Abstract
Long non-coding RNAs (lncRNAs) serve important roles in colorectal cancer. The aim of the present study was to investigate the expression and role of cervical carcinoma expressed PCNA regulatory (CCEPR) lncRNA in colorectal cancer progression. The results demonstrated that CCEPR expression was significantly higher in colorectal cancer tissues when compared with paired adjacent normal tissues. In addition, CCEPR expression was significantly higher in patients with advanced colorectal cancer (stage III/IV) than those with early-stage colorectal cancer (stage I/II). High CCEPR expression was significantly associated with poor differentiation, advanced clinical stage, positive lymph node metastasis and distant metastasis. Of particular note, patients with colorectal cancer exhibiting high CCEPR expression levels had shorter survival rates when compared with patients with low CCEPR expression. In vitro experiments demonstrated that the expression of CCEPR was increased in colorectal cancer cell lines when compared with a normal colon cell line. Knockdown of CCEPR significantly inhibited colorectal cancer cell proliferation, colony formation and cell cycle progression, as well as cell migration and invasion. Finally, silencing of CCEPR downregulated matrix metalloproteinase (MMP)-2 and MMP-9 expression and suppressed epithelial-mesenchymal transition in colorectal cancer cells. In conclusion, the results of the present study suggest that CCEPR may exert an oncogenic role in colorectal cancer, and CCEPR may be a promising molecular target for colorectal cancer treatment.
Keywords: long non-coding RNA, cervical carcinoma expressed PCNA regulatory lncRNA, colorectal cancer
Introduction
Colorectal cancer is one of the most common malignant tumours worldwide, and has the third highest incidence rates among all malignancies (1–3). Despite significant advances in surgical resection combined with radiochemotherapy, the prognosis of patients with advanced stage colorectal cancer remains poor with a five-year survival rate of <20% (4,5). This is thought to be mainly due to the rapid growth and widespread metastasis of the cancer. The molecular mechanisms underlying colorectal cancer growth and metastasis is currently unclear. Understanding the molecular mechanisms may facilitate the identification of novel and effective therapeutic targets (6).
Long non-coding RNAs (lncRNAs) are a class of RNAs (>200 nucleotides in length) with no protein-coding ability, that regulate gene expression through lncRNA-mRNA interactions, lncRNA-microRNA (miR) interactions and lncRNA-protein interactions (7–9). Recently, an increasing number of lncRNAs have been reported to serve key roles in various biological processes, including cell proliferation, colony formation, cell cycle progression, cell migration, cell invasion and epithelial-mesenchymal transition (EMT) (10–12). In addition, deregulation of various lncRNAs has been observed in several human malignancies (including colorectal cancer) and lncRNAs have been demonstrated to serve either oncogenic or tumour suppressive roles (10–13). For instance, expression of the lncRNA, HOX transcript antisense RNA, is significantly upregulated in colorectal cancer and regulates the progression and chemoresistance of colorectal cancer by modulating miR-203a-3p expression and the activity of the Wnt/β-catenin signalling pathway (5).
Cervical carcinoma expressed PCNA regulatory lncRNA (CCEPR) is an lncRNA localized to the 10q21.1 chromosomal region. CCEPR expression was initially identified to be significantly upregulated in cervical cancer and is associated with poor patient prognosis (14). Yang et al (15) reported that CCEPR promotes cervical cancer cell proliferation via the upregulation of proliferating cell nuclear antigen (PCNA) expression. In addition, CCEPR exerts oncogenic roles in gastric cancer, lung cancer, bladder cancer and liver cancer (16–19). Recently, Gaballah et al (20) identified that CCEPR expression is significantly upregulated in colorectal cancer, and its expression is positively correlated with the expression of phosphorylated (p)-ERK1/2, cyclooxygenase (COX)-2, cyclin D1 and PCNA (20). This suggests that CCEPR may be involved in colorectal cancer progression by modulating the ERK/COX-2 signalling pathway and cell proliferation activity. However, the detailed role of CCEPR during colorectal cancer progression remains currently unclear.
Matrix metalloproteinase (MMP)-2 and MMP-9 are members of the MMP gene family. They function as zinc-dependent enzymes that cleave extracellular matrix components (21). It has been demonstrated that MMP-2 and MMP-9 serve crucial roles in tumour cell migration and invasion (21). EMT is characterized by the transition of cells from an epithelial-like phenotype to a mesenchymal phenotype, which causes tumour cells to acquire invasive and migratory capacities (22–24). In addition, MMP-2 and MMP-9 have been implicated in the process of EMT (25). However, whether CCEPR affects the expression of MMP-2 and MMP-9 and the process of EMT in colorectal cancer has not yet been explored.
The aims of the present study were to evaluate the clinical significance of CCEPR expression in colorectal cancer, and to investigate the function of CCEPR in regulating the malignant phenotypes of colorectal cancer cells in vitro.
