (45) | 0% | 4.2% (2) |
| Ampicillin/sulbactam (10-10 µg) | 8.5% (4) | 0% | 91.4% (43) |
| Tobramycin (10 µg) | 87.2% (41) | 2.1% (1) | 10.6% (5) |
| Cefotaxime (30 µg) | 97.8% (46) | 0% | 2.1% (1) |
| Amoxicillin/clavulanic acid (20-10 µg) | 100% (47) | 0% | 0% |
| Tetracycline (30 µg) | 63.8% (30) | 4.2% (2) | 31.9% (15) |
| Piperacillin/tazobactam (100-10 µg) | 63.8% (30) | 31.9% (15) | 4.2% (2) |
| Amikacin (30 µg) | 91.4% (43) | 6.3% (3) | 2.1% (1) |
| Ciprofloxacin (5 µg) | 95.7% (45) | 2.1% (1) | 2.1% (1) |
| Ceftazidime (30 µg) | 97.8% (46) | 0% | 2.1% (1) |
| Gentamicin (10 µg) | 95.7% (45) | 2.1% (1) | 2.1% (1) |
| Cefepime (30 µg) | 93.6% (44) | 4.2% (2) | 2.1% (1) |
Biofilm formation was classified as weak, moderate, and strong. The results of tube test indicated that 64% of strains produced strong and moderate biofilm formation in these XDR isolates [Table 4].
Table 4.
Amounts of biofilm formation
| Tube method | Percentage of strains (n) in VAP strains | Percentage of strains (n) in burn strains |
|---|---|---|
| Weak | 37.5% (12) | 4.8% (7) |
| Moderate | 34.3% (11) | 34% (16) |
| High/strong | 28.1% (9) | 51% (24) |
Oxa-23 gene was identified in 62 (78%) of strains, as the most detected gene [Figure 1]. Vim, imp, oxa-48, NDM-1, and SPM-1 genes were not discovered in any of the strains.
Figure 1.
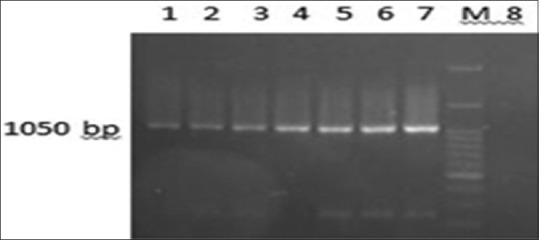
Oxa-23 gene. 1- 6: Positive strains, 7: Positive control, M: DNA ladder (DM2300), 8: Negative control
The results of RAPD PCR, as a molecular epidemiology method, showed 10 different bacterial clones in VAP strains and 12 clones in burns. However, 50% of the VAP isolates were related to two special clones, and 79% of burn isolates were associated with two specific clones.
Discussion
A. baumannii was identified as an opportunistic pathogen in recent years. The appearance of MDR and/or XDR strains of A. baumannii can be related to overusage or inappropriate usage of extended-spectrum antibiotics.[1,3] The presence of A. baumannii in hospital environment in long term can lead to its spread in hospital environment and antibiotic resistance gene transfer.[1,3] Resistance to broad-spectrum antibiotics can be due to carbapenemse-producing bacteria. On the other hand, the most prevalent carbapenemase genes are located in mobile genetic elements and can transfer to other bacteria.[1,3,8] Patients in intensive care unit are at high risk to get infected by nosocomial agents[4,5] and ultimately can lead to VAP and burn wounds infection.[4,5] Carbapenem resistant due to oxa type production has been increasingly noted in the recent studies.[1,8,15] Producing oxa-23 enzymes is one of the important mechanisms in carbapenem-resistant strains of A. baumannii. In this study, 78% of strains produced oxa-23 which can confirm the high prevalence of oxa-23 in A. baumannii. Azimi et al. in Iran in 2015 indicated that 80% of carbapenem-resistant A. baumannii isolated from burn patients have oxa-23 gene.[1] Similarly, Nowak et al. in Greece in 2017 reported that 80% oxa-23 produce A. baumannii isolated from VAP specimens.[15] The results of Royer et al.'s study in Brazil in 2015 showed that 100% of isolated carbapenem-resistant A. baumannii from VAP harbored oxa-23.[10] Another study from Iran by Mohammadi et al. in 2016 confirmed that all isolated carbapenem-resistant strains of A. baumannii isolated from VAP had oxa-23 gene.[9] The results of these studies[1,9,10,16] are similar to the recent research that confirmed the high prevalence of oxa-23 compared to other carbapenemase in carbapenem-resistant A. baumannii. The high rate of isolated XDR A. baumannii formVAP and burns is one of alarming result in this study. At least 64% and 78% of strains collected from burns and VAP, respectively, were resistant to 14-tested antibiotics. These findings are very considerable for control of nosocomial infection committees in hospitals. All of 79 isolates were XDR, and only 36% of them were categorized as a weak biofilm formation group. This result may indicate that biofilm formation can increase antibiotic resistance next to the other mechanisms of resistance. This was conducted by Krzyściak et al. in Netherland in 2017. The authors showed more antibiotic susceptibility in strong biofilm formation A. baumannii.[11] These results are not comparable to our findings. The increase of antibiotic resistance and the emergence of MDR strains due to decrease of antibiotic permeability because of biofilm formation can be an accepted reason. Molecular epidemiology confirmed two dominant clonal groups in VAP and also burn isolates that include the majority of strains in each group of specimens. The presence of genetic relationship in collected bacteria from VAP and burns may confirm nosocomial infection. The results of RAPD PCR determined the presence of two different predominant clonal groups in VAP and burns. This result can confirm the necessity of molecular epidemiology study in each hospital.
