Abstract
Adrenal glucocorticoid hormones are crucial for maintenance of homeostasis and adaptation to stress. They act via the mineralocorticoid receptors (MRs) and glucocorticoid receptors (GRs)—members of the family of nuclear receptors. MRs and GRs can mediate distinct, sometimes opposite, effects of glucocorticoids. Both receptor types can mediate nongenomic steroid effects, but they are best understood as ligand-activated transcription factors. MR and GR protein structure is similar; the receptors can form heterodimers on the DNA at glucocorticoid response elements (GREs), and they share a number of target genes. The transcriptional basis for opposite effects on cellular physiology remains largely unknown, in particular with respect to MR-selective gene transcription. In this review, we discuss proven and potential mechanisms of transcriptional specificity for MRs and GRs. These include unique GR binding to “negative GREs,” direct binding to other transcription factors, and binding to specific DNA sequences in conjunction with other transcription factors, as is the case for MRs and NeuroD proteins in the brain. MR- and GR-specific effects may also depend on specific interactions with transcriptional coregulators, downstream mediators of transcriptional receptor activity. Current data suggest that the relative importance of these mechanisms depends on the tissue and physiological context. Insight into these processes may not only allow a better understanding of homeostatic regulation but also the development of drugs that target specific aspects of disease.
Keywords: stress, adaptation, chromatin, nuclear receptor, DNA binding
Adrenal glucocorticoid hormones support physiology and are essential for the adaptive response to stress [1, 2]. The main glucocorticoid in humans is cortisol, whereas in rodents it is corticosterone [3]. An important target organ is the brain, where the action of these corticosteroids is mediated by the high-affinity mineralocorticoid receptor (MR) and the lower affinity glucocorticoid receptor (GR). Given its high affinity, MR is occupied at basal hormone levels, whereas GR is activated at the circadian peak of glucocorticoid secretion and during stress [4]. The dominant ligands of brain MRs are cortisol and corticosterone. Selective brain regions contain MR, which binds aldosterone to control physiology and behavior in relation to salt balance. This occurs in the nucleus of the solitary tract where MR-expressing neurons enzymatically convert glucocorticoids into the inactive form cortisone via 11β-hydroxysteroid dehydrogenase type 2, allowing access of aldosterone to the MR [5, 6].
GR is present in most brain regions and cell types, whereas MR is mainly expressed in limbic areas such as the hippocampus, amygdala, and prefrontal cortex [7]. GR and MR control a wide range of processes, ranging from neuronal differentiation [8] and excitability [9] to behavioral reactivity, mood, and cognition [2]—all processes that are needed to adapt to acute or chronic stress. MR activation during the early phases of acute stress is important in the appraisal process and memory retrieval; GR complements this by promoting memory consolidation and behavioral adaptation [2]. In addition to complementary actions of GR and MR, they can also exert opposing effects, even within the same cell type. This is best demonstrated by the excitability of hippocampal CA1 neurons, which are stimulated via MR and suppressed by GR activation [9]. However, differential effects of MR and GR are not limited to the brain; they also take place in the immune system, where MR-mediated proinflammatory effects contrast with the classical GR-mediated immune suppression [10], and in the heart [11].
The distinct effects of GR and MR in the brain are reflected in glucocorticoid effects on psychopathology. For instance, overactivation of GR is considered a risk factor for development of mood disorders. Patients with Cushing syndrome are exposed to excessively high cortisol levels and experience psychiatric symptoms such as personality changes, anxiety, irritability, and depressed mood. The use of a GR antagonist can relieve these symptoms [12–14]. On the other hand, low MR activity has been linked to psychiatric disorders. Observationally, in depression, schizophrenia, and bipolar disorder, there is decreased MR expression [15]. Genetically, MR gene variant haplotype 2 is known to enhance MR activity and is associated with lower risk of depression in women, indicating perhaps an interaction with sex steroids [16]. Clinically, a trial with an MR agonist as add-on treatment to antidepressant medication led to a faster antidepressant response in patients with major depressive disorder [17]. The protective effect of high levels of brain MR signaling may reflect direct effects on excitability in the limbic brain. It has also been proposed that lowered MR activation may lead to loss of its tonic inhibition of the hypothalamic-pituitary-adrenal (HPA) axis, causing chronically elevated cortisol levels. Subsequently, this high cortisol level increases the risk for major depressive disorder after a stressor [15, 18].
A recent argument for a direct effect of MR activation on mood comes from the use of synthetic glucocorticoid medication, which is known to cause adverse psychological, behavioral and cognitive effects [19, 20]. An example is the synthetic glucocorticoid dexamethasone, which is highly selective for the GR in vivo [21]. Dexamethasone strongly activates GR, which leads to suppressed HPA axis activity and reduced cortisol levels, thereby depleting MR of its ligand. The psychological adverse effects of dexamethasone may either be caused by GR overactivation, MR underactivation, or a disturbed balance in GR and MR activity [22]. A recent clinical trial demonstrated that psychological adverse effects and sleep-related difficulties caused by dexamethasone can be alleviated by reactivating MR with cortisol cotreatment [23]. This indicates that MR activation has an important role in stress-related psychopathology (i.e., in the context of high levels of GR activation).
Although it is established that MR and GR have differential intrinsic effects in the brain, it is unclear how these are established. In fact, MR and GR share not only basic mechanism of action but also target genes. We discuss proven and potential transcriptional mechanisms that can underlie transcriptional specificity for GR and MR.
1. Mechanisms of Action
MR and GR evolved from a common ancestral corticosteroid receptor gene; they share their basic overall nuclear receptor structure, with a central DNA-binding domain (DBD) and a c-terminal ligand-binding domain. These domains are similar enough to have overlap in, for example, ligand binding to the receptors and DNA binding by the receptors. Yet, some crucial mutations led to unique differences not only in ligand binding specificities but also in functionality [24–26].
A. Nongenomic Effects
Corticosteroids have well-documented, rapid effects that occur independently of DNA binding. Although they may be mediated via other mechanisms than binding to MR and GR [27], many of these effects in the brain are absent in MR/GR knockout mice [28, 29]. Nongenomic effects also occur via aldosterone-activated MR in the vasculature [30]. The MRs and GRs that mediate these rapid effects may be localized at or near the membrane (via unknown mechanisms). Nongenomic effects may also follow the dissociation of the receptors from their chaperones in the cytoplasm [31]. Of note, the pharmacology of the membrane-associated effects differs from the classical genomic effects in that higher hormone concentrations are needed to exert effects via membrane-associated receptors [28, 29]. For glucocorticoids secreted in response to stress, the nongenomic effects may support initial responses by rapid mediators such as noradrenalin, particularly with regard to MR-dependent effects [32, 33]. Interestingly, the nongenomic signaling does not simply precede genomic effects of the same episode of hormone secretion; it may also set the context for the genomic effects in a process called metaplasticity [34].
B. Glucocorticoid Response Elements
Genomic steroid receptor effects depend on at least three different processes: ligand binding, receptor association to the DNA, and interactions with other transcriptionally active proteins [35, 36]. As members of the nuclear receptor family, MR and GR translocate to the cell nucleus upon binding of hormone and can bind to glucocorticoid response elements (GREs) via the DBD. This occurs mainly in accessible chromatin, but at least GR can also pioneer the remodeling and opening of chromatin [37]. Dependence on accessible chromatin is one of the reasons that most putative GR-binding sequences are, in fact, not occupied by receptors and do not constitute actual GREs. Interactions of the receptors’ transcriptionally active proteins also determine whether a GRE sequence actually is a functional GRE [38]. The consequence of these interactions is that evolutionary conservation of GRE sequences is a predictor of functionality [39].