Materials and methods
Clinical tissue samples
The present study was approved by the Ethics Committee of the Hunan Provincial People's Hospital (Changsha, China). A total of 58 colorectal cancer tissues and paired adjacent normal tissues were collected from 58 patients with primary colorectal cancer admitted to Hunan Provincial People's Hospital between April 2012 and May 2013. These patients included 38 male and 20 female, from 33 years old to 75 years old. All patients provided written informed consent. The inclusion criteria were that all patients only had primary colorectal cancers. The exclusion criteria were colorectal cancer patients who had received chemotherapy or radiotherapy prior to undergoing surgery. The collected tissues were frozen using liquid nitrogen shortly after surgical resection, and stored at −80°C until experimentation. The total duration of follow-up was 5 years.
Cell culture and transfection
Human colorectal cancer cell lines, HT29, Caco2, SW480 and LS174T, and the normal human intestinal epithelial cell line, HIEC, were purchased from the American Type Culture Collection. Cells were cultured in DMEM (Thermo Fisher Scientific, Inc.) containing 10% FBS (Thermo Fisher Scientific, Inc.) and maintained in a humidified atmosphere at 37°C and 5% CO2. The HT29 and Caco-2 cell lines were established from primary adenocarcinomas of the colon. The SW480 and LS174T cell lines were established from a Duke's type B adenocarcinoma of the colon. HT29 and SW480 cells (5×106 cells per well) were seeded in six-well plates and transfected with 100 nM CCEPR small interfering (si) RNA (siCCEPR; Shanghai GenePharma Co., Ltd.) or 100 nM scrambled negative control (NC) siRNA (siNC; Shanghai GenePharma Co., Ltd.) using Lipofectamine 2000 (Thermo Fisher Scientific, Inc.) in accordance with the manufacturer's instructions. The target sequence of the CCEPR siRNA was 5′-CGAGGGCGAGCATGTTTGTTGTTTA-3′ (15). The NC siRNA sequences were 5′-TCAAGUCCACGACGACTTTG-3′. At 48-h post-transfection, the subsequent experiments were conducted.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from the tissue samples or cells using TRIzol reagent (Thermo Fisher Scientific, Inc.) in accordance with the manufacturer's instructions. Subsequently, 1 µg total RNA was used to perform RT-qPCR using a SuperScript III Platinum SYBR Green One-Step RT-qPCR kit (Thermo Fisher Scientific, Inc.) and the ABI 7500 real-time PCR system (Thermo Fisher Scientific, Inc.) in accordance with the manufacturer's instructions. The thermocycling conditions for reverse transcription were as follows: 16°C for 30 min, 42°C for 30 min and 85°C for 5 min. The thermocycling conditions for PCR were as follows: 95°C for 1 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 30 sec. Relative mRNA expression levels were calculated using the 2−ΔΔCq method (26) normalized to the internal reference gene GAPDH. The primer sequences were as follows: CCEPR forward, 5′-AAGGTCCCAGGATACTCGC-3′, and reverse, 5′-GTGTCGTGGACTGGCAAAAT-3′; GAPDH forward, 5′-ACAACTTTGGTATCGTGGAAGG-3′, and reverse, 5′-GCCATCACGCCACAGTTTC-3′. This assay was repeated 3 times.
Cell proliferation assay
Cell proliferation was examined using a Cell Counting kit-8 (CCK-8) assay (Thermo Fisher Scientific, Inc.). Briefly, HT29 and SW480 cells (10,000 cells/well) were seeded in 96-well plates and incubated at 37°C for 0, 24, 48 or 72 h. At the indicated time points, 10 µl CCK-8 reagent (Thermo Fisher Scientific, Inc.) was added to each well. The cells were subsequently incubated at 37°C for 2 h. The optical density at 450 nm was determined using a microplate reader (Bio-Rad Laboratories, Inc.).
Colony formation assay
Transfected HT29 and SW480 cells were seeded into six-well plates at a density of 200 cells/well. Following culture at 37°C for 14 days, the cells were washed with PBS (Thermo Fisher Scientific, Inc.) twice before they were stained with 0.5% crystal violet (Thermo Fisher Scientific, Inc.) at room temperature for 10 min. The number of colonies (containing >50 cells) were then counted under a light microscope (magnification, ×200).
Cell cycle analysis
Flow cytometry was used for cell cycle analysis. Briefly, transfected HT29 and SW480 cells were first fixed in 75% ethanol at 4°C overnight, and washed with PBS three times. The cells were then permeabilized using eBioscience™ Permeabilization Buffer (Thermo Fisher Scientific, Inc.) at 37°C for 30 min then stained with 500 µl propidium iodide solution (Thermo Fisher Scientific, Inc.) at 4°C for 30 min. The cell cycle distribution was determined using a FACScan flow cytometer (Becton, Dickinson and Company) and BD Accuri C6 system (32-bit) software version 1.0 (BD Biosciences).
Cell migration assay
A wound healing assay was performed to examine cell migration. Briefly, the transfected HT29 and SW480 cells were seeded into 12-well plates and incubated at 37°C without serum to ~90% confluence. Wounds were generated using a 100-µl pipette tip and the cells were then incubated at 37°C for 24 h. The wounds were photographed at 0 and 24-h using an inverted microscope (magnification, ×200; Olympus Corporation) and measured using ImageJ software version 1.8 (National Institutes of Health).
