Abstract
Previously, a number of microRNAs (miRNAs) have been reported to be dysregulated in cervical cancer, and dysregulated miRNAs may play crucial roles in the development and progression of cervical cancer. Hence, investigating the detailed roles of miRNAs that are aberrantly expressed in cervical cancer and the underlying molecular mechanisms is essential for early diagnosis and effective therapeutic approaches. miRNA-877 (miR-877) was found to be downregulated in hepatocellular carcinoma and renal cell carcinoma, and function as a tumor-suppressive miRNA. However, how miR-877 exerts an effect in cervical cancer progression and its underlying molecular mechanisms remains to be elucidated. In the current study, reverse transcription-quantitative PCR was performed to determine miR-877 expression in cervical cancer tissues and cell lines. The effects of miR-877 overexpression on cervical cancer cell proliferation and invasion were evaluated using MTT and Transwell cell invasion assays. In the present study, miR-877 was significantly downregulated in cervical cancer tissues and cell lines, and the decreased expression levels of miR-877 were significantly associated with increased International Federation of Gynecology and Obstetric stage as well as increased lymph node metastasis in patients with cervical cancer. Upregulation of miR-877 using miR-877 mimics resulted in the decreased proliferation and invasion of cervical cancer cells. Metastasis-associated in colon cancer-1 (MACC1) was assessed using bioinformatics analyses to determine whether it could be a potential target gene of miR-877, and the results were confirmed using a luciferase reporter assay. Furthermore, MACC1 was markedly upregulated in cervical cancer tissues, and its level was negatively correlated with the miR-877 level. Overexpression of miR-877 resulted in decreased expression levels of MACC1 in cervical cancer cells at both the mRNA and protein levels. In addition, the functional effects of MACC1 knockdown were similar to those induced by upregulated miR-877 in cervical cancer cells. MACC1 restored miR-877 overexpression-mediated suppression of cervical cancer cell proliferation and invasion. In conclusion, miR-877 may play an antitumor role in cervical cancer by directly targeting MACC1, which suggests that this miRNA may be a promising therapeutic target for the treatment of patients with such an aggressive gynecological cancer.
Keywords: microRNA-877, cervical cancer, proliferation, invasion, metastasis-associated in colon cancer-1
Introduction
Cervical cancer ranks third highest in incidence rate and fourth in cancer-associated mortality among women worldwide, with an estimated >500,000 new cases and 260,000 deaths caused by cervical cancer each year (1). In developing countries, the morbidity and mortality of cervical cancer has gradually increased, primarily due to the lack of screening programs, diagnosis methods and effective therapeutic methods (2). High-risk human papillomavirus (HPV) infection has been identified as a key attribute among the numerous risk factors associated with cervical cancer; however, HPV infection alone is inadequate to cause carcinogenesis (3). Extensive basic and clinical research has significantly facilitated the development of novel diagnostic and therapeutic techniques for patients with cervical cancer (4); unfortunately, the prognosis remains dissatisfactory, with an overall survival rate of ~30–50% for patients at an advanced stage of disease (5). Thus, further studies are critical in order to understand the molecular mechanisms that underlie cervical cancer occurrence and development in order to improve outcomes for patients with this malignancy.
MicroRNAs (miRNAs) are a large family of endogenous, non-coding, short RNA molecules that are ~22 nucleotides in length (6). miRNAs have been identified as critical gene regulators via directly binding to complementary sequences in the 3′-untranslated regions (3′-UTRs) of their target genes, resulting in translation suppression and/or mRNA degradation (7). It is well documented that one single miRNA is able to modulate numerous target genes, and ~30% of human protein-coding genes are predicted to be regulated by miRNAs (8,9). Previous studies focusing on miRNA expression revealed that a variety of miRNAs were dysregulated during cervical carcinogenesis and progression, such as miR-152 (10), miR-224 (11), miR-302 (12) and miR-1297 (13). Aberrantly expressed miRNAs can affect the aggressive behaviors of cervical cancer cells through acting as oncogenes or tumor suppressors (14). Therefore, a complete understanding of the abnormally expressed miRNAs in cervical cancer progression is important for the identification of potential therapeutic targets for treating patients with this disease.
Previous studies have indicated that miR-877 was downregulated in hepatocellular carcinoma (15,16) and renal cell carcinoma (17), and functioned as a tumor-suppressive miRNA. However, it has rarely been reported how miR-877 exerts an effect in cervical cancer progression and its underlying molecular mechanisms. Hence, the present study detected the expression levels of miR-877 in cervical cancer and determined its clinical significance. In addition, the effects of miR-877 in the malignant behaviors of human cervical cancer cells and involved mechanisms were investigated.
Materials and methods
Tissue collection
The present study was approved by the Ethics Committee of Jilin Cancer Hospital (Changchun, China). All patients enrolled in the present study provided written informed consent based on the principles of the Declaration of Helsinki. A total of 57 pairs of cervical cancer tissues and matched adjacent normal tissues (2 cm away from tumor tissues) were collected from patients (age range: 43–68 years old) in the Jilin Cancer Hospital between March 2015 and August 2017, and were confirmed as cervical cancer via analysis by a pathologist. Patients diagnosed as cervical cancer and had not been treated with radiotherapy or chemotherapy prior to surgery participated in the current study. All patients were divided into two groups, a low miR-877 expression group and a high miR-877 expression group, based on the median value of miR-877. All tissues were frozen immediately in liquid nitrogen following surgical resection and then stored at −80°C until further use.
Cell lines and culture
A total of four human cervical cancer cell lines (CaSki, HeLa, C-33A and SiHa) and a normal human cervix epithelial cell line (Ect1/E6E7) were purchased from the American Type Culture Collection. All cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% antibiotics (100 U/ml penicillin and 100 µg/ml streptomycin; all from Invitrogen; Thermo Fisher Scientific, Inc.). All cultures were maintained at 37°C in an incubator containing 5% CO2 until further use.