The DBD is 96% identical between MRs and GRs, and, therefore, they share the same GRE as their primary binding motif. GREs consist of inverted repeats to which the receptors bind as homo- or heterodimers [40, 41]. Recently there have been suggestions of higher-order complexes at the GR, with these complexes consisting of tetramers, and/or MRs that interact with DNA-bound GR, independent of the MR DBD [42, 43]. It seems that for GR, the homodimer binding to the GRE alone is not the final active form but rather it triggers tetramerization of GR, thereby adding an extra level of regulation [43]. MR interactions with the GRE-bound GRs may be a variation on this theme [42]. In addition, there can be cooperation between receptors that are bound to GREs and GRE half sites that are in close (functional) proximity [44, 45]. Binding to these GREs is normally linked to target gene activation rather than repression [24].
Once the agonist-bound receptors are bound to GREs on the DNA, they recruit coactivator and corepressor proteins that form the bridge to the transcription-initiation complexes [46]. The coregulator recruitment depends critically on the exact receptor conformation, which is determined by amino acid sequence, the ligand that is bound [47], and the DNA element to which the receptor is bound [48, 49]. The fact that coregulator interactions depend on the exact DNA-binding sequence predicts that specific coregulators are critical for specific sets of MR and GR target genes. Indeed, this is clearly the case, for example in relation to GR signaling in the brain: steroid receptor coactivator 1 (SRC-1) is a coregulator of GR and is necessary for the regulation of the Crh and Pomc genes, but not Fkbp5 [50, 51].
MR and GR recruit coregulators via two domains or activation functions: AF-1 is located in the intrinsically unstructured N-terminal part of the receptors [52], whereas AF-2 is formed in the highly structured ligand-binding domain [45]. Receptor-coregulator interactions via AF-2 can be studied using an in vitro peptide array called MARCoNI [53]. This array recapitulates ligand-induced interactions based on coregulator-derived peptide sequences: Agonists induce interactions with coactivator peptides, while antagonists displace the agonist, reduce coactivator interactions, and, in fact, may lead to recruitment of corepressors leading to active gene repression [46]. Many of these AF-2 interacting coregulators are shared between nuclear receptors, and this is certainly true for MR and GR [54].
C. Negative GREs
The GR is the only steroid receptor that can also bind to negative GREs (nGREs), at which two receptor molecules bind, but which does not require dimerization of these receptors [24, 55]. At the nGRE, the receptor adopts a different conformation, is SUMOylated in its N-terminal domain, and recruits corepressors such as NCOR and SMRT, rather than coactivators [24, 56]. Given that GR agonists do not induce such corepressor interactions in DNA-free protein-protein interaction assays [53, 57], it is more difficult to predict the behavior of different ligands at such nGREs. The MR lacks the ability to act via these nGREs, because this requires specific mutations from the ancestral steroid receptor that are unique to GR [24].
D. Protein-Protein Interactions
GR is well known to interact with transcription factors like AP-1 to repress transcription independent of direct binding to the DNA [58]. MR activation, in many instances, also leads to repression of transcription, as evidenced by transcriptomics studies, all outside the brain [11, 59]. Because nGREs are specific to monomeric GR binding, it seems likely that transrepression of genes via MR depends on interactions with other transcription factors. Interactions between MR and transcription factor SP-1 may be a case in point [37, 60]. The use of such tethering mechanisms is highly tissue specific. In a genome-wide characterization of DNA-binding sites for aldosterone-activated MR in a human kidney cell line, the majority of loci contained no apparent GRE but rather binding site motifs for transcription factors like EGR1, FOX, PAX5, and AP-1 [61]. In contrast, motif analysis of genomic MR binding in the rat hippocampus suggested exclusive binding to GRE-like sequences [62]. Also GR binding in the rat hippocampus, but not in cell lines, seems to be predominantly GRE dependent [63–65].
Thus, the cistromes of MR and GR in the hippocampus point to a predominance of GRE binding. However, corticosterone clearly suppresses the expression of many hippocampal genes [66, 67]. The part of the MR cistrome that might be associated with transrepression, as previously identified for individual genes [60, 68], therefore, is unclear. A caveat regarding the current experiments on the brain (and other tissue homogenates) is that binding events in specifically activated neurons (e.g., after learning tasks) may have been diluted beyond detection. On the other hand, results in cell lines may be relevant for mitotic cell populations, and obviously, by definition, are not in a physiological context. A full overview of all the cross-talk partners in different cells and tissues awaits dedicated experiments in different physiological settings.
Of note, although classical protein-protein transrepression depends on tethering of MR and GR, there are also hybrid mechanisms by which GR directly binds to DNA near transcription factors like AP-1 as a compound GRE [69, 70]. As with tethering mechanisms, the outcome of GR activation may be repressive or rather synergistic, depending on the exact nature and spacing of the interacting partners. Composite elements are important for recruitment of GR to the chromatin via transcription factor AP-1 [71]. Variations on this theme are the codependence of MR or GR binding to GREs in conjunction with other transcription factors that may require longer-range interactions on the DNA [38, 72].
2. Mechanisms for MR Specificity Over GR
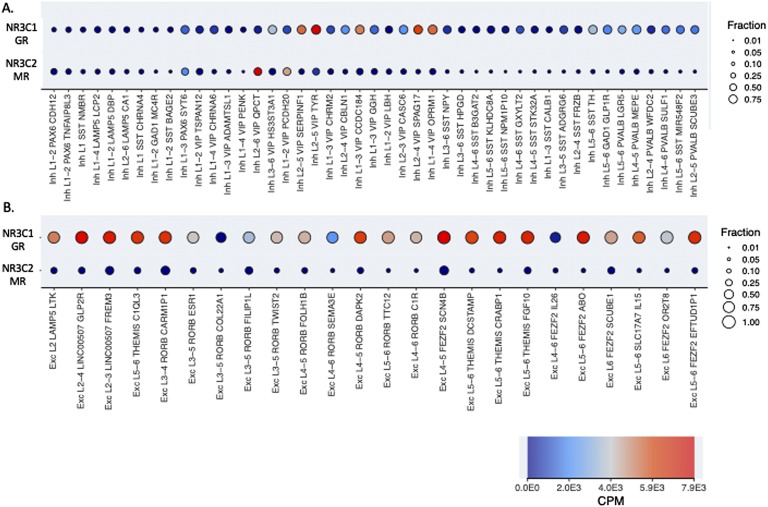
Foremost, the presence or absence of the receptors in a cell will determine whether cortisol will act via MR and/or GR. Differential effects of MR and GR activation in physiology may be explained in part by targeting of different cell types in the body that may or may not express either receptor. Single-cell sequencing will reveal, in the near future, the MR-to-GR ratio at the mRNA level for any cell type in the brain, as has been done already for the human temporal cortex, where 69 types of neurons were identified [73]. Figure 1 shows relative GR and MR expression in these cell types. For now, the current literature suggests there may be dominant MR presence in CA3 pyramidal cells in the hippocampus, even if GR is also present [74], and some GABA-ergic cortical neurons. MR-exclusive cells have not been proven, although one may expect that in neurons with aldosterone-selective MR, there should be very little GR activity.
Figure 1.
Expression of GR and MR in all individual 45 inhibitory and 24 excitatory neuronal cell types of the human temporal cortex. (A) GABA-ergic (‘inh’, or inhibitory) neurons show variable expression of GR and MR. Two GABA-ergic neurons show higher expression of MR than of GR: layer 2-6 VIP/OPCT positive neurons and layer 1-2 VIP/PCDH20 positive neurons. In most cell types, receptors are expressed at intermediate to low levels. (B) Glutamatergic (‘Exc’, or excitatory) neurons generally express high levels of GR and low levels of MR. Exonic expression is shown. Color codes for counts per million (CPM) transcripts on a linear scale, and size indicates the fraction per cell types with expression >1 CPM. Data and image are from http://celltypes.brain-map.org/rnaseq/human.
