Abstract
Background
At present, a number of long non-coding RNAs (lncRNAs) have been realized as the critical regulators of breast cancers. Current evidence indicates that dysregulation of UFC1 contributes to the tumorigenesis and progression of various types of human cancer. However, the roles of UFC1 in breast cancer are still unclear.
Material/Methods
Firstly, we measured the expression of UFC1 in breast cancer tissues and cells lines compared with corresponding controls. Then, cell functional assays were performed to determine the roles of UFC1 in breast cancer progression in vitro. Moreover, the correlation between UFC1 and miR-34a was determined by luciferase reporter assays. Further, the role of miR-34a in regulating biological function of breast cancer and its downstream target CXCL10 was applied by a series of functional assays.
Results
In present study, we found that UFC1 was highly expressed in breast tissue and cells lines compared with normal tissues and cell lines. Silenced UFC1 suppressed multiple biological activities of breast cancer cells, which also functioned as a miR-34a sponge in breast cancer. Furthermore, over-expressing miR-34a could prominently suppress cell growth, invasion, migration and inducing apoptosis in breast cancer cells. In addition, we verified that miR-34a was a target of CXCL10 by bioinformatics analysis and luciferase reporter assay.
Conclusions
LncRNA UFC1 regulated biological activity of breast cancer via miR-34a/CXCL10 axis, providing a novel diagnosis biomarker and potential therapeutic target for breast cancer.
MeSH Keywords: Breast Diseases; MicroRNAs; RNA, Long Noncoding
Background
As one of the most common life-threating diseases, breast cancer accounts for a large proportion of cancer-related mortality among women around the world [1]. According to recent studies, over 1 675 000 women were diagnosed of breast cancer, and over 500 000 women were die for this cancer each year [2,3]. However, the most common choice for treating breast cancer is chemotherapy, which is usually associated with a number of drawbacks as drug toxicity, accompanying serious infection and other organ damage [4]. Therefore, it is an emergency concern which needs to explore pathogenesis, as well as find more available diagnostic and therapeutic strategy for breast cancer.
Long non-coding RNAs (lncRNAs), are a group of RNAs which identified as more than 200 nucleotides in length with limited protein coding potential [5]. A large number of research studies have demonstrated that more than 16 000 lncRNAs in the human transcriptome were frequently spliced, polyadenylated, and principally transcribed by RNA polymerase II, which might play multitudinous roles in cancer cellular life circles as proliferation, metastasis, epithelial-mesenchymal transition (EMT), apoptosis, and differentiation [6]. MicroRNAs (miRNAs) are another kind of non-coding single stranded RNA (ncRNA) encoded by an endogenous gene, with length of about 22 nucleotides, involved in the transcriptional and post- transcriptional regulation of gene expression [7]. Recent evidence revealed that lncRNAs could interact with sponge miRNAs as a competitive endogenous RNAs (ceRNAs), which regulated miRNA-mediated target gene expression in transcriptional or post-transcriptional level [8]. The potential mechanisms of ceRNAs involved in oncogenesis, progression and metastasis of various human cancers were gradually clarified, for example, Liu et al. found that HOTAIR represented as a biomarker of poor prognosis in gastric cancer, which could promote cancer cell proliferation and migration by regulating miR-331-3p/HER2 axis [9]; Huang et al. showed that MIR100HG could facilitate cell proliferation and invasion in laryngeal squamous cell carcinoma via down-regulation of miR-204-5p [10].
Owing to deep microarrays application, numerous lncRNAs were considered to be abnormal expression in breast cancer, which could be broadly divided into 2 categories such as oncogenes or tumor suppressors [6]. The lncRNA UFC1, locating in chromosome 1q23.3, was first reported in hepatocellular carcinoma which was remarkably upregulated in cancer tissues and function as a tumor promoter [11]. As a newly discovered lncRNA, UFC1 was currently only explored in a small number of diseases including osteoarthritis [12], gastric cancer [13] and colon cancer[14], the functional role of which in breast cancer was still unclear.
In present study, we focused on exploring UFC1, which was upregulated in breast cancer tissues and cell lines. Several attempts were performed to detect the underlining roles of UFC1 in breast cancer cell proliferation, invasion, migration, EMT, and apoptosis. Our findings validated that UFC could modulate the biological function of breast cancer cells via miR-34a/CXCL10 axis, which might provide an effective therapeutic target for breast cancer.
