Abstract
A most interesting exception within the parasitic Apicomplexa is Nephromyces, an extracellular, probably mutualistic, endosymbiont found living inside molgulid ascidian tunicates (i.e., sea squirts). Even though Nephromyces is now known to be an apicomplexan, many other questions about its nature remain unanswered. To gain further insights into the biology and evolutionary history of this unusual apicomplexan, we aimed to 1) find the precise phylogenetic position of Nephromyces within the Apicomplexa, 2) search for the apicoplast genome of Nephromyces, and 3) infer the major metabolic pathways in the apicoplast of Nephromyces. To do this, we sequenced a metagenome and a metatranscriptome from the molgulid renal sac, the specialized habitat where Nephromyces thrives. Our phylogenetic analyses of conserved nucleus-encoded genes robustly suggest that Nephromyces is a novel lineage sister to the Hematozoa, which comprises both the Haemosporidia (e.g., Plasmodium) and the Piroplasmida (e.g., Babesia and Theileria). Furthermore, a survey of the renal sac metagenome revealed 13 small contigs that closely resemble the genomes of the nonphotosynthetic reduced plastids, or apicoplasts, of other apicomplexans. We show that these apicoplast genomes correspond to a diverse set of most closely related but genetically divergent Nephromyces lineages that co-inhabit a single tunicate host. In addition, the apicoplast of Nephromyces appears to have retained all biosynthetic pathways inferred to have been ancestral to parasitic apicomplexans. Our results shed light on the evolutionary history of the only probably mutualistic apicomplexan known, Nephromyces, and provide context for a better understanding of its life style and intricate symbiosis.
Keywords: sporozoa, endosymbiont, mutualism, plastome, co-infection, Nephromycida, Hematozoa, Lankesteria, gregarine
Introduction
Nephromyces is an endosymbiont found exclusively in molgulid ascidian tunicates (also known as sea squirts or sea grapes). Thus far, Nephromyces has been found in every adult molgulid individual sampled (i.e., there is a 100% infection rate in natural populations; Saffo 1981, 1982, 1991). Inside its molgulid hosts, Nephromyces inhabits the voluminous lumen of the so-called “renal sac,” a ductless, urate- and calcium oxalate-rich organ of uncertain function (Saffo 1978; Saffo and Lowenstam 1978; Saffo 1981). Inside the renal sac, Nephromyces appears to degrade abundant urate as a carbon and nitrogen source thanks to the conservation of peroxisomes and a highly expressed urate oxidase (Saffo 1991; Paight et al. 2019). The identity of the probably mutualistic Nephromyces, which appears to be the only eukaryotic symbiont within the renal sac, remained a phylogenetic enigma for more than a century since its discovery (Saffo 1981; Saffo et al. 2010). Nephromyces was first thought to be a community of microbes that included gregarines (de Lacaze-Duthiers 1874). Giard later thought these endosymbionts to be chytridiomycetes and called them “Nephromyces” (Giard 1888; Harant 1931). Subsequent biologists considered Nephromyces to be an unaffiliated “lower” fungus (Buchner 1930, 1965) or simply an eukaryote of unknown taxonomic affinities (incertae sedis; Cavalier-Smith 2003). Nephromyces was even placed next to the flagellate Telonema in the Opalozoa (Cavalier-Smith 1993a, 1993b). Such taxonomic confusion is not surprising given its unusual morphology and habitat, and complex life cycle (Saffo 1981; Saffo and Nelson 1983; Saffo 1991; Saffo et al. 2010).
