Abstract
Background
Hepatocellular carcinoma (HCC) is a commonly occurring liver malignancy. Its prognosis remains unsatisfactory. Accumulating evidence has revealed that exosomal microRNAs (miRNAs) act as biomarkers and play crucial roles in the advancement of HCC. The current study explored the biological role and fundamental mechanism of exosomal miR-744 in HCC.
Material/Methods
The serum exosomes of HCC patients were isolated by differential ultracentrifugation. MiR-744 expression in HCC tissues, cell lines and serum exosomes were detected by quantitative real-time polymerase chain reaction (qRT-PCR). EdU (5-ethynyl-2′-deoxyuridine) assay and Cell Counting Kit-8 (CCK-8) assay were conducted to show the impacts of miR-744 or exosomal miR-744 on proliferation and sorafenib resistance in HepG2 cells. The target of miR-744 was ascertained by regulating the level of miR-744 in HepG2 cells.
Results
MiR-744 is downregulated in HCC tissues and cell lines as well as in exosomes derived from patient serum and HepG2 cells. Additionally, downregulated miR-744 promotes HepG2 cell proliferation and inhibits the chemosensitivity of HepG2 cells to sorafenib. PAX2 was identified as the functional target of miR-744. Interestingly, miR-744 is decreased in exosomes derived from sorafenib-resistant HepG2 cells. Furthermore, when treated with the miR-744-enriched exosomes, the proliferation of HepG2 cells was significantly suppressed, and the sorafenib resistance was reduced.
Conclusions
MiR-744 has an imperative role in the propagation and chemoresistance of HCC. Serum exosomal miR-744 might act as a biomarker of HCC, and exosomal miR-744 might offer an innovative strategy for HCC treatment.
MeSH Keywords: Antineoplastic Agents; Carcinoma, Hepatocellular; Cell Proliferation; Exosomes; MicroRNAs
Background
Hepatocellular carcinoma (HCC) is the commonest tumor of the digestive tract and the sixth most commonly occurring malignancy of the whole body. In addition, HCC is the second most prominent cause of cancer-associated deaths universally [1]. Most HCC patients are identified at medium or progressive stages, with tumors approximately >2 cm. At that time, it is usually too late for superior treatment. Except for traditional chemotherapeutic medications, such as gemcitabine (GEM), oxaliplatin (OXA), and 5-fluorouracil (5-FU), the only effective treatments are transarterial chemoembolization (TACE) plus targeted treatment with the multikinase inhibitor sorafenib [2,3]. Even when advanced comprehensive therapy is applied, HCC patients still have a poor prognosis. This is mainly because HCC exhibits great resistance to normally preferred chemotherapeutic medicines [4]. Hence, the discovery of novel targets and the advancement of unique treatment tactics to inhibit chemoresistance are essential.
Exosomes are small extracellular vesicles discharged by almost all cell types including cultured cells. They play momentous roles in cell-to-cell communication because they carry DNA, mRNAs, microRNAs (miRNAs), proteins and additional molecules as cargo to recipient cells from donor cells and regulate their function [5]. A growing body of studies have reported that tumor cell-derived exosomes can stimulate tumor progression and act as effective biomarkers of the specific cancer [6]. Exosomes are the main transporters of miRNAs in serum. Recently, serum exosomes generated from chemoresistant cells have been indicated to deliver miRNAs and transfer the drug resistant property to sensitive cells [7,8]. Conversely, there are also many miRNAs that are decreased in chemoresistant cancer cell-derived exosomes. For instance, Yuwen and colleagues found that the miR-146a-5p level in serum exosomes is decreased, which indicates the cisplatin chemoresistance in non-small cell lung cancer [9]. MiR-770 was considerably diminished in chemoresistant tissues and could be transferred by exosomes [10]. Therefore, certain serum exosome miRNAs can be developed as effective biomarkers for the chemoresistance of specific cancers. More importantly, exosomes could be used to carry miRNAs to tumor cells and improve their sensitivity to chemotherapy.
Previous reports suggested that miR-744 was dysregulated in many cancers. It inhibits the expression of eEF1A2 and leads to the impedance of MCF7 cancer cell propagation [11]. MiR-744 was also found to be dysregulated in multiple myeloma, which was related to lower overall survival and the deterioration of myeloma patients [12]. In addition, the imperative role of miR-744 in HCC has received increasing attention. Decreased miR-744 has been demonstrated to promote the cell propagation of HCC [13]. However, whether serum exosomes can reflect the change in miR-744 in HCC patients is still unknown.
In the current study, we purified serum exosomes from HCC patients as well as exosomes from HepG2 cell culture media and analyzed the levels of miR-744. We established that miR-744 was reduced in serum exosomes and sorafenib-resistant HepG2 (HepG2-R) cell-derived exosomes. Furthermore, exosomes derived from miR-744-overexpressing HepG2 cells can deliver miR-744 into cells with low miR-744 levels and inhibit their proliferation and sorafenib resistance. Our study provides a potential strategy to suppress the growth of HCC with exosomes.