Material and Methods
Clinical specimens
We collected 76 matched breast ductal cancer tissue and non-cancer tissue from patients who underwent breast cancer surgery at First Affiliated Hospital of Gannan Medical University between December 2015 and October 2016. The exclusion criteria contained chemotherapy or radiotherapy. Tissue specimens were immediately frozen in liquid nitrogen at 170°C rapidly. All participants were approved by the Institute Research Ethics Committee of the First Affiliated Hospital of Gannan Medical University. All procedures were in accordance with the Helsinki Declaration and the institutional guidelines. In addition, all procedures provided written informed consent.
Cell culture and transfection
All the cell lines were provided by Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Wuhan, China), including normal breast cell line (HBL-100) and breast cancer cell lines (MDA-MB-231, SKBR-3,MDA-MB-453, BT-474, and MCF-7). And then, cells were cultured in RPMI 1640 (Gibco, USA) with 10% fetal bovine serum (FBS; Gibco, USA) at 37°C in a humidified atmosphere of 5% CO2.
For transfection, miR-34a mimics, miRNA control (miR-NC), UFC1 siRNA (si-UFC1) were all were designed and synthesized by GenePharma (Shanghai, China). All transfections were applied using Lipofectamine 3000 (Thermo Fisher Scientific, USA) according to the manufacturer’s protocols. After 48 hours of transfection, the cells were collected for subsequent experiments.
Quantitative real-time polymerase chain reaction (qRT-PCR)
The total RNA extracted from tissues and cells using TRIzol reagent (Invitrogen, USA) according to the manufacturer’s protocols. Reverse transcription was performed to generate the cDNA using Reverse Transcription Kit (Promega, USA), and then the expression level of RNAs was measured using SYBR Premix Ex Taq II (TaKaRa, Dalian, China) on the Roche Lightcycler 480 RT-PCR system (Roche Diagnostics, Switzerland). The following thermocycling conditions were applied: 40 cycles, including denaturation at 95°C for 30 seconds, primer annealing at 55°C for 30 seconds, and extension at 72°C for 5 minutes. The relative RNA expression changes were normalized to GAPDH or U6 and was determined by 2−ΔΔCt method. The primers are shown in Table 1.
Table 1.
Forward and reverse primers.
| Genes | Primer sequences |
|---|---|
| UFC1 | F: 5′TCCAACCTGAGTGACATAGCGA3′ |
| R: 5′CTGACCTCCAACTCCAACGAAT3′ | |
| CXCL10 | F: 5′CCGGAATTCGAGCCTACAGCAGAGGAACC3′ |
| R: 5′CCGCTCGAGTTTGATCCCCTCTGGTTTTA3′ | |
| Bcl-2 | F: 5′GCTCAGCCTGTGCCACCTG3′ |
| R: 5′CAGAGGTCGGCATGCAGGACTT3′ | |
| Bax | F: 5′TCCACCAAGAAGCTGAGCGAG3′ |
| R: 5′GTCCAGCCCATGATGGTTCT3′ | |
| E-cadherin | F: 5′GACGCGGACGATGATGTGAAC3′ |
| R: 5′TTGTACTGTTGTGGATTGAAG3′ | |
| N-cadherin | F: 5′CACCCAACATGTTTACAATCAACAATGAGAC3′ |
| R: 5′CTGCAGCAACAGTAAGGACAAACATCCTATT3′ | |
| Vimentin | F: 5′GGAAGAGAACTTTGCCGTTGAA3′ |
| R: 5′GTGACGAGCCATTTCCTCCTT3′ | |
| U6 | F: 5′CTCGCTTCGGCAGCACA3′ |
| R: 5′AACGCTTCACGAATTTGCGT3′ | |
| GAPDH | F: 5′AGAAGGCTGGGGCTCATTTG3′ |
| R: 5′AGGGGCCATCCACAGTCTTC3′ |
Cellular proliferative ability assays
Cell Counting Kit-8 (CCK-8) assay (Dodinjo, Japan) and colony formation assay were utilized to determined cell viability and colony formation ability respectively. For CCK-8 assay, transfected cells (1×103 cells/well) were seeded in 96-well plates, and then 10 μL CCK-8 solution was added into each well at 0, 24, 48, 72, 96, and 120 hours, and incubated for another 3 hours. The absorbance at 450 nm was measured by a microplate reader.
For colony formation assay, the transfected cells (5×102 cells/well) were seeded in 6-well plates and cultured for 14 days, then the colon cells were stained by 1% crystal violet and counted under a light microscope (Olympus Corporation, Japan).