Nephromyces, however, has ultimately been found to be an apicomplexan (Saffo et al. 2010). More precisely though, Nephromyces is a sporozoan; the Apicomplexa comprises the free-living Apicomonada (i.e., “chrompodellids”) and the ancestrally parasitic Sporozoa (e.g., gregarines, coccidians, and hematozoans) (Cavalier-Smith 2018; Muñoz-Gómez and Slamovits 2018). Phylogenetic trees inferred using the nuclear SSU rRNA gene (also called 18S “rDNA”) suggested that the closest relatives of Nephromyces are apicomplexans isolated from other, nonmolgulid ascidians: Cardiosporidium cionae, a parasite of the ascidian Ciona instestinalis, and an environmental sequence associated with the ascidian Halocynthia roretzi (Saffo et al. 2010). Saffo et al. (2010) suggested that the novel Nephromyces-Cardiosporidium clade was itself sister to piroplasmids (e.g., Babesia and Theileria); however, the precise phylogenetic placement of Nephromyces and Cardiosporidium remained unclear (Saffo et al. 2010). This uncertainty arose from the fact that 1) haemosporidians (e.g., Plasmodium) were excluded from the phylogenetic analyses due to their high degree of divergence (i.e., long branches), 2) statistical branch support for the clade uniting Nephromyces and allies with the piroplasmids was poor or barely moderate (0.89 posterior probability [PP] and 71% bootstrap support), and 3) the phylogenetic analysis relied on a single gene (nuclear SSU rRNA gene). More recently, Cavalier-Smith has inferred nuclear SSU rRNA gene trees using site-heterogeneous models and a broad taxon sampling (Cavalier-Smith 2014). He concluded that the Nephromycida (i.e., a taxon formally established by Cavalier-Smith in 1993 for Nephromyces and its close relatives) is sister to the Piroplasmida. However, support for the sisterhood of the Nephromycida and Piroplasmida was only low or moderate, and the Haemosporidia unexpectedly appeared to be unrelated to both the Nephromycida and the Piroplasmida (Cavalier-Smith 2014). The precise phylogenetic placement of Nephromyces, and the Nephromycida, thus remains to be determined.
Genomic and transcriptomic surveys of Nephromyces could shed light not only on the evolutionary history of this unusual apicomplexan, but also on its metabolism, symbiosis, life style, and general ecology (e.g., Paight et al. 2019). To gain further insights into the evolutionary history of the Nephromycida, and the Apicomplexa more generally, we have sequenced both a Nephromyces-enriched metagenome and metatranscriptome from the contents of the renal sac of two different single individuals of Molgula occidentalis (one of the host species for Nephromyces). Our diverse multi-gene phylogenetic analyses robustly show that the Nephromycida is sister to both the Haemospiridia and the Piroplasmida, or the Hematozoa. We also show that the apicoplast genome and metabolism of Nephromyces closely resembles those of other derived apicomplexans. On the basis of the finding of several diverse apicoplast genomes and nucleus-encoded gene orthologues, we argue that a disparate set of genetically divergent but most closely related lineages of Nephromyces can co-inhabit the renal sac of a single tunicate host.
Materials and Methods
Metagenome and Metatranscriptome Sequencing
Individuals of M. occidentalis were obtained from the Gulf Specimen Marine Laboratories, Inc. (http://www.gulfspecimen.org/; last accessed July 25, 2019) and dissected to remove their renal sacs whose contents were emptied into 2 ml tubes that were then quickly frozen in liquid nitrogen. Total DNA was isolated from the inside contents of a dissected renal sac of a single individual of M. occidentalis using CTAB/chloroform extraction. A sequencing library was made using the PrepX DNA Library Kit (Takara Bio, USA). The DNA library was sequenced (2 × 250 bp) in an Illumina MiSeq instrument (University of Rhode Island). Total RNA was isolated from the inside contents (which were mixed with RNAlater reagent; ThermoFisher Scientific) of a dissected renal sac of another single individual of M. occidentalis using the TRIzol reagent (ThermoFisher Scientific). A cDNA sequencing library was made using the TruSeq RNA Sample Prep Kit v2 (Illumina). The cDNA library was sequenced (2 × 300 bp) in a local Illumina MiSeq instrument (Centre for Comparative Genomic and Evolutionary Bioinformatics, Dalhousie University).
Metagenomic and Metatranscriptomic Analyses
Quality assessments of the libraries were done with FastQC v0.11.7. The Illumina short reads from the two sequenced DNA libraries were processed with Trimmomatic v0.32 (Bolger et al. 2014). Quality-filtered reads (43.99 GB) were assembled into a metagenome with SPAdes v3.11.1 (Bankevich et al. 2012). The assembled contigs were then processed with the Anvi’o v4 metagenomic workflow (Eren et al. 2015). In brief, the Anvi’o metagenomic workflow allowed us to map Illumina short reads to contigs with Bowtie2 to obtain coverage values, assign the taxonomy to each contig based on DIAMOND searches of predicted contig genes, and identify contigs in which a custom collection of 29 HMM profiles for apicoplast genes were overrepresented. Anvi-interactive super-imposed all these layers of information on the metagenome contigs which were later visually inspected to retrieve a candidate set of 13 contigs that most likely corresponded to apicoplast genomes. Each of the 13 Nephromyces lineages represented by an apicoplast contig was named after the contig number in the metagenome assembly. Apicoplast contigs were closed (circularized) with NOVOPlasty v2.6.2 (Dierckxsens et al. 2017) using divergent seeds and a closed reference; the two apicoplast genomes (Nephromyces 638 and 654) that have the lowest abundances in the renal sac metagenome are incomplete by missing either one or two of the rRNA-gene operons and could also not be circularized by NOVOPlasty. The apicoplast genomes identified were manually annotated in Artemis. ORFs larger than 50 amino acids were predicted, and BLAST searches against the NCBI GenBank non-redundant (nr) database were performed. tRNA genes were predicted using tRNA-scan-SE v2.0 using the “Mold and Protozoan Mito” genetic code and both the “Eukaryotic” and “Bacterial” mode. rRNA genes were annotated based on predictions made by RNAmmer v1.2 using the “Bacteria” setting.