Cell invasion assay
Cell invasion was examined using a 24-well Transwell chamber (8 mm pore size; Corning, Inc.) pre-coated with Matrigel for 1-h at room temperature (EMD Millipore; Merck KGaA). Transfected HT29 and SW480 cells (50,000 cells/well) were seeded in 300 µl serum-free DMEM to the upper chamber. A total of 500 µl DMEM containing 20% FBS was added into the bottom chamber. Following incubation at 37°C for 24 h, cells on the insert were carefully removed with a cotton-tipped swab. The cells that had invaded through the membrane were stained with 0.5% crystal violet at room temperature for 10 min and photographed under an inverted microscope (magnification, ×200).
Western blot analysis
Transfected HT29 and SW480 cells were lysed using radioimmunoprecipitation buffer (Beyotime Institute of Biotechnology) according to the manufacturer's protocols. A bicinchoninic acid protein assay kit (Pierce; Thermo Fisher Scientific, Inc.) was then used to determine the protein concentrations. Protein samples (60 µg/lane) were separated by 12% SDS-PAGE before they were transferred to polyvinylidene difluoride membranes (Beyotime Institute of Biotechnology). The membranes were blocked with 5% non-fat dry milk overnight at 4°C. After washing with PBS at room temperature three times, the membranes were incubated with rabbit anti-human E-cadherin (dilution, 1:200; cat. no. ab40772; Abcam), rabbit anti-human N-cadherin (dilution, 1:500; cat. no. ab18203; Abcam), rabbit anti-human vimentin (dilution, 1:200, cat. no. ab92547; Abcam), rabbit anti-human MMP-2 (dilution, 1:500, cat. no. ab181286; Abcam), rabbit anti-human MMP-9 (dilution, 1:200, cat. no. ab137867; Abcam), or rabbit anti-human GAPDH (dilution 1:200, cat. no. ab181602; Abcam) primary antibodies at room temperature for 4 h. After washing with PBS at room temperature three times, the membranes were then incubated with a horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (dilution, 1:5,000, cat. no. ab6721; Abcam) at room temperature for 30 min. Chemiluminescence was examined using SuperSignal™ West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. ImageJ software (v1.46; National Institutes of Health) was used for densitometry analysis with GAPDH as the loading control.
Statistical analysis
Data are presented as the mean ± standard deviation and were analysed using GraphPad Prism 3.0 software (GraphPad Software, Inc.). Differences between 2 groups were analysed using paired or unpaired Student's t-tests. Differences between multiple groups were analysed by one-way analysis of variance followed by a post hoc Tukey's test. The association between CCEPR expression and clinicopathological characteristics of colorectal cancer patients was analysed using chi-square tests. Survival analysis was performed using Kaplan-Meier survival curves and log-rank tests. P<0.05 was considered to indicate a statistically significant difference.
Results
Increased CCEPR expression is associated with colorectal cancer progression
In the present study, the expression of CCEPR in 58 primary colorectal cancer tissues and adjacent normal tissues was first examined. RT-qPCR results indicated that CCEPR expression was significantly upregulated in colorectal cancer tissues compared with adjacent normal tissues (Fig. 1A). In addition, CCEPR expression levels were significantly higher in advanced colorectal cancer tissues (III/IV) than in early-stage colorectal cancer tissues (I/II; Fig. 1B). The association between CCEPR expression and the clinicopathological characteristics of colorectal cancer patients was then determined. Using the median expression value of CCEPR as a cut-off value (2.71), patients were divided into low and high CCEPR expression groups. Statistical analysis of the results demonstrated that high CCEPR expression was significantly associated with poor differentiation, positive lymph node metastasis, distant metastasis and advanced clinical stage (Table I). This suggests that upregulation of CCEPR expression may serve a key role during colorectal cancer progression. Of particular note, patients with high CCEPR expression levels exhibited shorter survival rates than patients with low CCEPR expression levels over 5 years (Fig. 1C), suggesting that CCEPR expression may be a promising predictive marker for colorectal cancer prognosis.
Figure 1.
Upregulation of CCEPR is associated with colorectal cancer progression. (A) RT-qPCR analysis demonstrated that CCEPR is more highly upregulated in colorectal cancer tissues when compared with adjacent normal tissues (**P<0.01 vs. adjacent tissue). (B) RT-qPCR analysis demonstrated that CCEPR expression is higher in advanced colorectal cancer tissues (stage III/IV) than in early-stage colorectal cancer tissues (stage I/II; **P<0.01 vs. stage I/II). (C) Patients with colorectal cancer exhibiting with high CCEPR expression had shorter overall survival rates compared with those exhibiting low CCEPR expression. CCEPR, cervical carcinoma expressed PCNA regulatory lncRNA; lncRNA, long non-coding RNA; RT-qPCR, reverse transcription-quantitative PCR.
Table I.
Association between CCEPR expression and clinicopathological characteristics of patients with colorectal cancer.
| Variables | Cases (n=58) | Low CCEPR levels (n=33) | High CCEPR levels (n=25) | P-value |
|---|---|---|---|---|
| Age (years) | 0.384 | |||
| ≤55 | 31 | 16 | 15 | |
| >55 | 27 | 17 | 10 | |
| Sex | 0.832 | |||
| Male | 38 | 22 | 16 | |
| Female | 20 | 11 | 9 | |
| Differentiation | 0.032a | |||
| Well and moderately | 41 | 27 | 14 | |
| Poor | 17 | 6 | 11 | |
| Node metastasis | 0.043a | |||
| Present | 26 | 11 | 15 | |
| Absent | 32 | 22 | 10 | |
| Distant metastasis | 0.003a | |||
| Present | 9 | 1 | 8 | |
| Absent | 49 | 32 | 17 | |
| Clinical stage | 0.043a | |||
| I–II | 32 | 22 | 10 | |
| III–IV | 26 | 11 | 15 |
P<0.05. CCEPR, cervical carcinoma expressed PCNA regulatory lncRNA; lncRNA, long non-coding RNA.