Transwell migration and invasion assays
The ability of migration and invasion of cells were detected using a 24-well Transwell chamber (Corning Costar, USA) with an 8.0 μm pore size. The transfected cells were suspended in serum-free RPMI-1640 medium and inoculated onto the upper chamber not coated without Matrigel (BD Biosciences, USA) for migration assay, whereas Matrigel-coated chambers were used for the invasion assay, and then medium containing 10% FBS was added to the lower chambers. After 48-hour cultured, the migratory and invading cells in the lower chamber were fixed in 70% ethanol, stained with 0.1% crystal violet and photographed under a light microscope (Olympus Corporation, Japan).
Western blot
The protein samples from cells were acquired using RIPA lysis buffer (Beyotime, China), and the protein concentration was measured by a BCA protein assay kit (Beyotime, China). Next, the equal amounts of protein were separated on 10% SDS-PAGE and transferred to PVDF membrane (Millipore, USA) blocking with 5% non-fat milk. Where after, the membranes were incubated at 4°C overnight with primary antibodies against Bcl-2 (1: 1000, Abcam, UK), Bax(1: 1000, Abcam, UK), E-cadherin (1: 1000, Abcam, UK), N-cadherin (1: 1000, Abcam, UK), vimentin (1: 1000, Abcam, UK), CXCL10 (1: 1000, Abcam, UK) and GAPDH (1: 5000, Abcam, UK). The membranes were then incubated with horseradish peroxidase-conjugated secondary antibody (1: 5,000; Abcam) at room temperature for 2 hours. The signals of membranes were measured by enhanced chemiluminescence kit (ECL) (Millipore, USA).
Luciferase reporter assays
Simply, the 3′-UTR fragments of UFC-1 and CXCL10 containing miR-34a binding site were amplified by Shanghai GenePharma Co., Ltd. and cloned into the pmirGLO vector (Promega, USA). Then, the wild type of UFC-1 or mutant UFC-1 3′-UTR (UFC-1-WT or UFC-1-WT), as well as the wild type of CXCL10 or mutant CXCL10 3′-UTR (CXCL10-WT or CXCL10-WT) was constructed. Next, the constructed plasmids were co-transfected with miR-34a mimics or mimic-NC into HKE293 cells by using Lipofectamine 3000 (Invitrogen, USA) according to the manufacturer’s instructions. After 48 hours of transfection, luciferase activity was measured by the Dual-Luciferase Reporter System (Promega, USA).
Statistical analysis
All data are presented as the mean±standard deviation. Statistical analysis was performed by SPSS 25.0 (BMI, USA) through a Student’s t-test or ANOVA following LSD (least significant difference) post-hoc method. The survival rates of breast cancer patients were analyzed by The Kaplan-Meier method along with the log-rank test. Statistically significant were considered as P<0.05.
Results
UFC1 was highly expressed in breast cancer tissues
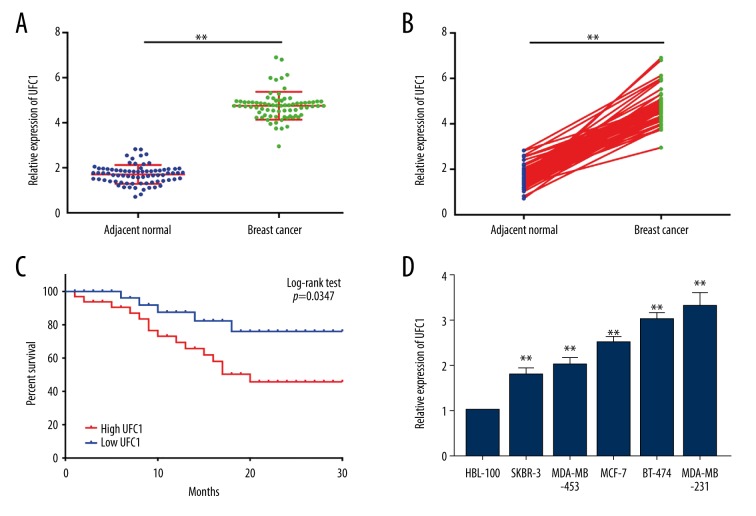
It has been shown that expressions of UFC1 between normal tissues and breast ductal cancer tissues were significantly distinct, whereas UFC1 expression within breast cancer tissues was remarkably beyond compared to that within paired adjacent normal tissues (Figure 1A, 1B). Moreover, Log-rank test and Kaplan-Meier analysis indicated patients with high expression of UFC1 had a worse prognosis than patients with low expression at 30 months follow-up (Figure 1C). Besides, the expression of UFC1 was also upregulated in human breast cancer cell lines (SKBR-3, BT-474, MDA-MB-231, MDA-MB-453, and MCF-7) when compared to normal breast cell line (HBL-100) (Figure 1D). Together, these data suggest that UFC1 expression was significantly increased in breast cancer which might be a novel biomarker for the diagnosis and prognosis of breast cancer.
