Abstract
Carotenoids are isoprenoid pigments synthesized by all photosynthetic organisms and many heterotrophic microorganisms. They are equipped with a conjugated double-bond system that builds the basis for their role in harvesting light energy and in protecting the cell from photo-oxidation. In addition, the carotenoids polyene makes them susceptible to oxidative cleavage, yielding carbonyl products called apocarotenoids. This oxidation can be catalyzed by carotenoid cleavage dioxygenases or triggered nonenzymatically by reactive oxygen species. The group of plant apocarotenoids includes important phytohormones, such as abscisic acid and strigolactones, and signaling molecules, such as β-cyclocitral. Abscisic acid is a key regulator of plant’s response to abiotic stress and is involved in different developmental processes, such as seed dormancy. Strigolactone is a main regulator of plant architecture and an important signaling molecule in the plant-rhizosphere communication. β-Cyclocitral, a volatile derived from β-carotene oxidation, mediates the response of cells to singlet oxygen stress. Besides these well-known examples, recent research unraveled novel apocarotenoid growth regulators and suggests the presence of yet unidentified ones. In this review, we describe the biosynthesis and biological functions of established regulatory apocarotenoids and touch on the recently identified anchorene and zaxinone, with emphasis on their role in plant growth, development, and stress response.
Keywords: apocarotenoids, abscisic acid (ABA), strigolactones (SLs), β-cyclocitral, zaxinone, anchorene
Introduction
Carotenoids are a large group of isoprenoid pigments, which includes more than 600 distinct compounds (Britton et al., 2012). Carotenoids are present in all clades of life; however, their synthesis is restricted to photosynthetic organisms and some nonphotosynthetic fungi and bacteria (Ruiz-Sola and Rodríguez-Concepción, 2012; Moise et al., 2014; Nisar et al., 2015). In plants, carotenoids are essential components of the photosynthetic apparatus where they act as photoprotective pigments and contribute to the light harvesting process. In addition, plant carotenoids confer their bright orange, reddish, or yellow colors to many fruits and flowers (DellaPenna and Pogson, 2006; Ahrazem et al., 2016a). Animals are not able to synthesize carotenoids de novo and rely on nutritional sources to cover their needs of these pigments that act as vitamin A precursor and antioxidants (Fraser and Bramley, 2004).
The electron-rich polyene system of carotenoids makes them susceptible to oxidation processes that break the carotenoid backbone. This cleavage reaction is catalyzed by carotenoid cleavage dioxygenases (CCDs), which build a ubiquitous family of non-heme iron enzymes, and leads to products called apocarotenoids (Giuliano et al., 2003; Hou et al., 2016). Apocarotenoids can also arise through non-enzymatic attack by reactive oxygen species (ROS). In both cases, apocarotenoids fulfill, with or without further enzymatic modifications, many important biological functions. For instance, these metabolites act as precursors of the phytohormones abscisic acid (ABA) and strigolactones (SLs), which are best known for their role in abiotic stress response and adapting plant architecture to nutrition availability, respectively (Jia et al., 2017). Other apocarotenoids are known as signaling molecules regulating cell response to oxidative stress or have been recently discovered as plant growth regulators. In this review, we describe the role of apocarotenoids in plant growth and development and address their biosynthesis and transport, focusing on ABA, SLs, and recently identified regulatory apocarotenoids.
General Aspects of Apocarotenoid Formation
The formation of apocarotenoids is initiated by oxidative cleavage reactions that can be catalyzed by enzymes, generally CCDs, or take place through exposure of carotenoids to ROS. β-Cyclocitral is an example for a carotenoid cleavage product that can be formed following both scenarios (Ramel et al., 2012b; Rodrigo et al., 2013). The formation of this β-carotene–derived volatile in photosynthetic tissues is triggered by a singlet oxygen 1O2 attack, which takes place in the photosystem II, particularly under high light stress (D’Alessandro et al., 2019), while in citrus fruits, it is catalyzed by a CCD (CCD4b) that cleaves β-carotene, β-cryptoxanthin, and zeaxanthin (Rodrigo et al., 2013). Although some CCDs show a quite relaxed substrate and site (double bond) specificity (Ilg et al., 2014), cleavage reactions catalyzed by these enzymes generally take place at certain double bond(s) in defined carotenoid/apocarotenoid substrate(s) (Walter and Strack, 2011; McQuinn et al., 2015). It should be also mentioned here that some bacterial and fungal members of the CCD family cleave biphenylic, stilbene substrates instead of carotenoids (Brefort et al., 2011).
The genome of Arabidopsis encodes nine members of the CCD family, which includes five 9-cis-epoxy carotenoid dioxygenases (NCED2, NCED3, NCED5, NCED6, and NCED9) and four, CCDs (CCD1, CCD4, CCD7, and CCD8). NCED enzymes catalyze the first steps in ABA biosynthesis by cleaving the (C11–C12) double bond in 9-cis-violaxanthin and 9′-cis-neoxanthin, which yields the ABA precursor xanthoxin (Tan et al., 2003). CCD1 enzymes have the ability to cleave several carotenoid and apocarotenoid substrates—linear, monocycle, and bicycle—at different positions along the carbon backbone (Schwartz et al., 2001; Vogel et al., 2008; Ilg et al., 2009; Ilg et al., 2014), which leads to volatile compounds responsible for aroma and flavor in different plant species. CCD4 is also involved in the formation of volatile compounds by cleaving carotenoids either at the (C7–C8) double bond in cryptoxanthin and zeaxanthin, as shown for Citrus CCD4b, or at the (C9–C10) double bond in bicyclic carotenoids, as shown for the Arabidopsis and potato CCD4 (Rubio-Moraga et al., 2014; Bruno et al., 2015; Bruno et al., 2016). CCD4 activity regulates carotenoid content in different plant tissues and produces the citrus pigment citraurin. Finally, the sequential action of CCD7 and CCD8 enzymes leads to the SL precursor carlactone. CCD7 cleaves 9-cis-β-carotene formed by the carotene isomerase DWARF27 from the corresponding all-trans-isomer at the (C9′–C10′) double bound to produce 9-cis-β-apo-10′-carotenal and β-ionone (Alder et al., 2012; Bruno et al., 2014; Bruno and Al-Babili, 2016; Haider et al., 2018). CCD8 action follows by converting 9-cis-β-apo-10′-carotenal into carlactone the substrate of CYP450 enzymes (711A1 clade) that produce different SLs (Abe et al., 2014; Zhang et al., 2014). A further CCD type (CCD2) has been identified in Crocus species, which cleaves the carotenoid chain at the (C7–C8) and (C7′–C8′) double bounds, converting zeaxanthin into crocetin dialdehyde, the precursor of crocin, a saffron pigment, and 3-hydroxy-cyclocitral (Frusciante et al., 2014; Ahrazem et al., 2016b). Very recently, a survey of 748 sequences of all CCDs genes from 69 plant species unraveled an overlooked plant CCD clade represented by the rice zaxinone synthase (ZAS) that cleaves the apocarotenoid apo-10′-zeaxanthinal at the (C13–C14) double bond to produce zaxinone, a novel growth regulator required for normal rice growth (Wang et al., 2019). Recently, the discovery of a new biosynthetic pathway for apocarotenoids was shown to be CCDs independent, through the action of TomLocX/LOX lipoxygenase (in tomato and Arabidopsis, respectively) (Gao et al., 2019).
Abscisic Acid
Abscisic acid is an essential carotenoid-derived plant hormone that coordinates plant response to stress stimuli and is involved in different developmental processes. In addition, ABA regulates water uptake (Dodd, 2013; Harris, 2015) and coordinates stomata closure to minimize water loss under drought condition (Merilo et al., 2015). Furthermore, this hormone controls seed maturation and is responsible for seed dormancy that prevents newly produced seeds from germination under unfavorable conditions (Tuan et al., 2018).
ABA Metabolism
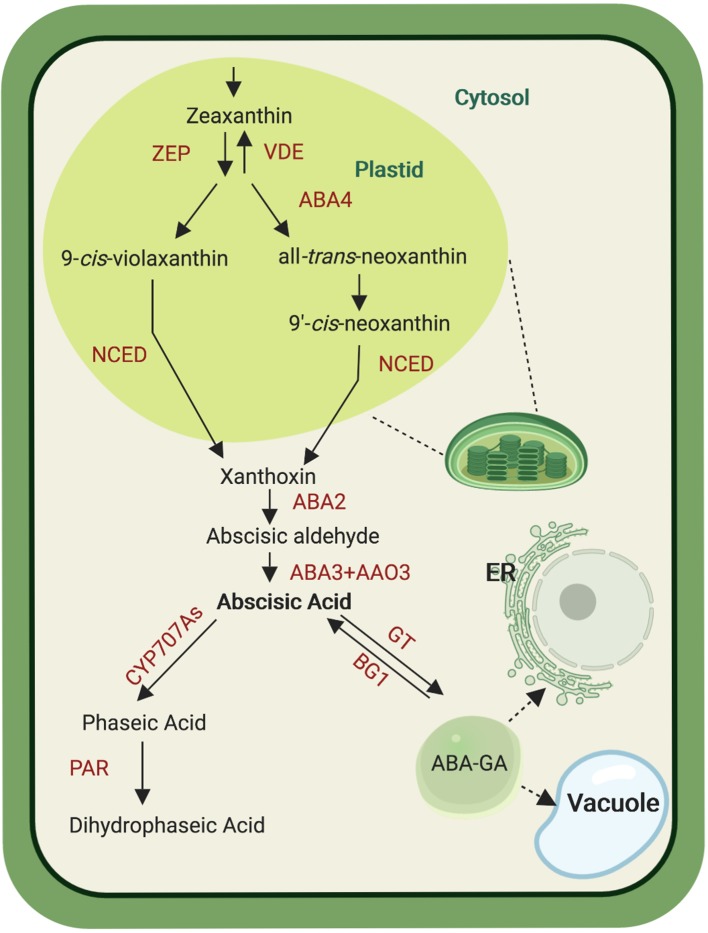
Plant ABA biosynthesis takes place in different cell compartments, as it starts in plastids and is finalized in the cytosol ( Figure 1 ). This metabolic process is initiated by cleaving C40 9-cis-epoxycarotenoids, i.e. 9-cis-violaxanthin and 9′-cis-neoxanthin, which represents the first committed and rate-limiting step in ABA biosynthesis (Finkelstein, 2013). Both xanthophylls, violaxanthin and neoxanthin, are formed from all-trans-zeaxanthin by two epoxidation steps catalyzed by zeaxanthin epoxidase, yielding all-trans-violaxanthin. These epoxidation reactions can be reversed by violaxanthin de-epoxidase. All-trans-violaxanthin is then converted into all-trans-neoxanthin in a less understood process that involves the protein ABA-deficient 4 (ABA4) in Arabidopsis (North et al., 2007). Both all-trans-xanthophylls are isomerized into the required cis-forms by yet unclear mechanism(s) (Dejonghe et al., 2018). The following cleavage reaction, catalyzed by NCED, takes place in plastids and yields the C15 compound xanthoxin and a C25-apocarotenoid. The former is then relocalized from plastids to the cytosol where it becomes available for the final two steps of the ABA biosynthesis. The latter consists of the C25-apocarotenoid conversion into abscisic aldehyde by the short-chain dehydrogenase/reductase-like enzyme ABA-deficient 2 (ABA2), followed by the oxidation to ABA by abscisic aldehyde oxidase (AAO) that belongs to a multienzyme family represented by four members in Arabidopsis (AAO1, AAO2, AAO3, and AAO4). Mutant analysis points to AAO3 as the major contributor to ABA formation, among these four enzymes (Seo et al., 2000). The AAO activity requires molybdenum cofactor that is formed by the sulfurase ABA3 (Iuchi et al., 2001; Hauser et al., 2017), which is supported by the ABA deficiency in aba3 loss of function mutants (Watanabe et al., 2018). Abscisic acid can be further metabolized by ABA-8′-hydroxylases, CYP enzymes of the 707A clade, which introduce a hydroxyl-group at the C-8′ position, yielding the instable 8′-OH-ABA that isomerizes spontaneously by internal cyclization to phaseic acid (PA). A soluble reductase converts PA to dihydrophaseic acid (DPA) by reducing the 4′-ketone group. In addition to the C-8′ position, ABA can be hydroxylated at the C-7′ and C-9′ positions. PA and DPA are considered as major ABA catabolites (Hauser et al., 2017; Ma et al., 2018). However, it was recently shown that PA acts also as a hormone in seed plants and can be recognized by some ABA receptors (Weng et al., 2016). Abscisic acid can also be further catabolized through the action of the glucosyltransferase subfamily ABA uridine diphosphate glucosyltransferases into ABA glucosyl esters (ABA-GE). ABA-GE accumulates in vacuoles and is considered as a storage or transport form of ABA, which can be cleaved back into ABA by β-glucosidases under dehydration stress (Lee et al., 2006; Xu et al., 2012; Ma et al., 2018).
Figure 1.