When MR and GR are coexpressed, they can differentially affect cellular physiology. This is exemplified by the classical U-shaped effect of corticosterone on excitability of hippocampal CA1 neuron. In these cells, MR activation stimulates excitability by suppressing the responsiveness to 5-HT1A receptor and calcium currents. Concomitant GR activation has the opposite effects on these cellular responses. In this way MR and GR mediate opposite effects of different doses of corticosterone on neuronal excitability [9].
At the mRNA level, the single-cell sequencing studies will help fine tune our understanding of where MR and GR can exert their effects in complex tissues like the brain [73]. At the protein level, knowing the relative abundance of MR and GR is complicated by technical issues (e.g., dependence on antibodies) and by the existence of MRs and GRs that differ in the length of their N-terminus because of alternative translation of their mRNAs [75, 76]. Also posttranslational modifications will affect the relative activity of the receptors [77].
There are several options as to how MR and GR differentially affect transcription, if they are both present in the same cell, including binding to specific GREs (potentially in conjunction with other transcription factors), differential binding to nGREs, differential interaction with other transcription factors, and differential interaction with downstream coregulators once the receptors have bound to the DNA. It is clear that binding to negative GREs is exclusive for GR [24]. It was also noted early on that GR is more potent than MR at transrepression of the transcription factor AP-1 [78]. Such effects may explain unique effects of cortisol via GR, such as anti-inflammatory effects, but MR also has intrinsic effects that differ from those mediated by GR [9, 10].
A. DNA Binding
In the rat hippocampus, we were able to make a direct comparison between whole-genome MR and GR binding in the same samples 45 minutes after treatment with a high dose of corticosterone. This revealed many DNA loci that were bound by either MR, GR, or both receptors. Motif analysis suggested that the GRE sequence was present in all bound DNA fragments, although the consensus sequence was slightly more degenerate for MR binding sites [62, 63].
Strikingly, the consensus binding site for NeuroD factors always co-occurred with the GRE for MR-specific binding sites but not for GR-specific binding sites. This motif was also reported for ∼15% of all GR binding sites in a separate study [64], and we suggest these loci represent sites where both MR and GR can bind either as homo- or as heterodimers [40]. Chromatin immunoprecipitation experiments confirmed the presence of the NeuroD2 protein, typically within 300 nucleotides of the MR binding site on hippocampal DNA. In forebrain-specific MR knockout mice, NeuroD2 was still present at these sites, suggesting that any functional directionality involves binding of NeuroD2 before MR [79]. Therefore, NeuroD factors may selectively allow MR binding to GREs in the principal neurons of the hippocampus. Complete absence of MR, in most cases, did not lead to GR binding at the shared MR and GR loci, nor did GR bind to specific MR loci in the MR-knockout mice. Competition between MR and GR for DNA binding, therefore, does not seem to be a dominant mechanism. However, in absence of MR, GR binding was increased at the Per1 locus [79]. This may be related to the fact that this particular GRE is a very high affinity GR binding site, given the very low EC50 value for glucocorticoid induction of the per1 gene [80].
How NeuroD would confer MR-specific DNA binding is not fully understood. The somewhat more degenerate GRE consensus for MR-selective loci may point to the necessity of NeuroD proteins as stabilizing factors for MR binding at these loci, but this remains to be proven. Structure-function analysis of MR and GR interactions with NeuroD in cell lines did not recapitulate the MR specificity: In transient transfections, NeuroD binding near a GRE could potentiate both MR- and GR-mediated transactivation. This may be explained by an incomplete chromatin context of transfected plasmids and/or reflects interactions of NeuroD factors with a third factor that is present in hippocampus but not in cultured cells. The notion of indirect interactions between MR and NeuroD factors is supported by additional studies in cell lines in which NeuroD proteins potentiated corticosterone-induced transcription of both N- and C-terminal truncations of MR and GR [62].
NeuroD proteins form a subfamily of bHLH transcription factors and have an important role in the terminal differentiation of particular neurons [81, 82]. Family members NeuroD1, 2, and 6 are expressed at substantial levels in different subfields of the adult mouse and rat hippocampus [62]. The link with neuronal end differentiation may explain the much more limited neuronal expression of MR compared with GR, in that MR would be specific only to particular differentiation programs. The intrinsic connection of MR binding sites to NeuroD factors suggest that via MR basal glucocorticoid levels are linked to the “neuron-ness” of such cells. This may relate to the early electrophysiological findings that showed increased excitability of the principal neurons in the hippocampus upon MR activation [9]. Of interest, NeuroD proteins have been found differentially expressed in postmortem brain tissue of depressed subjects [83].
NeuroD proteins are closely related to the MyoD family of transcription factors, which plays a role in the differentiation of muscle. MyoD factors can bind to a subset of NeuroD response elements and are somewhat better understood in terms of structure-function relationship [84]. MyoD family members may regulate gene expression either by direct transcriptional activation or by remodeling of the local chromatin [85]. In reporter studies, MR responses were affected only by the chromatin-modification function rather than the direct transcriptional activation. In contrast, GR-dependent transcription was potentiated by MyoD proteins via both mechanisms [79]. Knockout models for individual NeuroD factors may suffer from early death [86] and absence of particular neuronal populations [87], but on the other hand, they may show compensatory upregulation of family-member bHLH protein [88]. Nevertheless, in (inducible) models that show survival [86, 89], it will be very interesting to test MR functionality in the hippocampus.
As in the NeuroD study, GR seemed to be a much stronger transcription factor in reporter studies [44], but this seems at odds with powerful in vivo observations, both in terms of gene expression [90] and DNA binding [62]. Of note, in other cell types, functional interactions between MyoD and GR have been observed [91]. Thus, there seems to be a more general interaction mechanism between MR and/or GR and bHLH transcription factors that is dependent on yet different factors, given the cell specificity of the effects. For the hippocampal loci where MR and GR bind, NeuroD may help recruit MRs, also in the face of high hormone concentrations that would otherwise bias toward exclusive GR binding. Thus, on one hand, joint MR and GR occupancy may simply extend the dose-response curve for endogenous glucocorticoids and, on the other hand, fine tune the magnitude of transcriptional responses by, for example, heterodimerization.
B. Coregulators
It is clear that some coregulators are shared by MR, GR, and a host of other nuclear receptors, in particular for the AF-2 domain that is highly structured and similar between related nuclear receptors [53]. It is also clear that there is coregulator specificity, not only per receptor (in particular, via AF-1 [92]) but also per ligand. For example, coactivators may prefer aldosterone-bound MR to cortisol-bound MR [93, 94].
The final transcriptional outcome of glucocorticoid exposure depends on the type of ligand, type of receptor, the gene, and the cell type. This complexity is an uncomfortable fact if we want to understand and predict steroid action in particular conditions. One way to grasp what may happen in particular cell types is to use coexpression data of receptors and potential coregulators. This was done for all steroid receptors in the mouse brain, based on in situ hybridization data from the Allen Brain Institute [95]. The host of current and coming single-cell transcriptomes [73, 96] will allow us to link expression of receptors, their signaling partners, and their potential target genes, and, via “guilt-by-association,” pinpoint relevant interaction and predict steroid responses [97].
Ligand-selective coregulator recruitment is one of the mechanisms by which selective receptor modulators act, that is, ligands that combine agonism and antagonism of steroid receptors, dependent on the gene and tissue of interest [98]. Selective modulators induce alternative conformations of the receptors, allowing interactions with only a subset of coregulators [56, 99, 100]. Selective GR modulators have been pursued for a long time, primarily to separate anti-inflammatory effects from adverse effects of glucocorticoid therapy [101]. Selective MR modulators are of interest to block the hypokalemia that comes with MR antagonism aimed at the heart [102]. Conceptually, if stimulating brain MRs would be a therapeutic target [16, 17], selective MR modulation could prevent overactivation of the aldosterone-sensitive MRs regulating blood pressure and salt intake [5].