Figure 1.
UFC1 was highly expressed in breast tissues, indicating the poor prognosis of breast cancer patients. (A, B) RT-PCR was performed to test UFC1 expression in breast tissue specimens compared with adjacent non-tumor tissue. (C) The prognosis of breast cancer patients who had higher or lower UFC1 expression analyzed by Kaplan-Meier analysis and log-rank test. (D) The expression of UFC1 in human breast cancer cell lines (SKBR-3, BT-474,MDA-MB-231,MDA-MB-453, and MCF-7) as well as normal breast cell line (HBL-100). Data are presented as the mean±standard deviation. ** P<0.01 versus control group. RT-PCR – real-time polymerase chain reaction.
Silenced UFC1 repressed the logical activity breast cancer in vitro
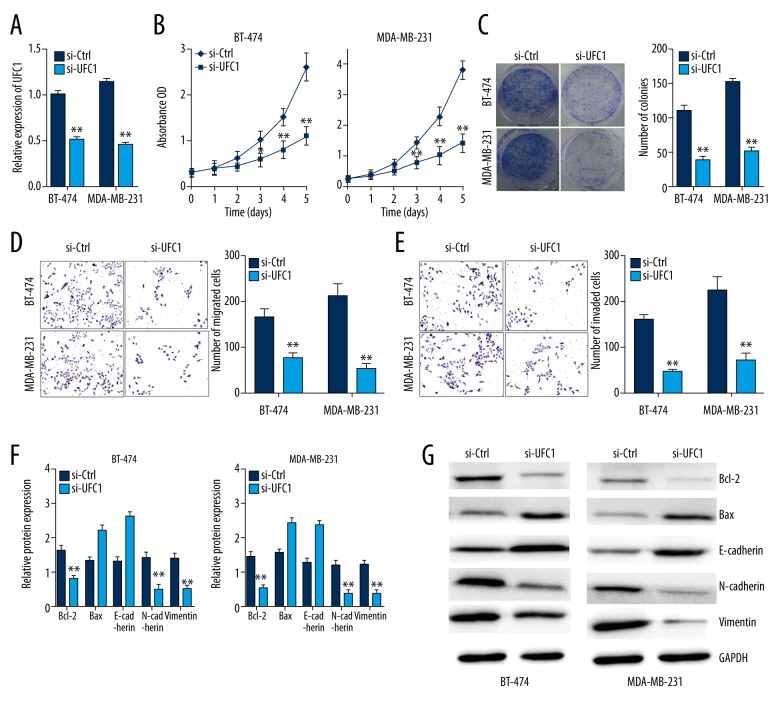
Based on the aforementioned results, we considered UFC1 might be involved in biological function of breast cancer, thus, a series of loss-of-functional experiments were performed using BT-474 and MDA-MB-231 cells lines to test our conjecture. First of all, we validated the efficiency of UFC1 siRNA on inhibiting UFC1 expression (Figure 2A). Then, CCK8 assay and colony formation assay were conducted to evaluate proliferation of breast cancer cells transfected with si-UFC1. Our results found that knockdown UFC1 could significantly suppress cell proliferation in BT-474 and MDA-MB-231 cells (Figure 2B, 2C). As shown in Figure 2D and 2E, knockdown of UFC1 also impaired migration and invasion abilities of breast cancer cells. Moreover, the apoptotic and EMT related gene expression as Bcl-2, Bax, E-cadherin, N-cadherin, vimentin were assessed in breast cancer cells transfected with si-UFC1 using RT-PCR and western blot. The results indicated that down-expression of UFC1 might inhibit EMT process and induce apoptosis in both BT-474 and MDA-MB-231 cells (Figure 2F, 2G). To sum up, silenced UFC1 might be capable of repress the proliferation, invasion, migration, EMT process and induce apoptosis of breast cancer in vitro.
Figure 2.
Silenced UFC1 repressed the biological activity breast cancer in vitro. (A) UFC1 expression in BT-474 and MDA-MB-231 cells transfected with si-UFC1 or si-NC was measured by RT-PCR. (B, C) CCK-8 assay and colony formation assay was conducted to evaluate the cell proliferation of breast cancer cells transfected with si-UFC1 or si-NC. (D, E) Transwell assay assessed the migration and invasion abilities of breast cancer cells transfected with si-UFC1 or si-NC. (F, G) The mRNA and protein expression of apoptotic and EMT related genes in breast cancer cells transfected with si-UFC1 or si-NC. Data are presented as the mean±standard deviation. ** P<0.01, * P<0.05 versus control group. NC – negative control; RT-PCR – real-time polymerase chain reaction; CCK-8 – Cell Counting Kit-8; EMT – epithelial-mesenchymal transition.