Material and Method
Patients and samples
Clinical tissue samples were acquired from HCC patients who underwent tumor resection at the First Affiliated Hospital of Xi’an Jiaotong University between August 2017 and June 2018. Tissues were obtained during operation and immediately preserved at −80°C. A total of 68 HCC tissue samples and 52 normal liver tissue samples were analyzed. Serum samples were acquired from 10 male patients who were diagnosed with HCC at our hospital. Ten healthy male volunteers aged 45 to 65 years were also recruited. None of the participants had undergone any preoperative treatment. No other underlying diseases were found in any of the participants. The serum or serum exosomal miRNAs in patients were collected prior to surgery. All samples were centrifuged immediately to eliminate cell debris. The acquired serum samples were kept at −80°C until further use. This study was approved by the Ethical Committee of the First Affiliated Hospital of Xi’an Jiaotong University. All participants signed informed consent.
Cell culture
The normal human liver cell line LO2 was acquired from the American Type Culture Collection (ATCC). Hepatocellular carcinoma cell lines HepG2 and MMC-7721 were acquired from the China Center for Type Culture Collection. Cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) complemented with 10% fetal bovine serum (FBS) and 100 IU/mL penicillin-streptomycin at 37°C with 5% CO2. To create the sorafenib-resistant cells (HepG2-R), HepG2 cells were treated with incremental concentrations of sorafenib (Selleck, Houston, TX, USA); starting with 0.5 μM, the concentration was doubled every 2 weeks until it reached 8 μM. Then, the HepG2-R cells were treated with 8 μM of sorafenib biweekly to maintain the resistant ability [14].
To obtain exosomes, cells were cultured in 15-cm dishes with 30 mL of the whole culture. After reaching 70% confluence, cells were washed with phosphate-buffered saline (PBS) and cultured in DMEM complemented with 10% exosome-depleted FBS for 48 hours. The exosome depleted FBS was obtained by ultracentrifugation at 100 000×g for 12 hours.
Cell transfection
HepG2 cells were transfected with 100 nM of miR-744 mimics, miR-744 inhibitor or negative control (RiboBio, Guangzhou, China) by Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). For PAX2 overexpression, HepG2 cells were transfected with 4 μg of plasmids (GenePharma, Shanghai, China) in 60 mm dishes. Then, 24 hours following transfection, the medium was discarded, fresh medium was added, and the cells were used for additional analyses. For exosome isolation, the medium was collected 48 hours after medium replacement.
Exosome isolation
Exosomes from serum samples and cell culture media were isolated by differential ultracentrifugation as reported previously [15]. Briefly, serum samples or cell culture media were acquired and centrifuged at 300×g for 10 minutes with subsequent centrifugation at 2000×g for 10 minutes. The supernatant was collected and centrifuged again at 10 000×g and 4°C for 30 minutes to eliminate large particles. Then, the supernatant was centrifuged at 100 000×g (Beckman Ti70) at 4°C for 70 minutes to gather the exosomes. The supernatant was discarded, and the pellets were resuspended in 12 mL of PBS for washing. Then, centrifugation at 100 000×g was conducted for 70 minutes to obtain the pure exosomes.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was acquired from HepG2 cells or tumor samples by TRIzol. Total serum RNA, including miRNA, was acquired by the miRNeasy Serum/Plasma Kit (Qiagen, Germany). Experiments were accomplished per the protocol. Total exosomal RNAs were obtained using the miRNeasy Mini Kit (Qiagen, Germany) based on the supplier’s instructions. Then, the RNA concentrations were detected by a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, USA). Afterward, the TaqMan microRNA Reverse Transcription Kit (Applied Biosystems, USA) was utilized to reverse transcribe the total RNA. All cDNA samples were either preserved at −20°C or used immediately for qRT-PCR.
The qRT-PCR for miR-744 was carried out on an Mx3005P Real-time PCR system (Stratagene, USA) along with SYBR® Premix Ex Taq™ (TaKaRa, China). Small nuclear RNA U6 (U6 snRNA) served as an internal control. The primers were purchased from RiboBio (Guangzhou, China) and were as follows:
miR-744 (F: 5′-ACACTCCAGCTGGGTGCGGGGCTAGGGCTAAC-3′ and
R: 5′-CTCAACTGGT GTCGTGGA-3′) and
U6 (F: 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and
R: 5′-CTCAACTGGTGTCGTGGA-3′).
Western blotting
Cells, tissue samples, and exosome pellets were lysed with radioimmunoprecipitation assay (RIPA) buffer (Beyotime Biotechnology, China). The concentrations of protein were detected by the bicinchoninic acid (BCA) method. Then, similar quantities of proteins were separated by 10% sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis (PAGE) and transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore, USA). After the blots were blocked with 1% bovine serum albumin (BSA) at room temperature for 2 hours, they were then incubated with primary antibodies (PAX2, Cell Signaling Technology (CST), USA; β-actin, Abcam, USA; FLOT1, 1: 1000, CST, USA; CD9 and CD81, SBI, USA) at 4°C overnight. Then, the blots were incubated in specific secondary antibodies at room temperature for 2 hours after washing. The blots were observed via Electrochemiluminescence (ECL)-associated fluorography (Millipore).
Transmission electron microscopy (TEM)
The separated exosome pellets were detected by transmission electron microscopy (TEM) as reported previously. In brief, using 1% glutaraldehyde to fix a drop of purified exosomes (around 10 μL) for 10 minute and then loaded onto carbon-coated copper grids. After staining with 1% uranyl acetate and drying, the copper grids were placed into the HT7700 TEM (Hitachi, Tokyo, Japan). Exosome images were captured, and the size was calculated.