Knockdown of CCEPR inhibits the growth of colorectal cancer cells
In vitro experiments were performed to investigate the function of CCEPR in colorectal cancer further. The expression levels of CCEPR were examined in several human colorectal cancer cell lines including HT29, Caco-2, SW480 and LS174T, as well as in the normal human intestinal epithelial cell line, HIEC. RT-qPCR results demonstrated that CCEPR expression was significantly increased in colorectal cancer cell lines compared with HIEC cells (Fig. 2A). HT29 and SW480 cells exhibited the highest CCEPR expression levels and were therefore selected for subsequent experiments. As CCEPR expression was observed to be significantly upregulated in colorectal cancer samples, CCEPR siRNA was transfected into HT29 and SW480 cells to reduce its expression levels. Following transfection, CCEPR levels were significantly decreased in the siCCEPR group compared with the siNC group (Fig. 2B). A CCK-8 assay was then performed to assess cell proliferation. The proliferation of the HT29 and SW480 cells in the siCCEPR group was significantly suppressed compared with cells in the siNC group (Fig. 2C and D). Thus, CCEPR may serve an oncogenic role in colorectal cancer growth. A colony formation assay was then performed to examine the effects of CCEPR downregulation on the colony formation capacity of colorectal cancer cells. The results indicated that the colony formation capacity of cells in the siCCEPR group was significantly inhibited when compared with cells in the siNC group (Fig. 2E). To further confirm these findings, flow cytometry was utilised to examine the effects of CCEPR downregulation on cell cycle progression in colorectal cancer cells. The results indicated that silencing CCEPR in HT29 and SW480 cells led to significant cell cycle arrest in the G1 stage (Fig. 2F and G). Therefore, these results demonstrated that knockdown of CCEPR inhibited the growth of colorectal cancer cells.
Figure 2.
Inhibition of CCEPR suppresses colorectal cancer cell growth in vitro. (A) CCEPR expression levels were significantly higher in colorectal cancer cell lines compared with the HIEC cell line (**P<0.01 vs. HIEC). (B) HT29 and SW480 cells were transfected with siNC or siCCEPR, and CCEPR levels were then examined using reverse transcription-quantitative PCR. The results demonstrated successful transfection efficiency. A cell counting kit-8 assay was used to determine cell proliferation in (C) HT29 and (D) SW480 cell lines. (E) Colony formation assays demonstrated that CCEPR silencing significantly reduced colony formation in HT29 and SW480 cells. The cell cycle distribution of (F) HT29 and (G) SW480 cells. **P<0.01 vs. siNC. CCEPR, cervical carcinoma expressed PCNA regulatory lncRNA, lncRNA, long non-coding RNA; si, small interfering RNA; NC, negative control; OD, optical density.
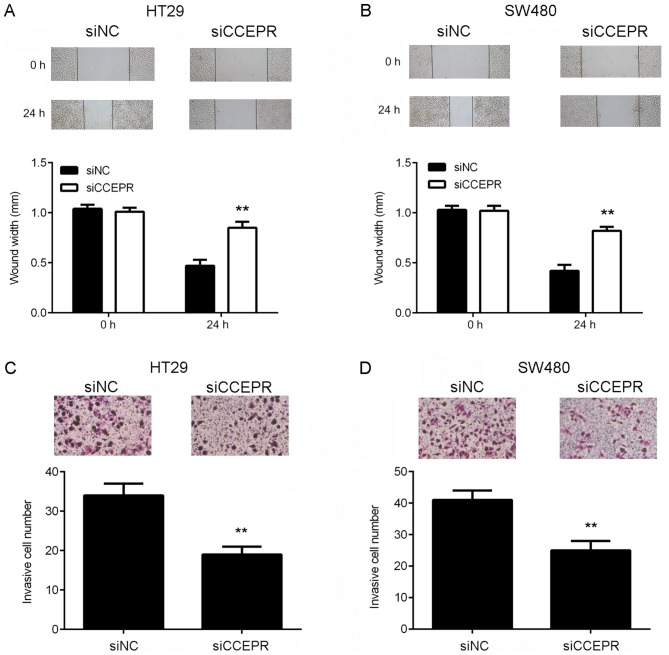
Knockdown of CCEPR suppresses the migration, invasion and EMT of colorectal cancer cells
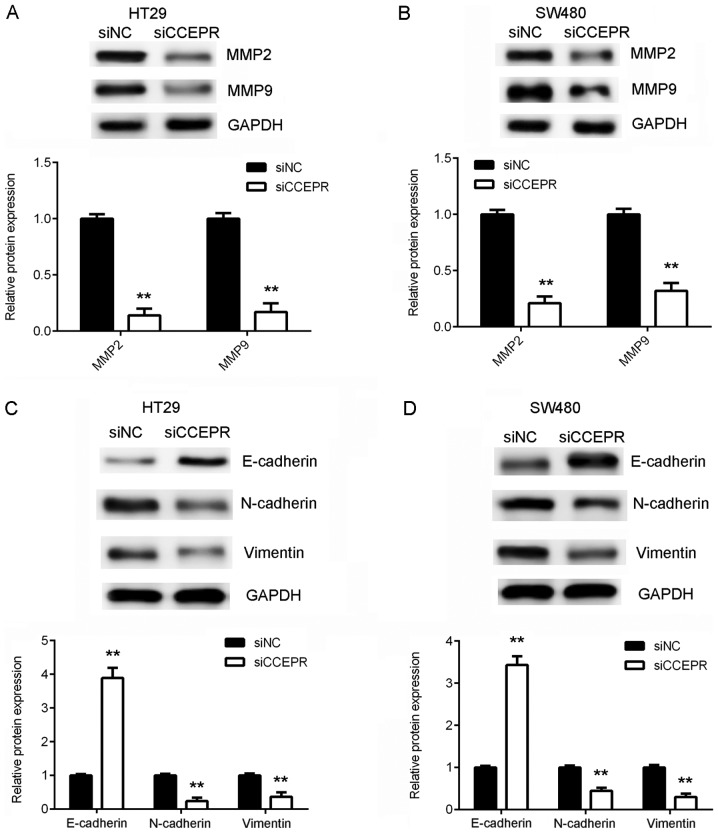
Following the cell growth experiments, the function of CCEPR in colorectal cancer metastasis in vitro was investigated. Wound healing and Transwell assays were performed to examine the effects of CCEPR downregulation on colorectal cancer cell migration and invasion, respectively. The results of the wound healing assay demonstrated that the migratory capacity of the HT29 and SW480 cells in the siCCEPR group was significantly attenuated when compared with the cells in the siNC group (Fig. 3A and B). In addition, the Transwell assay revealed that the number of invasive cells in the siCCEPR group was significantly decreased when compared