UFC1 acts as a miR-34a sponge in breast cancer
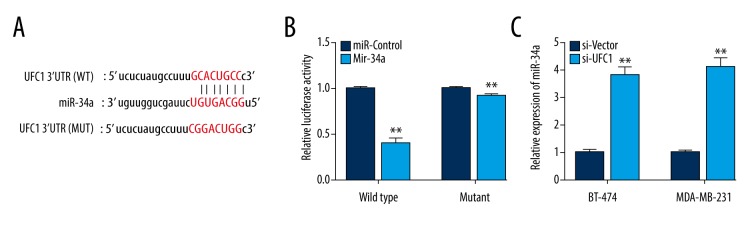
Increasing evidence indicated that lncRNAs could bind to specific miRNAs and activate the expression of its targeted genes competitively [6]. Therefore, we firstly predicted the potential binding sites of miR-34a in UFC1 3′UTR using bioinformatics analysis (Figure 3A). Then, luciferase assays were carried out to detect the relationship between UFC1 and miR-34a. As illustrated in Figure 3B, over-expression of miR-34a strikingly suppressed the luciferase activities of the UFC1-WT reporter vector but not UFC1-MUT reporter vector. Additionally, knockdown UFC1 could apparently promote miR-34a expression in breast cancer cells (Figure 3C). Taken together, our results indicated that UFC1 acted as a miR-34a sponge in breast cancer cells.
Figure 3.
UFC1 acts as a miR-34a sponge in breast cancer. (A) Putative binding sites of miR-34a within the UFC1 predicted by bioinformatics tools. (B) Luciferase reporter assay was performed to validate relationship between UFC1 and miR-34a. (C) The expression of miR-34a in BT-474 and MAD-MB-231 cells transfected with si-UFC1 or si-vector by using RT-PCR. Data are presented as the mean±standard deviation. ** P<0.01, * P<0.05 versus control group. NC – negative control.
MiR-34a over-expression inhibits biological activity of breast cancer cells by targeting CXCL10
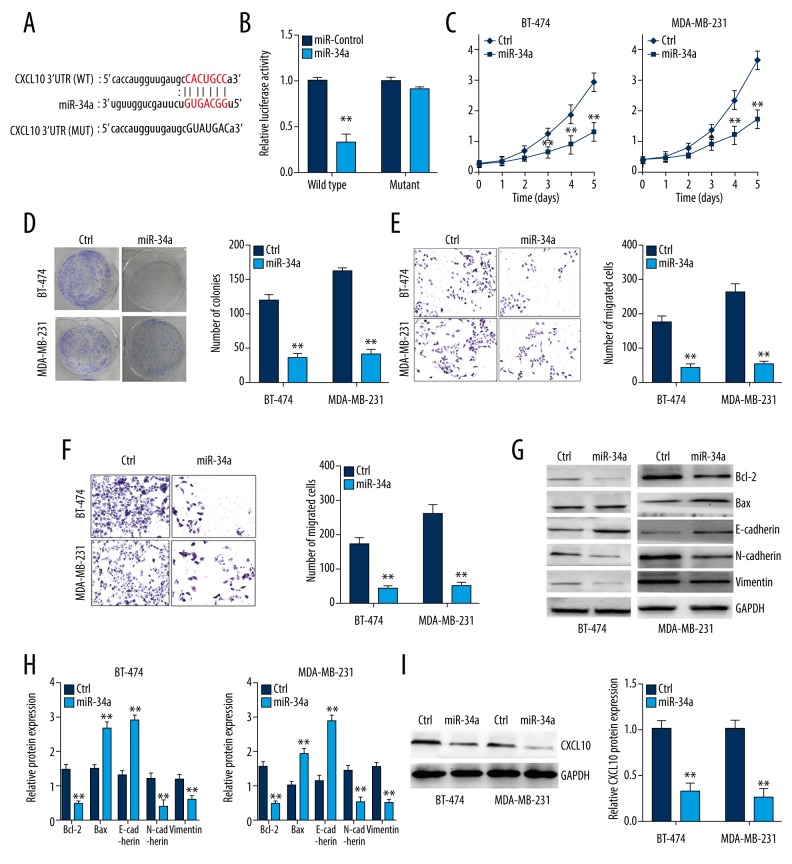
Through previous results, we supposed that miR-34a and UFC1 might form an RNA-induced silencing complex (RISC), where a downstream target of miR-34a was still unclear. Similarly, with the help of bioinformatics tools, we found complementary binding sites of CXCL10 mRNA 3′-UTR for miR-34a (Figure 4A). Besides, luciferase reporter assay showed that over-expression of miR-34a prominently suppressed the luciferase activities of the CXCL10-WT reporter vector but not CXCL10- MUT reporter vector (Figure 4B). To further estimate the function of miR-34a on the malignant behaviors of breast cancer, gain-of-functional experiments were conducted using BT-474 and MDA-MB-231 cells transfected with miR-34a mimics or control. As shown in Figure 4C and 4D, the cell viability of BT-474 and MDA-MB-231 cells was decreased when transaction with miR-34a mimics. Meanwhile, the migration and invasion ability of BT-474 and MDA-MB-231 cells transfected with miR-34a mimics were also suppressed (Figure 4E, 4F). Furthermore, our result indicated that upregulated miR-34a could inhibit EMT and apoptosis process in breast cancer by measuring apoptotic and EMT related gene expression (Figure 4G, 4H). In addition, the protein levels of CXCL10 in BT-474 and MDA-MB-231 cells transfected with miR-34a mimics were decreased, suggesting miR-34a might mediate pathological behaviors of breast cancer via its downstream target CXCL10 (Figure 4I). Taken together, our findings deemed that down-expression of UFC1 might regulate biologic function of breast cancer by targeting miR-34a/CXCL10 axis.
Figure 4.