Conclusion
The results of this study showed the high prevalence of MDR and XDR strains of A. baumannii as an important cause of health-care association infection. On the other hand, genetic relationship in XDR strains can confirm the nosocomial infection.
Limitation
In this study, 79 samples of A. baumannii (VAP: 32, burns: 47) were survived. The numbers of VAP isolates are few. VAP specimens in the selected educational hospital was low because it is very difficult to collect. On the other hand, because of budget limitation, we could not work on more burn specimens.
Financial support and sponsorship
Research reported in this publication was supported by Elite Researcher Grant Committee under award number (963355) from the National Institutes for Medical Research Development, Tehran, Iran.
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
Research reported in this publication was supported by Elite Researcher Grant Committee under award number (963355) from the National Institutes for Medical Research Development, Tehran, Iran.
References
- 1.Azimi L, Talebi M, Pourshafie MR, Owlia P, Rastegar Lari A. Characterization of carbapenemases in extensively drug resistance Acinetobacter baumannii in a burn care center in Iran. Int J Mol Cell Med. 2015;4:46–53. [PMC free article] [PubMed] [Google Scholar]
- 2.Owlia P, Azimi L, Gholami A, Asghari B, Lari AR. ESBL- and MBL-mediated resistance in Acinetobacter baumannii: A global threat to burn patients. Infez Med. 2012;20:182–7. [PubMed] [Google Scholar]
- 3.Azimi L, Motevallian A, Ebrahimzadeh Namvar A, Asghari B, Lari AR. Nosocomial infections in burned patients in Motahari hospital, Tehran, Iran. Dermatol Res Pract. 2011;2011:436952. doi: 10.1155/2011/436952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garnacho-Montero J, Ortiz-Leyba C, Fernández-Hinojosa E, Aldabó-Pallás T, Cayuela A, Marquez-Vácaro JA, et al. Acinetobacter baumannii ventilator-associated pneumonia: Epidemiological and clinical findings. Intensive Care Med. 2005;31:649–55. doi: 10.1007/s00134-005-2598-0. [DOI] [PubMed] [Google Scholar]
- 5.Tsakiridou E, Makris D, Daniil Z, Manoulakas E, Chatzipantazi V, Vlachos O, et al. Acinetobacter baumannii infection in prior ICU bed occupants is an independent risk factor for subsequent cases of ventilator-associated pneumonia. Biomed Res Int. 2014;2014:193516. doi: 10.1155/2014/193516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bose S, Khodke M, Basak S, Mallick SK. Detection of biofilm producing staphylococci: Need of the hour. J Clin Diagn Res. 2009;3:1915–20. [Google Scholar]
- 7.Babapour E, Haddadi A, Mirnejad R, Angaji SA, Amirmozafari N. Biofilm formation in clinical isolates of nosocomial Acinetobacter baumannii and its relationship with multidrug resistance. Asian Pac J Trop Biomed. 2016;6:528–33. [Google Scholar]
- 8.Liu S, Wang Y, Xu J, Li Y, Guo J, Ke Y, et al. Genome sequence of an OXA23-producing, carbapenem-resistant Acinetobacter baumannii strain of sequence type ST75. J Bacteriol. 2012;194:6000–1. doi: 10.1128/JB.01440-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohammadi F, Goudarzi H, Hashemi A, Yousefi Nojookambari N, Khoshnood S, Sabzehali F. Detection of iSabai in Acinetobacter baumannii strains carrying oxa genes isolated from Iranian burn patients. Arch Pediatr Infect Dis. 2017;5:e39307. [Google Scholar]
- 10.Royer S, Faria AL, Seki LM, Chagas TP, Campos PA, Batistão DW, et al. Spread of multidrug-resistant Acinetobacter baumannii and Pseudomonas aeruginosa clones in patients with ventilator-associated pneumonia in an adult intensive care unit at a university hospital. Braz J Infect Dis. 2015;19:350–7. doi: 10.1016/j.bjid.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krzyściak P, Chmielarczyk A, Pobiega M, Romaniszyn D, Wójkowska-Mach J. Acinetobacter baumannii isolated from hospital-acquired infection: Biofilm production and drug susceptibility. APMIS. 2017;125:1017–26. doi: 10.1111/apm.12739. [DOI] [PubMed] [Google Scholar]
- 12.Performance Standards for Antimicrobial Susceptibility Testing: Nineteenth Informational Supplement M100–S21. Wayne, PA: Clinical Laboratory Standards Institute; 2017. Clinical Laboratory Standards Institute. [Google Scholar]
- 13.Hassan A, Usman J, Kaleem F, Omair M, Khalid A, Iqbal M. Evaluation of different detection methods of biofilm formation in the clinical isolates. Braz J Infect Dis. 2011;15:305–11. [PubMed] [Google Scholar]
- 14.Higgins PG, Lehmann M, Wisplinghoff H, Seifert H. GyrB multiplex PCR to differentiate between Acinetobacter calcoaceticus and Acinetobacter genomic species 3. J Clin Microbiol. 2010;48:4592–4. doi: 10.1128/JCM.01765-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nowak P, Paluchowska P, Budak A. Distribution of blaOXA genes among carbapenem-resistant Acinetobacter baumannii nosocomial strains in Poland. New Microbiol. 2012;35:317–25. [PubMed] [Google Scholar]
- 16.Carr E, Eason H, Feng S, Hoogenraad A, Croome R, Soddell J, et al. RAPD-PCR typing of Acinetobacter isolates from activated sludge systems designed to remove phosphorus microbiologically. J Appl Microbiol. 2001;90:309–19. doi: 10.1046/j.1365-2672.2001.01245.x. [DOI] [PubMed] [Google Scholar]