Abscisic acid (ABA) biosynthesis pathway. De novo ABA biosynthesis begins in plastid, catalyzed by several enzymes (ZEP, zeaxanthin epoxidase; VDE, violaxanthin de-epoxidase; ABA4, ABA-deficient 4; NCED, nine-cis-apoxicarotenoid-dioxygenase ) that convert zeaxanthin into xanthoxin. After transport into the cytosol, xanthoxin is subjected to oxidation steps mediated by three enzymes (ABA2, ABA-deficient 2; AAO3, abscisic aldehyde oxidase and ABA3, ABA-deficient 3) giving rise to ABA. Abscisic acid can be modified by CYP707A enzymes that yield phaseic acid (PA). The latter is the substrate of the PA reductase (PAR) that forms dihydrophaseic acid. Alternatively, ABA is glucosylated by ABA glucosyltransferase (GT) into ABA-GT that can be transported into the vacuole and the endoplasmic reticulum (ER). Abscisic acid–GT can be converted back into ABA by the glycosyl hydrolase BG1. Created with ‘Biorender’.
ABA Signaling Pathways
Modulation of plant growth upon exposure to abiotic stress conditions (high salinity, drought, and low temperature) is coordinated by several hormones and proteins, including regulatory factors (Todaka et al., 2015). In Arabidopsis, the phytohormone ABA has been shown to be a key player in this process, by activating the ABA-dependent signaling pathway. This pathway consists of three major components: PYR (pyrabactin resistance)/PYL (PYR1-LIKE)/RCAR (regulatory component of ABA response) group of ABA receptors, PP2C (protein phosphatase 2C; negative regulator), and SnRK2 (sucrose nonfermenting 1–related protein kinase 2; positive regulator) (Todaka et al., 2015). Together, these components form an ABA sensing and signal transducing complex: in the absence of ABA, the negative regulator PP2C represses SnRK2 activity by dephosphorylating its kinase activating loop, while in the presence of ABA, the receptors PYR/PYL/RCAR5 and PP2C form a complex that prevents the PP2C-mediated dephosphorylation of SnRK2. The resulting activated SnRK2 phosphorylates and activates ABA-responsive element (ABRE)–binding protein/ABRE-binding factor (AREB/ABF) transcription factors, which bind to promoters of drought responsive genes and trigger plant stress response (Todaka et al., 2015; Wang et al., 2017; Xie et al., 2019). There is also an ABA-independent signaling pathway that regulates plant response to abiotic stress. Dehydration responsive element-binding (DREB) proteins are transcription factors induced by stress conditions, such as high salinity, drought, and heat stress (Todaka et al., 2015). Under high temperature stress, heat shock factor A1 and ABRE-binding proteins/ABRE binding factor 3 (AREB1/AREB2/ABF3) regulate DREB2A expression through the binding to ABRE and heat shock element motifs, in their promoter region, to enhance plant resistance to high temperature (Todaka et al., 2015; Xie et al., 2019).
ABA Regulates Seed Dormancy and Germination
Seed dormancy is a desirable trait allowing the uniform postharvest germination of seeds under favorable environmental conditions. In Arabidopsis seeds, ABA shows two peaks of accumulation, which arise during middle (25 days after pollination [DAP]) and late phases (35 DAP) of seed maturation (Kanno et al., 2010). In rice, it was shown that the accumulation of ABA, which occurs at early and middle stages of seed development (10–20 DAP), induces seed dormancy (Suzuki et al., 2000) and that the decrease of ABA level in seeds results in loss of dormancy. These changes in ABA concentration are determined by the expression of the ABA biosynthesis (NCED) and catabolic genes (CYP707A1and CYP707A2) ( Figure 2 ) (Du et al., 2015). The role of CYP707A in reducing ABA content and paving the way for seed germination was also observed in barley. It was shown that the increase in the expression level of HvABA8′OH, a barley CYP707A enzyme, correlated with a reduction of ABA content in ripe seeds. HvABA8′OH1 RNAi lines confirmed the role of this enzyme in ABA catabolism, as they showed enhanced seed dormancy (Gubler et al., 2008). But how does ABA regulate seed dormancy?
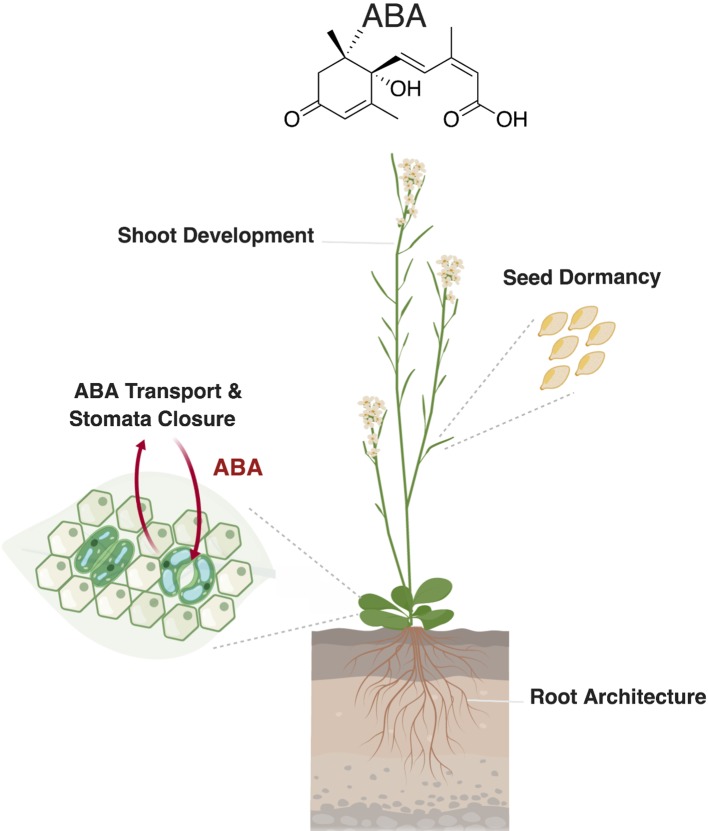
Figure 2.
Role of abscisic acid (ABA) in plant development and stress response. Abscisic acid regulates different developmental processes, such as seed dormancy and shoot and root development. In leaves, ABA transport to the guard cells triggers stomatal closure in response to biotic and abiotic stresses. Created with ‘Biorender’.
Gibberellic acid (GA) is an additional hormone that plays a key role in seed germination. Thus, this developmental process is regulated by a balance between ABA and GA, which act here as antagonists in signaling and biosynthesis pathways (Finkelstein et al., 2008; Née et al., 2017b). In Arabidopsis, DELLA proteins act as regulators of ABA and GA response, balancing between dormancy and germination by interacting with different genes. In the absence of GA, DELLAs interact with (i) XERICO, a RING-H2 zinc finger ubiquitin ligase, which enhances ABA accumulation, and (ii) ABA insensitive 5 (ABI5), an ABA-regulated transcription factor, leading to inhibition of seed germination/maintenance of seed dormancy (Zeng et al., 2014; Zeng et al., 2015). Previous studies identified another genetic element, called delay of germination 1 (DOG1), which regulates seed dormancy and germination in Arabidopsis, as it interferes with both ABA and GA. Two PP2C phosphatases, ABA-hypersensitive germination 1 (AHG1) and AHG3, have been identified interacting with DOG1 in imbibed and dry seeds and revealed to be essential for DOG1 function in controlling seed dormancy (Née et al., 2017a).
ABA Regulates Shoot Development
Several lines of evidence suggest a role of ABA in inhibiting shoot branching by maintaining axillary buds dormancy (reviewed in Barbier et al., 2019). Branched 1 (BRC1), a transcription factor of the Teosinte branched 1, Cycloidea, PCF1 (TCP) family, plays an important role in regulating bud outgrowth in response to plant hormones, nutrients availability, and light quality. BRC1 binds and activates three homeodomain leucine zipper (HD-ZIP) transcription factors, which stimulate NCED3 expression. The increase in NCED3 activity results in enhanced ABA level, indicating that ABA enhances bud dormancy downstream of BRC1 (Barbier et al., 2019). In addition, recent results show that ABA is involved in the regulation of shoot branching by light quality and intensity, as ABA accumulation and synthesis in buds negatively correlate with the red to far red-light ratio (R:FR) (Reddy et al., 2013; González-Grandío et al., 2017; Holalu and Finlayson, 2017). Abscisic acid also affects leaf and shoot growth through repressing ethylene production, which maintains leaf expansion and enhances shoot growth (Hussain et al., 2000). A further evidence for the role of ABA in shoot growth is provided by the ABA-deficient mutant line aba2-1 that shows shoot growth inhibition, a phenotype that can be rescued by exogenous application of this hormone (LeNoble et al., 2004) ( Figure 2 ).
Antagonistic or Synergistic Actions of ABA With Other Phytohormones on the Root System Development
Plants modulate their architecture in response to environmental stimuli. These adaptation processes include also alteration in the root system that shows high sensitivity to environmental changes, particularly to harsh growth conditions. In response to salt stress, ABA stimulates the elongation of primary and lateral roots (Harris, 2015). During drought stress, ABA promotes primary root elongation, allowing the roots to grow deeper to reach water (Hauser et al., 2017). Generally, in well-watered conditions, ABA is known as an inhibitor of root development (Sharp and LeNoble, 2002). However, exogenous ABA application shows that at low concentration (0.1 μM), ABA stimulates root growth, while it exerts the opposite effect when applied at high concentration (10 μM). Thus, ABA acts in a concentration-dependent manner, antagonistically or synergistically with other phytohormones, such as ethylene and auxin, to fine tune plant root architecture. Ethylene represses root growth under stress conditions by inhibiting root cell elongation (Le et al., 2001; Ruzicka et al., 2007; Li et al., 2017). It has been shown that the growth inhibitory effect observed upon application of high ABA concentrations is an ethylene-dependent process (Ghassemian et al., 2000), while the promoting effect of low ABA concentrations is ethylene independent (Li et al., 2017). Auxin was shown to regulate root development in response to both high and low ABA concentrations. Roots of auxin signaling and influx mutants (iaa7/axr2 and aux1, respectively) were less responsive to exogenous ABA regardless its concentration, indicating that both ABA effects are regulated by auxin. Additionally, Pin-formed 2 (PIN2), an important auxin efflux transporter expressed in the lateral root cap, is also involved in ABA response (Li et al., 2017).
Abscisic acid regulates root development also through the action of microRNAs (miRNAs), short single-stranded, small noncoding RNAs that play a major role in plant growth and development, as well as in hormone and stress response (Yan et al., 2017). In Populus euphratica, ABA modulates root growth through miRNA-mediated pathway, together with other hormones, such as GA and brassinosteroids (BRs) (Lian et al., 2018). For instance, peu-miRNA477 was shown to target the repressor of GA1-like (RGL1), a negative regulator of gibberellin response. The rgl1 mutant line showed a dwarf shoot phenotype with an increased lateral root biomass, revealing the importance of RGL1 in plant development. Interestingly, treatment of wild-type plants with exogenous ABA resulted in an increase in peu-miRNA477 expression level, in both shoots and roots, accompanied by a remarkable phenotype, closely related to that of rgl1 mutant: repressed shoot and promoted root growth. The second example links ABA with BR as BR-insensitive 1 (BRI1) gene was shown to mediate BR signaling response in root growth through interaction with BAK1 (BR-insensitive 1–associated receptor kinase 1 related). Exogenous ABA treatment led to a decrease in the level of peu-miRNA-n68 that targets BAK1, resulting in an induction in BAK1 expression and enhanced root growth (Lian et al., 2018).
ABA Transport and Stomata Closure
In higher plants, water is absorbed from soil by the root system and is transported to the shoot where it transpires. Abscisic acid regulates the water status in plants by regulating the closure of stomata that mediate uptake and release of oxygen and carbon dioxide for respiration and photosynthesis (Jia and Zhang, 2008; Manzi et al., 2016). Moreover, stomata play an important role in pathogen defense, as they are considered as a port of entry of pathogens including bacteria, fungi, and viruses. By regulating stomata aperture and inducing the expression of defense-related genes, ABA initiates plant’s defenses against pathogen attack (Lim et al., 2015). Under mild water limitation, when the soil starts to dry, ABA accumulates in root tissues leading to the decrease of stomatal conductance, while under drought stress, ABA accumulates in xylem. This distribution indicates that ABA is synthesized in roots, released to xylem vessels, and transported to the shoots. However, ABA is not only synthesized in roots, as ABA biosynthesis genes, e.g. NCED2, ABA2, AAO3, are also expressed at the vascular bundle in leaves where the guard cells are located at the epidermal layer, suggesting that ABA is transported between leaf tissues to elicit stomata response (Kuromori et al., 2018). In Arabidopsis, the two ATP-binding cassette (ABC) transporter proteins ABCG25 and ABCG40, which are localized at the plasma membrane in vascular tissues and guard cells, respectively, have been shown to modulate stomata closure and ABA sensitivity ( Figure 2 ) (Kang et al., 2010; Kuromori et al., 2010). Overexpression of AtABCG25 led to enhanced stomata response, supporting the hypothesis that ABA is synthesized in the vascular tissues and transported to the guard cells (Kuromori et al., 2016). The Arabidopsis nitrate transporter 1/peptide transporter family (NPF4.6) is a further ABA transporter that is expressed in vascular tissues, suggesting a complementary role with AtABCG25 and AtABCG40 in modulating the amount of transported ABA from vascular cells to guard cells (Kuromori et al., 2018). Finally, ATDTX50, a member of multidrug and toxin efflux transporter family, was identified as an ABA exporter localized at the plasma membrane and expressed in vascular tissues and guard cells (Zhang et al., 2014). The corresponding mutant line dtx50 showed retarded growth, which might be linked to high ABA sensitivity or high endogenous ABA level (Kuromori et al., 2018).