MiR-34a over-expression inhibits biological activity of breast cancer cells by targeting CXCL10. (A) Putative binding sites of miR-34a within the CXCL10 predicted by bioinformatics tools. (B) Luciferase reporter assay was performed to validate relationship between CXCL10 and miR-34a. (C, D) CCK-8 assay and colony formation assay was conducted to evaluate the cell proliferation of breast cancer cells transfected with miR-34a mimics or mimics-NC. (E, F) Transwell assay assessed the migration and invasion abilities of breast cancer cells transfected with miR-34a mimics or mimics-NC. (G, H) The mRNA and protein expression of apoptotic and EMT related genes in breast cancer cells transfected with miR-34a mimics or mimics-NC. (I) The protein levels of CXCL10 in BT-474 and MDA-MB-231 cells transfected with miR-34a mimics or mimics-NC. Data are presented as the mean±standard deviation. ** P<0.01, * P<0.05 versus control group. CCK-8 – Cell Counting Kit-8; NC – negative control; EMT – epithelial-mesenchymal transition.
Discussion
A great amount of evidence indicated that lncRNA participated in cellular processes included proliferation, apoptosis, migration, and cell invasion and their dysregulated expression has been confirmed in several cancers [15]. UFC1 expression has been reported upregulated in several cancer and serves as a new biomarker for the prognosis and diagnosis of many cancers as gastric cancer, colorectal cancer and cervical cancer [16–18], however, its role in breast tumors has not been reported. Our study firstly investigated that the expression of UFC1 was significantly elevated in breast cancer tissues and cell lines, which also closely related to the prognosis of the patients. Recently, Zhang et al. showed that UFC1 had a promoting role in gastric cancer progression, which could stimulate proliferation, invasion, migration and EMT in corresponding cell lines [13]. At same time, Yu et al. also demonstrated that UFC1 was upregulated in colorectal cancer tissues, knockdown of which could distinctly inhibit proliferation and promote apoptosis cancer cells [14]. On account of the aforementioned research, we subsequently performed a battery of experiments to explore underlying mechanism of lncRNA UFC1 in the biological function of breast cancer cells. Our study revealed that silenced UFC1 was able to repress the proliferation, invasion and migration, as well as resulting intrinsic apoptosis of breast cancer in vitro, indicating that UFC1 was critical for breast carcinogenesis and progression.
Current research suggests that lncRNAs act as competitive endogenous RNA (ceRNA), which sponge for corresponding miRNAs and eliminate the endogenous inhibition of targeted genes of these miRNAs [6]. In breast cancer, Ding et al. identified that MIF-AS1 modulated the cell proliferation, migration and EMT by regulating level of Homeobox B8 (HOXB8) via binding to miR-1249-3p [19]; Li et al. found that HOXC13-AS facilitated tumorigenesis and progression through regulating miR-497-5p/PTEN axis [20]. Our results found that UFC1 was highly conservatively binding to miR-34a, and illustrated UFC1 could suppress the breast cancer cells proliferation, invasion, and migration, inducing apoptosis via targeting miR-34a/CXCL10 axis.
