Abstract
Kruppel-like factor 4 (KLF4) has been implicated in a number of different types of cancer; however, the role of KLF4 in papillary thyroid cancer remains elusive. The present study aimed to investigate the role of KLF4 in papillary thyroid cancer and its potential underlying molecular mechanisms. The expression of KLF4 in thyroid tumor tissue and adjacent non-cancerous tissues were detected via immunohistochemistry and western blotting. The papillary thyroid cancer cell line, KTC1, was transfected with viruses carrying KLF4 overexpression vectors. The relative expression of KLF4, E-cadherin, N-cadherin, Vimentin, matrix metalloproteinase (MMP)2, MMP9 and collagen was detected via quantitative-PCR. The viability of KTC1 cells was detected using a cell counting kit-8 assay at 24, 48 and 72 h. Cell invasion was examined via a transwell invasion assay. Cell migration was examined via a scratch migration assay at 0 and 24 h. Compared with adjacent non-cancerous tissues, the expression of KLF4 was significantly lower in thyroid tumor tissues. The expression of KLF4 in KTC1 cells were significantly increased compared with the blank or negative control groups. The expression of N-cadherin, MMP2, MMP9 and collagen was significantly decreased in the KLF4 overexpression group. The viability of KTC1 cells was markedly decreased in KLF4 overexpression group at 24, 48 and 72 h when compared with the blank or negative control groups. The invasion of KTC1 cells in the KLF4 overexpression group was markedly decreased. Compared with the negative control group, the KTC1 cell migration in the KLF4 overexpression group was markedly decreased at 24 h. The expression of KLF4 was also significantly lower in thyroid tumor tissues. The cell viability, tumor invasion and migration ability and expression levels of N-cadherin, MMP2, MMP9 and collagen in papillary thyroid cancer cells were markedly decreased with KLF4 overexpression.
Keywords: thyroid cancer, KTC1, Kruppel-like factor 4, tumor invasion
Introduction
There were >3,000,000 cases of thyroid cancer worldwide in 2015 and 31,900 deaths occurred as a result in the same year (1). Thyroid cancer most commonly occurs between the ages of 35 and 65 years, with women affected more frequently (2). Papillary thyroid cancer is the most common type of thyroid cancer, accounting for 75–85% of all thyroid cancer cases (3). Its incidence has tripled between 1975 and 2012, with the incidence in 2012 being 14.9 per 100,000 individuals (4). Papillary thyroid cancer is the most common thyroid cancer in children, as well as in those who had previously received radiation therapy to the head and neck (5). It has also been observed in patients with a family history of syndromes, including multiple endocrine neoplasia type 2 and familial adenomatous polyposis (6).
Kruppel-like factor 4 (KLF4) has been implicated in a number of different types of cancer, such as gastric cancer (7), lung cancer (8) and colorectal cancer (9). MicroRNA (miR)-7 has been previously demonstrated to suppress the metastasis of breast cancer stem-like cells into the brain via KLF4 modulation (10). Epigenetic inactivation of KLF4 has been reported to be associated with the progression and early recurrence of urothelial cancer (11). The KLF4/Musashi 2 signaling pathway is also known to regulate the growth and metastasis of pancreatic cancer (12). Additionally, KLF4 was revealed to suppress the proliferation of estrogen-dependent breast cancer by inhibiting the transcriptional activity of estrogen receptor-α (13). miR-25 has been reported to enhance non-small cell lung cancer cell migration and invasion by inhibiting KLF4 via the extracellular signal-regulated kinase (ERK) signaling pathway (14). The progestin-induced suppression of miR-29 has been demonstrated to promote the dedifferentiation of breast cancer cells via KLF4 (15). However, the role of KLF4 in papillary thyroid cancer remains elusive.
E-cadherin is an adhesion molecule that suppresses the invasion of various tumor cells (16). N-cadherin is also a transmembrane protein that functions to mediate cell-cell adhesion (17). The co-expression of E-cadherin and Vimentin is associated with the invasion and metastasis of breast cancer (18). Additionally, matrix metalloproteinase (MMP) 2 was revealed to be suppressed by regulatory T cells and was determined to be involved the regulation of urinary bladder cancer cell invasion (19). The gene expression of MMP9 is regulated by epigenetic modifications in breast cancer (20). Collagen was also revealed to be elevated in the serum of patients with non-small cell lung cancer (21). However, how KLF4 affects the expression of E-cadherin, N-cadherin, Vimentin, MMP2, MMP9 and collagen in papillary thyroid cancer remains unclear.
Therefore, the present study aimed to investigate the role of KLF4 in papillary thyroid cancer and to determine potential underlying molecular mechanisms.
Materials and methods
Reagents and patients
Primers and probes, TRIzol® reagent, SuperScript III Reverse Transcriptase, SYBR qPCR mix kit was purchased from Invitrogen; Thermo Fisher Scientific, Inc. DMEM was purchased from Gibco (Thermo Fisher Scientific, Inc.).
Thyroid cancer tissues together with the adjacent non-tumor tissue were surgically removed from 8 patients who were admitted to Baoshan District Integrated Traditional Chinese and Western Medicine Hospital (Shanghai, China) between December 2016 and November 2017. The distance between adjacent non-tumor tissue and the boundary of the cancer tissue was ~1 cm. All patients were diagnosed with papillary thyroid cancer and their clinicopathological characteristics were recorded (Table I) (22,23). After the study was explained, all patients provided written informed consent. The present study was approved by the Institutional Research Board of Baoshan District Integrated Traditional Chinese and Western Medicine Hospital (Shanghai, China).
Table I.
Clinicopathological features of patients in the present study.
| Patient no. | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|
| Age (years) | 66 | 50 | 57 | 35 | 55 | 48 | 48 | 52 |
| Sex | F | F | F | M | M | F | F | F |
| TNM | T1aN0M0 | T1aN1M0 | T1N1M0 | T1aN1M0 | T1aN1M0 | T1aN0M0 | T1bN0M0 | T1bN0M0 |
| Tumor stage | I | III | III | I | III | I | I | I |
Immunohistochemistry (IHC)
Tumor tissues and adjacent non-tumor tissues were embedded in paraffin and cut into slides (5 µm thick). Slides were fixed in 4% paraformaldehyde for 10 min at room temperature, blocked with 5% bovine serum albumin (Bio-west, Inc.) at room temperature for 30 min and incubated with primary antibodies against KLF4 (1:500; cat. no. Bs-1064R; Bioss Antibodies, Inc.) at 4°C overnight. Following overnight incubation, slides were washed with PBS and incubated in the dark with FITC-conjugated goat anti-rabbit secondary antibodies (1:1,000; cat. no. ab6717; Abcam) at room temperature for 1 h. Slides were subsequently washed with PBS (3 times; each, 30 sec). Slides were prepared with mounting media and Antifades (Invitrogen; Thermo Fisher Scientific, Inc.) and observed using a fluorescence microscope (magnification, ×100). The mean intensity, calculated by multiplying the area (size) and average density of fluorescence, was evaluated using Image-Pro Plus software (version 7; Media Cybernetics, Inc.).