C. Specific and Shared Target Genes
The existence of unique and shared DNA-binding sites of MR and GR suggests that there will be unique and shared target genes. Transrepressed genes via GR in the activated immune system are a clear example, but this mechanism appears not to have a major role in the hippocampus [62–64]. The opposite effects of MR and GR on hippocampal excitability have biased the search for target genes toward genes that are uniquely regulated by one receptor type or even in opposite ways via MR and GR.
We used the hippocampal MR chromatin binding profile in the rat to identify MR-regulated genes. Because only ∼10% of DNA-bound steroid receptor can be directly linked to gene expression [103], we selected binding sites that were unique for MR located in intronic regions or within 5 kilobases of transcription start sites. Because MR also bound these loci in mouse hippocampal chromatin, we evaluated their expression in mice lacking MR expression in the forebrain [104]. mRNA of Jdp2, Nos1ap, and Supv3l1 was up to 50% less in the brains of the MR-knockout mice, suggesting that these genes are selective MR target genes [105]. Although this list is surely incomplete, and RNA sequencing should reveal other MR-dependent transcripts, the current set of likely MR-specific transcripts may be of use to probe functional MR activity in different paradigms of stress and/or steroid exposure.
It is important that we can attribute particular effects to either MR or GR and evaluate their relative activity in clinical states or experimental models. Having specific readouts for GR and MR activity is not trivial, because the GRE, as such, is shared by both receptors, and most glucocorticoid-induced mRNAs that are routinely used as readouts for glucocorticoid effects can be stimulated via both MR and GR. This includes mRNAs for Per1 [80, 106, 107], Sgk1 [90, 108], and GILZ [109, 110]. In fact, next to some genes associated with selective MR binding, basal levels of FKBP5 mRNA also were substantially downregulated in the hippocampus of forebrain-MR knockout mice [105]. That target genes are shared suggests (in well-studied cases) MR and GR cooperate to extend the cellular sensitivity for glucocorticoids over a range of three orders of magnitude. Functionally, the cooperative actions of MR and GR are perhaps made most clear by the fact that both MR and GR mediate negative feedback on the HPA axis. GR mediates negative feedback in pituitary and hypothalamus, and MR does so in the hippocampus [4, 111, 112]. The involvement of both receptor types means that negative feedback takes place in a gradual manner, from minor elevations basal trough levels to very high levels of hormones. The common regulation of genes like FKBP5 also merits attention. Often, FKBP5 function is considered in relation to GR functionality, and the (epi-)genetic variation in the FKBP5 gene in human disease is likewise being linked mainly or exclusively to GR [113, 114]. Although in some cases (perhaps peripheral blood) this may be justified, the contribution of MR to FKBP5 expression merits a less GR-centric view of this major transcriptional target of glucocorticoids. The cellular diversity in the brain calls for more refined experiments to understand which MR and GR target genes are joined, or rather are receptor specific, in which particular cell types.
3. Summary and Conclusions
The cortisol- and corticosterone-preferring brain MR plays an important role in regulation of stress responsiveness, adaption, and mood. It does so via nongenomic and genomic actions in interplay with GR. MR and GR share a number of target genes and may cooperate at the transcriptional level within the cell, as well as at the functional level. MR and GR can also mediate independent effects on gene expression and, in this way, have opposite effects on cellular physiology. MR-specific gene transcripts have been identified, and these seem to depend on a permissive effect of NeuroD factors for MR binding to GREs that mediate transactivation of target genes. More generically, the gene- and cell type–dependent effects of MR and GR on gene expression depend on interactions with several different types of proteins, including transcription factors and coregulators. The coexpression of these interacting partners can be assessed using single-cell sequencing data from repositories or experimental models. Such interactions may be selectively targeted with new receptor ligands, to better understand adaptation to stress, and for therapeutic purposes in stress-related disease.
Acknowledgments
Financial Support: This review is based on research projects funded to OCM by NWO-ALW (Grant 82302002) and ZON-MW, the Dutch Foundation for Health Research and Development (Grant 95105005), and COST project ADMIRE.
Additional Information
Disclosure Summary: O.C.M. receives research funding from Corcept Therapeutics.
Data Availability: Data sharing is not applicable to this article because no data sets were generated or analyzed during the current study.
Glossary
Abbreviations:
- DBD
DNA-binding domain
- GR
glucocorticoid receptor
- GRE
glucocorticoid response element
- HPA
hypothalamic-pituitary-adrenal
- MR
mineralocorticoid receptor
- nGRE
negative glucocorticoid response element
References and Notes
- 1. Sapolsky RM, Romero LM, Munck AU. How do glucocorticoids influence stress responses? Integrating permissive, suppressive, stimulatory, and preparative actions. Endocr Rev. 2000;21(1):55–89. [DOI] [PubMed] [Google Scholar]
- 2. de Kloet ER, Joëls M, Holsboer F. Stress and the brain: from adaptation to disease. Nat Rev Neurosci. 2005;6(6):463–475. [DOI] [PubMed] [Google Scholar]
- 3. Karssen AM, Meijer OC, van der Sandt IC, Lucassen PJ, de Lange EC, de Boer AG, de Kloet ER. Multidrug resistance P-glycoprotein hampers the access of cortisol but not of corticosterone to mouse and human brain. Endocrinology. 2001;142(6):2686–2694. [DOI] [PubMed] [Google Scholar]
- 4. Reul JM, van den Bosch FR, de Kloet ER. Relative occupation of type-I and type-II corticosteroid receptors in rat brain following stress and dexamethasone treatment: functional implications. J Endocrinol. 1987;115(3):459–467. [DOI] [PubMed] [Google Scholar]
- 5. Geerling JC, Loewy AD. Aldosterone in the brain. Am J Physiol Renal Physiol. 2009;297(3):F559–F576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gasparini S, Resch JM, Narayan SV, Peltekian L, Iverson GN, Karthik S, Geerling JC. Aldosterone-sensitive HSD2 neurons in mice. Brain Struct Funct. 2019;224(1):387–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Reul JM, de Kloet ER. Two receptor systems for corticosterone in rat brain: microdistribution and differential occupation. Endocrinology. 1985;117(6):2505–2511. [DOI] [PubMed] [Google Scholar]
- 8. Fitzsimons CP, van Hooijdonk LWA, Schouten M, Zalachoras I, Brinks V, Zheng T, Schouten TG, Saaltink DJ, Dijkmans T, Steindler DA, Verhaagen J, Verbeek FJ, Lucassen PJ, de Kloet ER, Meijer OC, Karst H, Joëls M, Oitzl MS, Vreugdenhil E. Knockdown of the glucocorticoid receptor alters functional integration of newborn neurons in the adult hippocampus and impairs fear-motivated behavior. Mol Psychiatry. 2013;18(9):993–1005. [DOI] [PubMed] [Google Scholar]