Cell viability assay
Cell Counting Kit-8 (CCK-8) detection kit (Beyotime Biotechnology, China) was applied to detect cell viability with a as instructed by the supplier. Cells were seeded in 48-well plates and treated with 1 μM of sorafenib when they reached 80% confluence. Then, 24 hours later or at the designated time points, the medium was replaced with 200 μL of fresh medium along with 10 μL of CCK-8. Following incubation for 2 hours, the absorbance was read at 450 nm.
EdU (5-ethynyl-2′-deoxyuridine) assay
Cell propagation was evaluated by a Cell-Light EdU DNA cell proliferation kit (RiboBio, China) according to the supplier’s instruction [16]. Concisely, cells were sowed in 96-well plates, and EdU (5-ethynyl-2′-deoxyuridine) medium was added for 2 hours. Then, the cells were fixed with 4% paraformaldehyde and neutralized with glycine. After penetrating with 0.5% Triton X-100, Apollo 643 dyeing solution was added for 30 minutes without light followed by washing with PBS containing 0.5% Triton X-100. DAPI (4′,6-diamidino-2-phenylindole) solution was used to stain the nucleus. Fluorescence microscopy was used to capture images. The fraction of EdU-positive cells was described as the proliferation rate.
Statistical analysis
The analysis was done with GraphPad Prism 8 (USA), and all data are mentioned in the form of the mean±standard deviation (SD). The t-test or paired t-test was implemented to compare the data between 2 groups. One-way analysis of variance (ANOVA) followed by a Tukey multiple comparison test was utilized for investigating multiple groups. P<0.05 was considered to be statistical difference.
Results
HCC tissues and cell lines express lower levels of miR-744
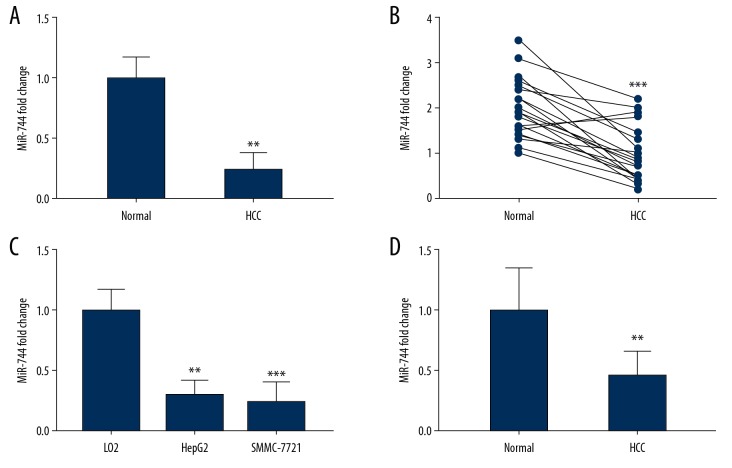
We examined miR-744 expression in 68 HCC tissue samples and 52 normal liver tissue samples. Our results showed an evident downregulation of miR-744 in HCC tissues (Figure 1A). Furthermore, we analyzed 21 paired HCC tissue samples and their adjacent normal tissues. Total RNA was extracted, and qRT-PCR was carried out to identify the levels of miR-744 in these tissues. We established that the miR-744 level was markedly decreased in HCC tissues compared with the adjacent normal tissues (Figure 1B). The HepG2 and SMMC-7721 cell lines were utilized to determine the levels of miR-744. The results showed that miR-744 was declined in the HCC cell lines (Figure 1C). Furthermore, ten healthy male volunteers and ten male HCC patients aged 45 to 65 were recruited. Serum was collected, and total RNA was extracted to determine the level of miR-744. The qRT-PCR assay suggested that the expression of miR-744 in healthy serum was greater than that in HCC serum (Figure 1D).
Figure 1.
The expression of miR-744 in HCC tissues, cell lines and the serum of HCC patients. (A) qRT-PCR investigation of miR-744 in 68 HCC tissues and 52 normal liver tissues. (B) The expression of miR-744 in 21 paired HCC tissues and their corresponding adjacent normal liver tissues. (C) qRT-PCR investigation of miR-744 in LO2, HepG2. and SMMC-7721 cell lines. (D) The serum of HCC patients and healthy individuals was collected, and the miR-744 level was identified by qRT-PCR. ** P<0.01, *** P<0.001. HCC – hepatocellular carcinoma; qRT-PCR – quantitative real-time polymerase chain reaction.
Exosomes derived from patients’ serum and HepG2 cells contain decreased levels of miR-744
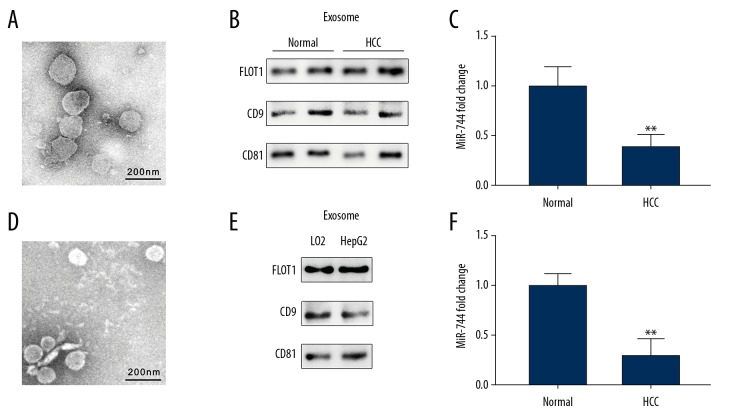
The serum exosomes of HCC patients and healthy volunteers were isolated by ultracentrifugation. TEM was used to identify the morphology of the exosomes, and they presented a classical shape (Figure 2A). In addition, exosome markers, such as CD9, CD81, and FLOT1, were detected by western blotting to examine the purification of the exosomes (Figure 2B). Total RNA was acquired from 10 μg of exosomes, and the miR-744 level was determined by qRT-PCR assay. The results showed serum exosomes from HCC patients were significantly reduced compared with those derived from healthy individuals (Figure 2C). Then, HepG2 and LO2 cells were cultured, and the exosomes in the culture media were isolated. Morphology and exosome markers were also detected (Figure 2D, 2E). Further study revealed that miR-744 levels were decreased in exosomes from the HepG2 culture medium compared to the LO2 culture medium (Figure 2F).