Western blot analysis
The protein expression of KLF4 in tumor tissues and adjacent non-cancerous tissues was detected via western blot analysis. In addition, the protein expression of N-cadherin, MMP2, MMP9 and collagen in KTC1 cells were detected via western blotting. Tissues were digested and lysed in lysis buffer (cat. no. P0013; Beyotime Institute of Biotechnology) at 4°C with inhibitors of phosphatase and protease (cat. no. P1045; Beyotime Institute of Biotechnology). The lysis mixture was centrifuged at 4°C for 10 min at 10,000 × g and the supernatant containing cellular proteins was utilized in the following experiments. Protein concentration was determined using a BCA kit. Proteins were separated via SDS-PAGE (10% gel; 40 µg loaded per lane; 120 V). Separated proteins were then transferred to PVDF membranes (100 V for 120 min; Beyotime Institute of Biotechnology), which were subsequently blocked with 5% non-fat milk at room temperature for 1 h. Membranes were then incubated with the following primary antibodies obtained from Abcam at 4°C overnight: Anti-KLF4, anti-N-cadherin (cat. no. ab18203), anti-MMP2 (cat. no. ab97779), anti-MMP9 (cat. no. ab228402), anti-collagen (cat. no. ab138492), anti-GAPDH (cat. no. ab181602) and anti-β-actin (cat. no. ab8227; all 1:1,000). Membranes were washed with Tris-buffered saline containing Tween 20 and incubated with horseradish peroxidase-conjugated goat anti-rabbit secondary antibodies (cat. no. ab6721; 1:2,000; Abcam) at room temperature for 1 h. Membranes were incubated in enhanced chemiluminescence solution (Beyotime Institute of Biotechnology). Images were captured on film (Beyotime Institute of Biotechnology) in a dark room. Experiments were repeated three times. The western blot images were quantified in greyscale using ImageJ software (version 1.5.2; National Institutes of Health).
Construction of recombinant plasmids and lentiviral packaging
cDNA sequence of KLF4 (NM_004235) was synthesized and subcloned into lentivirus vector pL6.3-CMV-GFPa1-IRES-MCS (Novobio) for lentivirus production. The KLF4 recombinant lentivirus vector, pL6.3-CMV-GFPa1-IRES-KLF4, was confirmed by Sanger sequencing (24).
Packaging mix (9 µg; Novobio) and KLF4 recombinant lentiviral plasmids (3 µg) were added into Opti-Minimum Essential Medium (Opti-MEM; Thermo Fisher Scientific Inc.) and mixed. Lipofectamine™ 2000 (36 µl; Thermo Fisher Scientific Inc.) was mixed with Opti-MEM (1.5 ml) and incubated at room temperature for 5 min. The plasmid solution and diluted Lipofectamine 2000 were then mixed and incubated at room temperature for 5 min. The mixture was added into a culture dish with 293 T cells (Novobio), and cells were cultured for 48 h. Cell supernatants were then collected, centrifuged at 1,500 × g for 10 min at room temperature and filtered. The lentivirus solution was then condensed via centrifugation at 50,000 × g for 2 h at 4°C and re-suspended in DMEM. KLF4 recombinant lentivirus was derived.
The human papillary thyroid carcinoma cell line KTC1 (3×105 cells/well in six-well plates) was transfected with the pL6.3-CMV-GFPa1-IRES-KLF4 and pL6.3-CMV-GFPa1-IRES-MCS (control). The transduction MOI was 30. Quantitative PCR (qPCR) was utilized to detect the efficiency of KLF4 overexpression after 48 h. There was no transfection in blank group.
Reverse transcription-quantitative PCR (RT-qPCR)
The expression of E-cadherin, N-cadherin, Vimentin, MMP2, MMP9 and collagen in KTC1 cells was detected via qPCR. Total RNA was extracted using TRIzol® reagent, according to the manufacturer's protocol. A universal cDNA synthesis kit (Invitrogen Thermo Fisher Scientific, Inc.) was utilized for reverse transcription at 42°C for 1 h. Each reaction contained 0.5 µl random hexamers primers (dN6; 0.2 µg/µl; Novobio) and 1 µl SuperScript III reverse transcriptase (200 U/µl). The specific primers used are listed in Table II. PCR was performed using a SYBR qPCR mix kit (Invitrogen; Thermo Fisher Scientific, Inc.). The PCR conditions were as follows: Pre-denaturation at 95°C for 2 min; 40 cycles of denaturation at 95°C for 10 sec annealing at 60°C for 30 sec and polymerization at 70°C for 45 sec. qPCR was performed using a CFX96 Touch™ Real-Time PCR Detection system (Bio-Rad Laboratories, Inc.). Gene expression was determined and normalized to β-actin. The primer used for rat β-actin was as follows: Forward, 5′-AGGGAAATCGTGCGTGAC-3′ and reverse, 5′-CGCTCATTGCCGATAGTG-3′. The 2−∆∆Cq method was utilized to measure PCR results (25).
Table II.
Primers for quantitative PCR.
| Primer | Sequences (5′ to 3′) |
|---|---|
| KLF4-F | TTCCCATCTCAAGGCACACC |
| KLF4-R | CATGTGTAAGGCGAGGTGGT |
| MMP2-F | GATACCCCTTTGACGGTAAGGA |
| MMP2-R | CCTTCTCCCAAGGTCCATAGC |
| MMP9-F | GTACTCGACCTGTACCAGCG |
| MMP9-R | TTCAGGGCGAGGACCATAGA |
| Collagen I-F | AGTGGTTTGGATGGTGCCAA |
| Collagen I-R | GCACCATCATTTCCACGAGC |
| Vimentin (VIM)-F | TGGACCAGCTAACCAACGAC |
| Vimentin (VIM)-R | GCCAGAGACGCATTGTCAAC |
| E-cadherin (CDH1)-F | TCATGAGTGTCCCCCGGTAT |
| E-cadherin (CDH1)-R | TCTTGAAGCGATTGCCCCAT |
| N-cadherin (CDH2)-F | TGACAATGACCCCACAGCTC |
| N-cadherin (CDH2)-R | GTCCTGCTCACCACCACTAC |
Cell viability assay
The viability of KTC1 cells was measured using a cell counting kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.) cell viability assay at 24, 48 and 72 h after KTC1 cells were transfected with the aforementioned viruses. CCK-8 solution was added to each well and incubated at 37°C for 4 h. The absorbance was subsequently measured using a microplate reader at 490 nm. Relative tumor cell viability rate was calculated by dividing the reading of each group at 24, 48 and 72 h by the baseline reading at 0 h. Experiments were repeated three times.
Transwell invasion assay
The membrane of the upper compartment was coated with Matrigel (1 g/l; 50 µl), which was allowed to solidify via incubation at 37°C for 1 h. KTC1 cell suspension (1×104 cells/ml, 200 µl) in 2% DMEM was added to the upper compartment of each Transwell insert, while 800 µl DMEM with 10% FBS (HyClone; GE Healthcare Life Sciences) was added to the lower compartment. Cells were incubated at 37°C for 24 h. Subsequently, 4% paraformaldehyde was utilized to fix cells on the microporous membrane at room temperature for 30 min. Cells on the lower side of the membrane were stained with 1% crystal violet at room temperature for 10 min and washed with PBS twice. Cells were then observed under Olympus IX50 fluorescent microscope (magnification, ×400; Olympus Corporation) and the number of cells that had transgressed through the membrane was counted. Relative tumor cell invasion was calculated by dividing the average number of cells that invaded through the membrane in the experimental groups by that in the blank group. Experiments were repeated three times.