- 9. Joëls M. Corticosteroid effects in the brain: U-shape it. Trends Pharmacol Sci. 2006;27(5):244–250. [DOI] [PubMed] [Google Scholar]
- 10. Chantong B, Kratschmar DV, Nashev LG, Balazs Z, Odermatt A. Mineralocorticoid and glucocorticoid receptors differentially regulate NF-kappaB activity and pro-inflammatory cytokine production in murine BV-2 microglial cells. J Neuroinflammation. 2012;9(1):260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oakley RH, Cruz-Topete D, He B, Foley JF, Myers PH, Xu X, Gomez-Sanchez CE, Chambon P, Willis MS, Cidlowski JA. Cardiomyocyte glucocorticoid and mineralocorticoid receptors directly and antagonistically regulate heart disease in mice. Sci Signal 2019;12(577):eaau9685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nieman LK, Chrousos GP, Kellner C, Spitz IM, Nisula BC, Cutler GB, Merriam GR, Bardin CW, Loriaux DL. Successful treatment of Cushing’s syndrome with the glucocorticoid antagonist RU 486. J Clin Endocrinol Metab. 1985;61(3):536–540. [DOI] [PubMed] [Google Scholar]
- 13. van der Lely AJ, Foeken K, van der Mast RC, Lamberts SW. Rapid reversal of acute psychosis in the Cushing syndrome with the cortisol-receptor antagonist mifepristone (RU 486). Ann Intern Med. 1991;114(2):143–144. [DOI] [PubMed] [Google Scholar]
- 14. Pereira AM, Tiemensma J, Romijn JA. Neuropsychiatric disorders in Cushing's syndrome. Neuroendocrinology 2010;92(Suppl 1):65–70. [DOI] [PubMed] [Google Scholar]
- 15. ter Heegde F, De Rijk RH, Vinkers CH. The brain mineralocorticoid receptor and stress resilience. Psychoneuroendocrinology. 2015;52:92–110. [DOI] [PubMed] [Google Scholar]
- 16. Klok MD, Giltay EJ, Van der Does AJW, Geleijnse JM, Antypa N, Penninx BWJH, de Geus EJC, Willemsen G, Boomsma DI, van Leeuwen N, Zitman FG, de Kloet ER, DeRijk RH. A common and functional mineralocorticoid receptor haplotype enhances optimism and protects against depression in females. Transl Psychiatry. 2011;1(12):e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Otte C, Hinkelmann K, Moritz S, Yassouridis A, Jahn H, Wiedemann K, Kellner M. Modulation of the mineralocorticoid receptor as add-on treatment in depression: a randomized, double-blind, placebo-controlled proof-of-concept study. J Psychiatr Res. 2010;44(6):339–346. [DOI] [PubMed] [Google Scholar]
- 18. de Kloet ER, Otte C, Kumsta R, Kok L, Hillegers MHJ, Hasselmann H, Kliegel D, Joëls M. Stress and depression: a crucial role of the mineralocorticoid receptor. J Neuroendocrinol. 2016;28(8). [DOI] [PubMed] [Google Scholar]
- 19. Fardet L, Petersen I, Nazareth I. Suicidal behavior and severe neuropsychiatric disorders following glucocorticoid therapy in primary care. Am J Psychiatry. 2012;169(5):491–497. [DOI] [PubMed] [Google Scholar]
- 20. Judd LL, Schettler PJ, Brown ES, Wolkowitz OM, Sternberg EM, Bender BG, Bulloch K, Cidlowski JA, de Kloet ER, Fardet L, Joëls M, Leung DYM, McEwen BS, Roozendaal B, Van Rossum EFC, Ahn J, Brown DW, Plitt A, Singh G. Adverse consequences of glucocorticoid medication: psychological, cognitive, and behavioral effects. Am J Psychiatry. 2014;171(10):1045–1051. [DOI] [PubMed] [Google Scholar]
- 21. Reul JM, Gesing A, Droste S, Stec IS, Weber A, Bachmann C, Bilang-Bleuel A, Holsboer F, Linthorst AC. The brain mineralocorticoid receptor: greedy for ligand, mysterious in function. Eur J Pharmacol. 2000;405(1-3):235–249. [DOI] [PubMed] [Google Scholar]
- 22. Meijer OC, de Kloet ER. A refill for the brain mineralocorticoid receptor: the benefit of cortisol add-on to dexamethasone therapy. Endocrinology. 2017;158(3):448–454. [DOI] [PubMed] [Google Scholar]
- 23. Warris LT, van den Heuvel-Eibrink MM, Aarsen FK, Pluijm SMF, Bierings MB, van den Bos C, Zwaan CM, Thygesen HH, Tissing WJE, Veening MA, Pieters R, van den Akker ELT. Hydrocortisone as an intervention for dexamethasone-induced adverse effects in pediatric patients with acute lymphoblastic leukemia: results of a double-blind, randomized controlled trial. J Clin Oncol. 2016;34(19):2287–2293. [DOI] [PubMed] [Google Scholar]
- 24. Hudson WH, Youn C, Ortlund EA. The structural basis of direct glucocorticoid-mediated transrepression. Nat Struct Mol Biol. 2013;20(1):53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Harms MJ, Thornton JW. Historical contingency and its biophysical basis in glucocorticoid receptor evolution. Nature. 2014;512(7513):203–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bridgham JT, Carroll SM, Thornton JW. Evolution of hormone-receptor complexity by molecular exploitation. Science. 2006;312(5770):97–101. [DOI] [PubMed] [Google Scholar]
- 27. Gasser PJ, Lowry CA. Organic cation transporter 3: a cellular mechanism underlying rapid, non-genomic glucocorticoid regulation of monoaminergic neurotransmission, physiology, and behavior. Horm Behav. 2018;104:173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Karst H, Berger S, Turiault M, Tronche F, Schütz G, Joëls M. Mineralocorticoid receptors are indispensable for nongenomic modulation of hippocampal glutamate transmission by corticosterone. Proc Natl Acad Sci USA. 2005;102(52):19204–19207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nahar J, Haam J, Chen C, Jiang Z, Glatzer NR, Muglia LJ, Dohanich GP, Herman JP, Tasker JG. Rapid nongenomic glucocorticoid actions in male mouse hypothalamic neuroendocrine cells are dependent on the nuclear glucocorticoid receptor. Endocrinology. 2015;156(8):2831–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dooley R, Harvey BJ, Thomas W. Non-genomic actions of aldosterone: from receptors and signals to membrane targets. Mol Cell Endocrinol. 2012;350(2):223–234. [DOI] [PubMed] [Google Scholar]
- 31. Gutièrrez-Mecinas M, Trollope AF, Collins A, Morfett H, Hesketh SA, Kersanté F, Reul JMHM. Long-lasting behavioral responses to stress involve a direct interaction of glucocorticoid receptors with ERK1/2-MSK1-Elk-1 signaling. Proc Natl Acad Sci USA. 2011;108(33):13806–13811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jiang C-L, Liu L, Tasker JG. Why do we need nongenomic glucocorticoid mechanisms? Front Neuroendocrinol. 2014;35(1):72–75. [DOI] [PubMed] [Google Scholar]
- 33. Joëls M, Sarabdjitsingh RA, Karst H. Unraveling the time domains of corticosteroid hormone influences on brain activity: rapid, slow, and chronic modes. Pharmacol Rev. 2012;64(4):901–938. [DOI] [PubMed] [Google Scholar]
- 34. Karst H, Berger S, Erdmann G, Schütz G, Joëls M. Metaplasticity of amygdalar responses to the stress hormone corticosterone. Proc Natl Acad Sci USA. 2010;107(32):14449–14454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Monczor F, Chatzopoulou A, Zappia CD, Houtman R, Meijer OC, Fitzsimons CP. A model of glucocorticoid receptor interaction with coregulators predicts transcriptional regulation of target genes. Front Pharmacol. 2019;10:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ong KM, Blackford JA Jr, Kagan BL, Simons SS Jr, Chow CC. A theoretical framework for gene induction and experimental comparisons. Proc Natl Acad Sci USA. 2010;107(15):7107–7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. John S, Sabo PJ, Thurman RE, Sung M-H, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43(3):264–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. So AY-L, Cooper SB, Feldman BJ, Manuchehri M, Yamamoto KR. Conservation analysis predicts in vivo occupancy of glucocorticoid receptor-binding sequences at glucocorticoid-induced genes. Proc Natl Acad Sci USA. 2008;105(15):5745–5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Datson NA, Polman JAE, de Jonge RT, van Boheemen PTM, van Maanen EMT, Welten J, McEwen BS, Meiland HC, Meijer OC. Specific regulatory motifs predict glucocorticoid responsiveness of hippocampal gene expression. Endocrinology. 2011;152(10):3749–3757. [DOI] [PubMed] [Google Scholar]