with the siNC group (Fig. 3C and D), indicating that knockdown of CCEPR suppressed colorectal cancer cell invasion. Taking the results presented thus far into consideration, the authors of the present study hypothesized that CCEPR may serve an oncogenic role in colorectal cancer metastasis. To further confirm this hypothesis, the protein expression levels of MMP-2 and MMP-9 were then analysed. The results demonstrated that knockdown of CCEPR significantly downregulated MMP-2 and MMP-9 expression in HT29 and SW480 cells (Fig. 4A and B). EMT is known to serve an essential role in cancer cell migration and invasion (10). Therefore, the effects of CCEPR downregulation on EMT in colorectal cancer cells was subsequently investigated. In HT29 and SW480 cells from the siCCEPR group, the protein expression levels of E-cadherin (an epithelial marker) were significantly increased, while the protein expression levels of N-cadherin and vimentin (mesenchymal markers) were reduced when compared with cells from the siNC group (Fig. 4C and D). This indicated that knockdown of CCEPR significantly inhibited EMT in colorectal cancer cells. Therefore, it was hypothesized that silencing CCEPR may inhibit colorectal cancer cell migration and invasion via suppression of EMT.
Figure 3.
Inhibition of CCEPR suppresses colorectal cancer cell migration and invasion. Cells were transfected with siCCEPR or siNC and wound healing assay analysis demonstrated that CCEPR knockdown significantly reduced cell migration in (A) HT29 and (B) SW480 cell lines. Transwell assay analysis demonstrated that CCEPR knockdown reduced cell invasion significantly in (C) HT29 and (D) SW480 cell lines. **P<0.01 vs. siNC. CCEPR, cervical carcinoma expressed PCNA regulatory lncRNA; lncRNA, long non-coding RNA; si, small interfering RNA; NC, negative control.
Figure 4.
Knockdown of CCEPR inhibits EMT in colorectal cancer cells. MMP-2 and MMP-9 protein expression was significantly decreased following CCEPR knockdown in (A) HT29 and (B) SW480 cell lines. E-cadherin protein expression was significantly increased, while N-cadherin and vimentin protein expression was significantly decreased, following CCEPR knockdown in (C) HT29 and (D) SW480 cell lines. **P<0.01 vs. siNC. CCEPR, cervical carcinoma expressed PCNA regulatory lncRNA; lncRNA, long non-coding RNA; EMT, epithelial-mesenchymal transition; MMP, matrix metalloproteinase; si, small interfering RNA; NC, negative control.
Discussion
The underlying molecular mechanisms of CCEPR in colorectal cancer progression is currently unknown. The present study observed that the expression of CCEPR was significantly higher in colorectal cancer tissues when compared with paired adjacent normal tissues. In addition, CCEPR expression was significantly higher in advanced colorectal cancer tissues (stage III/IV) than early-stage colorectal cancer tissues (stage I/II). High CCEPR expression was significantly associated with poor differentiation, advanced clinical stage, positive lymph node metastasis and distant metastasis. Of particular note, patients with colorectal cancer exhibiting high CCEPR expression had shorter survival rates when compared with patients exhibiting low CCEPR expression. Subsequent in vitro experiments determined that the expression of CCEPR was significantly higher in colorectal cancer cell lines when compared with a normal colon cell line. Knockdown of CCEPR significantly inhibited colorectal cancer cell proliferation, colony formation, cell cycle progression, cell migration and invasion. In addition, silencing of CCEPR also downregulated MMP-2 and MMP-9 expression, and suppressed EMT in colorectal cancer cells.