Figure 2.
Exosome isolation and miR-744 expression in HCC patient serum-derived exosomes and cell culture medium-derived exosomes. (A) Transmission electron microscopy image of exosomes extracted from the serum of HCC patients. (B) Representative western blotting bands of exosome protein markers in the exosomes extracted from the serum of HCC patients. (C) Exosomal miR-744 from the serum of HCC patients was evaluated by qRT-PCR. (D) Transmission electron microscopy image of exosomes extracted from the culture medium of HepG2 and LO2 cells. (E) Exosome protein markers of cell culture medium-derived exosomes were detected by western blotting. (F) Exosomal miR-744 of the cell culture medium was evaluated by qRT-PCR. ** P<0.01. HCC – hepatocellular carcinoma; qRT-PCR – quantitative real-time polymerase chain reaction.
PAX2 is upregulated in HCC tissue and was proven to be a target of miR-744
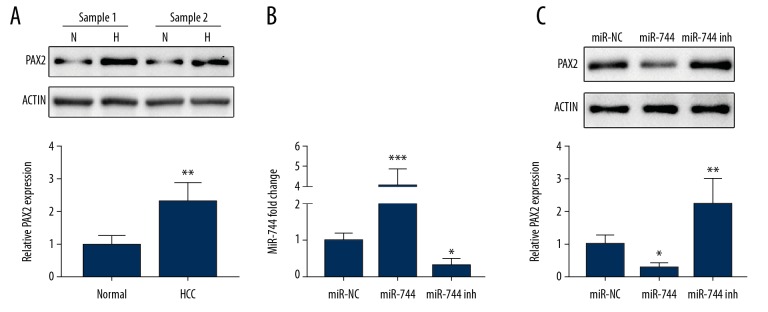
To investigate the role of miR-744 in HCC, we searched for the possible target of miR-744 using TargetScan software. After analysis and testing, we found that PAX2 was a target of miR-744. Western blotting showed a significant increase of PAX2 in HCC tissues compare to the adjoining regular tissues (Figure 3A). Moreover, HepG2 cells were transfected with plasmids of miR-744 mimic or inhibitor, and PAX2 was detected. PAX2 was downregulated by the miR-744 mimic and rose by the miR-744 inhibitor (Figure 3C). Figure 3B shows the transfection efficiency of miR-744 in HepG2 cells.
Figure 3.
The negative regulation of PAX2 by miR-744. (A) Western blotting shows the expression of PAX2 in 2 representative paired HCC tissues and their adjacent normal liver tissue samples (N for normal and H for HCC). (B) HepG2 cells were transfected with miR-NC, miR-744 mimic or miR-744 inhibitor plasmids, and miR-744 expression was detected by qRT-PCR. (C) After transfection with miR-NC, miR-744 mimic or miR-744 inhibitor plasmids, PAX2 expression in HepG2 cells was assessed. * P<0.05, ** P<0.01, *** P<0.001. HCC – hepatocellular carcinoma; qRT-PCR – quantitative real-time polymerase chain reaction.
MiR-744 regulates the proliferation and sorafenib sensitivity of HCC cells
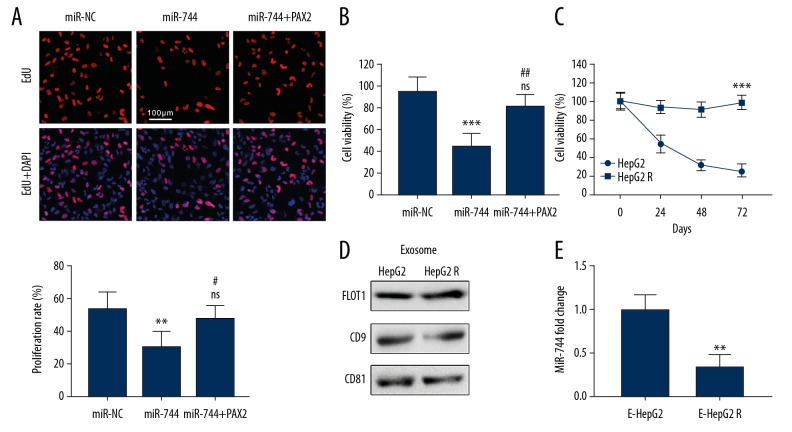
MiR-744 was proven to be declined in HCC tissues, but how it affects the development of HCC is still unknown. We transfected miR-744 mimic plasmids into HepG2 cells and detected the proliferation of the cells by EdU staining. Thirty-six hours after transfection, the proliferation rate of miR-744-overexpressing cells was reduced compared with that of the control cells (Figure 4A). The decrease in the proliferation rate was diminished if HepG2 cells were co-transfected with the miR-744 mimic and PAX2 plasmid (Figure 4A). In addition, the chemoresistance of HepG2 cell to sorafenib was also regulated by miR-744. A CCK-8 assay was implemented to evaluate cell viability after sorafenib administration. The overexpression of miR-744 promoted cell death when exposed to sorafenib (Figure 4B). Moreover, cells co-transfected with the miR-744 mimic and PAX2 plasmid exhibited no significant difference in cell viability compared to control cells (Figure 4B). Furthermore, we established HepG2-R cells and purified the exosomes from the culture medium (Figure 4C, 4D). Exosomal miR-744 was detected by qRT-PCR and was greatly decreased compared with that in normal HepG2 cells (Figure 4E).