Strigolactones
Strigolactones are carotenoid-derived plant phytohormones that regulate different aspects of plant development, including shoot branching, establishing of root architecture, and leaf senescence, and are involved in biotic and abiotic stress response (Xie et al., 2010; Al-Babili and Bouwmeester, 2015; Decker et al., 2017; Saeed et al., 2017; Jia et al., 2019b). However, SLs were originally identified as inducer of root parasitic plant seed germination of genera Striga, Orobanche, Alectra, and Phelipanche (Xie et al., 2010). These weeds are a major threat for global food security, causing enormous yield losses in yields of cereals and many other crops (Parker, 2009; Parker, 2012;Atera et al., 2011;Bouwmeester et al., 2003; Pennisi, 2015). Later on, it was shown that SLs act as a chemical signal released into the soil to attract arbuscular mycorrhizal (AM) fungi for establishing the beneficial AM symbiosis (Akiyama et al., 2005;Lopez-Raez et al., 2015). Approximately 80% of land plants form this symbiosis that provides the AM fungi (AMF) with photosynthetic products and the plant with water and important micronutrients with low mobility in soil, especially phosphorus (Bonfante and Genre, 2010; Gutjahr and Parniske, 2013; Lanfranco et al., 2017; Yoneyama et al., 2019); therefore, SL biosynthesis and release are induced upon phosphate deficiency (Yoneyama et al., 2012).
SL Biosynthesis
There are around 25 characterized natural SLs so far, which are classified based on their chemical structure in canonical and noncanonical SLs. Canonical SLs consist of a butenolide lactone ring (D ring) linked by enol ether bridge in R-configuration (2′R), to a tricyclic lactone (ABC ring). Canonical SLs are divided into strigol- and orobanchol-like SLs based on the stereochemistry of B-/C-ring junction, whereas in the strigol-type SLs the C-ring is in β-orientation (up; 8bS-configuration), while in orobanchol-type SLs, the C-ring is in α orientation (down; 8bR-configuration) (Jia et al., 2017). Noncanonical SLs contain the butenolide lactone ring (D ring) and the enol ether bridge in R-configuration, which are characteristic for all-natural SLs. However, they have, as a second moiety, different structures instead of the ABC ring. Examples for noncanonical SLs are methyl carlactonoate, heliolactone, zealactone and zeapyranolactone, and avenaol (Al-Babili and Bouwmeester, 2015; Jia et al., 2017; Jia et al., 2019b; Koltai and Prandi, 2019). The diversity of SLs is a result of the structural variability of noncanonical SLs and modifications of the A and B rings in canonical ones, which include methylation, hydroxylation, ketolation, and epoxidation (Jia et al., 2017; Wang and Bouwmeester, 2018; Jia et al., 2019b).
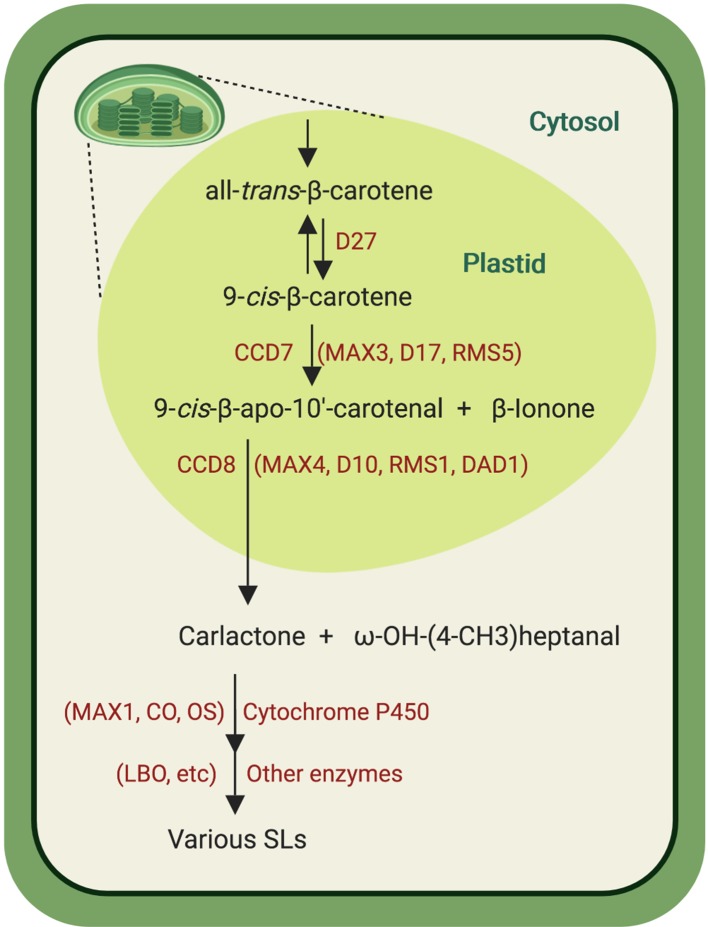
The similarity between the SL A-ring and the ionone ring in carotenoids led to the hypothesis that SLs might, like ABA, originate from carotenoids. This hypothesis was confirmed by investigating the SL release in different carotenoid-deficient mutants and through application of inhibitors of carotenoid biosynthesis (Matusova et al., 2005). However, SL biosynthesis remained elusive for many years because of lack of knowledge about the involved enzymes. The discovery that more branching/high tillering mutants from different plant species are SL deficient led to the breakthrough discovery of the SL hormonal function and provided candidate for biosynthesis enzymes and perception components, enabling the elucidation of major steps of SL biosynthesis and signal transduction. Strigolactone biosynthesis is initiated by the reversible conversion of the all-trans-β-carotene (C40) into 9-cis-β-carotene in the plastids, which is catalyzed by the 9-cis/trans-all-β-carotene isomerase Dwarf27 (D27) (Alder et al., 2012; Waters et al., 2012; Bruno and Al-Babili, 2016; Abuauf et al., 2018) ( Figure 3 ). The resulting 9-cis-β-carotene is cleaved by the carotenoid cleavage dioxygenase 7 (CCD7) (Arabidopsis MAX3, rice D17/HTD1, pea RMS5, and petunia DAD3), a stereospecific enzyme that converts only 9-cis-configured carotenoids into 9-cis-β-apo-10′-carotenal (C27) and β-ionone (C13) (Bruno et al., 2014). 9-cis-β-apo-10′-carotenal is the substrate of CCD8 (Arabidopsis MAX4, rice D10, pea RMS1, and petunia DAD1), an unusual CCD that catalyzes a combination of different reactions leading to the central SL biosynthesis intermediate carlactone (CL) and ω-OH-(4-CH3) heptanal (Alder et al., 2012; Bruno et al., 2017; Guillotin et al., 2017). Following its formation in plastids, CL is then exported into the cytosol where it is further converted by cytochrome P450 enzymes (CYP) of the 711 clade, e.g. the Arabidopsis MAX1 and the rice carlactone oxidase (Zhang et al., 2014). Recent investigation of the activity of CYP711 enzymes from different plant species allowed the classification of these enzymes into three types, types A1, A2, and A3 (Yoneyama et al., 2018). In Arabidopsis, CL is hydroxylated and oxidized at C19 into carlactonoic acid (CLA) by AtMAX1 (a CYP711A1 type). Carlactonoic acid is substrate of an unidentified methyltransferase that forms methyl carlactonoate (MeCLA) (Abe et al., 2014; Jia et al., 2017; Yoneyama et al., 2018). Recently, the enzyme lateral branching oxidoreductase (LBO), a member of the 2-oxoglutarate and FeII-dependent dioxygenase family, was identified as a further SL biosynthesis enzyme downstream of MAX1. In vitro studies indicated that LBO converts MeCLA into an unidentified product with an additional oxygen atom (Brewer et al., 2016). Rice genome encodes five MAX1 homologs (Os900, Os1400, Os1500, Os1900, Os5100). In vitro studies and transient expression in Nicotiana benthamiana suggest that Os900 (a CYP711A2 type) catalyzes the repeated oxygenation and ring closure in CL to yield 4-deoxyorobanchol (4DO), a canonical SL. Os1400, a rice CYP711A3 type enzyme, catalyzes the hydroxylation of 4DO into orobanchol (Sun et al., 2014). It was also shown that the three rice CYP711, Os1400, Os5100, and Os1900, can also convert CL into CLA, indicating that CLA is a common component in plant SL biosynthesis (Yoneyama et al., 2018). A recent study revealed that CCD7 and CCD8 can also utilize hydroxylated substrates, forming 3-OH-CL as a further product of the SL biosynthesis core pathway. 3-OH-CL might be the precursor of yet unidentified SLs, which contributes to SL diversity (Baz et al., 2018).
Figure 3.
Strigolactones biosynthesis pathway. The conversion of all-trans-β-carotene into carlactone is successively catalyzed by the cis/trans-β-carotene isomerase D27 that reversibly converts all-trans-β-carotene into 9-cis-β-carotene, which is subsequently cleaved by CCD7 into 9-cis-β-apo-10′-carotenal and β-ionone. The next enzyme, CCD8, transforms 9-cis-β-apo-10′-carotenal into carlactone and ω-OH-(4-CH3) heptanal. Carlactone is then oxidized, catalyzed by CYP711 enzymes [Arabidopsis MAX1, rice carlactone oxidase (CO), and orobanchol synthase (OS)] into carlactonoic acid and other SLs. After methylation, carlactonoic acid is oxidized by the oxidoreductase LBO into yet unidentified product. See text for abbreviations. Created with ‘Biorender’.
SL Transport
Grafting is frequently used to investigate hormone and metabolite fluxes between shoots and roots. Using this technique, it was shown that the more branching phenotype of SL biosynthesis mutants scions can be rescued by wild-type root stock, suggesting the presence of a root to shoot SL transport (Leyser, 2009; López-Ráez et al., 2010). Strigolactones are also released into the rhizosphere, which suggests the presence of a further transport system. In Petunia, this release was shown to be mediated by an ABC transporter protein pleiotropic drug resistance 1 (PDR1) (Kretzschmar et al., 2012), acting as a SL transporter. Pleiotropic drug resistance 1 is localized in the plasma membrane of the cortex cells in root tip and of the hypodermal passage cells, the passage points of the AMF. The latter localization indicates that PDR1 may guide the hyphae of AMF to the hypodermal passage cells (Jia et al., 2019b).
SL Signal Transduction
Strigolactone perception is initiated by binding to the SL receptor, Dwarf14 (D14), a α/β-hydrolase superfamily protein, which contains a conserved Ser-His-Asp catalytic triad required for SL hydrolysis and signal transduction (Hamiaux et al., 2012; Nakamura et al., 2013; Waters et al., 2015). In the absence of SLs, this receptor exhibits an open solvent-exposed ligand-binding pocket. Binding of the SL ligand in the pocket triggers its hydrolysis into the ABC ring and an intermediate D-ring moiety, which is followed by releasing the ABC ring and covalent binding of the D-ring to form the so-called covalently linked intermediate molecule (CLIM), accompanied by a conformational change of D14. These processes are critical for SL signal transduction that proceeds through binding to the F-box protein MAX2 (D3 in rice) and recruiting target expression repressors, such as the rice D53 or members of the Arabidopsis SMXL proteins for ubiquitination and proteasome-mediated degradation. (Saeed et al., 2017; Waters et al., 2017; Jia et al., 2019b). Most recently, structure-function studies of the rice D14-D3-D53 complex show that D3 possesses a C-terminal α-helix (CTH), which can occur in two conformation states, i.e. an engaged form and a dislodged form. The engaged form facilitates the binding of D3 and D14 in its CLIM form, whereas the dislodged form recognizes unmodified D14 and inhibits its enzymatic activity. The D3 CTH enables D14 to recruit D53 in SL presence, which then triggers SL hydrolysis. This structural plasticity of the SCFD3–D14 ubiquitin ligase suggests a model that explains how D3 coordinates SL perception and hydrolysis (Shabek et al., 2018).