An increasing number of lncRNAs have been reported to serve as important mediators in human cancers (27,28). Specific lncRNAs are significantly deregulated in colorectal cancer and have been demonstrated to promote tumour growth and metastasis. For instance, the growth arrest-specific 5 lncRNA is a prognostic marker in colorectal cancer that has been demonstrated to inhibit tumour cell proliferation, induce G0/G1 arrest and apoptosis (29). In addition, the small nucleolar RNA host gene 12 lncRNA promotes the growth and inhibits apoptosis of colorectal cancer cells (30). The present study observed that the expression levels of CCEPR were significantly increased in colorectal cancer tissues and cell lines when compared with adjacent normal tissues and non-tumour HIEC cells, respectively. Consistent with these observations, Gaballah et al (20) also reported that CCEPR expression was upregulated in 60 colorectal cancer tissues than in adjacent normal tissues. The current study also demonstrated that the expression levels of CCEPR were higher in advanced colorectal cancer tissues (stage III/IV) compared with early-stage colorectal cancer tissues (stage I/II). Further investigation demonstrated that high CCEPR expression was associated with poor differentiation, positive lymph node metastasis, distant metastasis and advanced clinical stage in colorectal cancer. Consistent with these observations, Gaballah et al (20) determined that increased CCEPR expression was associated with increased tumour size, poor differentiation, advanced Dukes' stage, positive lymph node involvement and vascular invasion. Taking the results of the present and previous studies into consideration, the authors hypothesize that increased CCEPR expression may therefore serve a role in colorectal cancer progression. In addition, the present study is the first to investigate whether CCEPR expression may predict the prognosis of patients with colorectal cancer. The results demonstrated that patients with colorectal cancer exhibiting high CCEPR expression levels had shorter survival times than patients with low CCEPR expression levels.
Based on the results observed in clinical samples, the present study performed further in vitro experiments to investigate the function of CCEPR in regulating the malignant phenotypes of colorectal cancer cells. As CCEPR expression was significantly upregulated in colorectal cancer samples, CCEPR siRNA was used to transfect HT29 and SW480 cells to downregulate its expression. The results of the in vitro experiments demonstrated that silencing CCEPR significantly suppressed colorectal cancer cell proliferation and colony formation capacity, potentially via inducing cell cycle arrest at the G1 stage. These results suggest that CCEPR serves an oncogenic role in colorectal cancer growth. In addition, inhibition of CCEPR was associated with a significant reduction in colorectal cancer cell migration and invasion when compared with cells expressing normal endogenous CCEPR levels. This suggests that CCEPR may promote colorectal cancer metastasis, which is consistent with the clinical findings. Similarly, Liao et al (17) demonstrated that CCEPR promotes the proliferation, metastasis and invasion of non-small cell lung cancer cells. Peng et al (18) reported that silencing of CCEPR significantly inhibits hepatocellular carcinoma cell growth and induces cell apoptosis.
EMT is characterized by loss of the epithelial phenotype and the acquisition of mesenchymal properties; a process that is essential for cancer cells to escape their original sites and gain invasion and migration capabilities (22,31). It has been widely reported that inhibiting EMT effectively suppresses cancer cell migration and invasion (22,31). Recent studies have implicated several lncRNAs in the regulation of EMT in colorectal cancer (32–34). For instance, the X-inactive specific transcript lncRNA promotes metastasis and modulates EMT in colorectal cancer (32). However, whether CCEPR regulates EMT in colorectal cancer cells currently remains unclear. In the present study, western blot analysis demonstrated that silencing of CCEPR in HT29 and SW480 cells resulted in significant upregulation of the epithelial marker, E-cadherin, but a significant downregulation in the expression of two mesenchymal markers, N-cadherin and vimentin, indicating that EMT was suppressed. Therefore, the inhibitory effects of CCEPR downregulation on colorectal cancer cell invasion and migration may be attributed to the inhibition of EMT.