Figure 4.
MiR-744 suppresses the proliferation and chemoresistance of HCC. (A) HepG2 cells were transfected with the miR-744 mimic or co-transfected with miR-744 and PAX2. An EdU labeling assay was utilized to show the impact of miR-744 on cell proliferation (* compared with miR-NC, # compared with miR-744, ns: no significant difference compared with miR-NC). (B) The effect of the indicated transfection on the sorafenib resistance of HepG2 cells; CCK-8 assay was performed to assess cell viability 24 hours after sorafenib treatment (* compared with miR-NC, # compared with miR-744, ns: no significant difference compared with miR-NC). (C) CCK-8 assay shows the sorafenib-resistant ability of the established HepG2-R cells and normal HepG2 cells. (D) Western blotting shows the exosome protein markers of the isolated exosomes derived from HepG2 and HepG2-R cell culture media. (E) The exosomal miR-744 levels in HepG2 and HepG2-R cell culture media were evaluated by qRT-PCR. * and # represent P<0.05, ** and ## represent P<0.01, and *** P<0.001. HCC – hepatocellular carcinoma; EdU – 5-ethynyl-2′-deoxyuridine; CCK-8 – Cell Counting Kit-8; qRT-PCR – quantitative real-time polymerase chain reaction.
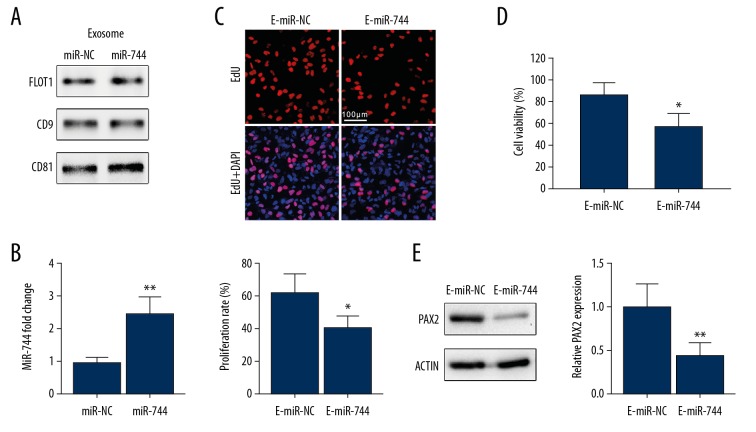
Exosomes overexpressing miR-744 inhibit the proliferation and chemoresistance of HCC cells
HepG2 cells were transfected with miR-744 mimic or control plasmids 72 hours before exosomes were purified from the culture medium. Western blotting showed the quantification of exosomes from each cell group (Figure 5A). We detected miR-744 level in the exosomes of each group by qRT-PCR, and an increase in the level of miR-744 was shown in HepG2 cell-derived exosomes (Figure 5B). Then, 20 μg of exosomes from each cell group was added to HepG2 cells, and proliferation was assessed. EdU staining indicated that exosomal miR-744 clearly hindered the propagation of HepG2 cells (Figure 5C). To examine the impact of miR-744-enriched exosomes on the chemoresistance of HCC cells, we treated HepG2-R cells with miR-744-enriched exosomes and negative control exosomes. Cell viability was analyzed by CCK-8 assay after sorafenib treatment. We found that HepG2-R cells treated with miR-744-overexpressing exosomes were more sensitive to sorafenib (Figure 5D). Western blotting indicated that PAX2 in HepG2-R cells was also downregulated by exosomal miR-744 (Figure 5E).
Figure 5.
MiR-744-enriched exosome-treated HepG2 cells showed inhibited proliferation and chemoresistance. (A) HepG2 cells were transfected with miR-NC or miR-744 mimic plasmids for 24 hours followed by 48 hours of culturing in fresh medium. Then, the exosomes of the culture medium were isolated. Western blotting shows the exosome protein markers of the isolated exosomes. (B) The miR-744 level of exosomes from the indicated cells was evaluated by qRT-PCR. (C) HepG2 cells were treated with the exosomes described in (A). An EdU labeling assay was utilized to show the proliferation rate. (D) HepG2-R cells were treated with the exosomes described in (A) for 24 hours. Then, CCK-8 assay was performed to assess cell viability 24 hours after sorafenib treatment. (E) PAX2 expression in HepG2-R cells treated with the specified exosomes was assessed. * P<0.05, ** P<0.01. qRT-PCR – quantitative real-time polymerase chain reaction; EdU – 5-ethynyl-2′-deoxyuridine; CCK-8 – Cell Counting Kit-8.