Scratch migration assay
A confluent monolayer of KTC1 cells was used in the scratch migration assay. A marker pen was used to draw a straight line at the back of plate. Pippet tips were utilized to draw scratch lines vertical to the straight line on the second day. PBS was used to wash cells three times and DMEM without serum was added. Images were taken under Olympus IX50 (magnification, ×400) and cultured in an incubator with 5% CO2 at 37°C. Images were then taken at 24 h and migration distances were calculated under the same field using Image-Pro Plus software. Migration distance at 24 h relative to 0 h was recorded for both negative control and overexpression groups. Then relative migration distances were normalized to the negative control group.
Statistical analysis
Experiments were repeated three times. Statistical data was analyzed using GraphPad Prism software (version 5.0; GraphPad Software Inc.). The results are presented as the mean ± standard deviation. Differences among more than three groups were compared by a one-way analysis of variance followed by the Bonferroni post-hoc test. Differences between two groups were compared using Student's unpaired t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of KLF4 is significantly lower in thyroid tumor tissue
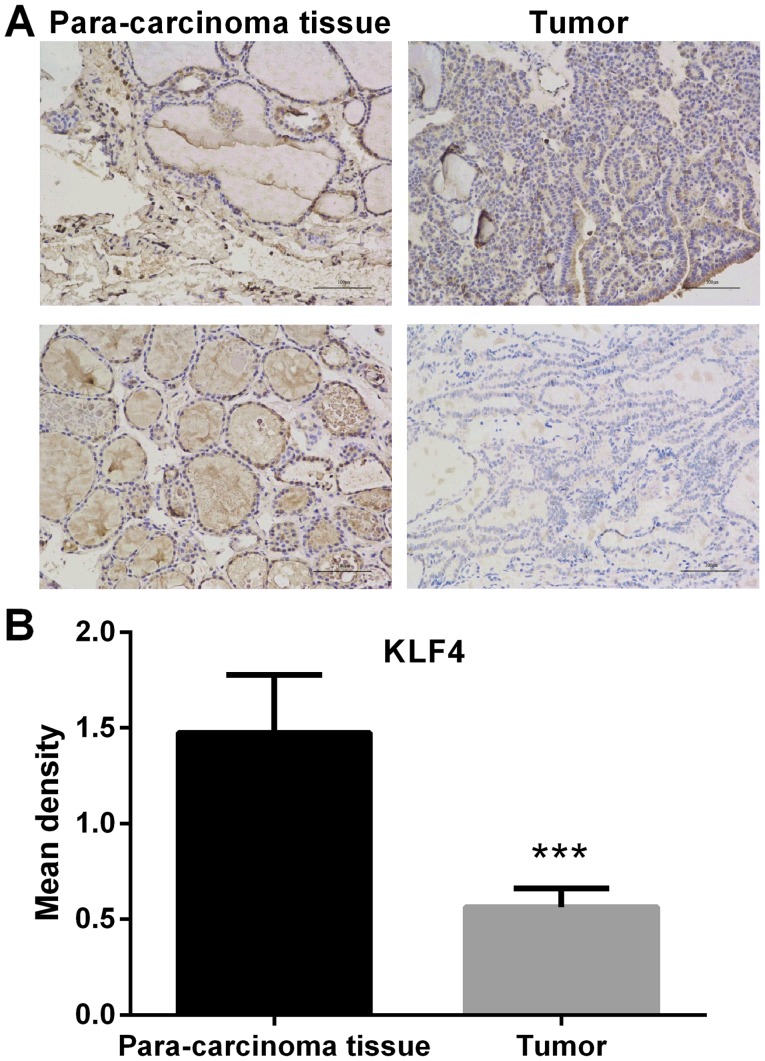
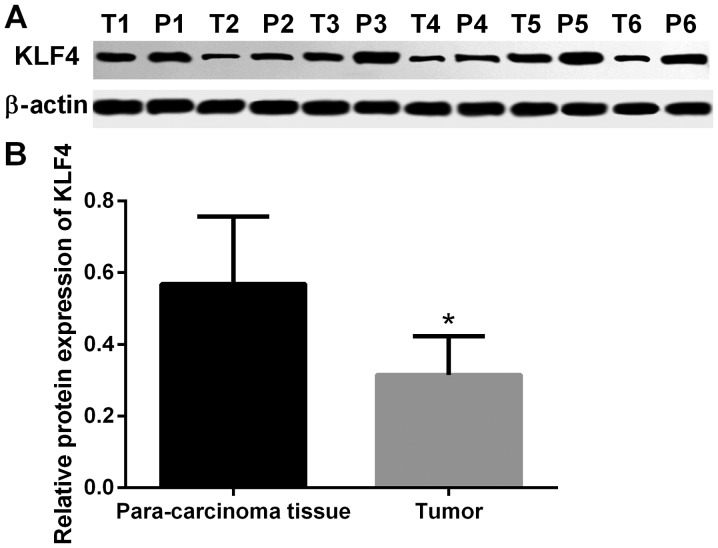
The expression of KLF4 in thyroid tumor tissue and paracarcinoma tissue was detected by IHC. In the microscopic image of KLF4 staining (Fig. 1), positive cell nucleus was stained by diaminobenzidine (DAB) and appeared as brown. Negative cell nucleus was stained by hematoxylin and appeared as blue. Compared with adjacent non-cancerous tissue, the protein expression of KLF4 was significantly lower in thyroid tumor tissue (P<0.001; Fig. 1). In addition, the protein expression of KLF4 in tumor and adjacent non-cancerous tissue was detected via western blot analysis. The protein expression of KLF4 was markedly lower in thyroid tumor tissue when compared with adjacent non-cancerous tissues (P<0.05; Fig. 2).
Figure 1.
IHC images of thyroid tumor tissue and paracarcinoma tissue. (A) Representative IHC image. Scale bar=50 µm. (B) Statistical analysis of immunohistochemistry results. Expression of KLF4 in thyroid tumor tissue and paracarcinoma tissue was detected by IHC. Compared with the adjacent non-cancerous tissues, the expression of KLF4 was significantly lower in thyroid tumor tissue (mean ± standard deviation; n=3 per group). ***P<0.001 vs. paracarcinoma tissue. IHC, immunohistochemistry; KLF4, Kruppel-like factor 4.
Figure 2.
Western blot images of thyroid tumor tissue and paracarcinoma tissue. (A) Results of western blotting and (B) subsequent statistical analysis. The protein expression of KLF4 in tumor tissue and adjacent non-tumor tissue was detected via western blotting. The expression of KLF4 was markedly lower in thyroid tumor tissue compared with paracarcinoma tissue (mean ± standard deviation; n=6 per group). *P<0.05 vs. paracarcinoma tissue. KLF4, Kruppel-like factor 4; T, tumor; P, paracarcinoma.
Confirmation of KLF4 overexpression
KTC1 cells were transfected with viruses carrying KLF4 overexpression vectors. The relative expression of KLF4 was detected via qPCR. The expression of KLF4 in KTC1 cells was significantly increased compared with the blank or negative control groups (P<0.001; Fig. 3).
Figure 3.