SL and Shoot Architecture
Strigolactone is one of the key hormones in determining shoot architecture. In fact, the discovery of SLs as a new plant hormone was achieved through characterizing a series of more branching or high tillering mutants in different species. These mutants include more axillary growth (max) in Arabidopsis (Sorefan et al., 2003; Booker et al., 2004; Booker et al., 2005; Stirnberg et al., 2007), ramosus (rms) in pea (Beveridge et al., 2000; Morris et al., 2001; Sorefan et al., 2003; Foo et al., 2013), dwarf (d)/high-tillering dwarf (htd) in rice (Ishikawa et al., 2005; Arite et al., 2007; Arite et al., 2009; Lin et al., 2009; Zhou et al., 2013), and decreased apical dominance (dad) in petunia (Snowden et al., 2005; Simons et al., 2007; Drummond et al., 2009; Drummond et al., 2012; Hamiaux et al., 2012). This branching phenotype was resolved at the molecular level through SL perception mechanism in vascular plants. As previously mentioned, SL perception requires α-β hydrolase D14 and MAX2 and leads to (i) the cleavage of SL and (ii) the proteasome-mediated degradation of D53 and SMXL6/7/8 in rice and Arabidopsis, respectively, in order to regulate shoot branching. Indeed, it was shown that the smxl6 smxl7 smxl8 triple mutants could restore the branching and SL level to wild type (Soundappan et al., 2015).
Auxin is a major regulator of shoot branching. Many lines of evidence demonstrate that SLs interact with auxin to regulate this developmental process (Leyser, 2009). It has been shown that SLs regulate auxin homeostasis by modulating polar auxin transport (PAT) that is supposed to determine axillary bud dormancy/release in Arabidopsis. On the other hand, auxin controls the expression of SL biosynthesis genes and hence determines SL biosynthesis. Moreover, the negative feedback exerted by SLs on their own biosynthesis is mediated by auxin. In Arabidopsis, it was proposed that the inhibitory effect of SLs on shoot branching is caused, at least partially, by a reduction of PAT from the axillary buds to shoots by down-regulating the expression of the auxin transporter, PIN1. In addition, local auxin transport between the PAT stream and surrounding tissues, which is called connective auxin transport, rather than PAT itself, was shown later to be essential for the initial growth of axillary buds initial growth, while PAT is essential for their sustainable growth in both in Arabidopsis and pea (Leyser, 2009; Barbier et al., 2019). Sucrose also plays an important role in regulating bud outgrowth, by interacting with plant hormones: sucrose induces the expression of auxin biosynthesis genes and promotes the accumulation of cytokinin accumulation, which triggers buds outgrowth (Barbier et al., 2019) ( Figure 4 ).
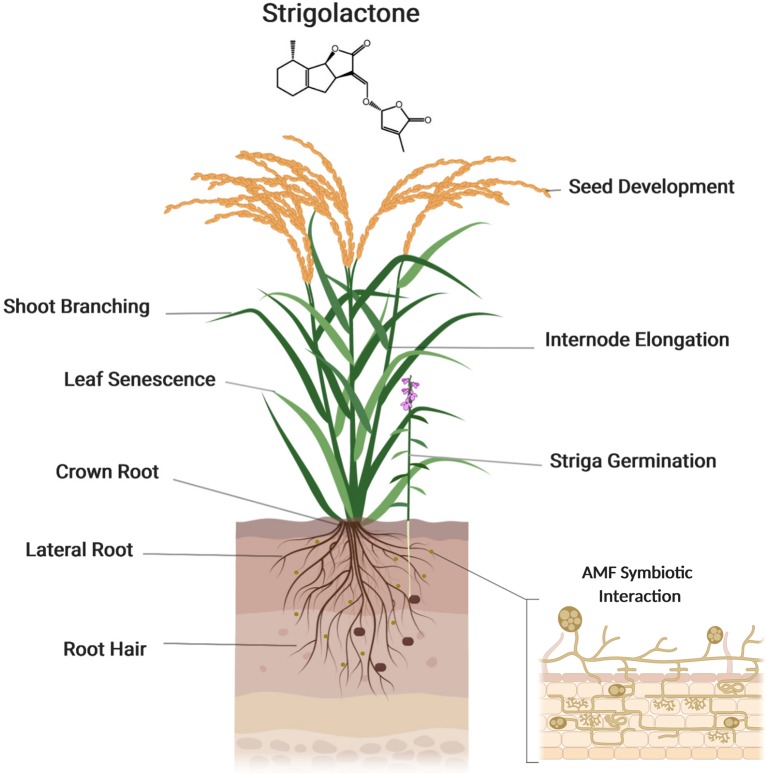
Figure 4.
Strigolactones (SLs) function in plant growth and development. Strigolactones regulate several developmental processes, such as shoot branching, seed development, internode elongation, and leaf senescence. Strigolactones are also a key regulator of root architecture, modulating the growth of crown and lateral roots and root hairs. Created with ‘Biorender’.
SLs in Root Development
In addition to shoot architecture, SLs play an important role in root development. The rice SL biosynthesis (d10, d17 and d27) and perception (d3 and d14) mutants exhibit shorter crown roots and contain a lower number of root meristem cells, compared to wild-type plant (Arite et al., 2012). In barley, the SL receptor mutant d14 mutant shows an increase in lateral root density and shorter seminal roots (Marzec et al., 2016). In Arabidopsis, treatment with the SL analog rac-GR24 leads to a reduction in the number and growth of lateral roots in a MAX2-dependent manner, while max2 mutant, but not SL biosynthesis mutants, shows a strong increase in lateral root density (Kapulnik et al., 2011a; Ruyter-Spira et al., 2011) due to SL insensitivity. SMXL6,7,8 might act downstream of MAX2, as the smxl6,7,8 triple mutants shows less lateral root density compared to wild type, while the smxl6,7,8 in max2 mutant background showed ∼50% decrease in lateral root density, suggesting that SMXLs are growth regulators thus repressed by SL signaling (Soundappan et al., 2015).
Strigolactones also regulate root hair elongation, as shown by treatment of tomato and Arabidopsis plants with rac-GR24. Confirming the specificity of this effect, application of rac-GR24 to Arabidopsis max2 mutant did not promote root hair elongation (Kapulnik et al., 2011a; Kapulnik et al., 2011b; Matthys et al., 2016). Strigolactones also inhibit adventitious root formation (Rasmussen et al., 2012). The effect of SLs on root development largely depends on auxin (Ruyter-Spira et al., 2011).
SLs in Leaf Senescence
Leaf senescence is a final phase of plant development. This process starts with the degradation of proteins and lipids into small-molecular-weight compounds and relocation of nutrients to younger cells or seeds (Guo and Gan, 2005). Leaf senescence is induced by several factors such as stress, aging, darkness, and plant hormones including SLs. Thus, Arabidopsis more axilliary branches 2/ORESARA 9 (MAX2/OREA9) gene was initially identified as a leaf senescence gene (Stirnberg et al., 2007). Mutation in the rice homolog D3 leads also to delayed leaf senescence (Yan et al., 2007), and a similar phenotype was observed in the corresponding petunia and Lotus japonicas mutant (Liu et al., 2013). Providing a further evidence for the positive role of SLs in leaf senescence, exogenous application of rac-GR24 accelerates leaf senescence in rice wild-type and SL biosynthesis mutants, i.e. d27, d17, and d10, but not in seedlings of the SL signaling mutants d14 and d13 (Ali et al., 2018). Similarly, the Arabidopsis SL-deficient and SL-insensitive mutants show delayed leaf senescence (Ueda and Kusaba, 2015).
Novel Carotenoid-Derived Signaling Molecules
In the last years, there has been large progress in identifying novel carotenoid-derived signaling molecules and regulatory metabolites. In the following, we touch on the exciting apocarotenoid volatile cyclocitral and its functions in regulating stress response and determining root architecture. We also briefly describe the recently discovered regulatory metabolites, anchorene, and zaxinone and their role in plant development.
β-Cyclocitral
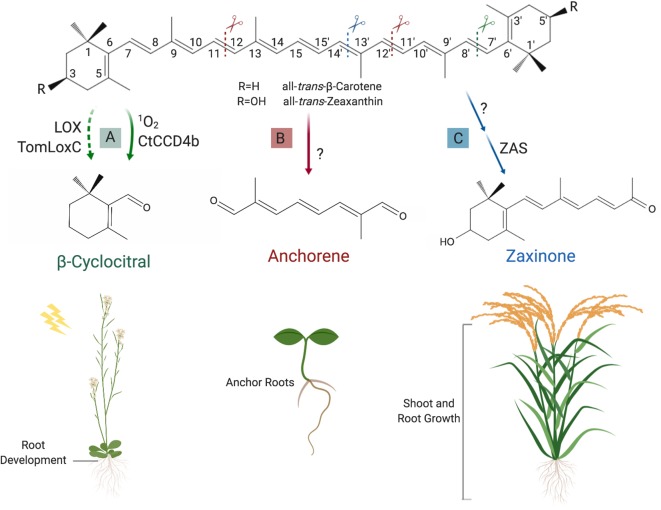
β-Cyclocitral is a β-carotene cleavage product that can be formed by enzymes from a subgroup of the CCD4 clade (Rodrigo et al., 2013) ( Figure 5A ) and, non-enzymatically, through attack by singlet oxygen (1O2) (Ramel et al., 2012a). Very recently, Gao et al. (2019) showed that the amount of β-cyclocitral (and other carotenoid-derived volatiles) was proportional to the LOX 13S-lipoxygenase expression level; a decrease of apocarotenoid content was observed in TomLocX antisense transgenic tomato and in AtLOX Arabidopsis mutant, compared to wild type. This study confirms previous in vitro observations where carotenoids were shown to act as a cosubstrate for these enzymes during the fatty acid oxidation (Hayward et al., 2017) and reveals the existence of an alternative route for apocarotenoid biosynthesis in planta. β-Cyclocitral has been detected in many plant species, including tomato (Tieman et al., 2012), rice (Hinge et al., 2016), parsley (Linde et al., 2016), tea (Ojha and Roy, 2018), grape (Chen et al., 2017), various trees (Gómez and Ledbetter, 1997; Tewari et al., 2015), and moss (Saritas et al., 2001). Studies in Arabidopsis showed that β-cyclocitral is a likely second messenger that conveys the 1O2 stress signal to the nucleus, altering the transcription of 1O2-regulated genes via methylene blue sensitivity protein, a small zinc finger protein (Ramel et al., 2012b; Shumbe et al., 2017). Similar activity was also reported for dihydroactinidiolide, a volatile resulting from the oxidation of β-ionone by 1O2 (Ramel et al., 2012b) (for a recent review, see D’Alessandro and Havaux, 2019). The transcriptome reprograming process, triggered by β-cyclocitral, increases the plant tolerance to photo-oxidative stress and initiates acclimation to high light conditions (Ramel et al., 2012b). Recently, it was shown that high light leads to a significant increase in the content of glycosylated β-cyclocitral (Mi et al., 2019b), which might be a mechanism for deactivation of this signaling molecule (D’Alessandro and Havaux, 2019).
Figure 5.
Formation and function of novel regulatory apocarotenoids. (A) β-Cyclocitral can be produced through cleavage of all-trans-β-carotene at the (C7′–C8′) double bound, which is catalyzed by CtCCD4b or 1O2 attack. Very recently, it was also shown that lipoxygenases, e.g. the tomato TomLoxC and the Arabidopsis LOX, can mediate the formation of this volatile, which acts as a growth regulator of roots and is involved in high light acclimation. (B) Anchorene, which can be formed by cleaving the (C11–C12) and (C11′–C12′) double bounds in all carotenoids downstream ζ-carotene in the carotenoid pathway, stimulates ANRs formation in Arabidopsis seedlings. (C) The recently discovered zaxinone is synthesized from all-trans-zeaxanthin (or all-trans-lutein) via the intermediate apo-10′-zeaxanthinal (not shown). The latter is the substrate of Zaxinone synthase (ZAS) that mediates the cleavage of the (C13–C14) double bond. Zaxinone is required for normal rice growth and development and is a negative regulator of SL biosynthesis. Created with ‘Biorender’.
A very recent study underpins the biological importance of β-cyclocitral by revealing its role as root growth regulator (Dickinson et al., 2019). Assuming that unknown apocarotenoid signals are required for root branching and lateral roots emergence, the authors used a targeted chemical genetic approach to identify apocarotenoids that enhance root branching. To increase the sensitivity of the roots by minimizing the level of endogenous apocarotenoids, they performed this screen in the presence of D15, an inhibitor of CCDs, at a concentration that reduces lateral roots emergence by 50% (Dickinson et al., 2019). Treatment with β-cyclocitral and dihydroactinidiolide led to significant increases in lateral root branching, with β-cyclocitral showing the strongest effect. In addition, application of β-cyclocitral in the absence of D15 also stimulated the growth of primary roots. Investigation of β-cyclocitral effect at cellular level showed that this compound promotes cell division in undifferentiated cells in meristems of lateral and primary roots. This effect is independent of ROS, auxin, and BR signaling pathways, as shown by using corresponding hormone-responsive marker lines and mutants.