Oligonucleotides, plasmids and transfection
Cells were plated in six-well plates at a density of 5×106 cells per well. Cells were transfected with miR-877 mimics (Shanghai GenePharma Co., Ltd.) to increase miR-877 expression. The small interfering RNA (siRNA) targeting MACC1 (MACC1 siRNA; Guangzhou RiboBio Co., Ltd.) was introduced into cells to knockdown endogenous MACC1 expression. miRNA mimics negative control (miR-NC) and scrambled negative control siRNA (NC siRNA) were used as the control for miR-877 mimics and MACC1 siRNA transfection, respectively. The miR-877 mimics sequence was 5′-GUAGAGGAGAUGGCGCAGGG-3′ and the miR-NC sequence was 5′-UAUGCACUCCUGAAGGGCUCGC-3′. The MACC1 siRNA sequence was 5′-AAGAUUGGACUUGUACACUGC-3′ and the NC siRNA sequence was 5′-UUCUCCGAACGUGUCACGUTT-3′. MACC1 overexpression plasmid pCMV-MACC1 and empty pCMV plasmid were provided by the Chinese Academy of Sciences. Cells were transfected with miRNA mimics (100 pmol), siRNA (100 pmol) or plasmid (4 µg) using Lipofectamine™ 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and a Transwell cell invasion assay was performed at 24 h post-transfection. MTT assay and western blot analysis was performed at 24 h and 72 h after transfection, respectively.
RT-qPCR
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was used to isolate total RNA from tissue samples or cells. All-in-One™ miRNA RT-qPCR Detection kit (GeneCopoeia, Inc.) was used to measure miR-877 expression. The thermocycling conditions were as follows: 95°C for 10 min, 45 cycles of denaturation at 95°C for 15 sec and annealing/elongation at 60°C for 15 sec. To quantify MACC1 mRNA expression, reverse transcription was performed to produce cDNA from total RNA using a PrimeScript RT reagent kit (Takara Biotechnology Co., Ltd.). The thermocycling conditions for reverse transcription was as follows: 37°C for 15 min and 85°C for 5 sec. The synthesized cDNA was then subjected to MACC1 mRNA expression detection using a SYBR Premix Ex Taq™ (Takara Biotechnology Co., Ltd.). The thermocycling conditions for qPCR were performed as follows: 5 min at 95°C, followed by 40 cycles of 95°C for 30 sec and 65°C for 45 sec. U6 small nuclear RNA and GAPDH served as the internal references for miR-877 and MACC1 mRNA levels, respectively. The relative gene expression was analyzed and normalized using the 2−ΔΔCq method (18). The primers were designed as follows: miR-877 forward, ‘5-GTAGAGGAGATGGCGCAGGG-3′ and reverse, 5′-CAGTGCGTGTCGTGGAGT-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′; MACC1 forward, 5′-CACAACTTGCGGAGGTCAC-3′ and reverse, 5′-AAGCTGTGGGGTTTTTCC-3′; and GAPDH forward, 5′-CGGAGTCAACGGATTTGGTCGTAT-3′ and reverse 5′-AGCCTTCTCCATGGTGGTGAAGAC-3′.
MTT assay
Transfected cells were harvested and seeded into 96-well plates with a density of 3×103 cells/well. Cells were incubated at 37°C in an incubator supplied with 5% CO2 for 0, 24, 48 and 72 h. MTT assay was applied to determine cellular proliferation at indicated time points by adding 20 µl MTT solution (5 mg/ml; Sigma-Aldrich; Merck KGaA) into each well. After 4 h of incubation at 37°C, the culture medium was carefully removed and 150 µl of DMSO (Sigma-Aldrich; Merck KGaA) was added into each well in order to dissolve the formazan crystals. Finally, the absorbance of each sample was detected at a wavelength of 490 nm using a plate reader (Bio-Rad Laboratories, Inc.).
Transwell cell invasion assay
The invasive ability was evaluated using 24-well Transwell filters (8 micrometers pore size) that were pre-coated with Matrigel (both from BD Biosciences). After 48 h of transfection, cells were collected, suspended in FBS-free DMEM and then inoculated into the upper compartments of each filter (5×104 cells per filter). DMEM with 20% FBS was added into the lower compartments as a chemoattractant. Following incubation of the cells for 24 h at 37°C, non-invaded cells remaining on the upper surface of the membranes were removed with a cotton swab. The invasive cells were fixed in 4% paraformaldehyde at room temperature for 30 min and stained with 0.5% crystal violet at room temperature for 30 min. The number of invasive cells was acquired from five randomly selected areas under a light microscope (magnification, ×200).
Bioinformatics prediction of miR-877 targets
Target gene prediction software, including microRNA.org (August 2010; Release Last Update: 2010-11-01; http://www.microrna.org/microrna/) and TargetScan (Release 7.2: March 2018; http://www.targetscan.org/vert_71/), was used to search for the potential targets of miR-877.
Luciferase reporter assay
The wild-type (wt) and mutant (mut) of MACC1 3′-UTR was amplified by Shanghai GenePharma Co., Ltd., and cloned into the pMIR-REPORT miRNA Expression Reporter vector (Ambion; Thermo Fisher Scientific, Inc.) generating the pMIR-MACC1-3′-UTR wt and pMIR-MACC1-3′-UTR mut, respectively. Cells were plated in 24-well plates with a density of 1.0×105 cells per well and co-transfected with wt or mut luciferase plasmid and miR-877 mimics or miR-NC using Lipofectamine™ 2000. After a 48-h incubation, transfected cells were harvested and assessed for luciferase activity using a dual-luciferase reporter assay system (Promega Corporation), according to the manufacturer's protocol. The firefly luciferase activity was normalized to that of Renilla luciferase activity.