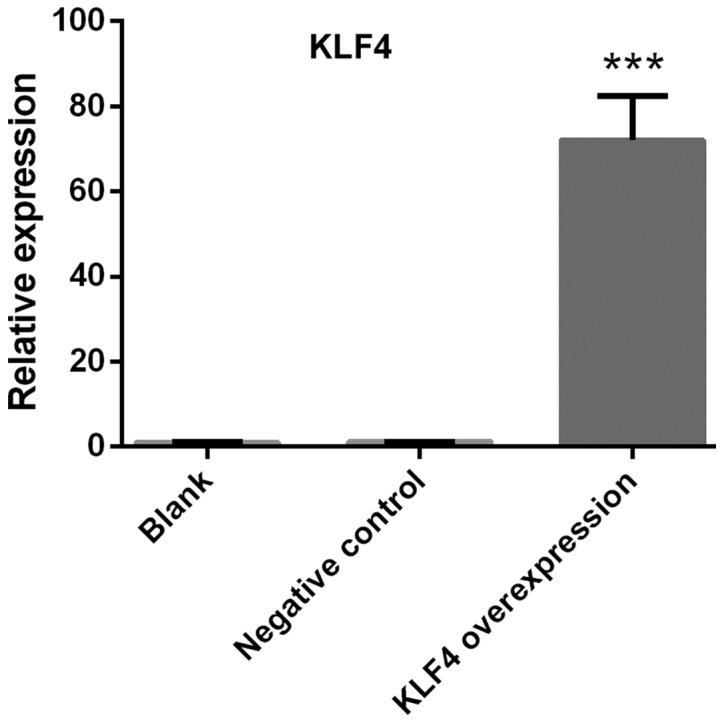
Confirmation of KLF4 overexpression. Papillary thyroid cancer KTC1 cells were transfected with viruses carrying KLF4 overexpression vectors. The relative expression of KLF4 was detected via quantitative PCR. The expression of KLF4 in KTC1 cells was significantly increased compared with the blank or negative control groups (mean ± standard deviation; n=3 per group). ***P<0.001 compared with the blank or negative control groups.
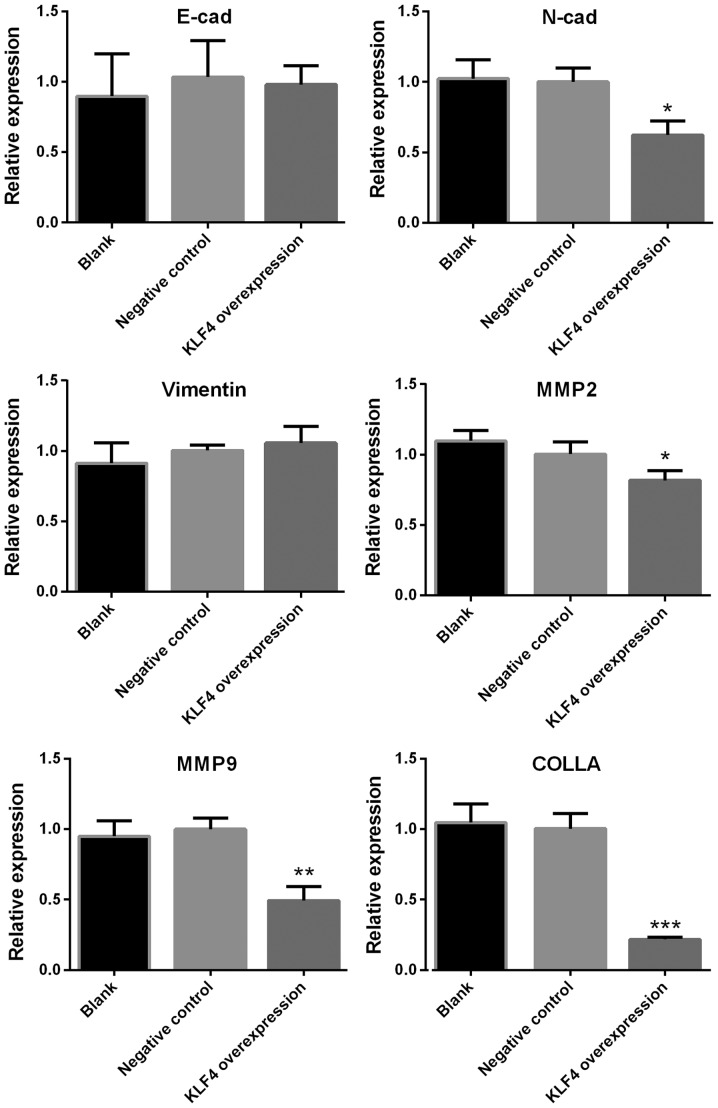
Expression of N-cadherin, MMP2, MMP9 and collagen is significantly decreased in the KLF4 overexpression group
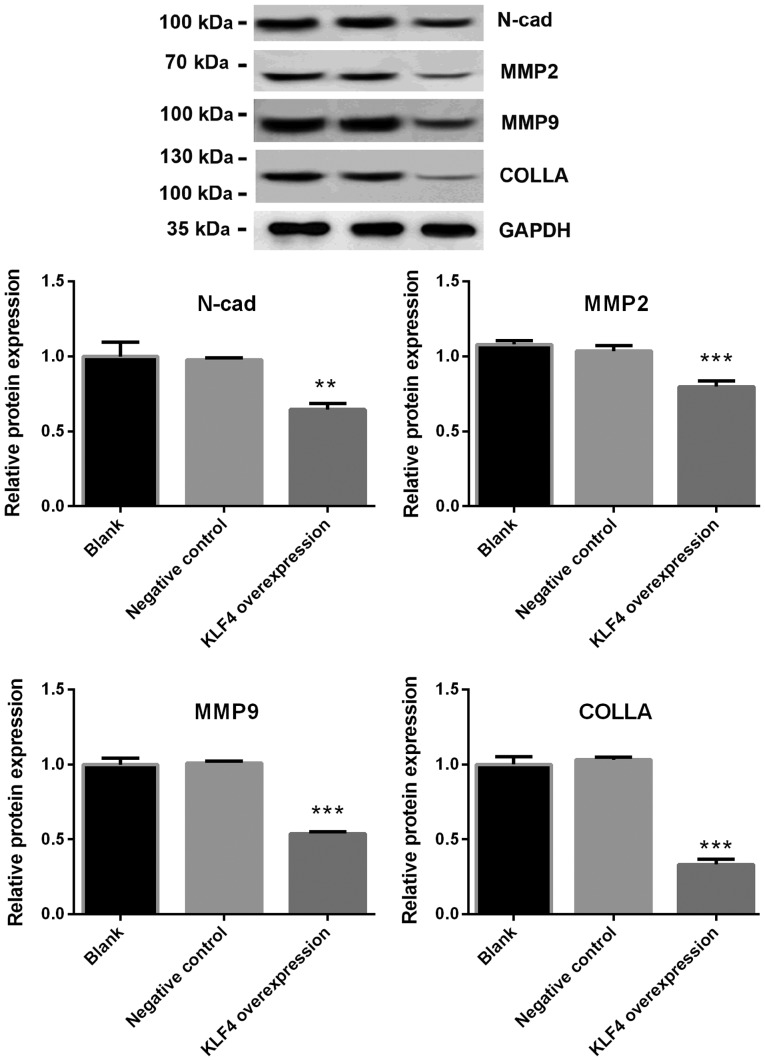
The mRNA levels of E-cadherin, N-cadherin, Vimentin, MMP2, MMP9 and collagen in KTC1 cells were detected via qPCR. The protein expression of N-cadherin, MMP2, MMP9 and collagen in KTC1 cells were confirmed by western blotting. Among all the genes screened, the mRNA expression of N-cadherin, MMP2, MMP9 and collagen were significantly decreased in the KLF4 overexpression group when compared with the blank or negative control groups (P<0.05 for N-cadherin and MMP2; P<0.01 for MMP9; P<0.001 for collagen; Fig. 4). No significant differences were observed in the levels of E-cadherin and Vimentin (Fig. 4). In addition, the protein expression of N-cadherin, MMP2, MMP9 and collagen were significantly decreased in the KLF4 overexpression group when compared with blank or negative control group (P<0.01 for N-cadherin; P<0.001 for MMP-2, MMP-9 and collagen; Fig. 5).