- 40. Liu W, Wang J, Sauter NK, Pearce D. Steroid receptor heterodimerization demonstrated in vitro and in vivo. Proc Natl Acad Sci USA. 1995;92(26):12480–12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mifsud KR, Reul JMHM. Acute stress enhances heterodimerization and binding of corticosteroid receptors at glucocorticoid target genes in the hippocampus. Proc Natl Acad Sci USA. 2016;113(40):11336–11341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rivers CA, Rogers MF, Stubbs FE, Conway-Campbell BL, Lightman SL, Pooley JR. Glucocorticoid receptor-tethered mineralocorticoid receptors increase glucocorticoid-induced transcriptional responses. Endocrinology. 2019;160(5):1044–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Presman DM, Ganguly S, Schiltz RL, Johnson TA, Karpova TS, Hager GL. DNA binding triggers tetramerization of the glucocorticoid receptor in live cells. Proc Natl Acad Sci USA. 2016;113(29):8236–8241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Adams M, Meijer OC, Wang J, Bhargava A, Pearce D. Homodimerization of the glucocorticoid receptor is not essential for response element binding: activation of the phenylethanolamine N-methyltransferase gene by dimerization-defective mutants. Mol Endocrinol. 2003;17(12):2583–2592. [DOI] [PubMed] [Google Scholar]
- 45. Liu W, Wang J, Yu G, Pearce D. Steroid receptor transcriptional synergy is potentiated by disruption of the DNA-binding domain dimer interface. Mol Endocrinol. 1996;10(11):1399–1406. [DOI] [PubMed] [Google Scholar]
- 46. O’Malley BW. Coregulators: from whence came these “master genes”. Mol Endocrinol. 2007;21(5):1009–1013. [DOI] [PubMed] [Google Scholar]
- 47. Atucha E, Zalachoras I, van den Heuvel JK, van Weert LTCM, Melchers D, Mol IM, Belanoff JK, Houtman R, Hunt H, Roozendaal B, Meijer OC. A mixed glucocorticoid/mineralocorticoid selective modulator with dominant antagonism in the male rat brain. Endocrinology. 2015;156(11):4105–4114. [DOI] [PubMed] [Google Scholar]
- 48. Meijsing SH, Pufall MA, So AY-L, Bates DL, Chen L, Yamamoto KR. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324(5925):407–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Weikum ER, Knuesel MT, Ortlund EA, Yamamoto KR. Glucocorticoid receptor control of transcription: precision and plasticity via allostery. Nat Rev Mol Cell Biol. 2017;18(3):159–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lachize S, Apostolakis EM, van der Laan S, Tijssen AMI, Xu J, de Kloet ER, Meijer OC. Steroid receptor coactivator-1 is necessary for regulation of corticotropin-releasing hormone by chronic stress and glucocorticoids. Proc Natl Acad Sci USA. 2009;106(19):8038–8042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zalachoras I, Verhoeve SL, Toonen LJ, van Weert LTCM, van Vlodrop AM, Mol IM, Meelis W, de Kloet ER, Meijer OC. Isoform switching of steroid receptor co-activator-1 attenuates glucocorticoid-induced anxiogenic amygdala CRH expression. Mol Psychiatry. 2016;21(12):1733–1739. [DOI] [PubMed] [Google Scholar]
- 52. Fischer K, Kelly SM, Watt K, Price NC, McEwan IJ. Conformation of the mineralocorticoid receptor N-terminal domain: evidence for induced and stable structure. Mol Endocrinol. 2010;24(10):1935–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Desmet SJ, Dejager L, Clarisse D, Thommis J, Melchers D, Bastiaensen N, Ruijtenbeek R, Beck IM, Libert C, Houtman R, Meijer OC, De Bosscher K. Cofactor profiling of the glucocorticoid receptor from a cellular environment. Methods Mol. Biol. 2014;1204:83–94. [DOI] [PubMed] [Google Scholar]
- 54. Broekema MF, Hollman DAA, Koppen A, van den Ham H-J, Melchers D, Pijnenburg D, Ruijtenbeek R, van Mil SWC, Houtman R, Kalkhoven E. Profiling of 3696 nuclear receptor-coregulator interactions: a resource for biological and clinical discovery. Endocrinology. 2018;159(6):2397–2407. [DOI] [PubMed] [Google Scholar]
- 55. Surjit M, Ganti KP, Mukherji A, Ye T, Hua G, Metzger D, Li M, Chambon P. Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell. 2011;145(2):224–241. [DOI] [PubMed] [Google Scholar]
- 56. Hua G, Paulen L, Chambon P. GR SUMOylation and formation of an SUMO-SMRT/NCoR1-HDAC3 repressing complex is mandatory for GC-induced IR nGRE-mediated transrepression. Proc Natl Acad Sci USA. 2016;113(5):E626–E634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zalachoras I, Houtman R, Atucha E, Devos R, Tijssen AMI, Hu P, Lockey PM, Datson NA, Belanoff JK, Lucassen PJ, Joëls M, de Kloet ER, Roozendaal B, Hunt H, Meijer OC. Differential targeting of brain stress circuits with a selective glucocorticoid receptor modulator. Proc Natl Acad Sci USA. 2013;110(19):7910–7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yang-Yen HF, Chambard JC, Sun YL, Smeal T, Schmidt TJ, Drouin J, Karin M. Transcriptional interference between c-Jun and the glucocorticoid receptor: mutual inhibition of DNA binding due to direct protein-protein interaction. Cell. 1990;62(6):1205–1215. [DOI] [PubMed] [Google Scholar]
- 59. Robert-Nicoud M, Flahaut M, Elalouf JM, Nicod M, Salinas M, Bens M, Doucet A, Wincker P, Artiguenave F, Horisberger JD, Vandewalle A, Rossier BC, Firsov D. Transcriptome of a mouse kidney cortical collecting duct cell line: effects of aldosterone and vasopressin. Proc Natl Acad Sci USA. 2001;98(5):2712–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Meinel S, Ruhs S, Schumann K, Strätz N, Trenkmann K, Schreier B, Grosse I, Keilwagen J, Gekle M, Grossmann C. Mineralocorticoid receptor interaction with SP1 generates a new response element for pathophysiologically relevant gene expression. Nucleic Acids Res. 2013;41(17):8045–8060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Meijer OC, Williamson A, Dallman MF, Pearce D. Transcriptional repression of the 5-HT1A receptor promoter by corticosterone via mineralocorticoid receptors depends on the cellular context. J Neuroendocrinol. 2000;12(3):245–254. [DOI] [PubMed] [Google Scholar]
- 62. Le Billan F, Khan JA, Lamribet K, Viengchareun S, Bouligand J, Fagart J, Lombès M. Cistrome of the aldosterone-activated mineralocorticoid receptor in human renal cells. FASEB J. 2015;29(9):3977–3989. [DOI] [PubMed] [Google Scholar]
- 63. van Weert LTCM, Buurstede JC, Mahfouz A, Braakhuis PSM, Polman JAE, Sips HCM, Roozendaal B, Balog J, de Kloet ER, Datson NA, Meijer OC. NeuroD factors discriminate mineralocorticoid from glucocorticoid receptor dna binding in the male rat brain. Endocrinology. 2017;158(5):1511–1522. [DOI] [PubMed] [Google Scholar]