Western blot analysis
Western blot analysis was applied to detect MACC1 protein expression. Total protein was isolated from cultured cells or homogenized tissues using a cold radioimmunoprecipitation assay buffer (Shanghai Qcbio Science & Technologies Co., Ltd.). Total protein was quantified according to the protocol of a Bicinchoninic Acid Protein Assay kit (Bio-Rad Laboratories, Inc.). An equal mass of proteins (20 µg) were separated by SDS-PAGE (10% gel), blotted onto PVDF membranes (EMD Millipore) and blocked at room temperature in Tris-buffered saline containing 0.1% Tween-20 (TBST) supplemented with 5% dried skimmed milk for 2 h. Subsequently, the membranes were incubated with primary antibodies overnight at 4°C followed by incubation with horseradish peroxidase-conjugated goat anti-rabbit secondary antibodies (1:5,000; catalog no. ab6721; Abcam) at room temperature for 2 h. Following extensive washing with TBST, an Enhanced Chemiluminescence (ECL) Western blotting kit (Pierce; Thermo Fisher Scientific, Inc.) was used to visualize the immune complex on the PVDF membranes. The primary antibodies used in the present study were as follows: Rabbit anti-human MACC1 antibody (1:1,000; catalog no. ab106579) and rabbit anti-human GAPDH antibody (1:1,000; catalog no. ab128915; both from Abcam). GAPDH was used as an internal control. Quantity One software (version 4.62; Bio-Rad Laboratories, Inc.) was utilized to analyze the protein signals.
Statistical analysis
All assays were repeated at least three times. Data are presented as the mean ± standard deviation and were analyzed using SPSS software (version 17.0; SPSS Inc.). Differences between groups were determined using Student's t-tests or one-way analysis of variance (ANOVA). Student-Newman-Keuls (SNK) was used as the post hoc analysis following ANOVA. The association between the clinicopathological characteristics of the patients with cervical cancer and miR-877 or MACC1 expression was assessed with χ2 test. Spearman's correlation analysis was used to evaluate the correlation between miR-877 and MACC1 mRNA expression levels in cervical cancer tissues. P<0.05 was considered to indicate a statistically significant result.
Results
miR-877 is downregulated in cervical cancer tissues and cell lines
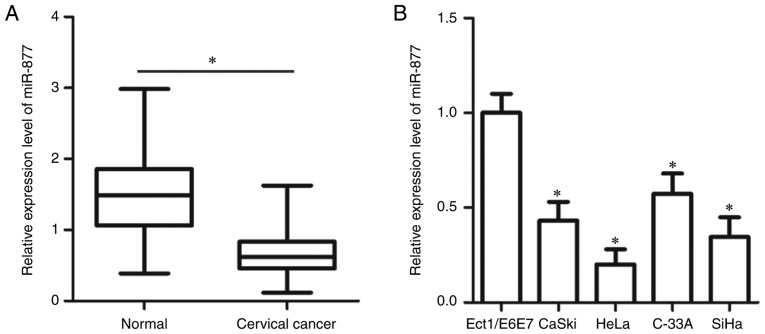
To determine the expression patterns of miR-877 in cervical cancer, RT-qPCR was utilized to measure miR-877 expression in 57 pairs of cervical cancer tissues and matched adjacent normal tissues. The expression level of miR-877 was lower in cervical cancer tissues when compared with the adjacent normal tissues (P<0.05; Fig. 1A). In addition, miR-877 expression was determined in a panel of human cervical cancer cell lines, including CaSki, HeLa, C-33A and SiHa. A normal human cervix epithelial cell line Ect1/E6E7 was used as the control. The data obtained from RT-qPCR revealed that miR-877 expression was decreased in all four cervical cancer cell lines compared with in Ect1/E6E7 (P<0.05; Fig. 1B).
Figure 1.
miR-877 is expressed at low levels in cervical cancer tissues and cell lines. (A) RT-qPCR was used to analyze the expression levels of miR-877 in 57 pairs of cervical cancer tissues and matched adjacent normal tissues. *P<0.05. (B) Expression levels of miR-877 were determined in four cervical cancer cell lines (CaSki, HeLa, C-33A and SiHa) and a normal human cervix epithelial cell line Ect1/E6E7 via RT-qPCR. *P<0.05 vs. Ect1/E6E7. miR, microRNA; RT-qPCR, reverse transcription-quantitative PCR.
To evaluate the clinical value of miR-877 in cervical cancer, all patients enrolled in the present study were divided into two groups, a low miR-877 expression group and a high miR-877 expression group, based on the median value of miR-877 (0.62). As presented in Table I, low miR-877 expression was significantly correlated with the International Federation of Gynecology and Obstetric (FIGO) stage (19) (P=0.017) and lymph node metastasis (P=0.007), but not with age (P=0.395), tumor size (P=0.417), HPV infection (P=0.514), vascular involvement (P=0.530) or myometrium invasion (P=0.175). These results suggest that downregulation of miR-877 may be involved in the malignant progression of cervical cancer.
Table I.