Quality assessments of the cDNA libraries were done with FastQC v0.11.7. The Illumina short reads from the two sequenced cDNA libraries were processed with Trimmomatic v0.32. Quality-filtered reads (6.92 GB) were assembled into a metatranscriptome with Trinity v2.6.6 (Haas et al. 2013). A proteome was predicted from the assembled metatranscriptome with TransDecoder v5.0.2 (Haas et al. 2013), and annotated with Trinotate v3.1.1. The completeness of the metatranscriptome was assessed with BUSCO v3.0.2 using the eukaryote marker gene set. Reciprocal BLAST searches were done to find orthologs of plastid-derived enzymes against the Nephromyces-enriched metatranscriptome using Toxoplasma gondii and Plasmodium falciparum queries. BLAST searches against the NCBI GenBank nr database using the candidate orthologs from Nephromyces were also done to corroborate their apicomplexan affiliation. For the UROS enzyme, HMMER searches were done using the HEM4 Pfam profile HMM (F02602) because of the high degree of divergence observed among apicomplexans. BLAST searches against a local database that comprises 97 eukaryotes and 85 bacteria were done to retrieve homologs for each plastid-derived enzyme. The homologs for each plastid-derived enzyme were aligned with MAFFT v7.310 (L-INS-i) (Katoh et al. 2005), trimmed with trimAl v1.4.rev15 (-automated1) (Capella-Gutiérrez et al. 2009), and their trees were then inferred with IQ-TREE v1.6.5 and the LG4X model (Nguyen et al. 2015; Hoang et al. 2018). Signal peptides of plastid-targeted enzymes were predicted with SignalP v4.1 and TargetP v1.1. Transit peptides were predicted with ChloroP v1.1 after removing the signal peptides predicted by either SignalP v4.1 or TargetP v1.1 with a custom Perl script. FASI-like transcripts of Nephromyces were identified by BLASTp searches using T.gondii’s FASI as a query and then annotated with the NCBI Conserved Domain Search.
Phylogenetic Analyses
A data set comprising 90 conserved eukaryotic proteins from Janouškovec et al. (2015) (which was itself a subset of Burki et al. 2012) was used (see supplementary table S1, Supplementary Material online for a functional annotation of these genes). This data set was chosen because it included the largest sampling available for apicomonad species, or also known as “chrompodellids,” and because it minimized missing data for these taxa. Several species were added to this data set, including Nephromyces endosymbiont of M. occidentalis, the gregarines Lankesteria abbotti, Gregarina niphandrodes, and Ascogregarina taiwanensis. Some sporozoan species, found in NCBI Genbank and EuPathDB, were added to the data set to increase sampling within the Haemosporidia, Piroplasmida and Coccidia (see supplementary table S2, Supplementary Material online for data sources). To add the new taxa, T.gondii genes were used as queries to search our database. All best hits with an E-value lower than 1E-10 were kept and added to the Janouškovec et al. (2015) unaligned and untrimmed gene sets. E-values and scores were appended to each sequence name so to choose orthologs in the single gene trees much more easily. Single gene files were aligned with MAFFT v7.310 (L-INS-i) (Katoh et al. 2005) and their corresponding trees were later inferred with IQ-TREE v1.6.5 (Nguyen et al. 2015; Hoang et al. 2018) and