- 64. Polman JAE, de Kloet ER, Datson NA. Two populations of glucocorticoid receptor-binding sites in the male rat hippocampal genome. Endocrinology. 2013;154(5):1832–1844. [DOI] [PubMed] [Google Scholar]
- 65. Pooley JR, Flynn BP, Grøntved L, Baek S, Guertin MJ, Kershaw YM, Birnie MT, Pellatt A, Rivers CA, Schiltz RL, Hager GL, Lightman SL, Conway-Campbell BL. Genome-wide identification of basic helix-loop helix and NF-1 motifs underlying GR binding sites in male rat hippocampus. Endocrinology. 2017;158(5):1486–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Datson NA, van den Oever JME, Korobko OB, Magarinos AM, de Kloet ER, McEwen BS. Previous history of chronic stress changes the transcriptional response to glucocorticoid challenge in the dentate gyrus region of the male rat hippocampus. Endocrinology. 2013;154(9):3261–3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bhargava A, Meijer OC, Dallman MF, Pearce D. Plasma membrane calcium pump isoform 1 gene expression is repressed by corticosterone and stress in rat hippocampus. J Neurosci. 2000;20(9):3129–3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ou XM, Storring JM, Kushwaha N, Albert PR. Heterodimerization of mineralocorticoid and glucocorticoid receptors at a novel negative response element of the 5-HT1A receptor gene. J Biol Chem. 2001;276(17):14299–14307. [DOI] [PubMed] [Google Scholar]
- 69. Diamond MI, Miner JN, Yoshinaga SK, Yamamoto KR. Transcription factor interactions: selectors of positive or negative regulation from a single DNA element. Science. 1990;249(4974):1266–1272. [DOI] [PubMed] [Google Scholar]
- 70. Pearce D, Matsui W, Miner JN, Yamamoto KR. Glucocorticoid receptor transcriptional activity determined by spacing of receptor and nonreceptor DNA sites. J Biol Chem. 1998;273(46):30081–30085. [DOI] [PubMed] [Google Scholar]
- 71. Biddie SC, John S, Sabo PJ, Thurman RE, Johnson TA, Schiltz RL, Miranda TB, Sung M-H, Trump S, Lightman SL, Vinson C, Stamatoyannopoulos JA, Hager GL. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol Cell. 2011;43(1):145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. O’Hara B, Alvarez de la Rosa D, Rajendran VM. Multiple mineralocorticoid response elements localized in different introns regulate intermediate conductance K+ (Kcnn4) channel expression in the rat distal colon. PLoS One. 2014;9(6):e98695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hodge RD, Bakken TE, Miller JA, Smith KA, Barkan ER, Graybuck LT, Close JL, Long B, Penn O, Yao Z, Eggermont J, Höllt T, Levi BP, Shehata SI, Aevermann B, Beller A, Bertagnolli D, Brouner K, Casper T, Cobbs C, Dalley R, Dee N, Ding S-L, Ellenbogen RG, Fong O, Garren E, Goldy J, Gwinn RP, Hirschstein D, Keene CD, Keshk M, Ko AL, Lathia K, Mahfouz A, Maltzer Z, McGraw M, Nguyen TN, Nyhus J, Ojemann JG, Oldre A, Parry S, Reynolds S, Rimorin C, Shapovalova NV, Somasundaram S, Szafer A, Thomsen ER, Tieu M, Scheuermann RH, Yuste R, Sunkin SM, Lelieveldt B, Feng D, Ng L, Bernard A, Hawrylycz M, Phillips J, Tasic B, Zeng H, Jones AR, Koch C, Lein ES. Conserved cell types with divergent features between human and mouse cortex [published online ahead of print August 5, 2018]. bioRxiv. www.biorxiv.org/content/10.1101/384826v1. [Google Scholar]
- 74. Sarabdjitsingh RA, Meijer OC, de Kloet ER. Specificity of glucocorticoid receptor primary antibodies for analysis of receptor localization patterns in cultured cells and rat hippocampus. Brain Res. 2010;1331:1–11. [DOI] [PubMed] [Google Scholar]
- 75. Lu NZ, Cidlowski JA. Translational regulatory mechanisms generate N-terminal glucocorticoid receptor isoforms with unique transcriptional target genes. Mol Cell. 2005;18(3):331–342. [DOI] [PubMed] [Google Scholar]
- 76. Faresse N. Post-translational modifications of the mineralocorticoid receptor: how to dress the receptor according to the circumstances? J Steroid Biochem Mol Biol. 2014;143:334–342. [DOI] [PubMed] [Google Scholar]
- 77. Vandevyver S, Dejager L, Libert C. Comprehensive overview of the structure and regulation of the glucocorticoid receptor. Endocr Rev. 2014;35(4):671–693. [DOI] [PubMed] [Google Scholar]
- 78. Pearce D, Yamamoto KR. Mineralocorticoid and glucocorticoid receptor activities distinguished by nonreceptor factors at a composite response element. Science. 1993;259(5098):1161–1165. [DOI] [PubMed] [Google Scholar]
- 79. van Weert LTCM, Buurstede JC, Sips HCM, Mol IM, Puri T, Damsteegt R, Roozendaal B, Sarabdjitsingh RA, Meijer OC. Mechanistic insights in NeuroD potentiation of mineralocorticoid receptor signaling. Int J Mol Sci. 2019;20(7):e1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Reddy TE, Gertz J, Crawford GE, Garabedian MJ, Myers RM. The hypersensitive glucocorticoid response specifically regulates period 1 and expression of circadian genes. Mol Cell Biol. 2012;32(18):3756–3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Bormuth I, Yan K, Yonemasu T, Gummert M, Zhang M, Wichert S, Grishina O, Pieper A, Zhang W, Goebbels S, Tarabykin V, Nave K-A, Schwab MH. Neuronal basic helix-loop-helix proteins Neurod2/6 regulate cortical commissure formation before midline interactions. J Neurosci. 2013;33(2):641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Schwab MH, Bartholomae A, Heimrich B, Feldmeyer D, Druffel-Augustin S, Goebbels S, Naya FJ, Zhao S, Frotscher M, Tsai MJ, Nave KA. Neuronal basic helix-loop-helix proteins (NEX and BETA2/Neuro D) regulate terminal granule cell differentiation in the hippocampus. J Neurosci. 2000;20(10):3714–3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Labonté B, Engmann O, Purushothaman I, Menard C, Wang J, Tan C, Scarpa JR, Moy G, Loh YE, Cahill M, Lorsch ZS, Hamilton PJ, Calipari ES, Hodes GE, Issler O, Kronman H, Pfau M, Obradovic ALJ, Dong Y, Neve RL, Russo S, Kazarskis A, Tamminga C, Mechawar N, Turecki G, Zhang B, Shen L, Nestler EJ. Sex-specific transcriptional signatures in human depression [published correction appears in Nat Med. 2018;24(4):525]. Nat Med. 2017;23(9):1102–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Fong AP, Yao Z, Zhong JW, Johnson NM, Farr GH III, Maves L, Tapscott SJ. Conversion of MyoD to a neurogenic factor: binding site specificity determines lineage. Cell Reports. 2015;10(12):1937–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Conerly ML, Yao Z, Zhong JW, Groudine M, Tapscott SJ. Distinct activities of Myf5 and MyoD indicate separate roles in skeletal muscle lineage specification and differentiation. Dev Cell. 2016;36(4):375–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Huang H-P, Chu K, Nemoz-Gaillard E, Elberg D, Tsai M-J. Neogenesis of beta-cells in adult BETA2/NeuroD-deficient mice. Mol Endocrinol. 2002;16(3):541–551. [DOI] [PubMed] [Google Scholar]