Application of β-cyclocitral to tomato and rice seedlings demonstrated that this apocarotenoid is a conserved root growth regulator across plant species. The positive effect of β-cyclocitral was also observed under salt stress conditions: applied to salt-stressed rice roots rescued the negative effect of salt on root depth and showed in salty soil a positive effect on the vigor of rice plants (Dickinson et al., 2019).
Anchorene
Carotenoids can be targeted by repeated oxidative cleavage, which yields dialdehyde products (diapocarotenoids), besides monocarbonyl apocarotenoids. However, carotenoid-derived dialdehydes have attracted little attention because of their instability that makes their detection in biological systems and the identification of their possible biological activity difficult. Indeed, diapocarotenoids were mainly investigated because of their role as precursor of crocin and bixin, two pigments accumulated in saffron stigma and bixa seeds. (Giuliano et al., 2003; Frusciante et al., 2014; Demurtas et al., 2018). Recently, Jia et al. established a screening system to identify known and predicted diapocarotenoids involved in plant development, by assaying alterations in Arabidopsis roots. This approach led to the discovery of the presumed diapocarotenoid anchorene as a novel carotenoid-derived bioactive that promotes the development of anchor roots (ANRs) (Jia et al., 2019a), a less investigated type of Arabidopsis roots, which develop from the collet region situated at the root hypocotyl junction (Lucas et al., 2011). Although it is still unclear how it is formed, anchorene (C10) is a natural metabolite, as confirmed by LC-MS analysis (Jia et al., 2019a; Mi et al., 2019a), and its structure indicates that it can be produced by cleaving the (C11–C12) and (C11′–C12′) double bound from all carotenoids downstream of ζ-carotene in the carotenoid biosynthesis pathway ( Figure 5B ). In Arabidopsis, application of anchorene induces ANR formation. The regulation of ANR by anchorene is highly specific to its chemical structure, as application of a structural isomer did not show an effect on ANR development, and altering anchorene functional group(s) to alcohol, acid, or acid-ethyl ester resulted in a loss of activity (Jia et al., 2019a). The usage of auxin reporter lines and an auxin transport inhibitor, together with transcriptome analysis, showed that anchorene’s effect on ANR development is caused by modulating auxin homeostasis. Indicating their role in increasing nitrogen uptake, Jia et al. showed that ANR formation is triggered by nitrogen deficiency. Interestingly, lack of nitrogen led also to an increase in anchorene content, pointing to a possible function in regulating plant’s response to this adverse growth condition (Jia et al., 2019a).
Zaxinone
A recent study unraveled a sixth new clade in the plant CCD family, which is present in many plant species but not in Arabidopsis or other members of Brassicacea. In vitro characterization of a rice representative of this clade showed that it cleaves apo-10′-zeaxanthinal (3-OH-β-apo-10′-carotenal, C27) at the (C13–C14) double bound, resulting in a C18-ketone and an instable dialdehyde (C9) (Wang et al., 2019). Wang et al. called the C18 apocarotenoid zaxinone and, accordingly, the corresponding enzyme ZAS ( Figure 5C ). They also confirmed that zaxinone is a natural metabolite present in several plant species, including rice (Wang et al., 2019; see also Mi et al., 2018). Analysis of a corresponding zas mutant in comparison to wild type showed that it has similar zaxinone content in shoots but contained less amounts of this compound in roots, the tissue with the highest ZAS expression levels. These results confirm the enzymatic activity of ZAS, but also demonstrate that zaxinone can be formed via a route(s) that does not involve ZAS. The lower zaxinone content in zas roots mutant was accompanied by a reduced crown root length and number, a decrease in root and shoot biomass, and a lower tiller and panicle number. Taking into consideration that low tillering phenotype might be a result of increased SL biosynthesis, Wang et al. determined SL content in root tissues and exudates and found that zas mutant produced and released higher amounts of SLs. Accordingly, zas root exudates were more active in inducing Striga germination, compared to wild type. Exogenous application of zaxinone rescued the root phenotypes in zas mutant and increased root growth in wild-type seedlings, suggesting that this metabolite is growth promoting and required for normal rice growth and development. Moreover, zaxinone’s application led to a decrease in SL content and release, demonstrating that zaxinone is a negative regulator of SL biosynthesis. Expression analysis of SL biosynthetic genes showed that zaxinone acts at transcript level. Interestingly, application of zaxinone to a Striga susceptible rice variety alleviated the infestation by this root parasitic plant, indicating an application potential of zaxinone in combating Striga, besides the possibility of employing it to promote rice growth (Wang et al., 2019).
Conclusion
Plant growth and development, as well as response to environmental changes and stress factors, are regulated by a set of hormones and signaling molecules, which includes metabolites originating from carotenoids. The initial step in the formation of these carotenoid-derived signals is an oxidation step that yields shortened, carbonyl-group containing molecules called apocarotenoids. Some apocarotenoids can immediately act as signaling molecules, e.g. β-cyclocitral, or growth regulators, e.g. zaxinone and anchorene. Further modifications of the primary cleavage products lead to the known hormones ABA and SLs. It can be expected that future work will unravel new carotenoid-derived regulatory metabolites and that carotenoid metabolism will remain an intriguing and important research field in plant science, connecting photosynthesis, the primary metabolic process, with plant growth and development.
Author Contributions
This review was written by AF and JB. SA-B supervised the writing of the review and was involved in its further editing and revision. MZ contributed to the editing and organization of the review.
Funding
This work was supported by baseline funding and Competitive Research Grant (CRG2017) given by King Abdullah University of Science and Technology (KAUST) to SA-B, and by the Deutsche Forschungsgemeinschaft (DFG, German research Foundation) under Germany’s Excellence Strategy EXC 2048/1-390686111.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Kun-Peng Jia, Dr. Jianing Mi, and Dr. Imran Haider for valuable comments.
References
- Abe S., Sado A., Tanaka K., Kisugi T., Asami K., Ota S., et al. (2014). Carlactone is converted to carlactonoic acid by MAX1 in Arabidopsis and its methyl ester can directly interact with AtD14 in vitro. Proc. Natl. Acad. Sci. 111 (50), 18084–18089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abuauf H., Haider I., Jia K.-P., Ablazov A., Mi J., Blilou I., et al. (2018). The Arabidopsis DWARF27 gene encodes an all-trans-/9-cis-β-carotene isomerase and is induced by auxin, abscisic acid and phosphate deficiency. Plant Sci. 277, 33–42. 10.1016/j.plantsci.2018.06.024 [DOI] [PubMed] [Google Scholar]
- Ahrazem O., Gómez-Gómez L., Rodrigo M., Avalos J., Limón M. (2016. a). Carotenoid cleavage oxygenases from microbes and photosynthetic organisms: features and functions. Int. J. Mol. Sci. 17 (11), 1781. 10.3390/ijms17111781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrazem O., Rubio-Moraga A., Berman J., Capell T., Christou P., Zhu C., et al. (2016. b). The carotenoid cleavage dioxygenase CCD2 catalysing the synthesis of crocetin in spring crocuses and saffron is a plastidial enzyme. New Phytol. 209 (2), 650–663. 10.1111/nph.13609 [DOI] [PubMed] [Google Scholar]
- Al-Babili S., Bouwmeester H. J. (2015). Strigolactones, a novel carotenoid-derived plant hormone. Annu. Rev. Plant Biol. 66, 161–186. 10.1146/annurev-arplant-043014-114759 [DOI] [PubMed] [Google Scholar]
- Alder A., Jamil M., Marzorati M., Bruno M., Vermathen M., Bigler P., et al. (2012). The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 335 (6074), 1348–1351. 10.1126/science.1218094 [DOI] [PubMed] [Google Scholar]
- Ali A., Gao X., Guo Y. (2018). Initiation, progression, and genetic manipulation of leaf senescence. In Plant senescence (Springer) (New York, NY: Humana Press; ) 1744, 9–31. 10.1007/978-1-4939-7672-0_2 [DOI] [PubMed] [Google Scholar]
- Arite T., Iwata H., Ohshima K., Maekawa M., Nakajima M., Kojima M., et al. (2007). DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J. 51 (6), 1019–1029. 10.1111/j.1365-313X.2007.03210.x [DOI] [PubMed] [Google Scholar]
- Arite T., Kameoka H., Kyozuka J. (2012). Strigolactone positively controls crown root elongation in rice. J. Plant Growth Regul. 31 (2), 165–172. 10.1007/s00344-011-9228-6 [DOI] [Google Scholar]
- Arite T., Umehara M., Ishikawa S., Hanada A., Maekawa M., Yamaguchi S., et al. (2009). d14, A strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. 50 (8), 1416–1424. 10.1093/pcp/pcp091 [DOI] [PubMed] [Google Scholar]
- Akiyama K., Matsuzaki K. I., Hayashi H. (2005). Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435 (7043), 824. 10.1038/nature03608 [DOI] [PubMed] [Google Scholar]
- Atera E. A., Itoh K., Onyango J. C. (2011). Evaluation of ecologies and severity of Striga weed on rice in sub-Saharan Africa. Agric. Biol. J. N. Am. 2 (5), 752–760. 10.5251/abjna.2011.2.5.752.760 [DOI] [Google Scholar]
- Barbier F. F., Dun E. A., Kerr S. C., Chabikwa T. G., Beveridge C. A. (2019). An update on the signals controlling shoot branching. Trends Plant Sci. 24 (3), 220–236. 10.1016/j.tplants.2018.12.001 [DOI] [PubMed] [Google Scholar]
- Baz L., Mori N., Mi J., Jamil M., Kountche B. A., Guo X., et al. (2018). 3-Hydroxycarlactone, a novel product of the strigolactone biosynthesis core pathway. Mol. Plant 11, 1312–1314. 10.1016/j.molp.2018.06.008 [DOI] [PubMed] [Google Scholar]
- Beveridge C. A., Symons G. M., Turnbull C. G. (2000). Auxin inhibition of decapitation-induced branching is dependent on graft-transmissible signals regulated by genes Rms1 and Rms2 . Plant Physiol. 123 (2), 689–698. 10.1104/pp.123.2.689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfante P., Genre A. (2010). Mechanisms underlying beneficial plant–fungus interactions in mycorrhizal symbiosis. Nat. Commun. 1, 48. 10.1038/ncomms1046 [DOI] [PubMed] [Google Scholar]
- Booker J., Auldridge M., Wills S., McCarty D., Klee H., Leyser O. (2004). MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr. Biol. 14 (14), 1232–1238. 10.1016/j.cub.2004.06.061 [DOI] [PubMed] [Google Scholar]
- Booker J., Sieberer T., Wright W., Williamson L., Willett B., Stirnberg P., et al. (2005). MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Dev. Cell 8 (3), 443–449. 10.1016/j.devcel.2005.01.009 [DOI] [PubMed] [Google Scholar]
- Bouwmeester H. J., Matusova R., Zhongkui S., Beale M. H. (2003). Secondary metabolite signalling in host–parasitic plant interactions. Curr. Opin. Plant Biol. 6 (4), 358–364. 10.1016/S1369-5266(03)00065-7 [DOI] [PubMed] [Google Scholar]
- Brefort T., Scherzinger D., Limón M. C., Estrada A. F., Trautmann D., Mengel C., et al. (2011). Cleavage of resveratrol in fungi: characterization of the enzyme Rco1 from Ustilago maydis. Fungal Genet. Biol. 48 (2), 132–143. 10.1016/j.fgb.2010.10.009 [DOI] [PubMed] [Google Scholar]
- Brewer P. B., Yoneyama K., Filardo F., Meyers E., Scaffidi A., Frickey T., et al. (2016). Lateral branching oxidoreductase acts in the final stages of strigolactone biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. 113 (22), 6301–6306. 10.1073/pnas.1601729113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton G., Liaaen-Jensen S., Pfander H., editors. (2012). Carotenoids: handbook. Birkhäuser: Springer Basel AG. [Google Scholar]