Association between miR-877 and clinicopathological characteristics of patients with cervical cancer.
| miR-877 expression level | |||
|---|---|---|---|
| Characteristic | Low | High | P-value |
| Age, years | 0.395 | ||
| <45 | 11 | 7 | |
| ≥45 | 18 | 21 | |
| Tumor size, cm | 0.417 | ||
| <4 | 16 | 19 | |
| ≥4 | 13 | 9 | |
| HPV | 0.514 | ||
| Negative | 12 | 14 | |
| Positive | 17 | 14 | |
| Vascular involvement | 0.530 | ||
| Yes | 8 | 5 | |
| No | 21 | 23 | |
| FIGO stage | 0.017 | ||
| I–II | 9 | 18 | |
| III–IV | 20 | 10 | |
| Myometrium invasion | 0.175 | ||
| <1/2 | 15 | 20 | |
| ≥1/2 | 14 | 8 | |
| Lymph node metastasis | 0.007 | ||
| No | 11 | 21 | |
| Yes | 18 | 7 | |
miR, microRNA; HPV, human papillomavirus; FIGO, International Federation of Gynecology and Obstetrics.
miR-877 has a suppressive effect on the proliferation and invasion of cervical cancer cells
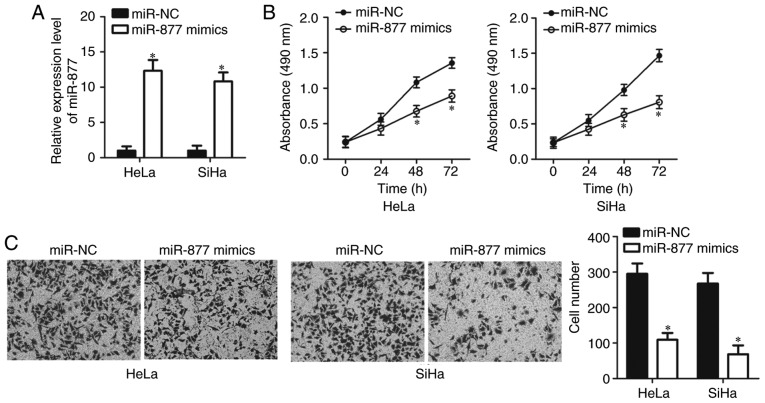
HeLa and SiHa cell lines exhibited relatively lower miR-877 expression levels out of all the cervical cancer cell lines (Fig. 1B), therefore the two cell lines were selected for subsequent functional assays. In order to investigate the potential functions of miR-877 in the malignant phenotypes of cervical cancer, HeLa and SiHa cells were transfected with miR-877 mimics or miR-NC. The results of the RT-qPCR confirmed that miR-877 was notably upregulated in miR-877 mimic-transfected HeLa and SiHa cells compared with the cells transfected with miR-NC (P<0.05; Fig. 2A). The impact of miR-877 upregulation on cell proliferation in cervical cancer was examined using MTT assay. miR-877-overexpressing HeLa and SiHa cells demonstrated a decreased proliferative ability when compared with that in the miR-NC group (P<0.05; Fig. 2B). The role of miR-877 in the regulation of cervical cancer cell invasion was then determined. Data from the transwell cell invasion assay revealed that increased expression levels of miR-877 markedly inhibited the invasion of HeLa and SiHa cells (P<0.05; Fig. 2C). These results implied that miR-877 may play a tumor suppressive role in the development of cervical cancer.
Figure 2.
miR-877 overexpression prohibits HeLa and SiHa cell proliferation and invasion. (A) miR-877 mimics or miR-NC were introduced into HeLa and SiHa cells. Following transfection, reverse transcription-quantitative PCR was performed to evaluate miR-877 expression. (B) The MTT assay was used to evaluate the cell proliferation in HeLa and SiHa cells following miR-877 mimic or miR-NC transfection. (C) Impact of miR-877 upregulation on the invasion of HeLa and SiHa cells was examined through transwell cell invasion assay (magnification, ×200). *P<0.05 vs. miR-NC. miR, microRNA; NC, negative control.
miR-877 inhibits MACC1 expression by directly targeting its 3′-UTR in cervical cancer cells
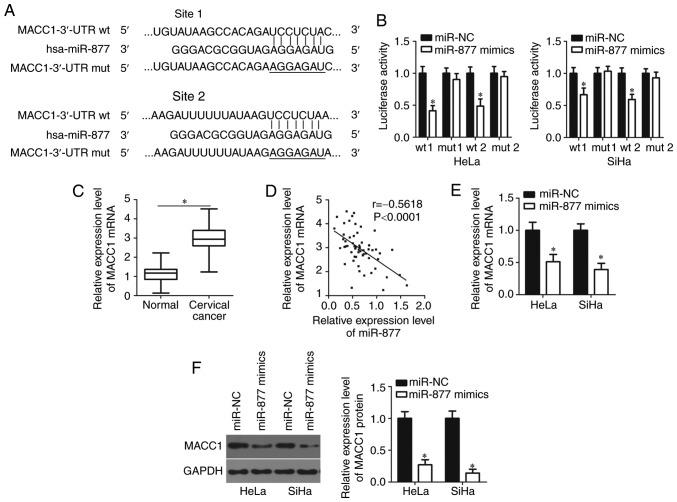
To decipher the mechanisms underlying how miR-877 affects cervical cancer progression, bioinformatics analyses were performed using microRNA.org and TargetScan to search for the potential target of miR-877. Two highly conserved putative binding sites were observed in 3,470–3,476 bp and 5,492–5,499 bp of MACC1 3′-UTR (Fig. 3A). MACC1 was selected for further identification as this gene has been widely reported as implicated in cervical cancer carcinogenesis and progression (20–25). Luciferase reporter assay was applied to confirm whether the 3′-UTR of MACC1 could be directly targeted by miR-877 in cervical cancer cells. In HeLa and SiHa cells co-transfected with pMIR-MACC1-3′-UTR wt (site 1 and 2) and miR-877 mimics, luciferase activity was significantly decreased compared with that in cells co-transfected with miR-NC and pMIR-MACC1-3′-UTR wt (P<0.05; site 1 and 2); however, no obvious influence was indicated on the luciferase activity of the plasmid harboring mutant miR-877 binding site (site 1 and 2; Fig. 3B).
Figure 3.