Discussion
Two main factors contributing to HCC-related deaths are the lack of early diagnostic biomarkers and the chemoresistance to drugs [17]. In this study, we identified exosomal miR-744 from serum as a biomarker of HCC. We found that miR-744 levels in the serum exosomes of HCC patients were lower than those in non-HCC patients. Interestingly, the level of miR-744 was considerably lower in exosomes derived from sorafenib-resistant HCC cells. Furthermore, we confirmed PAX2 as a direct as well as functional target of miR-744 in HCC cells. Moreover, exosomal miR-744 can be developed as a new therapeutic approach for HCC, as it suppresses the propagation and chemoresistance of HCC by inhibiting the expression of PAX2.
Exosomes originate from internal multivesicular bodies in almost all cells [5]. They can be discharged into peripheral blood and extensively spread in human body fluids, such as saliva, urine, blood, and breast milk. Exosomes have critical functions in intercellular interactions through the transport of biological cargoes, comprising proteins, mRNAs, and miRNAs [18]. This is mainly due to their stable status in blood, as they play a protective role against degradation by enzymes [19]. Exosomal miRNAs have been studied as valuable biomarkers of many diseases, especially cancers [20,21]. There are several advantages of developing exosomal miRNAs as biomarkers for cancer. Exosomes can be conveniently collected from serum, and the analysis of miRNAs is simple. Examining exosomal miRNAs is non-invasive and might be utilized as an early cancer screening method. Numerous circulating microRNAs have been described as candidates for biomarkers related to HCC in preceding studies. In TACE-treated HCC patients, serum exosomal miRNA-122 and miRNA-21 have been documented as prognostic biomarkers [22]. Tang and his colleagues confirmed that miR-9-3p in exosome represses HBGF-5 manifestation and is a useful biomarker in HCC [23]. Serum exosomal miR-125b is a new predictive indicator for HCC [24]. Qu Z et al. reported the use of exosomal miR-665 as a new nominally aggressive biomarker for HCC diagnosis as well as prognosis [25]. In the current study, the serum levels of miR-744 were investigated in patients with HCC. A previous study showed a decrease in miR-744 in HCC tissues [26], and we established that miR-744 was also reduced in the serum of HCC patients. Furthermore, we obtained exosomes from the serum of HCC patients and from HCC cell lines and detected the miR-744 level; the results showed low levels of miR-744 compared to those from normal persons and cell lines. This finding indicated that serum exosomal miR-744 could be a potential candidate HCC biomarker.
Though the expression of miR-744 in HCC has been investigated in a few studies, its key function remains unclear. The role of miR-744 in other cancers has been reported in many studies. Previous evidence has shown that miR-744 is a proto-oncogene in several malignant tumors, for example breast cancer, prostate cancer, and nasopharyngeal carcinoma [27–29]. However, it was proved to be a cancer suppressor in numerous other cancers, especially tumors of the digestive system. MiR-744 prevents tumor cell propagation as well as incursion of gastric cancer by targeting brain derived neurotrophic factor [30]. MiR-744 also prevents cellular propagation as well as incursion of colorectal cancer [31]. Another report indicated that that lesser miR-744 was an autonomous projector of inferior prognosis of HCC patients [26]. Nevertheless, its underlying mechanism in HCC and whether the change in miR-744 is detectable in serum exosomes is still unknown.
In our work, we proved that serum exosomal miR-744 is downregulated in HCC tissues. In addition, we identified that a direct target of miR-744 is PAX2, which has been indicated to regulate the chemotherapy response in several cancers [32–34]. In HCC, PAX2 is in high expression level in cancer tissues compared with adjacent normal tissues, which is contrary to the expression of miR-744. Interestingly, the predicted binding site of miR-744 was found in PAX2 mRNA by TargetScan software. To further confirm that PAX2 is a direct target of miR-744, we artificially manipulated miR-744 levels in HepG2 cells. PAX2 was markedly inhibited when cells were transfected with the miR-744 mimic and accumulated when miR-744 was decreased. This result proves that PAX2 can be effectively regulated by miR-744 in HCC. PAX2 exerts a powerful antiapoptotic effect in kidney cancer and ovarian cancer by activating the expression of some apoptosis inhibitory genes [35,36]. It also promotes the proliferation of cancer cells by re-entering them into the mitotic cycle. Hence, PAX2 plays a significant role in weakening the outcomes of chemotherapy. By inhibiting miR-744, the expression of PAX2 is increased in HCC cells, thus promoting proliferation and sorafenib resistance.
Resistance to chemotherapy is one of the main obstacles to improving the prognosis of HCC patients. Previous studies have shown that chemoresistant cancer cells are enhanced in exosomes that might function as genetic modulators [37,38]. The resistant-cell population might be capable of extending their resistance qualities to other cells by their distinctive exosomes [39]. Therefore, we can capture these distinctive exosomes from the circulation to predict the resistance characteristics of cancer cells and select more effective strategies. We found that the decreased miR-744 level in serum exosomes indicates the sorafenib resistance of HCC cells. Moreover, when we treated HCC cells with miR-744-enriched exosomes, the sensitivity of HCC cells to sorafenib was markedly augmented and so was the proliferation ability. This decrease was consistent with the decrease in PAX2 levels.