- Bruno M., Al-Babili S. (2016). On the substrate specificity of the rice strigolactone biosynthesis enzyme DWARF27. Planta 243 (6), 1429–1440. 10.1007/s00425-016-2487-5 [DOI] [PubMed] [Google Scholar]
- Bruno M., Beyer P., Al-Babili S. (2015). The potato carotenoid cleavage dioxygenase 4 catalyzes a single cleavage of β-ionone ring-containing carotenes and non-epoxidated xanthophylls. Arch. Biochem. Biophys. 572, 126–133. 10.1016/j.abb.2015.02.011 [DOI] [PubMed] [Google Scholar]
- Bruno M., Hofmann M., Vermathen M., Alder A., Beyer P., Al-Babili S. (2014). On the substrate- and stereospecificity of the plant carotenoid cleavage dioxygenase 7. FEBS Letters 588 (9), 1802–1807. 10.1016/j.febslet.2014.03.041 [DOI] [PubMed] [Google Scholar]
- Bruno M., Koschmieder J., Wuest F., Schaub P., Fehling-Kaschek M., Timmer J., et al. (2016). Enzymatic study on AtCCD4 and AtCCD7 and their potential to form acyclic regulatory metabolites. J. Exp. Bot. 67 (21), 5993–6005. 10.1093/jxb/erw356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno M., Vermathen M., Alder A., Wüst F., Schaub P., van der Steen R., et al. (2017). Insights into the formation of carlactone from in-depth analysis of the CCD8-catalyzed reactions. FEBS Letters 591 (5), 792–800. 10.1002/1873-3468.12593 [DOI] [PubMed] [Google Scholar]
- Chen W.-K., Yu K.-J., Liu B., Lan Y.-B., Sun R.-Z., Li Q., et al. (2017). Comparison of transcriptional expression patterns of carotenoid metabolism in ‘Cabernet Sauvignon’ grapes from two regions with distinct climate. J. Plant Physiol. 213, 75–86. 10.1016/j.jplph.2017.03.001 [DOI] [PubMed] [Google Scholar]
- D’Alessandro S., Havaux M. (2019). Sensing β-carotene oxidation in photosystem II to master plant stress tolerance. New Phytol. 223 (4), 1776–1783. 10.1111/nph.15924 [DOI] [PubMed] [Google Scholar]
- D’Alessandro S., Mizokami Y., Legeret B., Havaux M. (2019). The Apocarotenoid β-Cyclocitric Acid Elicits Drought Tolerance in Plants. iScience, 19, 461–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker E. L., Alder A., Hunn S., Ferguson J., Lehtonen M. T., Scheler B., et al. (2017). Strigolactone biosynthesis is evolutionarily conserved, regulated by phosphate starvation and contributes to resistance against phytopathogenic fungi in a moss, Physcomitrella patens. New Phytol. 216 (2), 455–468. 10.1111/nph.14506 [DOI] [PubMed] [Google Scholar]
- Dejonghe W., Okamoto M., Cutler S. R. (2018). Small molecule probes of ABA biosynthesis and signaling. Plant Cell Physiol. 59 (8), 1490–1499. 10.1093/pcp/pcy126 [DOI] [PubMed] [Google Scholar]
- DellaPenna D., Pogson B. J. (2006). Vitamin synthesis in plants: tocopherols and carotenoids. Annu. Rev. Plant Biol. 57, 711–738. 10.1146/annurev.arplant.56.032604.144301 [DOI] [PubMed] [Google Scholar]
- Demurtas O. C., Frusciante S., Ferrante P., Diretto G., Azad N. H., Pietrella M., et al. (2018). Candidate enzymes for saffron crocin biosynthesis are localized in multiple cellular compartments. Plant Physiol. 177 (3), 990–1006. 10.1104/17.01815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson A. J., Lehner K., Mi J., Jia K.-P., Mijar M., Dinneny J., et al. (2019). β-Cyclocitral is a conserved root growth regulator. Proc. Natl. Acad. Sci. 116 (21), 10563–10567. 10.1073/pnas.1821445116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd I. C. (2013). Abscisic acid and stomatal closure: a hydraulic conductance conundrum? New Phytol. 197 (1), 6–8. 10.1111/nph.12052 [DOI] [PubMed] [Google Scholar]
- Drummond R. S., Martínez-Sánchez N. M., Janssen B. J., Templeton K. R., Simons J. L., Quinn B. D., et al. (2009). Petunia hybrida carotenoid cleavage dioxygenase7 is involved in the production of negative and positive branching signals in petunia. Plant Physiol. 151 (4), 1867–1877. 10.1104/pp.109.146720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond R. S., Sheehan H., Simons J. L., Martínez-Sánchez N. M., Turner R. M., Putterill J., et al. (2012). The expression of petunia strigolactone pathway genes is altered as part of the endogenous developmental program. Front. Plant Sci. 2, 115. 10.3389/fpls.2011.00115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W., Cheng J., Cheng Y., Wang L., He Y., Wang Z., et al. (2015). Physiological characteristics and related gene expression of after-ripening on seed dormancy release in rice. Plant Biol. 17 (6), 1156–1164. 10.1111/plb.12371 [DOI] [PubMed] [Google Scholar]
- Finkelstein R. (2013). Abscisic acid synthesis and response. Arabidopsis Book Am. Soc. Plant Biol. 11, e0166. 10.1199/tab.0166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R., Reeves W., Ariizumi T., Steber C. (2008). Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 59, 387–415. 10.1146/annurev.arplant.59.032607.092740 [DOI] [PubMed] [Google Scholar]
- Foo E., Yoneyama K., Hugill C. J., Quittenden L. J., Reid J. B. (2013). Strigolactones and the regulation of pea symbioses in response to nitrate and phosphate deficiency. Mol. Plant 6 (1), 76–87. 10.1093/mp/sss115 [DOI] [PubMed] [Google Scholar]
- Fraser P. D., Bramley P. M. (2004). The biosynthesis and nutritional uses of carotenoids. Prog. Lipid Res. 43 (3), 228–265. 10.1016/j.plipres.2003.10.002 [DOI] [PubMed] [Google Scholar]
- Frusciante S., Diretto G., Bruno M., Ferrante P., Pietrella M., Prado-Cabrero A., et al. (2014). Novel carotenoid cleavage dioxygenase catalyzes the first dedicated step in saffron crocin biosynthesis. Proc. Natl. Acad. Sci. 111 (33), 12246–12251. 10.1073/pnas.1404629111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghassemian M., Nambara E., Cutler S., Kawaide H., Kamiya Y., McCourt P. (2000). Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell 12 (7), 1117–1126. 10.1105/tpc.12.7.1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Gonda I., Sun H., Ma Q., Bao K., Tieman D. M., et al. (2019). The tomato pan-genome uncovers new genes and a rare allele regulating fruit flavor. Nat. Genet. 51 (6), 1044. 10.1038/s41588-019-0410-2 [DOI] [PubMed] [Google Scholar]
- Giuliano G., Al-Babili S., von Lintig J. (2003). Carotenoid oxygenases: cleave it or leave it. Trends Plant Sci. 8 (4), 145–149. 10.1016/S1360-1385(03)00053-0 [DOI] [PubMed] [Google Scholar]
- Gómez E., Ledbetter C. A. (1997). Development of volatile compounds during fruit maturation: characterization of apricot and plum× apricot hybrids. J. Sci. Food Agric. 74 (4), 541–546. [DOI] [Google Scholar]
- González-Grandío E., Pajoro A., Franco-Zorrilla J. M., Tarancón C., Immink R. G., Cubas P. (2017). Abscisic acid signaling is controlled by a BRANCHED1/HD-ZIP I cascade in Arabidopsis axillary buds. Proc. Natl. Acad. Sci 114 (2), E245– E254. 10.1073/pnas.1613199114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F., Hughes T., Waterhouse P., Jacobsen J. (2008). Regulation of dormancy in barley by blue light and after-ripening: effects on abscisic acid and gibberellin metabolism. Plant Physiol. 147 (2), 886–896. 10.1104/pp.107.115469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillotin B., Etemadi M., Audran C., Bouzayen M., Bécard G., Combier J. P. (2017). Sl-IAA27 regulates strigolactone biosynthesis and mycorrhization in tomato (var. MicroTom). New Phytol. 213 (3), 1124–1132. 10.1111/nph.14246 [DOI] [PubMed] [Google Scholar]
- Guo Y., Gan S. (2005). Leaf senescence: signals, execution, and regulation. Curr. Top. Dev. Biol. 71, 83–112. 10.1016/S0070-2153(05)71003-6 [DOI] [PubMed] [Google Scholar]
- Gutjahr C., Parniske M. (2013). Cell and developmental biology of arbuscular mycorrhiza symbiosis. Annu. Rev. Cell Dev. Biol. 29, 593–617. 10.1146/annurev-cellbio-101512-122413 [DOI] [PubMed] [Google Scholar]
- Haider I., Andreo-Jimenez B., Bruno M., Bimbo A., Floková K., Abuauf H., et al. (2018). The interaction of strigolactones with abscisic acid during the drought response in rice. J. Exp. Bot. 69 (9), 2403–2414. 10.1093/jxb/ery089 [DOI] [PubMed] [Google Scholar]
- Hamiaux C., Drummond R. S., Janssen B. J., Ledger S. E., Cooney J. M., Newcomb R. D., et al. (2012). DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr. Biol. 22 (21), 2032–2036. 10.1016/j.cub.2012.08.007 [DOI] [PubMed] [Google Scholar]
- Harris J. (2015). Abscisic acid: hidden architect of root system structure. Plants 4 (3), 548–572. 10.3390/plants4030548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser F., Li Z., Waadt R., Schroeder J. I. (2017). SnapShot: abscisic acid signaling. Cell 171 (7), 1708–1708. e1700. 10.1016/j.cell.2017.11.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinge V., Patil H., Nadaf A. (2016). Comparative characterization of aroma volatiles and related gene expression analysis at vegetative and mature stages in basmati and non-basmati rice (Oryza sativa L.) cultivars. Appl. Biochem. Biotechnol. 178 (4), 619–639. 10.1007/s12010-015-1898-2 [DOI] [PubMed] [Google Scholar]
- Hayward S., Cilliers T., Swart P. (2017). Lipoxygenases: from isolation to application. Compr. Rev. Food Sci. F. 16 (1), 199–211. 10.1111/1541-4337.12239 [DOI] [PubMed] [Google Scholar]
- Holalu S. V., Finlayson S. A. (2017). The ratio of red light to far red light alters Arabidopsis axillary bud growth and abscisic acid signalling before stem auxin changes. J. Exp. Bot. 68 (5), 943–952. 10.1093/jxb/erw479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X., Rivers J., León P., McQuinn R. P., Pogson B. J. (2016). Synthesis and function of apocarotenoid signals in plants. Trends Plant Sci. 21 (9), 792–803. 10.1016/j.tplants.2016.06.001 [DOI] [PubMed] [Google Scholar]
- Hussain A., Black C., Taylor I., Roberts J. (2000). Does an antagonistic relationship between ABA and ethylene mediate shoot growth when tomato (Lycopersicon esculentum Mill.) plants encounter compacted soil? Plant Cell Environ. 23 (11), 1217–1226. 10.1046/j.1365-3040.2000.00639.x [DOI] [Google Scholar]
- Ilg A., Beyer P., Al-Babili S. (2009). Characterization of the rice carotenoid cleavage dioxygenase 1 reveals a novel route for geranial biosynthesis. FEBS J. 276 (3), 736–747. 10.1111/j.1742-4658.2008.06820.x [DOI] [PubMed] [Google Scholar]
- Ilg A., Bruno M., Beyer P., Al-Babili S. (2014). Tomato carotenoid cleavage dioxygenases 1A and 1B: relaxed double bond specificity leads to a plenitude of dialdehydes, mono-apocarotenoids and isoprenoid volatiles. FEBS Open Bio. 4, 584–593. 10.1016/j.fob.2014.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa S., Maekawa M., Arite T., Onishi K., Takamure I., Kyozuka J. (2005). Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol. 46 (1), 79–86. 10.1093/pcp/pci022 [DOI] [PubMed] [Google Scholar]
- Iuchi S., Kobayashi M., Taji T., Naramoto M., Seki M., Kato T., et al. (2001). Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 27 (4), 325–333. 10.1046/j.1365-313x.2001.01096.x [DOI] [PubMed] [Google Scholar]
- Jia K., Dickinson A. J., Mi J., Cui G., Kharbatia N. M., Guo X., et al. (2019. a). Anchorene is an endogenous diapocarotenoid required for anchor root formation in Arabidopsis. Sci. Adv. in press. 10.1101/496737 [DOI] [PMC free article] [PubMed]
- Jia K.-P., Baz L., Al-Babili S. (2017). From carotenoids to strigolactones. J. Exp. Bot. 69 (9), 2189–2204. 10.1093/jxb/erx476 [DOI] [PubMed] [Google Scholar]
- Jia K.-P., Li C., Bouwmeester H. J., Al-Babili S. (2019. b). “Strigolactone biosynthesis and signal transduction,” in Strigolactones—biology and applications (Cham: Springer; ), 1–45. 10.1007/978-3-030-12153-2_1 [DOI] [Google Scholar]
- Jia W., Zhang J. (2008). Stomatal movements and long-distance signaling in plants. Plant Signal. Behav. 3 (10), 772–777. 10.4161/psb.3.10.6294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Hwang J.-U., Lee M., Kim Y.-Y., Assmann S. M., Martinoia E., et al. (2010). PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc. Natl. Acad. Sci. 107 (5), 2355–2360. 10.1073/pnas.0909222107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y., Jikumaru Y., Hanada A., Nambara E., Abrams S. R., Kamiya Y., et al. (2010). Comprehensive hormone profiling in developing Arabidopsis seeds: examination of the site of ABA biosynthesis, ABA transport and hormone interactions. Plant Cell Physiol. 51 (12), 1988–2001. 10.1093/pcp/pcq158 [DOI] [PubMed] [Google Scholar]