Figure 4.
mRNA expression of N-cadherin, MMP2, MMP9 and collagen was significantly decreased in the KLF4 overexpression group. mRNA expression of E-cadherin, N-cadherin, Vimentin, MMP2, MMP9 and collagen in KTC1 cells were detected via quantitative PCR. Among all genes screened, the expression levels of N-cadherin, MMP2, MMP9 and collagen were significantly decreased in the KLF4 overexpression group, as compared with the blank or negative control groups (mean ± standard deviation; n=3 per group). *P<0.05, **P<0.01 and ***P<0.001 compared with the control or negative control group. MMP, matrix metalloproteinase; KLF4, Kruppel-like factor 4; COLLA, collagen; cad, cadherin.
Figure 5.
Protein expression of N-cadherin, MMP2, MMP9 and collagen was significantly decreased in the KLF4 overexpression group. Protein expression of E-cadherin, N-cadherin, Vimentin, MMP2, MMP9 and collagen in KTC1 cells were detected via western blotting. The protein expression of N-cadherin, MMP2, MMP9 and collagen was significantly decreased in the KLF4 overexpression group compared with the blank or negative control groups (mean ± standard deviation; n=3 per group). **P<0.01 and ***P<0.001 vs. the control or negative control groups. MMP, matrix metalloproteinase; KLF4, Kruppel-like factor 4; COLLA, collagen; N-cad, N-cadherin.
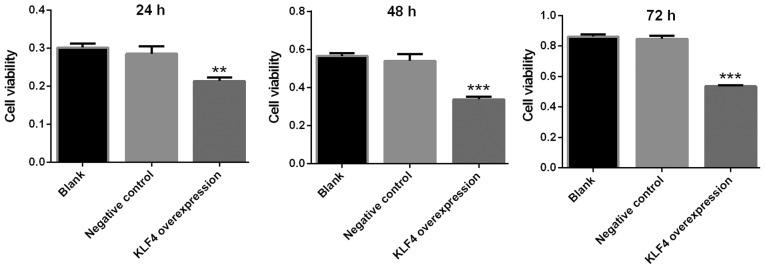
Viability of papillary thyroid cancer cells is markedly decreased in the KLF4 overexpression group
The viability of KTC1 cells was detected via a CCK-8 assay at 24, 48 and 72 h. The viability of KTC1 cells was markedly decreased in the KLF4 overexpression group at 24, 48 and 72 h when compared with the blank or negative control group (P<0.01 for 24 h; P<0.001 for 48 and 72 h; Fig. 6).
Figure 6.
Viability of papillary thyroid cancer cells was markedly decreased in the KLF4 overexpression group. The viability of KTC1 cells was detected using a cell counting kit-8 assay at 24, 48 and 72 h. The viability of KTC1 cells was markedly decreased in the KLF4 overexpression group at 24, 48 and 72 h when compared with the blank or negative control groups (mean ± standard deviation; n=3 per group). **P<0.01 and ***P<0.001 vs. the blank or negative control groups. KLF4, Kruppel-like factor 4.
Invasion of papillary thyroid cancer cells in the KLF4 overexpression group is significantly decreased
Cell invasion was investigated using a transwell invasion assay. Compared with the negative control group, the invasion of KTC1 cells in the KLF4 overexpression group were significantly decreased (P<0.01; Fig. 7).
Figure 7.
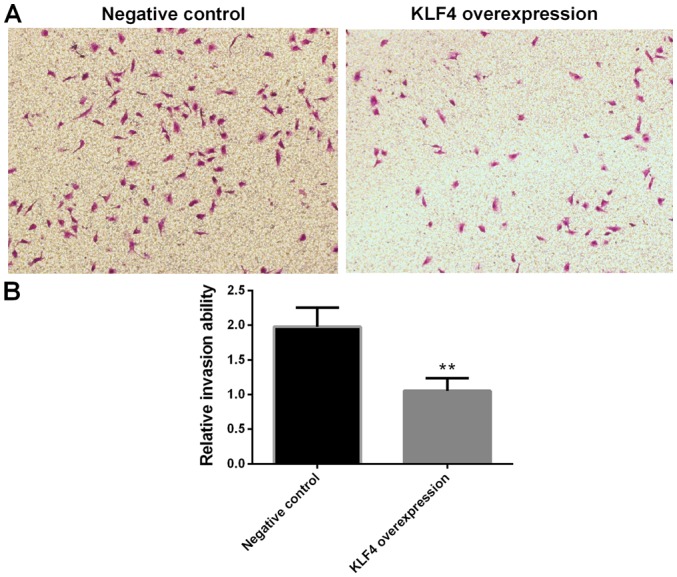
Invasion of papillary thyroid cancer cells in the KLF4 overexpression group was markedly decreased. (A) Representative image of transwell invasion assay (magnification, ×100). (B) Statistical analysis of the transwell invasion assay. Cell invasion ability was assessed using a transwell invasion assay. Compared with the negative control group, the invasion ability of KTC1 cells in the KLF4 overexpression group was markedly decreased (mean ± standard deviation; n=3 per group). **P<0.01 vs. the negative control group. KLF4, Kruppel-like factor 4.