MACC1 is a direct target of miR-877 in cervical cancer cells. (A) The two predicted binding sites of miR-877 in the 3′-UTR of MACC1. The mutant binding sites were produced in the seed region of the MACC1 3′-UTR by mutating 7 nucleotides that were targeted by miR-877. (B) The luciferase reporter assay was performed in HeLa and SiHa cells that were co-transfected with miR-877 mimics or miR-NC and luciferase reporter plasmid carrying the wt or mut miR-877 binding site. *P<0.05 vs. miR-NC. (C) Detection of MACC1 mRNA expression in 57 pairs of cervical cancer tissues and matched adjacent normal tissues was performed via RT-qPCR. *P<0.05. (D) Spearman's correlation analysis was used to determine the association between the expression levels of miR-877 and MACC1 mRNA in cervical cancer tissues. r=−0.5618, P<0.0001. MACC1 mRNA and protein expression in miR-877 mimic- or miR-NC-transfected HeLa and SiHa cells was detected via (E) RT-qPCR and (F) western blot analysis, respectively. *P<0.05 vs. miR-NC. MACC1, metastasis-associated in colon cancer 1; miR, microRNA; 3′-UTR, 3′-untranslated region; wt, wild type; mut, mutant; RT-qPCR, reverse transcription-quantitative PCR; NC, negative control.
The MACC1 mRNA expression levels in cervical cancer tissues through RT-qPCR. The expression levels of MACC1 mRNA were revealed to be markedly upregulated in cervical cancer tissues compared with that in adjacent normal tissues (P<0.05; Fig. 3C). The association between the mRNA level of MACC1 and the clinicopathological factors of patients with cervical cancer was examined. As presented in Table II, an increased MACC1 expression level was significantly associated with the increased FIGO stage (P=0.047), increased myometrium invasion (P=0.002) and increased lymph node metastasis (P=0.022). Notably, an inverse correlation was validated between miR-877 and MACC1 mRNA levels in cervical cancer tissues via Spearman's correlation analysis (P<0.0001; r=−0.5618; Fig. 3D). Furthermore, RT-qPCR and western blot analyses were performed in order to investigate whether miR-877 could regulate endogenous MACC1 expression levels in cervical cancer cells. The data revealed that ectopic miR-877 expression in HeLa and SiHa cells could decrease MACC1 expression at both the mRNA (P<0.05; Fig. 3E) and protein (P<0.05; Fig. 3F) levels. The results demonstrated that MACC1 was a direct target gene of miR-877 in cervical cancer cells.
Table II.
Association between MACC1 mRNA level and clinicopathological characteristics of patients with cervical cancer.
| MACC1 mRNA level | |||
|---|---|---|---|
| Characteristic | High | Low | P-value |
| Age, years | 0.570 | ||
| <45 | 8 | 10 | |
| ≥45 | 21 | 19 | |
| Tumor size, cm | 0.127 | ||
| <4 | 15 | 20 | |
| ≥4 | 14 | 8 | |
| HPV | 0.470 | ||
| Negative | 10 | 16 | |
| Positive | 19 | 12 | |
| Vascular involvement | 0.698 | ||
| Yes | 6 | 7 | |
| No | 23 | 21 | |
| FIGO stage | 0.047 | ||
| I–II | 10 | 17 | |
| III–IV | 19 | 11 | |
| Myometrium invasion | 0.002 | ||
| <1/2 | 12 | 23 | |
| ≥1/2 | 17 | 5 | |
| Lymph node metastasis | 0.022 | ||
| No | 12 | 20 | |
| Yes | 17 | 8 | |
MACC1, metastasis-associated in colon cancer 1; HPV, human papillomavirus; FIGO, International Federation of Gynecology and Obstetrics.
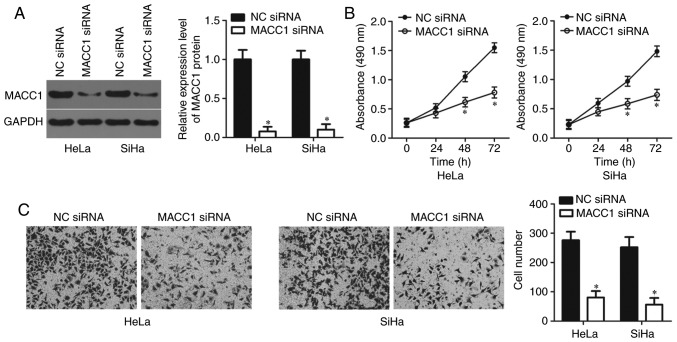
Knockdown of MACC1 restricts the proliferation and invasion of cervical cancer cells
In order to investigate the specific roles of MACC1 in cervical cancer, HeLa and SiHa cells were transfected with MACC1 siRNA with the aim of knocking down endogenous MACC1 expression. Transfection with NC siRNA served as the control group. Western blot analysis confirmed that the protein level of MACC1 was notably downregulated in HeLa and SiHa cells following MACC1 siRNA transfection (P<0.05; Fig. 4A). In addition, the results of the MTT and transwell cell invasion assays revealed that inhibition of MACC1 led to the decreased proliferative (P<0.05; Fig. 4B) and invasive (P<0.05; Fig. 4C) abilities of HeLa and SiHa cells, which was similar to the results demonstrated by miR-877 upregulation. Therefore, these results further suggest MACC1 as a functional downstream target of miR-877 in cervical cancer cells.
Figure 4.
MACC1 inhibition suppresses the proliferation and invasion of HeLa and SiHa cells. (A) MACC1 siRNA or NC siRNA was transfected into HeLa and SiHa cells. Following transfection, western blot analysis was used to measure MACC1 protein expression. (B) MTT and (C) transwell cell invasion assays (magnification, ×200) were performed to determine the proliferation and invasion abilities of HeLa and SiHa cells transfected with MACC1 siRNA or NC siRNA. *P<0.05 vs. NC siRNA. MACC1, metastasis-associated in colon cancer 1; siRNA, small interfering RNA; NC, negative control.