the LG4X model. Paralogs, pseudoparalogs and extremely divergent orthologs were manually removed after inspecting the single gene trees. A diverse set of apicomplexan orthologs most closely related to each other (i.e., a clade) were found for each gene in the Nephromyces-enriched renal sac metatranscriptome. We opted for reproducibly selecting the least divergent, or the shortest-branching, ortholog that also had a comparatively high BLAST score, or low E-value, within the apicomplexan clade as a representative sequence for Nephromyces. This constitutes the best compromise in the absence of a single-cell or single-species genome. Final single gene files comprising only orthologs were aligned with MAFFT v7.310 (L-INS-i) (Katoh et al. 2005), trimmed with trimAl v1.4.rev15 (-automated1) (Capella-Gutiérrez et al. 2009) and concatenated with SequenceMatrix (Vaidya et al. 2011). The data set was filtered using PhyloMCOA (de Vienne et al. 2012) to remove outlier sequences (distance = “nodal”; bvalue = 70); no taxon or gene outliers were detected by PhyloMCOA. The final data set comprised 45 taxa and 90 nucleus-encoded genes (24,754 amino acid sites; see supplementary table S2, Supplementary Material online for the percentage of missing data per taxon). Another data set comprising 35 apicoplast-encoded genes and a set of 56 taxa (6,403 amino acid sites) was assembled based on our own apicoplast genome reannotations. All apicomplexans for which complete or partial plastids had been sequenced as of June 2018 were included. Single gene files were aligned with MAFFT-homologs v7.310 (Katoh et al. 2005), trimmed with trimAl v1.4.rev15 (-automated1) (Capella-Gutiérrez et al. 2009) and concatenated with SequenceMatrix (Vaidya et al. 2011).
Phylogenetic analyses were performed under the maximum likelihood and Bayesian frameworks. Maximum likelihood analyses were done in IQ-TREE v1.6.5 (Nguyen et al. 2015; Hoang et al. 2018) using the LG+PMSF(C60)+F+G4 model which depended on a guide tree inferred with the LG+C60+F+G4 model (Wang et al. 2018). This model best fits the data according to the Akaike information criterion (AIC), the corrected Akaike information criterion (AICc), and the Bayesian information criterion (BIC) (see supplementary table S3, Supplementary Material online). Bayesian analyses were done in PhyloBayes MPI v1.7 using the CAT-GTR+G4 model (Lartillot and Philippe 2004; Lartillot et al. 2013). PhyloBayes MCMC chains were run for ∼11,000 cycles until convergence between the chains was achieved and the largest discrepancy was <0.1 (0.02). Sample trees were summarized into a Bayesian consensus tree from two PhyloBayes MCMC chains using a burn-in of 500 trees and subsampling every 10 trees (2,116 trees). The data set was recoded into the four-character state scheme SR4 to minimize saturation (Susko and Roger 2007). The recoded data set was analyzed in IQTREE v1.6.5 using the GTR+C60(SR4)+F+G4 model, and in PhyloBayes MPI v1.7 using the CAT-GTR+G4 model. The C60(SR4) model is an adaptation of the empirical mixture C60 model to four-character states; it is obtained by adding the frequencies of all amino acids that belong to each bin in the four-character state scheme SR4. Site removal based on site rates (-wsr option in IQTREE) was done using the script SiteStripper v1.03 (Verbruggen 2018).