As a member of miRNA, miR-34 had been realized that abnormal regulation in a great diversity of cancers, which was ascertained to be directly regulated by the tumor suppressor p53 and acted as a tumor suppressor [21]. Numerous studies reported that miR-34 was downregulated in breast cancer tissues, leading to inhibit biological function of cancer cells, for instance, Rui et al. showed that miR-34a might suppress breast cancer cell proliferation and invasion by partly targeting Notch1 [22]; Bayrakta et al. declared that miR-34a played as an tumor suppressor role in triple-negative breast cancer by dual-targeting of FOXM1/eEF2K signaling axis [23]. Therefore, from our study, it was suggested that UFC1/miR-34a axis served as a vital role in pathological mechanism of breast tumor. Similar to our findings, Zhang et al. and Cao et al. reported that UFC1 promoted proliferation and reduces apoptosis via controlling miR-34a expression in and chondrocytes and hepatocellular cancer cells respectively [11,12], suggesting that UFC1/miR-34a axis based gene therapy might be a potential therapeutic strategy in osteoarthritis and cancers.
CXCL10, the ligand of chemokine (C-X-C motif) receptor CXCR3, which played an important role in promoting chemotactic activity, inducing cells apoptosis, proliferation and angiogenesis [24–26]. Numerous studies indicated that CXCL10 served as a key role in tumorigenesis and progression of breast cancer, for example, Mulligan et al. reported that breast cancer functional screening serum chemokine spectrum showed that the HR-independent increased chemokine CXCL10 when compared to healthy controls [27]; Bu et al. showed that CXCL10 mediates metastatic breast cancer by activation of microglia to induce bone pain [28]. Meanwhile, a group of studies found that CXCL10 and miR-34a might be involved in metastatic potential of breast cancer at same time, due to strengthen EMT process and enlarged proportion of cancer stem-like cell phenotype in vitro [29]. More fascinatingly, Xu et el. indicated that miR-34a could alleviate breast tumorigenesis and progression by inhibition of the TLR signaling pathway via CXCL10 [30]. Similar to the previous study, we found that UFC1 had a promoting role in breast cancer progression by acting as a miR-34a sponge and then regulated expression of CXCL10, that means CXCL10 might also have a great influence on the development and progression of breast cancers.
Conclusions
Our data indicated that UFC1 was high expressed in breast cancer and regulated biological activity of breast cancer via miR-34a/CXCL10 axis, which would provide a potential target for therapy and a novel biomarker for diagnosis to breast cancer.
Footnotes
Source of support: Departmental sources
References
- 1.Froehlich K, Schmidt A, Heger JI, et al. Breast cancer, placenta and pregnancy. Eur J Cancer. 2019;115:68–78. doi: 10.1016/j.ejca.2019.03.021. [DOI] [PubMed] [Google Scholar]
- 2.Van Grembergen O, Bizet M, de Bony EJ, et al. Portraying breast cancers with long noncoding RNAs. Sci Adv. 2016;2:e1600220. doi: 10.1126/sciadv.1600220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 4.Damon LE, Cadman EC. The metabolic basis for combination chemotherapy. Pharmacol Ther. 1988;38:73–127. doi: 10.1016/0163-7258(88)90103-9. [DOI] [PubMed] [Google Scholar]
- 5.Xiong T, Li J, Chen F, Zhang F. PCAT-1: A novel oncogenic long non-coding rna in human cancers. Int J Biol Sci. 2019;15:847–56. doi: 10.7150/ijbs.30970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–58. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu XH, Sun M, Nie FQ, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92. doi: 10.1186/1476-4598-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang Y, Zhang C, Zhou Y. LncRNA MIR100HG promotes cancer cell proliferation, migration and invasion in laryngeal squamous cell carcinoma through the downregulation of miR-204-5p. Onco Targets Ther. 2019;12:2967–73. doi: 10.2147/OTT.S202528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao C, Sun J, Zhang D, et al. The long intergenic noncoding RNA UFC1, a target of MicroRNA 34a, interacts with the mRNA stabilizing protein HuR to increase levels of beta-catenin in HCC cells. Gastroenterology. 2015;148:415–26 e418. doi: 10.1053/j.gastro.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 12.Zhang G, Wu Y, Xu D, Yan X. Long noncoding RNA UFC1 promotes proliferation of chondrocyte in osteoarthritis by acting as a sponge for miR-34a. DNA Cell Biol. 2016;35:691–95. doi: 10.1089/dna.2016.3397. [DOI] [PubMed] [Google Scholar]