In conclusion, the results of the present study demonstrated that CCEPR is upregulated in colorectal cancer and may be associated with colorectal cancer progression, as well as poor prognosis in patients. In addition, CCEPR may serve an oncogenic role in regulating the malignant phenotypes of colorectal cancer cells. Therefore, CCEPR may be a promising therapeutic target for colorectal cancer treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
ZF collected clinical tissues. LZ designed the study and wrote the manuscript. YD and ZS performed experiments and the statistical analyses. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Hunan Provincial People's Hospital, Changsha, China. All patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Cusimano A, Balasus D, Azzolina A, Augello G, Emma MR, Di Sano C, Gramignoli R, Strom SC, McCubrey JA, Montalto G, Cervello M. Oleocanthal exerts antitumor effects on human liver and colon cancer cells through ROS generation. Int J Oncol. 2017;51:533–544. doi: 10.3892/ijo.2017.4049. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 4.Munera JO, Sundaram N, Rankin SA, Hill D, Watson C, Mahe M, Vallance JE, Shroyer NF, Sinagoga KL, Zarzoso-Lacoste A, et al. Differentiation of human pluripotent stem cells into colonic organoids via transient activation of BMP signaling. Cell Stem Cell. 2019;24:829. doi: 10.1016/j.stem.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiao Z, Qu Z, Chen Z, Fang Z, Zhou K, Huang Z, Guo X, Zhang Y. LncRNA HOTAIR is a prognostic biomarker for the proliferation and chemoresistance of colorectal Cancer via MiR-203a-3p-Mediated Wnt/ss-catenin signaling pathway. Cell Physiol Biochem. 2018;46:1275–1285. doi: 10.1159/000489110. [DOI] [PubMed] [Google Scholar]
- 6.Xie B, Deng Z, Pan Y, Fu C, Fan S, Tao Y, Zhou J, Xiao D. Post-transcriptional regulation DPC4 gene by miR-190 in colorectal cancer cells. J Cancer Res Ther. 2018;14:838–843. doi: 10.4103/jcrt.JCRT_577_17. [DOI] [PubMed] [Google Scholar]
- 7.Yu SY, Dong B, Tang L, Zhou SH. LncRNA MALAT1 sponges miR-133 to promote NLRP3 inflammasome expression in ischemia-reperfusion injured heart. Int J Cardiol. 2018;254:50. doi: 10.1016/j.ijcard.2017.10.071. [DOI] [PubMed] [Google Scholar]
- 8.Xu R, Zhu X, Chen F, Huang C, Ai K, Wu H, Zhang L, Zhao X. LncRNA XIST/miR-200c regulates the stemness properties and tumourigenicity of human bladder cancer stem cell-like cells. Cancer Cell Int. 2018;18:41. doi: 10.1186/s12935-018-0540-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiong W, Huang C, Deng H, Jian C, Zen C, Ye K, Zhong Z, Zhao X, Zhu L. Oncogenic non-coding RNA NEAT1 promotes the prostate cancer cell growth through the SRC3/IGF1R/AKT pathway. Int J Biochem Cell Biol. 2018;94:125–132. doi: 10.1016/j.biocel.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 10.Luo J, Chen J, Li H, Yang Y, Yun H, Yang S, Mao X. LncRNA UCA1 promotes the invasion and EMT of bladder cancer cells by regulating the miR-143/HMGB1 pathway. Oncol Lett. 2017;14:5556–5562. doi: 10.3892/ol.2017.6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu H, Zheng T, Yu J, Zhou L, Wang L. LncRNA XIST accelerates cervical cancer progression via upregulating Fus through competitively binding with miR-200a. Biomed Pharmacother. 2018;105:789–797. doi: 10.1016/j.biopha.2018.05.053. [DOI] [PubMed] [Google Scholar]
- 12.Zhang H, Lu W. LncRNA SNHG12 regulates gastric cancer progression by acting as a molecular sponge of miR320. Mol Med Rep. 2018;17:2743–2749. doi: 10.3892/mmr.2017.8143. [DOI] [PubMed] [Google Scholar]
- 13.Liu K, Yao H, Wen Y, Zhao H, Zhou N, Lei S, Xiong L. Functional role of a long non-coding RNA LIFR-AS1/miR-29a/TNFAIP3 axis in colorectal cancer resistance to pohotodynamic therapy. Biochim Biophys Acta. 2018;1864:2871–2880. doi: 10.1016/j.bbadis.2018.05.020. [DOI] [PubMed] [Google Scholar]
- 14.Chen Y, Wang CX, Sun XX, Wang C, Liu TF, Wang DJ. Long non-coding RNA CCHE1 overexpression predicts a poor prognosis for cervical cancer. Eur Rev Med Pharmacol Sci. 2017;21:479–483. [PubMed] [Google Scholar]
- 15.Yang M, Zhai X, Xia B, Wang Y, Lou G. Long noncoding RNA CCHE1 promotes cervical cancer cell proliferation via upregulating PCNA. Tumour Biol. 2015;36:7615–7622. doi: 10.1007/s13277-015-3465-4. [DOI] [PubMed] [Google Scholar]
- 16.Xu G, Zhang Y, Li N, Zhang JB, Xu R. LncRNA CCHE1 in the proliferation and apoptosis of gastric cancer cells. Eur Rev Med Pharmacol Sci. 2018;22:2631–2637. doi: 10.26355/eurrev_201805_14957. [DOI] [PubMed] [Google Scholar]
- 17.Liao Y, Cheng S, Xiang J, Luo C. lncRNA CCHE1 increased proliferation, metastasis and invasion of non-small lung cancer cells and predicted poor survival in non-small lung cancer patients. Eur Rev Med Pharmacol Sci. 2018;22:1686–1692. doi: 10.26355/eurrev_201803_14581. [DOI] [PubMed] [Google Scholar]