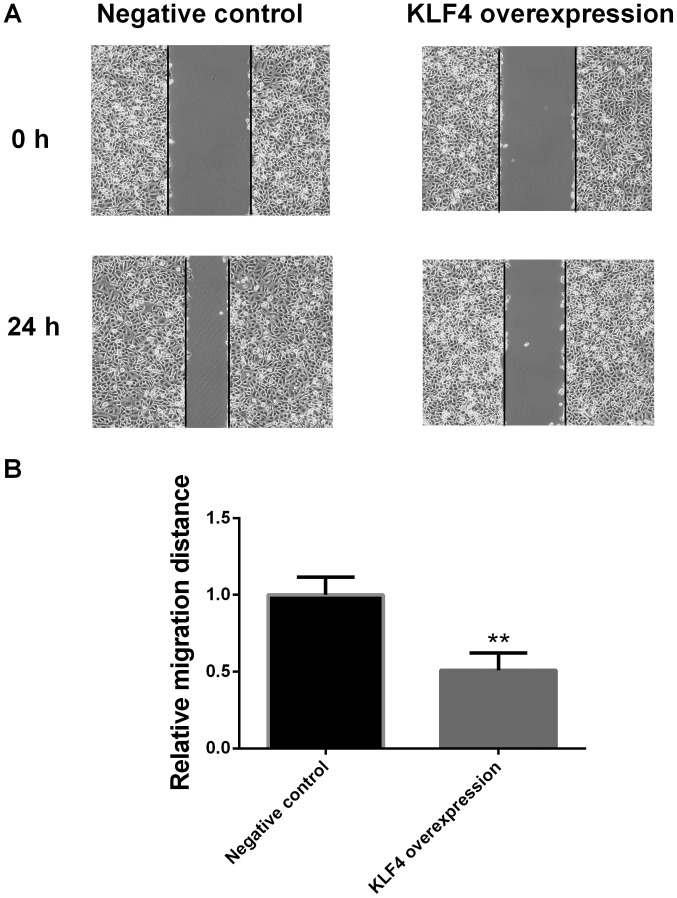
Migration of papillary thyroid cancer cells in the KLF4 overexpression group is significantly decreased at 24 h
Cell migration ability was examined via a scratch migration assay at 0 and 24 h. Compared with the negative control group, the migration of KTC1 cells in the KLF4 overexpression group was significantly decreased at 24 h (P<0.01; Fig. 8).
Figure 8.
Migration ability of papillary thyroid cancer cells in the KLF4 overexpression group was markedly decreased at 24 h. (A) Representative image and (B) statistical analysis of the scratch migration assay. Cell migration was examined via a scratch migration assay at 0 and 24 h. Compared with the negative control group, the migration of KTC1 cells in the KLF4 overexpression group was markedly decreased at 24 h (mean ± standard deviation; n=3 per group). **P<0.01 vs. the negative control group. KLF4, Kruppel-like factor 4.
Discussion
The present study demonstrated that the expression of KLF4 was significantly lower in thyroid tumor tissue compared with adjacent non-cancerous tissue. The viability, invasion and migration of cells, and the expression of N-cadherin, MMP2, MMP9 and collagen in papillary thyroid cancer cells were markedly decreased following KLF4 overexpression.
KLF4 has been reported to inhibit the proliferation of colorectal cancer cells via NMYC downstream-regulated gene 2 (26). Furthermore, KLF4 suppressed estrogen-dependent breast cancer growth by inhibiting the transcriptional activity of the estrogen receptor (27). KLF4 has also been demonstrated to inhibit the invasion of lung cancer cells by suppressing secreted protein acidic and cysteine rich gene expression (27). F-box protein-32 suppressed the tumorigenesis of breast cancer by targeting KLF4 for proteasomal degradation (28). In addition, the long non-coding RNA small nucleolar RNA host gene 5/miR-32 axis was revealed to regulate gastric cancer cell proliferation and migration by targeting KLF4 (29). KLF4 and KLF5 regulated the proliferation, apoptosis and invasion of esophageal cancer cells (30). Meanwhile, KLF4 was revealed to regulate adult lung tumor-initiating cells and repress K-Ras-mediated lung cancer (31). To the best of our knowledge, the present study revealed for the first time that the expression of KLF4 was significantly lower in thyroid tumor tissue when compared with adjacent non-cancerous tissues, and the viability, tumor invasion and migration of papillary thyroid cancer cells were significantly decreased following the overexpression of KLF4. These results may broaden the current understanding of the properties of KLF4, and shed light on possible therapeutic treatment of papillary thyroid cancer.
The present study revealed that the expression of N-cadherin, MMP2, MMP9 and collagen in papillary thyroid cancer cells were significantly decreased when KLF4 was overexpressed. N-cadherin expression in breast cancer has been demonstrated to be associated with an aggressive histological variant of invasive micropapillary carcinoma (32). N-cadherin, as a novel prognostic biomarker, was reported to drive the malignant progression of colorectal cancer (33). N-cadherin expression was also associated with enhanced invasion in erlotinib-resistant lung cancer cell lines (34). In addition, Trop2 has been indicated to enhance the invasion of thyroid cancer by inducing MMP2 via ERK and Janus kinase pathways (35). miR-29c suppressed the adhesion of lung cancer cell to the extracellular matrix, as well as metastasis, by targeting MMP2 (36). The association between the MMP2-1306 C/T polymorphism and prostate cancer susceptibility was revealed in a meta-analysis based on 3,906 subjects (37). Meanwhile, the downregulation of hepatoma-derived growth factor inhibited the migration and invasion of prostate cancer cells by suppressing MMP2 and MMP9 (38). miR-133b was reported to inhibit the cell growth, migration and invasion by targeting MMP9 in non-small cell lung cancer (39). Furthermore, the selective targeting of collagen IV in the cancer cell microenvironment was revealed to decrease tumor burden (40). Losartan loaded liposomes improved the antitumor efficacy of liposomal paclitaxel via the inhibition of collagen in breast cancer (41). In the present study, the significantly decreased expression of N-cadherin, MMP2, MMP9 and collagen in papillary thyroid cancer cells following KLF4 overexpression may impair the adhesion of thyroid cancer cells to the extracellular matrix, thus disrupting tumor invasion and migration.
Other factors may also account for the effects of KLF4 overexpression on tumor invasion and migration. KLF4 inhibited tumor growth and metastasis by targeting miR-31 in human hepatocellular carcinoma (42). Podocalyxin-like (PODXL) promoted the metastasis of gastric cancer, whereas KLF4 downregulated PODXL and prevented metastasis (43). miR-543 was also revealed to promote colorectal cancer proliferation and metastasis by targeting KLF4 (44). In addition, KLF4-mediated suppression of CD44 signaling decreased the stemness and metastasis of pancreatic cancer (45). Further studies are required to elucidate whether the aforementioned factors modulate the anti-metastasis effects of KLF4 in papillary thyroid cancer.
There are certain limitations to the present study. Normal thyroid cell lines were not used and there was no in vivo study. However, the present study detected the expression levels of KLF4 in human thyroid tumor tissue and adjacent normal tissues via IHC and western blotting. The experimental results in human tissues were consistent; therefore, similar experiments in normal thyroid cell lines or animals were not performed, which may or may not reflect the real situations in human.