Conclusions
In brief, we showed that miR-744 expression is downregulated in the serum exosomes and tissues of HCC patients. Furthermore, PAX2 was demonstrated to be a direct target of miR-744, which is increased in HCC tissues. Furthermore, diminished miR-744 facilitates the propagation and drug resistance of HCC cells. Importantly, when treated with miR-744-enriched exosomes, the propagation and drug resistance of HCC cells were significantly suppressed. These findings may be beneficial for providing a valuable biomarker for HCC and predicting drug resistance in HCC patients. Exosomal miR-744 may be an underlying effective therapeutic target for HCC patients.
Footnotes
Source of support: This study was supported by the Scientific and Technological Development Research Project Foundation from Shaanxi Province (No. 2015SF-057)
Conflicts of interest
None.
References
- 1.Bieze M, Klümpen HJ, Verheij J, et al. Diagnostic accuracy of (18) F-methylcholine positron emission tomography/computed tomography for intra- and extrahepatic hepatocellular carcinoma. Hepatology. 2014;59(3):996–1006. doi: 10.1002/hep.26781. [DOI] [PubMed] [Google Scholar]
- 2.Llovet JM, Villanueva A, Lachenmayer A, Finn RS. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat Rev Clin Oncol. 2015;12(7):408–24. doi: 10.1038/nrclinonc.2015.103. [DOI] [PubMed] [Google Scholar]
- 3.Liu D, Khong PL, Gao Y, et al. Radiation dosimetry of whole-body dual-tracer 18F-FDG and 11C-acetate PET/CT for hepatocellular carcinoma. J Nucl Med. 2016;57(6):907–12. doi: 10.2967/jnumed.115.165944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niu L, Liu L, Yang S, et al. New insights into sorafenib resistance in hepatocellular carcinoma: Responsible mechanisms and promising strategies. Biochim Biophys Acta Rev Cancer. 2017;1868(2):564–70. doi: 10.1016/j.bbcan.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Hessvik NP, Llorente A. Current knowledge on exosome biogenesis and release. Cell Mol Life Sci. 2018;75(2):193–208. doi: 10.1007/s00018-017-2595-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whiteside TL. Tumor-derived exosomes and their role in cancer progression. Adv Clin Chem. 2016;74:103–41. doi: 10.1016/bs.acc.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santos JC, Lima NDS, Sarian L, et al. Exosome-mediated breast cancer chemoresistance via miR-155 transfer. Sci Rep. 2018;8(1):829. doi: 10.1038/s41598-018-19339-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang M, Qiu R, Yu S, et al. Paclitaxel-resistant gastric cancer MGC-803 cells promote epithelial-to-mesenchymal transition and chemoresistance in paclitaxel-sensitive cells via exosomal delivery of miR-155-5p. Int J Oncol. 2019;54(1):326–38. doi: 10.3892/ijo.2018.4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuwen DL, Sheng BB, Liu J, et al. MiR-146a-5p level in serum exosomes predicts therapeutic effect of cisplatin in non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2017;21(11):2650–58. [PubMed] [Google Scholar]
- 10.Li Y, Liang Y, Sang Y, et al. MiR-770 suppresses the chemo-resistance and metastasis of triple negative breast cancer via direct targeting of STMN1. Cell Death Dis. 2018;9(1):14. doi: 10.1038/s41419-017-0030-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vislovukh A, Kratassiouk G, Porto E, et al. Proto-oncogenic isoform A2 of eukaryotic translation elongation factor eEF1 is a target of miR-663 and miR-744. Br J Cancer. 2013;108(11):2304–11. doi: 10.1038/bjc.2013.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kubiczkova L, Kryukov F, Slaby O, et al. Circulating serum microRNAs as novel diagnostic and prognostic biomarkers for multiple myeloma and monoclonal gammopathy of undetermined significance. Haematologica. 2014;99(3):511–18. doi: 10.3324/haematol.2013.093500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin F, Ding R, Zheng S, et al. Decrease expression of microRNA-744 promotes cell proliferation by targeting c-Myc in human hepatocellular carcinoma. Cancer Cell Int. 2014;14:58. doi: 10.1186/1475-2867-14-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han P, Li H, Jiang X, et al. Dual inhibition of Akt and c-Met as a second-line therapy following acquired resistance to sorafenib in hepatocellular carcinoma cells. Mol Oncol. 2017;11(3):320–34. doi: 10.1002/1878-0261.12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu F, Jiang W, Zhou L, Chen Z. Circulating exosomal miR-17-5p and miR-92a-3p predict pathologic stage and grade of colorectal cancer. Transl Oncol. 2018;11(2):221–32. doi: 10.1016/j.tranon.2017.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu J, Tian X, Chang J, et al. RUNX3 inhibits the proliferation and metastasis of gastric cancer through regulating miR-182/HOXA9. Biomed Pharmacother. 2017;96:782–91. doi: 10.1016/j.biopha.2017.08.144. [DOI] [PubMed] [Google Scholar]