Results and Discussion
Nephromyces Represents a Novel Apicomplexan Lineage that is Probably Sister to the Hematozoa
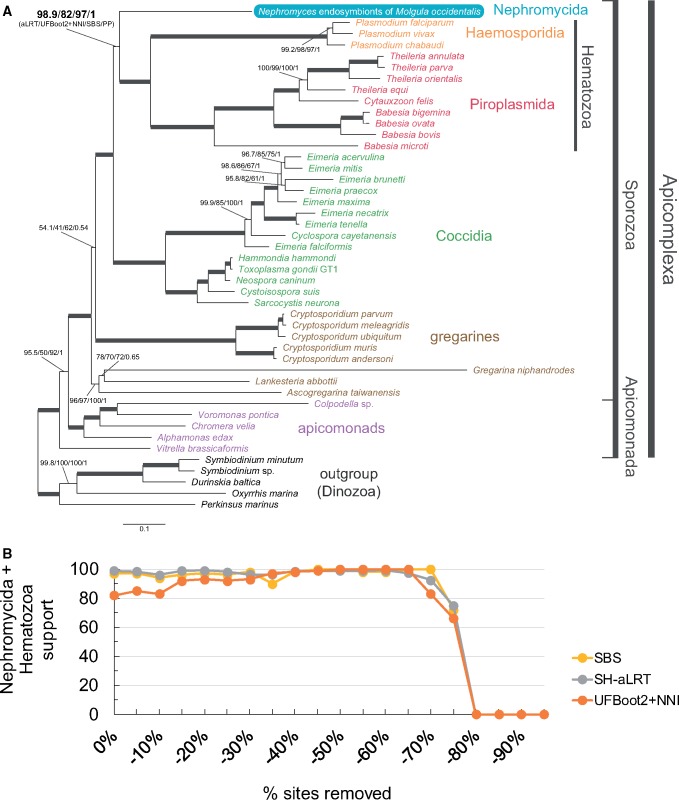
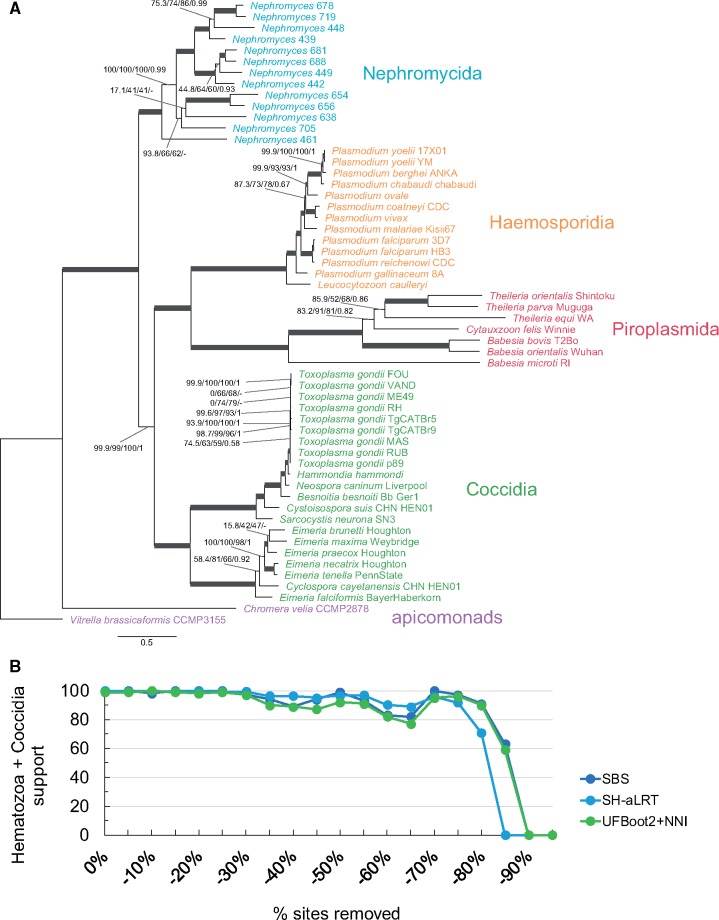
In an attempt to robustly resolve the placement of Nephromyces in the Apicomplexa tree, we sequenced a cDNA library (called a metatranscriptome; see below) from the renal sac of M. occidentalis. We expanded the data set used by Janouškovec et al. (2015), that comprises 90 nucleus-encoded conserved genes (24,754 amino acid sites; see supplementary table S1, Supplementary Material online), by including Nephromyces and three gregarines, and some other sporozoans (45 taxa in total; see Materials and Methods and supplementary table S2, Supplementary Material online). We then performed phylogenetic analyses using the most sophisticated and best-fitting site-heterogeneous models currently available, namely LG+PMSF(C60)+F+G4 in IQ-TREE (Nguyen et al. 2015; Wang et al. 2018) and CAT-GTR+G4 in PhyloBayes (Lartillot and Philippe 2004; Lartillot et al. 2013), both of which are capable of accounting for compositional heterogeneity across sites. Both a maximum-likelihood and a Bayesian consensus tree indicate that a community of Nephromyces lineages isolated from M. occidentalis is sister to both the Piroplasmida and the Haemosporidia (collectively called the Hematozoa; fig. 1A). This phylogenetic placement for the Nephromycida receives high statistical branch support (98.9% Shimodaira-Hasegawa approximate Likelihood Ratio Test, SH-aLRT; 82% UltraFast Bootstrap 2 with NNI optimization, UFBoot2+NNI; 97% nonparametric Standard BootStrap, SBS; and 1.0 PP; see fig. 1A), and is robust across different analyses (e.g., fig. 1 and supplementary figs. S1–S3, Supplementary Material online). For example, phylogenetic trees inferred from simpler site-homogeneous models also support the sister relationship between the Nephromycida and the Hematozoa (see supplementary figs. S1 and S2, Supplementary Material online). Moreover, support for the sister relationship between the Nephromycida and the Hematozoa remains high and reaches the highest support (SBS, SH-aLRT, UFBoot2+NNI) after 45–65% of the fastest-evolving sites are removed under both simpler site-homogeneous and more complex site-heterogeneous models (supplementary fig. S1B, Supplementary Material online and fig. 1B, respectively). Similarly, when the data set is recoded into the SR4 four-character state alphabet to minimize saturation (Susko and Roger 2007), the Nephromycida remains sister to the Hematozoa (supplementary fig. S3, Supplementary Material online). These results confirm that Nephromyces represents a novel lineage within the Sporozoa (fig. 1 and supplementary figs. S1–S3, Supplementary Material online), are in agreement with the SSU rRNA gene phylograms of Saffo et al. (2010), and suggest that future taxonomic schemes for the Apicomplexa should elevate the Nephromycida to a higher rank (contraCavalier-Smith 2014). Future efforts to include single isolates of Nephromyces and its sister group, C.cionae, will further test the phylogenetic sisterhood of the Nephromycida and the Hematozoa.
Fig. 1.