- 87. Miyata T, Maeda T, Lee JE. NeuroD is required for differentiation of the granule cells in the cerebellum and hippocampus. Genes Dev. 1999;13(13):1647–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Cho J-H, Klein WH, Tsai M-J. Compensational regulation of bHLH transcription factors in the postnatal development of BETA2/NeuroD1-null retina. Mech Dev. 2007;124(7-8):543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Brulet R, Zhu J, Aktar M, Hsieh J, Cho K-O. Mice with conditional NeuroD1 knockout display reduced aberrant hippocampal neurogenesis but no change in epileptic seizures. Exp Neurol. 2017;293:190–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Chen SY, Bhargava A, Mastroberardino L, Meijer OC, Wang J, Buse P, Firestone GL, Verrey F, Pearce D. Epithelial sodium channel regulated by aldosterone-induced protein sgk. Proc Natl Acad Sci USA. 1999;96(5):2514–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Oakley RH, Busillo JM, Cidlowski JA. Cross-talk between the glucocorticoid receptor and MyoD family inhibitor domain-containing protein provides a new mechanism for generating tissue-specific responses to glucocorticoids. J Biol Chem. 2017;292(14):5825–5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Pascual-Le Tallec L, Simone F, Viengchareun S, Meduri G, Thirman MJ, Lombès M. The elongation factor ELL (eleven-nineteen lysine-rich leukemia) is a selective coregulator for steroid receptor functions. Mol Endocrinol. 2005;19(5):1158–1169. [DOI] [PubMed] [Google Scholar]
- 93. Rogerson FM, Yao Y-Z, Young MJ, Fuller PJ. Identification and characterization of a ligand-selective mineralocorticoid receptor coactivator. FASEB J. 2014;28(10):4200–4210. [DOI] [PubMed] [Google Scholar]
- 94. Fuller PJ, Yang J, Young MJ. 30 Years of the mineralocorticoid receptor: coregulators as mediators of mineralocorticoid receptor signalling diversity. J Endocrinol. 2017;234(1):T23–T34. [DOI] [PubMed] [Google Scholar]
- 95. Mahfouz A, Lelieveldt BPF, Grefhorst A, van Weert LTCM, Mol IM, Sips HCM, van den Heuvel JK, Datson NA, Visser JA, Reinders MJT, Meijer OC. Genome-wide coexpression of steroid receptors in the mouse brain: Identifying signaling pathways and functionally coordinated regions. Proc Natl Acad Sci USA. 2016;113(10):2738–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Chen R, Wu X, Jiang L, Zhang Y. Single-cell RNA-Seq reveals hypothalamic cell diversity. Cell Reports. 2017;18(13):3227–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Chachlaki K, Prévot V. Coexpression profiles reveal hidden gene networks. Proc Natl Acad Sci USA. 2016;113(10):2563–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. De Bosscher K, Beck IM, Ratman D, Berghe WV, Libert C. Activation of the glucocorticoid receptor in acute inflammation: the SEDIGRAM concept. Trends Pharmacol Sci. 2016;37(1):4–16. [DOI] [PubMed] [Google Scholar]
- 99. Koorneef LL, van den Heuvel JK, Kroon J, Boon MR, ’t Hoen PAC, Hettne KM, van de Velde NM, Kolenbrander KB, Streefland TCM, Mol IM, Sips HCM, Kielbasa SM, Mei H, Belanoff JK, Pereira AM, Oosterveer MH, Hunt H, Rensen PCN, Meijer OC. Selective glucocorticoid receptor modulation prevents and reverses nonalcoholic fatty liver disease in male mice. Endocrinology. 2018;159(12):3925–3936. [DOI] [PubMed] [Google Scholar]
- 100. Meijer OC, Koorneef LL, Kroon J. Glucocorticoid receptor modulators. Ann Endocrinol (Paris). 2018;79(3):107–111. [DOI] [PubMed] [Google Scholar]
- 101. Coghlan MJ, Jacobson PB, Lane B, Nakane M, Lin CW, Elmore SW, Kym PR, Luly JR, Carter GW, Turner R, Tyree CM, Hu J, Elgort M, Rosen J, Miner JN. A novel antiinflammatory maintains glucocorticoid efficacy with reduced side effects. Mol Endocrinol. 2003;17(5):860–869. [DOI] [PubMed] [Google Scholar]
- 102. Grune J, Beyhoff N, Smeir E, Chudek R, Blumrich A, Ban Z, Brix S, Betz IR, Schupp M, Foryst-Ludwig A, Klopfleisch R, Stawowy P, Houtman R, Kolkhof P, Kintscher U. Selective mineralocorticoid receptor cofactor modulation as molecular basis for finerenone’s antifibrotic activity. Hypertension. 2018;71(4):599–608. [DOI] [PubMed] [Google Scholar]
- 103. Starick SR, Ibn-Salem J, Jurk M, Hernandez C, Love MI, Chung H-R, Vingron M, Thomas-Chollier M, Meijsing SH. ChIP-exo signal associated with DNA-binding motifs provides insight into the genomic binding of the glucocorticoid receptor and cooperating transcription factors. Genome Res. 2015;25(6):825–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Berger S, Wolfer DP, Selbach O, Alter H, Erdmann G, Reichardt HM, Chepkova AN, Welzl H, Haas HL, Lipp H-P, Schütz G. Loss of the limbic mineralocorticoid receptor impairs behavioral plasticity. Proc Natl Acad Sci USA. 2006;103(1):195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. van Weert LTCM, Buurstede JC, Sips HCM, Vettorazzi S, Mol IM, Prekovic S, Zwart W, Schmidt MV, Roozendaal B, Tuckermann J, Sarabdjitsingh RA, Meijer OC. Identification of mineralocorticoid receptor target genes in the mouse hippocampus. J Neuroendocrinol. 2019;e12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Le Billan F, Amazit L, Bleakley K, Xue Q-Y, Pussard E, Lhadj C, Kolkhof P, Viengchareun S, Fagart J, Lombès M. Corticosteroid receptors adopt distinct cyclical transcriptional signatures. FASEB J. 2018;32(10):5626–5639. [DOI] [PubMed] [Google Scholar]
- 107. Fletcher EK, Kanki M, Morgan J, Ray DW, Delbridge L, Fuller PJ, Clyne CD, Young MJ. Cardiomyocyte transcription is controlled by combined MR and circadian clock signalling. J Endocrinol. 2019;241(1):17–29. [DOI] [PubMed] [Google Scholar]
- 108. Webster MK, Goya L, Ge Y, Maiyar AC, Firestone GL. Characterization of sgk, a novel member of the serine/threonine protein kinase gene family which is transcriptionally induced by glucocorticoids and serum. Mol Cell Biol. 1993;13(4):2031–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Soundararajan R, Zhang TT, Wang J, Vandewalle A, Pearce D. A novel role for glucocorticoid-induced leucine zipper protein in epithelial sodium channel-mediated sodium transport. J Biol Chem. 2005;280(48):39970–39981. [DOI] [PubMed] [Google Scholar]
- 110. D’Adamio F, Zollo O, Moraca R, Ayroldi E, Bruscoli S, Bartoli A, Cannarile L, Migliorati G, Riccardi C. A new dexamethasone-induced gene of the leucine zipper family protects T lymphocytes from TCR/CD3-activated cell death. Immunity. 1997;7(6):803–812. [DOI] [PubMed] [Google Scholar]
- 111. Bradbury MJ, Akana SF, Cascio CS, Levin N, Jacobson L, Dallman MF. Regulation of basal ACTH secretion by corticosterone is mediated by both type I (MR) and type II (GR) receptors in rat brain. J Steroid Biochem Mol Biol. 1991;40(1-3):133–142. [DOI] [PubMed] [Google Scholar]
- 112. van Haarst AD, Oitzl MS, de Kloet ER. Facilitation of feedback inhibition through blockade of glucocorticoid receptors in the hippocampus. Neurochem Res. 1997;22(11):1323–1328. [DOI] [PubMed] [Google Scholar]
- 113. Klengel T, Mehta D, Anacker C, Rex-Haffner M, Pruessner JC, Pariante CM, Pace TWW, Mercer KB, Mayberg HS, Bradley B, Nemeroff CB, Holsboer F, Heim CM, Ressler KJ, Rein T, Binder EB. Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nat Neurosci. 2013;16(1):33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Yehuda R, Daskalakis NP, Desarnaud F, Makotkine I, Lehrner AL, Koch E, Flory JD, Buxbaum JD, Meaney MJ, Bierer LM. Epigenetic biomarkers as predictors and correlates of symptom improvement following psychotherapy in combat veterans with PTSD. Front Psychiatry. 2013;4:118. [DOI] [PMC free article] [PubMed] [Google Scholar]