- 17.Dutta R, Mahato RI. Recent advances in hepatocellular carcinoma therapy. Pharmacol Ther. 2017;173:106–17. doi: 10.1016/j.pharmthera.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farooqi AA, Desai NN, Qureshi MZ, et al. Exosome biogenesis, bioactivities and functions as new delivery systems of natural compounds. Biotechnol Adv. 2018;36(1):328–34. doi: 10.1016/j.biotechadv.2017.12.010. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–84. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valadi H, Ekström K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–59. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 21.Pant S, Hilton H, Burczynski ME. The multifaceted exosome: Biogenesis, role in normal and aberrant cellular function, and frontiers for pharmacological and biomarker opportunities. Biochem Pharmacol. 2012;83(11):1484–94. doi: 10.1016/j.bcp.2011.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suehiro T, Miyaaki H, Kanda Y, et al. Serum exosomal microRNA-122 and microRNA-21 as predictive biomarkers in transarterial chemoembolization-treated hepatocellular carcinoma patients. Oncol Lett. 2018;16(3):3267–73. doi: 10.3892/ol.2018.8991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang J, Li Y, Liu K, et al. Exosomal miR-9-3p suppresses HBGF-5 expression and is a functional biomarker in hepatocellular carcinoma. Minerva Med. 2018;109(1):15–23. doi: 10.23736/S0026-4806.17.05167-9. [DOI] [PubMed] [Google Scholar]
- 24.Liu W, Hu J, Zhou K, et al. Serum exosomal miR-125b is a novel prognostic marker for hepatocellular carcinoma. Onco Targets Ther. 2017;10:3843–51. doi: 10.2147/OTT.S140062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qu Z, Wu J, Wu J, et al. Exosomal miR-665 as a novel minimally invasive biomarker for hepatocellular carcinoma diagnosis and prognosis. Oncotarget. 2017;8(46):80666–78. doi: 10.18632/oncotarget.20881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan YL, Bai ZG, Zou WL, et al. MiR-744 is a potential prognostic marker in patients with hepatocellular carcinoma. Clin Res Hepatol Gastroenterol. 2015;39(3):359–65. doi: 10.1016/j.clinre.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 27.Yu Q, Zhang F, Du Z, Xiang Y. Up-regulation of serum miR-744 predicts poor prognosis in patients with nasopharyngeal carcinoma. Int J Clin Exp Med. 2015;8(8):13296–302. [PMC free article] [PubMed] [Google Scholar]
- 28.Chen X, Lu P, Wang DD, et al. The role of miRNAs in drug resistance and prognosis of breast cancer formalin-fixed paraffin-embedded tissues. Gene. 2016;595(2):221–26. doi: 10.1016/j.gene.2016.10.015. [DOI] [PubMed] [Google Scholar]
- 29.Zhang M, Li H, Zhang Y, Li H. Oncogenic miR-744 promotes prostate cancer growth through direct targeting of LKB1. Oncol Lett. 2019;17(2):2257–65. doi: 10.3892/ol.2018.9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu AJ, Fu LN, Wu HX, et al. MicroRNA-744 inhibits tumor cell proliferation and invasion of gastric cancer via targeting brain-derived neurotrophic factor. Mol Med Rep. 2017;16(4):5055–61. doi: 10.3892/mmr.2017.7167. [DOI] [PubMed] [Google Scholar]
- 31.Shen J, Li M. MicroRNA-744 inhibits cellular proliferation and invasion of colorectal cancer by directly targeting oncogene Notch1. Oncol Res. 2018 doi: 10.3727/096504018X15188747585738. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hurtado A, Holmes KA, Geistlinger TR, et al. Regulation of ERBB2 by oestrogen receptor-PAX2 determines response to tamoxifen. Nature. 2008;456(7222):663–66. doi: 10.1038/nature07483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hardy LR, Salvi A, Burdette JE. UnPAXing the divergent roles of PAX2 and PAX8 in high-grade serous ovarian cancer. Cancers (Basel) 2018;10(8) doi: 10.3390/cancers10080262. pii: E262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun P, Song Y, Liu D, et al. Potential role of the HOXD8 transcription factor in cisplatin resistance and tumour metastasis in advanced epithelial ovarian cancer. Sci Rep. 2018;8(1):13483. doi: 10.1038/s41598-018-31030-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hueber PA, Iglesias D, Chu LL, et al. In vivo validation of PAX2 as a target for renal cancer therapy. Cancer Lett. 2008;265(1):148–55. doi: 10.1016/j.canlet.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 36.Lin Z, Zhao J, Wang X, et al. Overexpression of microRNA-497 suppresses cell proliferation and induces apoptosis through targeting paired box 2 in human ovarian cancer. Oncol Rep. 2016;36(4):2101–7. doi: 10.3892/or.2016.5012. [DOI] [PubMed] [Google Scholar]
- 37.Bigagli E, Cinci L, D’Ambrosio M, Luceri C. Transcriptomic characterization, chemosensitivity and regulatory effects of exosomes in spontaneous EMT/MET transitions of breast cancer cells. Cancer Genomics Proteomics. 2019;16(3):163–73. doi: 10.21873/cgp.20122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu T, Zhang X, Du L, et al. Exosome-transmitted miR-128-3p increase chemosensitivity of oxaliplatin-resistant colorectal cancer. Mol Cancer. 2019;18(1):43. doi: 10.1186/s12943-019-0981-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Kanlikilicer P, Bayraktar R, Denizli M, et al. Exosomal miRNA confers chemo resistance via targeting Cav1/p-gp/M2-type macrophage axis in ovarian cancer. EBioMedicine. 2018;38:100–12. doi: 10.1016/j.ebiom.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]