MACC1 restores miR-877 overexpression-mediated suppression of cervical cancer cell proliferation and invasion
To verify whether MACC1 mediates the inhibitory effects of miR-877 in cervical cancer cells, the miR-877-overexpression HeLa and SiHa cells were further transfected with MACC1 overexpression plasmid pCMV-MACC1 or empty pCMV plasmid. Firstly, RT-qPCR analysis confirmed that transfection with pCMV-MACC1 significantly increased the expression level of MACC1 in HeLa and SiHa cells (P<0.05; Fig. 5A). Following transfection, the western blot analysis indicated that the expression levels of MACC1 protein were markedly decreased in miR-877 overexpression-HeLa and SiHa cells; however, the decreased MACC1 protein expression was almost recovered following pCMV-MACC1 co-transfection (P<0.05; Fig. 5B). Furthermore, MTT and transwell cell invasion assays demonstrated that MACC1 reintroduction reversed the inhibitory effects of miR-877 overexpression on the proliferation (P<0.05; Fig. 5C) and invasion (P<0.05; Fig. 5D) of HeLa and SiHa cells. These results suggest that MACC1 inhibition is required for the suppressive effects of miR-877 on the malignant phenotypes of cervical cancer cells.
Figure 5.
MACC1 abolishes the miR-877-mediated inhibition of proliferation and invasion in cervical cancer cells. (A) RT-qPCR was performed to determine the expression of MACC1 mRNA in HeLa and SiHa cells after transfection with pCMV-MACC1 or empty pCMV plasmid. *P<0.05 vs. pCMV. (B) miR-877 mimics were co-transfected with pCMV-MACC1 or empty pCMV plasmids into HeLa and SiHa cells. Following transfection, western blot analysis was performed to examine the protein level of MACC1. (C) Proliferative and (D) invasive capacities of HeLa and SiHa cells co-transfected with miR-877 mimics and pCMV-MACC1 or empty pCMV plasmids were investigated using MTT and transwell cell invasion (magnification, ×200) assays, respectively. *P<0.05 vs. miR-NC. #P<0.05 vs. miR-877 mimics+pCMV. MACC1, metastasis-associated in colon cancer; miR, microRNA; NC, negative control.
Discussion
Dysregulation of miRNAs has been widely reported in cervical cancer, and dysregulated miRNAs play crucial roles in cervical cancer carcinogenesis and progression (26–29). miRNAs are able to regulate all major cancer-associated biological behaviors in cervical cancer, including differentiation, proliferation, the cell cycle, apoptosis, migration and metastasis (30–32). Hence, investigating the detailed roles of miRNAs that are aberrantly expressed in cervical cancer and the involved molecular mechanisms is essential for early diagnosis and effective therapeutic approaches. To the best of our knowledge, the present study detected the miR-877 expression levels in cervical cancer and determined its clinical value for the first time. More importantly, the functional roles and molecular mechanisms responsible for the action of miR-877 in development of cervical cancer were investigated.
A number of studies have reported that miR-877 was downregulated in hepatocellular carcinoma (15,16). Decreased miR-877 expression was significantly associated with the histologic grade and the Tumor-Node-Metastasis stage of patients with hepatocellular carcinoma (15). Patients with hepatocellular carcinoma that exhibit low expression levels of miR-877 had shorter overall survival and disease-free survival rates (15). Furthermore, miR-877 was validated as an independent biomarker for predicting the poor prognosis of patients with hepatocellular carcinoma (15). Low expression levels of miR-877 expression were also revealed in the blood and tissues of patients with renal cell carcinoma (17). However, the expression status of miR-877 in cervical cancer remains unknown. In the present study, RT-qPCR was used to measure miR-877 expression in both cervical cancer tissues and cell lines. The data revealed that miR-877 was downregulated in cervical cancer, and the downregulation of miR-877 was positively associated with the FIGO stage and lymph node metastasis. These results suggest that miR-877 may emerge as a potential biomarker for the diagnosis of patients with the aforementioned human cancer types.
The dysregulation of miR-877 has been validated as an important driver for human cancer progression. For example, returning miR-877 expression levels to the norm inhibited hepatocellular carcinoma cell proliferation, colony formation, migration and invasion, as well as improving the chemosensitivity to paclitaxel (15,16). In renal cell carcinoma, miR-877 restoration suppressed the cellular proliferative and migratory capacities of renal cell carcinoma in vitro (17). However, the specific roles of miR-877 in the progression and development of cervical cancer remain uncertain. To elucidate these roles, MTT and transwell cell invasion assays were performed, which revealed that the overexpression of miR-877 inhibited the proliferation and invasion of cervical cancer cells. These observations suggest that miR-877 may be considered as a potential therapeutic target for treating patients with these specific types of human cancer.
Several genes, including cyclin-dependent kinase 14 (15), Forkhead box protein M1 (16) and eukaryotic elongation factor-2 kinase (17), have been verified as direct targets of miR-877. The mechanisms underlying the tumor-suppressive roles of miR-877 in cervical cancer were investigated in the present study. MACC1 was identified to be a direct target gene of miR-877 in cervical cancer cells.
MACC1, located on human chromosome 7 (7p21.1), was previously reported as overexpressed in multiple types of human cancer, such as glioma (33), breast cancer (34), hepatocellular carcinoma (35) and renal cell carcinoma (36). MACC1 was also observed as highly expressed in cervical cancer tissues and cell lines. Increased MACC1 expression levels were significantly correlated with the FIGO stage, pelvic lymph node metastasis and recurrence (20,21). Patients with cervical cancer with high MACC1 expression levels exhibited shorter overall survival times than those patients with low MACC1 expression (21). Furthermore, multivariate analyses validated MACC1 as an independent biomarker for predicting the overall survival of patients with cervical cancer (21). MACC1 has a role in oncogenic activity in the carcinogenesis and development of cervical cancer, and regulated a variety of aggressive behaviors (22–25). Herein, it was demonstrated that miR-877 was able to directly target MACC1 and inhibit the malignant progression of cervical cancer. Hence, the miR-877/MACC1 axis may represent a potential therapeutic target for patients with cervical cancer.