In conclusion, the present study demonstrated that the expression of KLF4 was significantly lower in thyroid tumor tissue. The cell viability, tumor invasion and migration, and expression levels of N-cadherin, MMP2, MMP9 and collagen in papillary thyroid cancer cells were markedly decreased with the overexpression of KLF4. Although further research is required to elucidate the underlying molecular mechanisms, the present study may provide the foundations for future therapeutic measures targeting papillary thyroid cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Baoshan District Science and Technology Commission Fund (grant no. 16-E-10), and Baoshan District Integrated Traditional Chinese and Western Medicine Hospital Fund (grant no. 201603).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
QW, LL and JX designed the experiments. QW and YC carried out the experiments. YC performed the statistical analyses. LL and JX gave advice on how to design and carry out experiments. QW wrote the manuscript, which was revised by the other authors. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Institutional Research Board of Baoshan District Integrated Traditional Chinese and Western Medicine Hospital. Each participant provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.GBD 2015 Disease and Injury Incidence and Prevalence Collaborators: Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388:1545–1602. doi: 10.1016/S0140-6736(16)31678-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen J, Hou H, Chen H, Luo Y, He Y, Zhang L, Zhang Y, Liu H, Zhang F, Liu Y, et al. Identification of β-hydroxybutyrate as a potential biomarker for female papillary thyroid cancer. Bioanalysis. 2019;11:461–470. doi: 10.4155/bio-2018-0273. [DOI] [PubMed] [Google Scholar]
- 3.James BC, Timsina L, Graham R, Angelos P, Haggstrom DA. Changes in total thyroidectomy versus thyroid lobectomy for papillary thyroid cancer during the past 15 years. Surgery. 2019;166:41–47. doi: 10.1016/j.surg.2019.01.007. [DOI] [PubMed] [Google Scholar]
- 4.Galuppini F, Pennelli G, Rugge M. The rising incidence of papillary thyroid cancer: More cancers or more assessments? Indian J Cancer. 2019;56:183–184. doi: 10.4103/ijc.IJC_483_18. [DOI] [PubMed] [Google Scholar]
- 5.Kazaure HS, Roman SA, Sosa JA. Aggressive variants of papillary thyroid cancer: Incidence, characteristics and predictors of survival among 43,738 patients. Ann Surg Oncol. 2011;19:1874–1880. doi: 10.1245/s10434-011-2129-x. [DOI] [PubMed] [Google Scholar]
- 6.Richards ML. Thyroid cancer genetics: Multiple endocrine neoplasia type 2, non-medullary familial thyroid cancer, and familial syndromes associated with thyroid cancer. Surg Oncol Clin N Am. 2009;18(viii):39–52. doi: 10.1016/j.soc.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 7.Zhu M, Zhang N, He S. Transcription factor KLF4 modulates microRNA-106a that targets Smad7 in gastric cancer. Pathol Res Pract. 2019:152467. doi: 10.1016/j.prp.2019.152467. [DOI] [PubMed] [Google Scholar]
- 8.Zhuan B, Lu Y, Chen Q, Zhao X, Li P, Yuan Q, Yang Z. Overexpression of the long noncoding RNA TRHDE-AS1 inhibits the progression of lung cancer via the miRNA-103/KLF4 axis. J Cell Biochem. 2019 May 22; doi: 10.1002/jcb.29029. (Epub ahead of print). doi: 10.1002/jcb.29029. [DOI] [PubMed] [Google Scholar]
- 9.Yang VW, Liu Y, Kim J, Shroyer KR, Bialkowska AB. Increased genetic instability and accelerated progression of colitis-associated colorectal cancer through intestinal epithelium-specific deletion of Klf4. Mol Cancer Res. 2019;17:165–176. doi: 10.1158/1541-7786.MCR-18-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okuda H, Xing F, Pandey PR, Sharma S, Watabe M, Pai SK, Mo YY, Iiizumi-Gairani M, Hirota S, Liu Y, et al. miR-7 suppresses brain metastasis of breast cancer stem-like cells by modulating KLF4. Cancer Res. 2013;73:1434–1444. doi: 10.1158/0008-5472.CAN-12-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Wang J, Xiao W, Xia D, Lang B, Wang T, Guo X, Hu Z, Ye Z, Xu H. Epigenetic inactivation of KLF4 is associated with urothelial cancer progression and early recurrence. J Urol. 2013;191:493–501. doi: 10.1016/j.juro.2013.08.087. [DOI] [PubMed] [Google Scholar]
- 12.Guo K, Cui J, Quan M, Xie D, Jia Z, Wei D, Wang L, Gao Y, Ma Q, Xie K. The novel KLF4/MSI2 signaling pathway regulates growth and metastasis of pancreatic cancer. Clin Cancer Res. 2017;23:687–696. doi: 10.1158/1078-0432.CCR-16-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akaogi K, Nakajima Y, Ito I, Kawasaki S, Oie SH, Murayama A, Kimura K, Yanagisawa J. KLF4 suppresses estrogen-dependent breast cancer growth by inhibiting the transcriptional activity of ERalpha. Oncogene. 2009;28:2894–2902. doi: 10.1038/onc.2009.151. [DOI] [PubMed] [Google Scholar]
- 14.Ding X, Zhong T, Jiang L, Huang J, Xia Y, Hu R. miR-25 enhances cell migration and invasion in non-small-cell lung cancer cells via ERK signaling pathway by inhibiting KLF4. Mol Med Rep. 2018;17:7005–7016. doi: 10.3892/mmr.2018.8772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cittelly DM, Finlay-Schultz J, Howe EN, Spoelstra NS, Axlund SD, Hendricks P, Jacobsen BM, Sartorius CA, Richer JK. Progestin suppression of miR-29 potentiates dedifferentiation of breast cancer cells via KLF4. Oncogene. 2012;32:2555–2564. doi: 10.1038/onc.2012.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong AS, Gumbiner BM. Adhesion-independent mechanism for suppression of tumor cell invasion by E-cadherin. J Cell Biol. 2003;161:1191–1203. doi: 10.1083/jcb.200212033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makrigiannakis A, Coukos G, Blaschuk O, Coutifaris C. Follicular atresia and luteolysis. Evidence of a role for N-cadherin. Ann N Y Acad Sci. 2000;900:46–55. doi: 10.1111/j.1749-6632.2000.tb06215.x. [DOI] [PubMed] [Google Scholar]
- 18.Yamashita N, Tokunaga E, Iimori M, Inoue Y, Tanaka K, Kitao H, Saeki H, Oki E, Maehara Y. Epithelial paradox: Clinical significance of coexpression of E-cadherin and vimentin with regard to invasion and metastasis of breast cancer. Clin Breast Cancer. 2018;18:e1003–e1009. doi: 10.1016/j.clbc.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 19.Winerdal ME, Krantz D, Hartana CA, Zirakzadeh AA, Linton L, Bergman EA, Rosenblatt R, Vasko J, Alamdari F, Hansson J, et al. Urinary bladder cancer Tregs suppress MMP2 and potentially regulate invasiveness. Cancer Immunol Res. 2018;6:528–538. doi: 10.1158/2326-6066.CIR-17-0466. [DOI] [PubMed] [Google Scholar]