- Kapulnik Y., Delaux P.-M., Resnick N., Mayzlish-Gati E., Wininger S., Bhattacharya C., et al. (2011. a). Strigolactones affect lateral root formation and root-hair elongation in Arabidopsis. Planta 233 (1), 209–216. 10.1007/s00425-010-1310-y [DOI] [PubMed] [Google Scholar]
- Kapulnik Y., Resnick N., Mayzlish-Gati E., Kaplan Y., Wininger S., Hershenhorn J., et al. (2011. b). Strigolactones interact with ethylene and auxin in regulating root-hair elongation in Arabidopsis. J. Exp. Bot. 62 (8), 2915–2924. 10.1093/jxb/erq464 [DOI] [PubMed] [Google Scholar]
- Koltai H., Prandi C. editors. (2019). Strigolactones-Biology and Applications. Switzerland: Springer Nature Switzerland AG. 10.1007/978-3-030-12153-2 [DOI] [Google Scholar]
- Kretzschmar T., Kohlen W., Sasse J., Borghi L., Schlegel M., Bachelier J. B., et al. (2012). A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature 483 (7389), 341. 10.1038/nature10873 [DOI] [PubMed] [Google Scholar]
- Kuromori T., Fujita M., Urano K., Tanabata T., Sugimoto E., Shinozaki K. (2016). Overexpression of AtABCG25 enhances the abscisic acid signal in guard cells and improves plant water use efficiency. Plant Sci. 251, 75–81. 10.1016/j.plantsci.2016.02.019 [DOI] [PubMed] [Google Scholar]
- Kuromori T., Miyaji T., Yabuuchi H., Shimizu H., Sugimoto E., Kamiya A., et al. (2010). ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc. Natl. Acad. Sci. 107 (5), 2361–2366. 10.1073/pnas.0912516107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromori T., Seo M., Shinozaki K. (2018). ABA transport and plant water stress responses. Trends Plant Sci. 23 (6), 513–522. 10.1016/j.tplants.2018.04.001 [DOI] [PubMed] [Google Scholar]
- Lanfranco L., Fiorilli V., Venice F., Bonfante P. (2017). Strigolactones cross the kingdoms: plants, fungi, and bacteria in the arbuscular mycorrhizal symbiosis. J. Exp. Bot. 69 (9), 2175–2188. 10.1093/jxb/erx432 [DOI] [PubMed] [Google Scholar]
- Le J., Vandenbussche F., Van Der Straeten D., Verbelen J.-P. (2001). In the early response of Arabidopsis roots to ethylene, cell elongation is up-and down-regulated and uncoupled from differentiation. Plant Physiol. 125 (2), 519–522. 10.1104/pp.125.2.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. H., Piao H. L., Kim H.-Y., Choi S. M., Jiang F., Hartung W., et al. (2006). Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell 126 (6), 1109–1120. 10.1016/j.cell.2006.07.034 [DOI] [PubMed] [Google Scholar]
- LeNoble M. E., Spollen W. G., Sharp R. E. (2004). Maintenance of shoot growth by endogenous ABA: genetic assessment of the involvement of ethylene suppression. J. Exp. Bot. 55 (395), 237–.245. 10.1093/jxb/erh031 [DOI] [PubMed] [Google Scholar]
- Leyser O. (2009). The control of shoot branching: an example of plant information processing. Plant Cell Environ. 32 (6), 694–703. 10.1111/j.1365-3040.2009.01930.x [DOI] [PubMed] [Google Scholar]
- Li X., Chen L., Forde B. G., Davies W. J. (2017). The biphasic root growth response to abscisic acid in Arabidopsis involves interaction with ethylene and auxin signalling pathways. Front. Plant Sci. 8, 1493. 10.3389/fpls.2017.01493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian C., Yao K., Duan H., Li Q., Liu C., Yin W., et al. (2018). Exploration of ABA responsive miRNAs reveals a new hormone signaling crosstalk pathway regulating root growth of Populus euphratica. Int. J. Mol. Sci. 19 (5), 1481. 10.3390/ijms19051481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C., Baek W., Jung J., Kim J.-H., Lee S. (2015). Function of ABA in stomatal defense against biotic and drought stresses. Int. J. Mol. Sci. 16 (7), 15251–15270. 10.3390/ijms160715251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Wang R., Qian Q., Yan M., Meng X., Fu Z., et al. (2009). DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 21 (5), 1512–1525. 10.1105/tpc.109.065987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linde G., Gazim Z., Cardoso B., Jorge L., Tešević V., Glamoćlija J., et al. (2016). Antifungal and antibacterial activities of Petroselinum crispum essential oil. Genet. Mol. Res. 15 (3), 1–11. 10.4238/gmr.15038538 [DOI] [PubMed] [Google Scholar]
- Liu J., Novero M., Charnikhova T., Ferrandino A., Schubert A., Ruyter-Spira C., et al. (2013). Carotenoid cleavage dioxygenase 7 modulates plant growth, reproduction, senescence, and determinate nodulation in the model legume Lotus japonicus . J. Exp. Bot. 64 (7), 1967–1981. 10.1093/jxb/ert056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Ráez J. A., Kohlen W., Charnikhova T., Mulder P., Undas A. K., Sergeant M. J., et al. (2010). Does abscisic acid affect strigolactone biosynthesis? New Phytol. 187 (2), 343–354. 10.1111/j.1469-8137.2010.03291.x [DOI] [PubMed] [Google Scholar]
- Lopez-Raez J. A., Fernández I., García J. M., Berrio E., Bonfante P., Walter M. H., et al. (2015). Differential spatio-temporal expression of carotenoid cleavage dioxygenases regulates apocarotenoid fluxes during AM symbiosis. Plant Sci. 230, 59–69. 10.1016/j.plantsci.2014.10.010 [DOI] [PubMed] [Google Scholar]
- Lucas M., Swarup R., Paponov I. A., Swarup K., Casimiro I., Lake D., et al. (2011). Short-root regulates primary, lateral, and adventitious root development in Arabidopsis. Plant Physiol. 155 (1), 384–398. 10.1104/pp.110.165126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Cao J., He J., Chen Q., Li X., Yang Y. (2018). Molecular mechanism for the regulation of ABA homeostasis during plant development and stress responses. Int. J. Mol. Sci. 19 (11), p.3643. 10.3390/ijms19113643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzi M., Lado J., Rodrigo M. J., Arbona V., Gómez-Cadenas A. (2016). ABA accumulation in water-stressed Citrus roots does not rely on carotenoid content in this organ. Plant Sci. 252, 151–161. 10.1016/j.plantsci.2016.07.017 [DOI] [PubMed] [Google Scholar]
- Marzec M., Gruszka D., Tylec P., Szarejko I. (2016). Identification and functional analysis of the HvD14 gene involved in strigolactone signaling in Hordeum vulgare. Physiol. Plant 158 (3), 341–355. 10.1111/ppl.12460 [DOI] [PubMed] [Google Scholar]
- Matthys C., Walton A., Struk S., Stes E., Boyer F.-D., Gevaert K., et al. (2016). The whats, the wheres and the hows of strigolactone action in the roots. Planta 243 (6), 1327–1337. 10.1007/s00425-016-2483-9 [DOI] [PubMed] [Google Scholar]
- Matusova R., Rani K., Verstappen F. W., Franssen M. C., Beale M. H., Bouwmeester H. J. (2005). The strigolactone germination stimulants of the plant-parasitic Striga and Orobanche spp. are derived from the carotenoid pathway. Plant Physiol. 139 (2), 920–934. 10.1104/pp.105.061382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuinn R. P., Giovannoni J. J., Pogson B. J. (2015). More than meets the eye: from carotenoid biosynthesis, to new insights into apocarotenoid signaling. Curr. Opin. Plant Biol. 27, 172–179. 10.1016/j.pbi.2015.06.020 [DOI] [PubMed] [Google Scholar]
- Merilo E., Jalakas P., Laanemets K., Mohammadi O., Hõrak H., Kollist H., et al. (2015). Abscisic acid transport and homeostasis in the context of stomatal regulation. Mol. Plant 8 (9), 1321–1333. 10.1016/j.molp.2015.06.006 [DOI] [PubMed] [Google Scholar]
- Mi J., Jia K. P., Wang J. Y., Al-Babili S. (2018). A rapid LC-MS method for qualitative and quantitative profiling of plant apocarotenoids. Anal. Chim. Acta 1035, 87–95. 10.1016/j.aca.2018.07.002 [DOI] [PubMed] [Google Scholar]
- Mi J., Jia K. P., Balakrishna A., Feng Q., Al-Babili S. (2019. a). A highly sensitive SPE-derivatization-UHPLC-MS approach for quantitative profiling of carotenoid-derived dialdehydes from vegetables. J. Agric. Food Chem. 67, 5899–5907 10.1021/acs.jafc.9b01749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi J., Jia K.-P., Balakrishna A., Wang J. Y., Al-Babili S. (2019. b). An LC-MS profiling method reveals a route for apocarotene glycosylation and shows its induction by high light stress in Arabidopsis. Analyst 144 (4), 1197–1204. 10.1039/C8AN02143K [DOI] [PubMed] [Google Scholar]
- Moise A. R., Al-Babili S., Wurtzel E. T. (2014). Mechanistic aspects of carotenoid biosynthesis. Chem. Rev. 114 (1), 164–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris S. E., Turnbull C. G., Murfet I. C., Beveridge C. A. (2001). Mutational analysis of branching in pea. Evidence that Rms1 and Rms5 regulate the same novel signal. Plant Physiol. 126 (3), 1205–1213. 10.1104/pp.126.3.1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H., Xue Y.-L., Miyakawa T., Hou F., Qin H.-M., Fukui K., et al. (2013). Molecular mechanism of strigolactone perception by DWARF14. Nat. Commun. 4, 2613. 10.1038/ncomms3613 [DOI] [PubMed] [Google Scholar]
- Née G., Kramer K., Nakabayashi K., Yuan B., Xiang Y., Miatton E., et al. (2017. a). Delay of germination1 requires PP2C phosphatases of the ABA signalling pathway to control seed dormancy. Nat. Commun. 8 (1), 72. 10.1038/s41467-017-00113-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Née G., Xiang Y., Soppe W. J. (2017. b). The release of dormancy, a wake-up call for seeds to germinate. Curr. Opin. Plant Biol. 35, 8–14. 10.1016/j.pbi.2016.09.002 [DOI] [PubMed] [Google Scholar]
- Nisar N., Li L., Lu S., Khin N. C., Pogson B. J. (2015). Carotenoid metabolism in plants. Mol. Plant 8 (1), 68–82. 10.1016/j.molp.2014.12.007 [DOI] [PubMed] [Google Scholar]
- North H. M., Almeida A. D., Boutin J. P., Frey A., To A., Botran L., et al. (2007). The Arabidopsis ABA-deficient mutant aba4 demonstrates that the major route for stress-induced ABA accumulation is via neoxanthin isomers. Plant J. 50 (5), 810–824. 10.1111/j.1365-313X.2007.03094.x [DOI] [PubMed] [Google Scholar]
- Ojha P. K., Roy K. (2018). PLS regression-based chemometric modeling of odorant properties of diverse chemical constituents of black tea and coffee. RSC Adv. 8 (5), 2293–2304. 10.1039/C7RA12914A [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker C. (2009). Observations on the current status of Orobanche and Striga problems worldwide. Pest Manag. Sci. Formerly Pestic. Sci. 65 (5), 453–459. 10.1002/ps.1713 [DOI] [PubMed] [Google Scholar]
- Parker C. (2012). Parasitic weeds: a world challenge. Weed Sci. 60 (2), 269–276. 10.1614/WS-D-11-00068.1 [DOI] [Google Scholar]
- Pennisi E. (2015). How crop-killing witchweed senses its victims. Science 350 (6257), 146–147. 10.1126/science.350.6257.146 [DOI] [PubMed] [Google Scholar]
- Ramel F., Birtic S., Cuiné S., Triantaphylidès C., Ravanat J.-L., Havaux M. (2012. a). Chemical quenching of singlet oxygen by carotenoids in plants. Plant Physiol. 158 (3), 1267–1278. 10.1104/pp.111.182394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramel F., Birtic S., Ginies C., Soubigou-Taconnat L., Triantaphylidès C., Havaux M. (2012. b). Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc .Natl. Acad. Sci. 109 (14), 5535–5540. 10.1073/pnas.1115982109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen A., Beveridge C. A., Geelen D. (2012). Inhibition of strigolactones promotes adventitious root formation. Plant Signal Behav. 7 (6), 694–697. 10.4161/psb.20224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy S. K., Holalu S. V., Casal J. J., Finlayson S. A. (2013). Abscisic acid regulates axillary bud outgrowth responses to the ratio of red to far-red light. Plant Physiol. 163 (2), 1047–1058. 10.1104/pp.113.221895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo M. J., Alquézar B., Alós E., Medina V., Carmona L., Bruno M., et al. (2013). A novel carotenoid cleavage activity involved in the biosynthesis of Citrus fruit-specific apocarotenoid pigments. J. Exp. Bot. 64 (14), 4461–4478. 10.1093/jxb/ert260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Moraga A., Rambla J. L., Fernández-de-Carmen A., Trapero-Mozos A., Ahrazem O., Orzáez D., et al. (2014). New target carotenoids for CCD4 enzymes are revealed with the characterization of a novel stress-induced carotenoid cleavage dioxygenase gene from Crocus sativus . Plant Mol. Biol. 86 (4–5), 555–569. 10.1007/s11103-014-0250-5 [DOI] [PubMed] [Google Scholar]