—Nephromyces, and the Nephromycida, branch within the Apicomplexa as sister to the Hematozoa. Nephromyces represents a chimeric taxon assembled from several most closely related lineages found inside the renal sac of a single Molgula occidentalis host. (A) A maximum-likelihood tree for the Apicomplexa inferred by IQ-TREE using the site-heterogeneous LG+PMSF(C60)+F+G4 model on 90 nucleus-encoded genes (24,754 amino acid sites). Branch support values are 100% SH-aLRT, 100% UFBoot2+NNI, 100% SBS, and 1.0 PP unless annotated. (B) Progressive removal of the fastest-evolving sites in increments of 5% and variation of SBS, SH-aLRT, and UFBoot2+NNI branch support values under the site-heterogeneous LG+PMSF(C60)+F+G4 model for the branch that united the Nephromycida and the Hematozoa.
There are two controversial deep branches across our analyses. First is the branch that unites the Cryptosporidium clade with all other sporozoan groups to the exclusion of the other three gregarines (fig. 1A and supplementary figs. S2 and S3, Supplementary Material online). The sister relationship between Cryptosporidium and all other sporozoans tends to increase on average, although inconsistently, as the fastest-evolving sites are removed but is lost after 55% of these have been removed (supplementary fig. S4, Supplementary Material online). Even though Cryptosporidium is known to be most closely related to gregarines based on SSU rRNA gene trees (e.g., Cavalier-Smith 2014; Simdyanov et al. 2017), ultrastructure, and life history (Aldeyarbi and Karanis 2016; Ryan et al. 2016; Thompson et al. 2016), our study does not contradict this hypothesis (i.e., support for the branch that unites Cryptosporidium with all other sporozoans is not consistently high across analyses). Second is the branch that unites apicomonads with all other apicomplexans to the exclusion of Vitrella brassicaformis (fig. 1 and supplementary figs. S2 and S3, Supplementary Material online). Janouškovec et al., (2015), who used the same multi-gene set but fewer taxa, recovered the monophyly of apicomonads. Otherwise, SSU rRNA gene trees are ambiguous in regard to the monophyly of apicomonads (Cavalier-Smith 2014; Janouškovec et al. 2015; Kwong et al. 2019). Our analyses with simpler evolutionary models (e.g., LG4X+F) recover apicomonads as monophyletic (supplementary fig. S1A, Supplementary Material online), but more sophisticated models (e.g., LG+PMSF(C60)+F+G4 and Bayesian CAT-GTR+G4 on the untreated data set and GTR+C60(SR4)+F+G4 on the SR4-recoded data set; see fig. 1 and supplementary fig. S3, Supplementary Material online, respectively) recover apicomonads as paraphyletic (fig. 1 and supplementary fig. S3, Supplementary Material online). Furthermore, the sister relationship between V.brassicaformis and all other apicomplexans (i.e., apicomonad paraphyly) becomes nearly fully supported after 10% of the fastest-evolving sites have been removed (supplementary fig. S4, Supplementary Material online). The monophyly of apicomonads (as well as that of both gregarines and Cryptosporidium) is lost when fast-evolving sites are also removed under the simpler site-homogeneous LG4X+F model (see supplementary fig. S5, Supplementary Material online). This suggests that the monophyly of apicomonads (and perhaps also that of gregarines and Cryptosporidium) might be artefactual and result from long-branch attraction (e.g., supplementary fig. S1A, Supplementary Material online). Our diverse analyses have therefore uncovered conflicting signals for both the monophyly of apicomonads and gregarines sensu lato (i.e., including Cryptosporidium).