- 18.Peng W, Fan H. Long noncoding RNA CCHE1 indicates a poor prognosis of hepatocellular carcinoma and promotes carcinogenesis via activation of the ERK/MAPK pathway. Biomed Pharmacother. 2016;83:450–455. doi: 10.1016/j.biopha.2016.06.056. [DOI] [PubMed] [Google Scholar]
- 19.Zhan Y, Li Y, Guan B, Chen X, Chen Z, He A, He S, Gong Y, Peng D, Liu Y, et al. Increased expression of long non-coding RNA CCEPR is associated with poor prognosis and promotes tumorigenesis in urothelial bladder carcinoma. Oncotarget. 2017;8:44326–44334. doi: 10.18632/oncotarget.17872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaballah HH, Gaber RA, Elrashidy MA, Elshahat DA, Hablus MA, Ebeid AM. Expression of long non-coding RNA CCHE1 in colorectal carcinoma: Correlations with clinicopathological features and ERK/COX-2 pathway. Mol Biol Rep. 2019;46:657–667. doi: 10.1007/s11033-018-4521-0. [DOI] [PubMed] [Google Scholar]
- 21.Pietruszewska W, Bojanowska-Pozniak K, Kobos J. Matrix metalloproteinases MMP1, MMP2, MMP9 and their tissue inhibitors TIMP1, TIMP2, TIMP3 in head and neck cancer: An immunohistochemical study. Otolaryngol Pol. 2016;70:32–43. doi: 10.5604/00306657.1202546. [DOI] [PubMed] [Google Scholar]
- 22.Wu Y, Sarkissyan M, Vadgama JV. Epithelial-mesenchymal transition and breast cancer. J Clin Med. 2016;5(pii):E13. doi: 10.3390/jcm5020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang Z, Song Q, Zeng R, Li J, Li J, Lin X, Chen X, Zhang J, Zheng Y. MicroRNA-218 inhibits EMT, migration and invasion by targeting SFMBT1 and DCUN1D1 in cervical cancer. Oncotarget. 2016;7:45622–45636. doi: 10.18632/oncotarget.9850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barrette K, Van Kelst S, Wouters J, Marasigan V, Fieuws S, Agostinis P, van den Oord J, Garmyn M. Epithelial-mesenchymal transition during invasion of cutaneous squamous cell carcinoma is paralleled by AKT activation. Br J Dermatol. 2014;171:1014–1021. doi: 10.1111/bjd.12967. [DOI] [PubMed] [Google Scholar]
- 25.Yang F, Yu N, Wang H, Zhang C, Zhang Z, Li Y, Li D, Yan L, Liu H, Xu Z. Downregulated expression of hepatoma-derived growth factor inhibits migration and invasion of prostate cancer cells by suppressing epithelial-mesenchymal transition and MMP2, MMP9. PLoS One. 2018;13:e0190725. doi: 10.1371/journal.pone.0190725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Zhu L, Yang N, Du G, Li C, Liu G, Liu S, Xu Y, Di Y, Pan W, Li X. LncRNA CRNDE promotes the epithelial-mesenchymal transition of hepatocellular carcinoma cells via enhancing the Wnt/β-catenin signaling pathway. J Cell Biochem. 2018 doi: 10.1002/jcb.26762. (Epub ahead of prin) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Y, Chen Y, Ding W, Hua Z, Wang L, Zhu Y, Qian H, Dai T. LncRNA UCA1 impacts cell proliferation, invasion, and migration of pancreatic cancer through regulating miR-96/FOXO3. IUBMB Life. 2018;70:276–290. doi: 10.1002/iub.1699. [DOI] [PubMed] [Google Scholar]
- 29.Yang Y, Shen Z, Yan Y, Wang B, Zhang J, Shen C, Li T, Ye C, Gao Z, Peng G, et al. Long non-coding RNA GAS5 inhibits cell proliferation, induces G0/G1 arrest and apoptosis, and functions as a prognostic marker in colorectal cancer. Oncol Lett. 2017;13:3151–3158. doi: 10.3892/ol.2017.5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang JZ, Xu CL, Wu H, Shen SJ. LncRNA SNHG12 promotes cell growth and inhibits cell apoptosis in colorectal cancer cells. Braz J Med Biol Res. 2017;50:e6079. doi: 10.1590/1414-431x20176079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith BN, Bhowmick NA. Role of EMT in metastasis and therapy resistance. J Clin Med. 2016;5(pii):E17. doi: 10.3390/jcm5020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen DL, Chen LZ, Lu YX, Zhang DS, Zeng ZL, Pan ZZ, Huang P, Wang FH, Li YH, Ju HQ, Xu RH. Long noncoding RNA XIST expedites metastasis and modulates epithelial-mesenchymal transition in colorectal cancer. Cell Death Dis. 2017;8:e3011. doi: 10.1038/cddis.2017.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tao Y, Han T, Zhang T, Ma C, Sun C. LncRNA CHRF-induced miR-489 loss promotes metastasis of colorectal cancer via TWIST1/EMT signaling pathway. Oncotarget. 2017;8:36410–36422. doi: 10.18632/oncotarget.16850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang X, Liu W, Xu X, Zhu J, Wu Y, Zhao K, He S, Li M, Wu Y, Zhang S, et al. Downregulation of long noncoding RNA UCA1 enhances the radiosensitivity and inhibits migration via suppression of epithelialmesenchymal transition in colorectal cancer cells. Oncol Rep. 2018;40:1554–1564. doi: 10.3892/or.2018.6573. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.