In conclusion, the present study indicated that miR-877 was downregulated in cervical cancer. Decreased miR-877 expression was significantly associated with the increased FIGO stage and lymph node metastasis. miR-877 upregulation restricted the proliferation and invasion of cervical cancer cells by directly targeting MACC1. The observations of the present study suggest that miR-877 may be a potential target for the therapy of cervical cancer, and provide a new technique for the prevention and treatment of patients with this disease. The present study was not without limitations; the association between miR-877 and prognosis of patients with cervical cancer was not investigated. Future investigations should collect the prognosis data and examine the association between miR-877 and prognosis of patients with cervical cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Youth Foundation of Jilin Province Science and Technology Development Plan (grant no. 20160520145JH).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
BS and FM designed the present study and performed the statistical analyses. FM, JO, JL and XL performed the RT-qPCR, the western blot analysis and the MTT assay. Transwell cell invasion and luciferase reporter assays were performed by YM, LY and PD. BS wrote the article. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Jilin Cancer Hospital (Changchun, China). All patients enrolled in the present study provided written informed consent based on the principles of the Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Su SY, Huang JY, Ho CC, Liaw YP. Evidence for cervical cancer mortality with screening program in Taiwan, 1981–2010: Age-period-cohort model. BMC Public Health. 2013;13:13. doi: 10.1186/1471-2458-13-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hildesheim A, Wang SS. Host and viral genetics and risk of cervical cancer: A review. Virus Res. 2002;89:229–240. doi: 10.1016/S0168-1702(02)00191-0. [DOI] [PubMed] [Google Scholar]
- 4.Sakuragi N. Up-to-date management of lymph node metastasis and the role of tailored lymphadenectomy in cervical cancer. Int J Clin Oncol. 2007;12:165–175. doi: 10.1007/s10147-007-0661-2. [DOI] [PubMed] [Google Scholar]
- 5.Mayr NA, Huang Z, Wang JZ, Lo SS, Fan JM, Grecula JC, Sammet S, Sammet CL, Jia G, Zhang J, et al. Characterizing tumor heterogeneity with functional imaging and quantifying high-risk tumor volume for early prediction of treatment outcome: Cervical cancer as a model. Int J Radiat Oncol Biol Phys. 2012;83:972–979. doi: 10.1016/j.ijrobp.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aigner A. MicroRNAs (miRNAs) in cancer invasion and metastasis: Therapeutic approaches based on metastasis-related miRNAs. J Mol Med (Berl) 2011;89:445–457. doi: 10.1007/s00109-010-0716-0. [DOI] [PubMed] [Google Scholar]
- 9.Cho WC. MicroRNAs: Potential biomarkers for cancer diagnosis, prognosis and targets for therapy. Int J Biochem Cell Biol. 2010;42:1273–1281. doi: 10.1016/j.biocel.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 10.Zhang H, Lu Y, Wang S, Sheng X, Zhang S. MicroRNA-152 acts as a tumor suppressor microRNA by inhibiting Kruppel-like factor 5 in human cervical cancer. Oncol Res. 2019;27:335–340. doi: 10.3727/096504018X15252202178408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu LM, Wang WW, Qi R, Leng TG, Zhang XL. MicroRNA-224 inhibition prevents progression of cervical carcinoma by targeting PTX3. J Cell Biochem. 2018;119:10278–10290. doi: 10.1002/jcb.27370. [DOI] [PubMed] [Google Scholar]
- 12.Jiang Y, Hou R, Li S, Li S, Dang G. MicroRNA-302 inhibits cell migration and invasion in cervical cancer by targeting DCUN1D1. Exp Ther Med. 2018;16:1000–1008. doi: 10.3892/etm.2018.6223. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Chen Z, Zhang M, Qiao Y, Yang J, Yin Q. MicroRNA-1297 contributes to the progression of human cervical carcinoma through PTEN. Artif Cells Nanomed Biotechnol 46 (Sup2) 2018:S1120–S1126. doi: 10.1080/21691401.2018.1479711. [DOI] [PubMed] [Google Scholar]
- 14.Srivastava SK, Ahmad A, Zubair H, Miree O, Singh S, Rocconi RP, Scalici J, Singh AP. MicroRNAs in gynecological cancers: Small molecules with big implications. Cancer Lett. 2017;407:123–138. doi: 10.1016/j.canlet.2017.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan TH, Qiu C, Sun J, Li WH. MiR-877-5p suppresses cell growth, migration and invasion by targeting cyclin dependent kinase 14 and predicts prognosis in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2018;22:3038–3046. doi: 10.26355/eurrev_201805_15061. [DOI] [PubMed] [Google Scholar]
- 16.Huang X, Qin J, Lu S. Up-regulation of miR-877 induced by paclitaxel inhibits hepatocellular carcinoma cell proliferation though targeting FOXM1. Int J Clin Exp Pathol. 2015;8:1515–1524. [PMC free article] [PubMed] [Google Scholar]