- 20.Klassen LMB, Chequin A, Manica GCM, Biembengut IV, Toledo MB, Baura VA, de O Pedrosa F, Ramos EAS, Costa FF, de Souza EM, Klassen G. MMP9 gene expression regulation by intragenic epigenetic modifications in breast cancer. Gene. 2018;642:461–466. doi: 10.1016/j.gene.2017.11.054. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen SH, Willumsen N, Brix S, Sun S, Manon-Jensen T, Karsdal M, Genovese F. Tumstatin, a matrikine derived from collagen type IVα3, is elevated in serum from patients with non-small cell lung cancer. Transl Oncol. 2018;11:528–534. doi: 10.1016/j.tranon.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujimori M, Sakauchi G. TNM classification-thyroid cancer. Gan No Rinsho. 1967;13:320–322. (In Japanese) [PubMed] [Google Scholar]
- 23.Haugen BR, Alexander EK, Bible KC, Doherty GM, Mandel SJ, Nikiforov YE, Pacini F, Randolph GW, Sawka AM, Schlumberger M, et al. 2015 American thyroid association management guidelines for adult patients with thyroid nodules and differentiated thyroid cancer. Thyroid. 2016;26:1–33. doi: 10.1089/thy.2015.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mancini P, Bonanno Ferraro G, Iaconelli M, Suffredini E, Valdazo-González B, Della Libera S, Divizia M, La Rosa G. Molecular characterization of human Sapovirus in untreated sewage in Italy by amplicon-based Sanger and next-generation sequencing. J Appl Microbiol. 2019;126:324–331. doi: 10.1111/jam.14129. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Ma Y, Wu L, Liu X, Xu Y, Shi W, Liang Y, Yao L, Zheng J, Zhang J. KLF4 inhibits colorectal cancer cell proliferation dependent on NDRG2 signaling. Oncol Rep. 2017;38:975–984. doi: 10.3892/or.2017.5736. [DOI] [PubMed] [Google Scholar]
- 27.Zhou Y, Hofstetter WL, He Y, Hu W, Pataer A, Wang L, Wang J, Zhou Y, Yu L, Fang B, Swisher SG. KLF4 inhibition of lung cancer cell invasion by suppression of SPARC expression. Cancer Biol Ther. 2010;9:507–513. doi: 10.4161/cbt.9.7.11106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou H, Liu Y, Zhu R, Ding F, Wan Y, Li Y, Liu Z. FBXO32 suppresses breast cancer tumorigenesis through targeting KLF4 to proteasomal degradation. Oncogene. 2017;36:3312–3321. doi: 10.1038/onc.2016.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao L, Han T, Li Y, Sun J, Zhang S, Liu Y, Shan B, Zheng D, Shi J. The lncRNA SNHG5/miR-32 axis regulates gastric cancer cell proliferation and migration by targeting KLF4. FASEB J. 2016;31:893–903. doi: 10.1096/fj.201600994R. [DOI] [PubMed] [Google Scholar]
- 30.Yang Y, Goldstein BG, Chao HH, Katz JP. KLF4 and KLF5 regulate proliferation, apoptosis and invasion in esophageal cancer cells. Cancer Biol Ther. 2005;4:1216–1221. doi: 10.4161/cbt.4.11.2090. [DOI] [PubMed] [Google Scholar]
- 31.Yu T, Chen X, Zhang W, Liu J, Avdiushko R, Napier DL, Liu AX, Neltner JM, Wang C, Cohen D, Liu C. KLF4 regulates adult lung tumor-initiating cells and represses K-Ras-mediated lung cancer. Cell Death Differ. 2015;23:207–215. doi: 10.1038/cdd.2015.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagi C, Guttman M, Jaffer S, Qiao R, Keren R, Triana A, Li M, Godbold J, Bleiweiss IJ, Hazan RB. N-cadherin expression in breast cancer: Correlation with an aggressive histologic variant-invasive micropapillary carcinoma. Breast Cancer Res Treat. 2005;94:225–235. doi: 10.1007/s10549-005-7727-5. [DOI] [PubMed] [Google Scholar]
- 33.Yan X, Yan L, Liu S, Shan Z, Tian Y, Jin Z. N-cadherin, a novel prognostic biomarker, drives malignant progression of colorectal cancer. Mol Med Rep. 2015;12:2999–3006. doi: 10.3892/mmr.2015.3687. [DOI] [PubMed] [Google Scholar]
- 34.Zhang X, Liu G, Kang Y, Dong Z, Qian Q, Ma X. N-cadherin expression is associated with acquisition of EMT phenotype and with enhanced invasion in erlotinib-resistant lung cancer cell lines. PLoS One. 2013;8:e57692. doi: 10.1371/journal.pone.0057692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guan H, Guo Z, Liang W, Li H, Wei G, Xu L, Xiao H, Li Y. Trop2 enhances invasion of thyroid cancer by inducing MMP2 through ERK and JNK pathways. BMC Cancer. 2017;17:486. doi: 10.1186/s12885-017-3475-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H, Zhu Y, Zhao M, Wu C, Zhang P, Tang L, Zhang H, Chen X, Yang Y, Liu G. miRNA-29c suppresses lung cancer cell adhesion to extracellular matrix and metastasis by targeting integrin β1 and matrix metalloproteinase 2 (MMP2) PLoS One. 2013;8:e70192. doi: 10.1371/journal.pone.0070192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang K, Chen X, Zhou J, Yang C, Zhang M, Chao M, Zhang L, Liang C. Association between MMP2-1306 C/T polymorphism and prostate cancer susceptibility: A meta-analysis based on 3906 subjects. Oncotarget. 2017;8:45020–45029. doi: 10.18632/oncotarget.16972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang F, Yu N, Wang H, Zhang C, Zhang Z, Li Y, Li D, Yan L, Liu H, Xu Z. Downregulated expression of hepatoma-derived growth factor inhibits migration and invasion of prostate cancer cells by suppressing epithelial-mesenchymal transition and MMP2, MMP9. PLoS One. 2018;13:e0190725. doi: 10.1371/journal.pone.0190725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhen Y, Liu J, Huang Y, Wang Y, Li W, Wu J. miR-133b inhibits cell growth, migration, and invasion by targeting MMP9 in non-small cell lung cancer. Oncol Res. 2016;25:1109–1116. doi: 10.3727/096504016X14800889609439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Revert F, Revert-Ros F, Blasco R, Artigot A, López-Pascual E, Gozalbo-Rovira R, Ventura I, Gutiérrez-Carbonell E, Roda N, Ruíz-Sanchis D, et al. Selective targeting of collagen IV in the cancer cell microenvironment reduces tumor burden. Oncotarget. 2018;9:11020–11045. doi: 10.18632/oncotarget.24280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xia T, He Q, Shi K, Wang Y, Yu Q, Zhang L, Zhang Q, Gao H, Ma L, Liu J. Losartan loaded liposomes improve the antitumor efficacy of liposomal paclitaxel modified with pH sensitive peptides by inhibition of collagen in breast cancer. Pharm Dev Technol. 2016;23:13–21. doi: 10.1080/10837450.2016.1265553. [DOI] [PubMed] [Google Scholar]
- 42.Tian C, Yao S, Liu L, Ding Y, Ye Q, Dong X, Gao Y, Yang N, Li Q. Klf4 inhibits tumor growth and metastasis by targeting microRNA-31 in human hepatocellular carcinoma. Int J Mol Med. 2016;39:47–56. doi: 10.3892/ijmm.2016.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang J, Zhu Z, Wu H, Yu Z, Rong Z, Luo Z, Xu Y, Huang K, Qiu Z, Huang C. PODXL, negatively regulated by KLF4, promotes the EMT and metastasis and serves as a novel prognostic indicator of gastric cancer. Gastric Cancer. 2019;22:48–59. doi: 10.1007/s10120-018-0833-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhai F, Cao C, Zhang L, Zhang J. miR-543 promotes colorectal cancer proliferation and metastasis by targeting KLF4. Oncotarget. 2017;8:59246–59256. doi: 10.18632/oncotarget.19495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan Y, Li Z, Kong X, Jia Z, Zuo X, Gagea M, Huang S, Wei D, Xie K. KLF4-mediated suppression of CD44 signaling negatively impacts pancreatic cancer stemness and metastasis. Cancer Res. 2016;76:2419–2431. doi: 10.1158/0008-5472.CAN-15-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.