- Ruiz-Sola M.Á., Rodríguez-Concepción M. (2012). Carotenoid biosynthesis in Arabidopsis: a colorful pathway. Arabidopsis Book 10, e0158. 10.1199/tab.0158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruyter-Spira C., Kohlen W., Charnikhova T., van Zeijl A., van Bezouwen L., de Ruijter N., et al. (2011). Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: another belowground role for strigolactones? Plant Physiol. 155 (2), 721–734. 10.1104/pp.110.166645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka K., Ljung K., Vanneste S., Podhorská R., Beeckman T., Friml J., et al. (2007). Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19 (7), 2197–2212. 10.1105/tpc.107.052126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed W., Naseem S., Ali Z. (2017). Strigolactones biosynthesis and their role in abiotic stress resilience in plants: a critical review. Front. Plant Sci. 8, 1487. 10.3389/fpls.2017.01487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saritas Y., Sonwa M. M., Iznaguen H., König W. A., Muhle H., Mues R. (2001). Volatile constituents in mosses (Musci). Phytochemistry 57 (3), 443–457. 10.1016/S0031-9422(01)00069-3 [DOI] [PubMed] [Google Scholar]
- Schwartz S. H., Qin X., Zeevaart J. A. (2001). Characterization of a novel carotenoid cleavage dioxygenase from plants. J. Biol. Chem. 276 (27), 25208–25211. 10.1074/jbc.M102146200 [DOI] [PubMed] [Google Scholar]
- Seo M., Koiwai H., Akaba S., Komano T., Oritani T., Kamiya Y., et al. (2000). Abscisic aldehyde oxidase in leaves of Arabidopsis thaliana. Plant J. 23 (4), 481–488. 10.1046/j.1365-313x.2000.00812.x [DOI] [PubMed] [Google Scholar]
- Shabek N., Ticchiarelli F., Mao H., Hinds T. R., Leyser O., Zheng N. (2018). Structural plasticity of D3–D14 ubiquitin ligase in strigolactone signalling. Nature 563 (7733), 652. 10.1038/s41586-018-0743-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp R. E., LeNoble M. E. (2002). ABA, ethylene and the control of shoot and root growth under water stress. J. Exp. Bot. 53 (366), 33–37. 10.1093/jxb/53.366.33 [DOI] [PubMed] [Google Scholar]
- Shumbe L., D’Alessandro S., Shao N., Chevalier A., Ksas B., Bock R., et al. (2017). Methylene blue sensitivity 1 (MBS1) is required for acclimation of Arabidopsis to singlet oxygen and acts downstream of β-cyclocitral. Plant Cell Environ. 40 (2), 216–226. 10.1111/pce.12856 [DOI] [PubMed] [Google Scholar]
- Simons J. L., Napoli C. A., Janssen B. J., Plummer K. M., Snowden K. C. (2007). Analysis of the decreased apical dominance genes of petunia in the control of axillary branching. Plant Physiol. 143 (2), 697–706. 10.1104/pp.106.087957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden K. C., Simkin A. J., Janssen B. J., Templeton K. R., Loucas H. M., Simons J. L., et al. (2005). The decreased apical dominance1/Petunia hybrida carotenoid cleavage dioxygenase8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. Plant Cell 17 (3), 746–759. 10.1105/tpc.104.027714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorefan K., Booker J., Haurogné K., Goussot M., Bainbridge K., Foo E., et al. (2003). MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev. 17 (12), 1469–1474. 10.1101/gad.256603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soundappan I., Bennett T., Morffy N., Liang Y., Stanga J. P., Abbas A., et al. (2015). SMAX1-LIKE/D53 family members enable distinct MAX2-dependent responses to strigolactones and karrikins in Arabidopsis. Plant Cell 27 (11), 3143–3159. 10.1105/tpc.15.00562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg P., Furner I. J., Ottoline Leyser H. (2007). MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J. 50 (1), 80–94. 10.1111/j.1365-313X.2007.03032.x [DOI] [PubMed] [Google Scholar]
- Sun H., Tao J., Liu S., Huang S., Chen S., Xie X., et al. (2014). Strigolactones are involved in phosphate-and nitrate-deficiency–induced root development and auxin transport in rice. J. Exp. Bot. 65 (22), 6735–6746. 10.1093/jxb/eru029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Matsuura T., Kawakami N., Noda K. (2000). Accumulation and leakage of abscisic acid during embryo development and seed dormancy in wheat. Plant Growth Regul. 30 (3), 253–260. 10.1023/A:1006320614530 [DOI] [Google Scholar]
- Tan B. C., Joseph L. M., Deng W. T., Liu L., Li Q. B., Cline K., et al. (2003). Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 35 (1), 44–56. 10.1046/j.1365-313X.2003.01786.x [DOI] [PubMed] [Google Scholar]
- Tewari G., Mohan B., Kishor K., Tewari L. M., Nailwal° T. K. (2015). Volatile constituents of Ginkgo biloba L. leaves from Kumaun. J. Indian Chem. Soc. 92, 1583–1586. 10.1080/0972060X.2014.958567 [DOI] [Google Scholar]
- Tieman D., Bliss P., McIntyre L. M., Blandon-Ubeda A., Bies D., Odabasi A. Z., et al. (2012). The chemical interactions underlying tomato flavor preferences. Curr. Biol. 22 (11), 1035–1039. 10.1016/j.cub.2012.04.016 [DOI] [PubMed] [Google Scholar]
- Todaka D., Shinozaki K., Yamaguchi-Shinozaki K. (2015). Recent advances in the dissection of drought-stress regulatory networks and strategies for development of drought-tolerant transgenic rice plants. Front. Plant Sci. 6, 84. 10.3389/fpls.2015.00084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuan P. A., Kumar R., Rehal P. K., Toora P. K., Ayele B. T. (2018). Molecular mechanisms underlying abscisic acid/gibberellin balance in the control of seed dormancy and germination in cereals. Front. Plant Sci. 9, 668. 10.3389/fpls.2018.00668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H., Kusaba M. (2015). Strigolactone regulates leaf senescence in concert with ethylene in Arabidopsis. Plant Physiol. 169 (1), 138–147. 10.1104/pp.15.00325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J. T., Tan B.-C., McCarty D. R., Klee H. J. (2008). The carotenoid cleavage dioxygenase 1 enzyme has broad substrate specificity, cleaving multiple carotenoids at two different bond positions. J. Biol. Chem. 283 (17), 11364–11373. 10.1074/jbc.M710106200 [DOI] [PubMed] [Google Scholar]
- Walter M. H., Strack D. (2011). Carotenoids and their cleavage products: biosynthesis and functions. Nat. Prod. Rep. 28 (4), 663–692. 10.1039/c0np00036a [DOI] [PubMed] [Google Scholar]
- Wang J. Y., Haider I., Jamil M., Fiorilli V., Saito Y., Mi J., et al. (2019). The apocarotenoid metabolite zaxinone regulates growth and strigolactone biosynthesis in rice. Nat. Commun. 10 (1), 810. 10.1038/s41467-019-08461-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Li C., Wu Z., Jia Y., Wang H., Sun S., et al. (2017). Abscisic acid regulates auxin homeostasis in rice root tips to promote root hair elongation. Front. Plant Sci. 8, 1121. 10.3389/fpls.2017.01121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Bouwmeester H. J. (2018). Structural diversity in the strigolactones. J. Exp. Bot. 69 (9), 2219–2230. 10.1093/jxb/ery091 [DOI] [PubMed] [Google Scholar]
- Watanabe S., Sato M., Sawada Y., Tanaka M., Matsui A., Kanno Y., et al. (2018). Arabidopsis molybdenum cofactor sulfurase ABA3 contributes to anthocyanin accumulation and oxidative stress tolerance in ABA-dependent and independent ways. Sci. Rep. 8 (1), 16592. 10.1038/s41598-018-34862-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters M. T., Brewer P. B., Bussell J. D., Smith S. M., Beveridge C. A. (2012). The Arabidopsis ortholog of rice DWARF27 acts upstream of MAX1 in the control of plant development by strigolactones. Plant Physiol. 159 (3), 1073–1085. 10.1104/pp.112.196253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters M. T., Gutjahr C., Bennett T., Nelson D. C. (2017). Strigolactone signaling and evolution. Annu. Rev. Plant Biol. 68, 291–322. 10.1146/annurev-arplant-042916-040925 [DOI] [PubMed] [Google Scholar]
- Waters M. T., Scaffidi A., Moulin S. L., Sun Y. K., Flematti G. R., Smith S. M. (2015). A Selaginella moellendorffii ortholog of karrikin insensitive2 functions in Arabidopsis development but cannot mediate responses to karrikins or strigolactones. Plant Cell 27 (7), 1925–1944. 10.1105/tpc.15.00146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng J.-K., Ye M., Li B., Noel J. P. (2016). Co-evolution of hormone metabolism and signaling networks expands plant adaptive plasticity. Cell 166 (4), 881–893. 10.1016/j.cell.2016.06.027 [DOI] [PubMed] [Google Scholar]
- Xie X., Yoneyama K., Yoneyama K. (2010). The strigolactone story. Annu. Rev. Phytopathol. 48, 93–117. 10.1146/annurev-phyto-073009-114453 [DOI] [PubMed] [Google Scholar]
- Xie Z., Nolan T. M., Jiang H., Yin Y. (2019). AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis . Front. Plant Sci. 10, p228. 10.3389/fpls.2019.00228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z.-Y., Lee K. H., Dong T., Jeong J. C., Jin J. B., Kanno Y., et al. (2012). A vacuolar β-glucosidase homolog that possesses glucose-conjugated abscisic acid hydrolyzing activity plays an important role in osmotic stress responses in Arabidopsis. Plant Cell 24 (5), 2184–2199. 10.1105/tpc.112.095935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H., Saika H., Maekawa M., Takamure I., Tsutsumi N., Kyozuka J. et al. (2007). Rice tillering dwarf mutant dwarf3 has increased leaf longevity during darkness-induced senescence or hydrogen peroxide-induced cell death. Genes Genet. Syst. 82(4), 361–366. [DOI] [PubMed] [Google Scholar]
- Yan J., Wang P., Wang B., Hsu C.-C., Tang K., Zhang H., et al. (2017). The SnRK2 kinases modulate miRNA accumulation in Arabidopsis. PLoS Genet. 13 (4), e1006753. 10.1371/journal.pgen.1006753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama K., Xie X., Kim H. I., Kisugi T., Nomura T., Sekimoto H., et al. (2012). How do nitrogen and phosphorus deficiencies affect strigolactone production and exudation? Planta 235 (6), 1197–1207. 10.1007/s00425-011-1568-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama K., Mori N., Sato T., Yoda A., Xie X., Okamoto M., et al. (2018). Conversion of carlactone to carlactonoic acid is a conserved function of MAX 1 homologs in strigolactone biosynthesis. New Phytol. 218 (4), 1522–1533. 10.1111/nph.15055 [DOI] [PubMed] [Google Scholar]
- Yoneyama K., Xie X., Yoneyama K., Nomura T., Takahashi I., Asami T., et al. (2019). Regulation of biosynthesis, perception, and functions of strigolactones for promoting arbuscular mycorrhizal symbiosis and managing root parasitic weeds. Pest Manag. Sci. 75 (9), 2353–2339. 10.1002/ps.5401 [DOI] [PubMed] [Google Scholar]
- Zeng D. E., Hou P., Xiao F., Liu Y. (2014). Overexpressing a novel RING-H2 finger protein gene, OsRHP1, enhances drought and salt tolerance in rice (Oryza sativa L.). J. Plant Biol. 57 (6), 357–365. 10.1007/s12374-013-0481-z [DOI] [Google Scholar]
- Zeng D. E., Hou P., Xiao F., Liu Y. (2015). Overexpression of Arabidopsis XERICO gene confers enhanced drought and salt stress tolerance in rice (Oryza sativa L.). J. Plant Biochem. Biotechnol. 24 (1), 56–64. 10.1007/s13562-013-0236-4 [DOI] [Google Scholar]
- Zhang Y., Van Dijk A. D., Scaffidi A., Flematti G. R., Hofmann M., Charnikhova T., et al. (2014). Rice cytochrome P450 MAX1 homologs catalyze distinct steps in strigolactone biosynthesis. Nat. Chem. Biol. 10 (12), 1028. 10.1038/nchembio.1660 [DOI] [PubMed] [Google Scholar]
- Zhou G., Xu D., Xu D., Zhang M. (2013). Southern rice black-streaked dwarf virus: a white-backed planthopper-transmitted fijivirus threatening rice production in Asia. Front. Microbiol. 4, 270. 10.3389/fmicb.2013.00270 [DOI] [PMC free article] [PubMed] [Google Scholar]