- 17.Shi Q, Xu X, Liu Q, Luo F, Shi J, He X. MicroRNA-877 acts as a tumor suppressor by directly targeting eEF2K in renal cell carcinoma. Oncol Lett. 2016;11:1474–1480. doi: 10.3892/ol.2015.4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Berek JS, Matsuo K, Grubbs BH, Gaffney DK, Lee SI, Kilcoyne A, Cheon GJ, Yoo CW, Li L, Shao Y, et al. Multidisciplinary perspectives on newly revised 2018 FIGO staging of cancer of the cervix uteri. J Gynecol Oncol. 2019;30:e40. doi: 10.3802/jgo.2019.30.e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou X, Xu CJ, Wang JX, Dai T, Ye YP, Cui YM, Liao WT, Wu XL, Ou JP. Metastasis-associated in colon cancer-1 associates with poor prognosis and promotes cell invasion and angiogenesis in human cervical Cancer. Int J Gynecol Cancer. 2015;25:1353–1363. doi: 10.1097/IGC.0000000000000524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo L, Lu W, Zhang X, Luo D, Zhang H. Metastasis-associated colon cancer-1 is a novel prognostic marker for cervical cancer. Int J Clin Exp Pathol. 2014;7:4150–4155. [PMC free article] [PubMed] [Google Scholar]
- 22.Chai H, Yang Y. Effects of MACC1 siRNA on biological behaviors of HeLa. Arch Gynecol Obstet. 2014;289:1271–1280. doi: 10.1007/s00404-013-3126-z. [DOI] [PubMed] [Google Scholar]
- 23.Chen XP, Ren XP, Lan JY, Chen YG, Shen ZJ. Analysis of HGF, MACC1, C-met and apoptosis-related genes in cervical carcinoma mice. Mol Biol Rep. 2014;41:1247–1256. doi: 10.1007/s11033-013-2969-5. [DOI] [PubMed] [Google Scholar]
- 24.Hua F, Xia Y, Wang H, Chen R, Ren Y, Yang J, Liang W. Effects of small interfering RNA silencing MACC-1 expression on cell proliferation, cell cycle and invasion ability of cervical cancer SiHa cells. Zhonghua Zhong Liu Za Zhi. 2014;36:496–500. (In Chinese) [PubMed] [Google Scholar]
- 25.Hua FF, Liu SS, Zhu LH, Wang YH, Liang X, Ma N, Shi HR. MiRNA-338-3p regulates cervical cancer cells proliferation by targeting MACC1 through MAPK signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21:5342–5352. doi: 10.26355/eurrev_201712_13919. [DOI] [PubMed] [Google Scholar]
- 26.Wang F, Li B, Xie X. The roles and clinical significance of microRNAs in cervical cancer. Histol Histopathol. 2016;31:131–139. [PubMed] [Google Scholar]
- 27.Servín-González LS, Granados-López AJ, López JA. Families of microRNAs expressed in clusters regulate cell signaling in cervical cancer. Int J Mol Sci. 2015;16:12773–12790. doi: 10.3390/ijms160612773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Díaz-González Sdel M, Deas J, Benítez-Boijseauneau O, Gómez-Cerón C, Bermúdez-Morales VH, Rodríguez-Dorantes M, Pérez-Plasencia C, Peralta-Zaragoza O. Utility of microRNAs and siRNAs in cervical carcinogenesis. Biomed Res Int. 2015;2015:374924. doi: 10.1155/2015/374924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pedroza-Torres A, López-Urrutia E, Garcia-Castillo V, Jacobo-Herrera N, Herrera LA, Peralta-Zaragoza O, López-Camarillo C, De Leon DC, Fernández-Retana J, Cerna-Cortés JF, Pérez-Plasencia C. MicroRNAs in cervical cancer: Evidences for a miRNA profile deregulated by HPV and its impact on radio-resistance. Molecules. 2014;19:6263–6281. doi: 10.3390/molecules19056263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li JH, Zhang Z, Du MZ, Guan YC, Yao JN, Yu HY, Wang BJ, Wang XL, Wu SL, Li Z. microRNA-141-3p fosters the growth, invasion, and tumorigenesis of cervical cancer cells by targeting FOXA2. Arch Biochem Biophys. 2018;657:23–30. doi: 10.1016/j.abb.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 31.Shang A, Zhou C, Bian G, Chen W, Lu W, Wang W, Li D. miR-381-3p restrains cervical cancer progression by downregulating FGF7. J Cell Biochem. 2019;120:778–789. doi: 10.1002/jcb.27438. [DOI] [PubMed] [Google Scholar]
- 32.Cai N, Hu L, Xie Y, Gao JH, Zhai W, Wang L, Jin QJ, Qin CY, Qiang R. MiR-17-5p promotes cervical cancer cell proliferation and metastasis by targeting transforming growth factor-β receptor 2. Eur Rev Med Pharmacol Sci. 2018;22:1899–1906. doi: 10.26355/eurrev_201804_14712. [DOI] [PubMed] [Google Scholar]
- 33.Yang T, Kong B, Kuang YQ, Cheng L, Gu JW, Zhang JH, Shu HF, Yu SX, He WQ, Xing XM, Huang HD. Overexpression of MACC1 protein and its clinical implications in patients with glioma. Tumour Biol. 2014;35:815–819. doi: 10.1007/s13277-013-1112-5. [DOI] [PubMed] [Google Scholar]
- 34.Huang Y, Zhang H, Cai J, Fang L, Wu J, Ye C, Zhu X, Li M. Overexpression of MACC1 and Its significance in human breast cancer progression. Cell Biosci. 2013;3:16. doi: 10.1186/2045-3701-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun DW, Zhang YY, Qi Y, Liu GQ, Chen YG, Ma J, Lv GY. Prognostic and clinicopathological significance of MACC1 expression in hepatocellular carcinoma patients: A meta-analysis. Int J Clin Exp Med. 2015;8:4769–4777. [PMC free article] [PubMed] [Google Scholar]
- 36.Jin Z, Xu N, Guo K, Xu P, Li P, Zhang Y, Li X, Zheng S, Liu C, Xu A, Huang P. Increased expression of metastasis-associated in colon cancer-1 in renal cell carcinoma is associated with poor prognosis. Int J Clin Exp Pathol. 2015;8:3857–3863. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.