- 13.Zhang X, Liang W, Liu J, et al. Long non-coding RNA UFC1 promotes gastric cancer progression by regulating miR-498/Lin28b. J Exp Clin Cancer Res. 2018;37:134. doi: 10.1186/s13046-018-0803-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu T, Shan TD, Li JY, et al. Knockdown of linc-UFC1 suppresses proliferation and induces apoptosis of colorectal cancer. Cell Death Dis. 2016;7:e2228. doi: 10.1038/cddis.2016.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan JH, Yang F, Wang F, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–81. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 16.Xi J, Feng J, Zeng S, Huang P. Long noncoding RNA UFC1 is activated by E2F1 and exerts oncogenic properties by functioning as a ceRNA of FOXP3. Cancer Med. 2018 doi: 10.1002/cam4.1556. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Batista PJ, Chang HY. Long noncoding RNAs: Cellular address codes in development and disease. Cell. 2013;152:1298–307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aalijahan H, Ghorbian S. Long non-coding RNAs and cervical cancer. Exp Mol Pathol. 2019;106:7–16. doi: 10.1016/j.yexmp.2018.11.010. [DOI] [PubMed] [Google Scholar]
- 19.Ding J, Wu W, Yang J, Wu M. Long non-coding RNA MIF-AS1 promotes breast cancer cell proliferation, migration and EMT process through regulating miR-1249-3p/HOXB8 axis. Pathol Res Pract. 2019;215:152376. doi: 10.1016/j.prp.2019.03.005. [DOI] [PubMed] [Google Scholar]
- 20.Li X, Wang Q, Rui Y, et al. HOXC13-AS promotes breast cancer cell growth through regulating miR-497-5p/PTEN axis. J Cell Physiol. 2019 doi: 10.1002/jcp.28800. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 21.Zhang L, Liao Y, Tang L. MicroRNA-34 family: A potential tumor suppressor and therapeutic candidate in cancer. J Exp Clin Cancer Res. 2019;38:53. doi: 10.1186/s13046-019-1059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rui X, Zhao H, Xiao X, et al. MicroRNA-34a suppresses breast cancer cell proliferation and invasion by targeting Notch1. Exp Ther Med. 2018;16:4387–92. doi: 10.3892/etm.2018.6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bayraktar R, Ivan C, Bayraktar E, et al. Dual suppressive effect of miR-34a on the FOXM1/eEF2-kinase axis regulates triple-negative breast cancer growth and invasion. Clin Cancer Res. 2018;24:4225–41. doi: 10.1158/1078-0432.CCR-17-1959. [DOI] [PubMed] [Google Scholar]
- 24.Liu M, Guo S, Hibbert JM, et al. CXCL10/IP-10 in infectious diseases pathogenesis and potential therapeutic implications. Cytokine Growth Factor Rev. 2011;22:121–30. doi: 10.1016/j.cytogfr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giuliani N, Bonomini S, Romagnani P, et al. CXCR3 and its binding chemokines in myeloma cells: expression of isoforms and potential relationships with myeloma cell proliferation and survival. Haematologica. 2006;91:1489–97. [PubMed] [Google Scholar]
- 26.Neville LF, Mathiak G, Bagasra O. The immunobiology of interferon-gamma inducible protein 10 kD (IP-10): A novel, pleiotropic member of the C-X-C chemokine superfamily. Cytokine Growth Factor Rev. 1997;8:207–19. doi: 10.1016/s1359-6101(97)00015-4. [DOI] [PubMed] [Google Scholar]
- 27.Mulligan AM, Raitman I, Feeley L, et al. Tumoral lymphocytic infiltration and expression of the chemokine CXCL10 in breast cancers from the Ontario Familial Breast Cancer Registry. Clin Cancer Res. 2013;19:336–46. doi: 10.1158/1078-0432.CCR-11-3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bu H, Shu B, Gao F, et al. Spinal IFN-gamma-induced protein-10 (CXCL10) mediates metastatic breast cancer-induced bone pain by activation of microglia in rat models. Breast Cancer Res Treat. 2014;143:255–63. doi: 10.1007/s10549-013-2807-4. [DOI] [PubMed] [Google Scholar]
- 29.Morata-Tarifa C, Jimenez G, Garcia MA, et al. Low adherent cancer cell subpopulations are enriched in tumorigenic and metastatic epithelial-to-mesenchymal transition-induced cancer stem-like cells. Sci Rep. 2016;6:18772. doi: 10.1038/srep18772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu M, Li D, Yang C, Ji JS. MicroRNA-34a inhibition of the TLR signaling pathway via CXCL10 suppresses breast cancer cell invasion and migration. Cell Physiol Biochem. 2018;46:1286–304. doi: 10.1159/000489111. [DOI] [PubMed] [Google Scholar]