The apicomplexan origin of Nephromyces helps explain some of its features, such as sporozoite and spore stages, tubular mitochondrial cristae, micropores in the filamentous trophozoites, and rhoptries or microneme vesicles in the sporozoites (Saffo 1990, 1991; Saffo et al. 2010). The predominantly extracellular life cycle of Nephromyces inside a marine invertebrate, and the gigantism of its purely extracellular filamentous trophozoites (trophic stage; up to 500 µm in length; Saffo and Nelson 1983) are reminiscent of some gregarines (most gregarines are ambicellular though; Cavalier-Smith 2014, and see Leander 2008). However, Nephromyces appears to have a more derived origin within the Sporozoa (fig. 1 and supplementary figs. S1–S3, Supplementary Material online). Our trees show that Nephromyces is phylogenetically embedded in a group of sporozoans (e.g., hematozoans and coccidians) that reproduce primarily intracellularly and do not have large extracellular trophic stages. Indeed, the sister group of Nephromyces, namely C.cionae, is an intracellular parasite of tunicate blood cells (i.e., haemocytes) and appears to not develop the conspicuous filamentous stages of Nephromyces (Ciancio et al. 2008). The above may suggest that Nephromyces convergently evolved extracellular gigantism when it invaded a novel and specialized habitat inside its host, the urate-rich renal sac—the colonization of the renal sac by Nephromyces might have been facilitated by its bacterial endosymbionts (Saffo 1990; Saffo et al. 2010), which could have freed Nephromyces from some its ancestral auxotrophies, as seen in some trypanosomatids (Harmer et al. 2018). Nephromyces may thus have followed the same convergent evolutionary path that some gregarines followed toward extracellular gigantism after having evolved from a presumed intracellular sporozoan cenancestor (Cavalier-Smith and Chao 2004; Cavalier-Smith 2014).
Nephromyces Possess an Apicoplast with a Reduced Genome
The derived phylogenetic placement of Nephromyces as sister to the Hematozoa would imply that Nephromyces has retained apicoplasts, as do hematozoans and coccidians. Indeed, organelles quite similar to apicoplasts (i.e., multimembraned vesicles) have been observed in micrographs of C.cionae, Nephromyces’ sister group (Ciancio et al. 2008). However, structures resembling apicoplasts have not yet been definitely identified in Nephromyces (Saffo 1990; Saffo et al. 2010). It is thus unknown whether Nephromyces has retained an apicoplast, or has lost it independently as it has happened in Cryptosporidium and presumably in G.niphandrodes (Zhu et al. 2000; Toso and Omoto 2007).
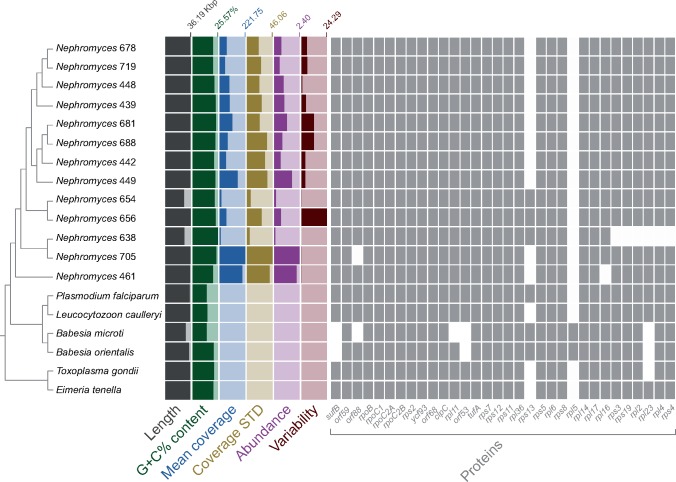
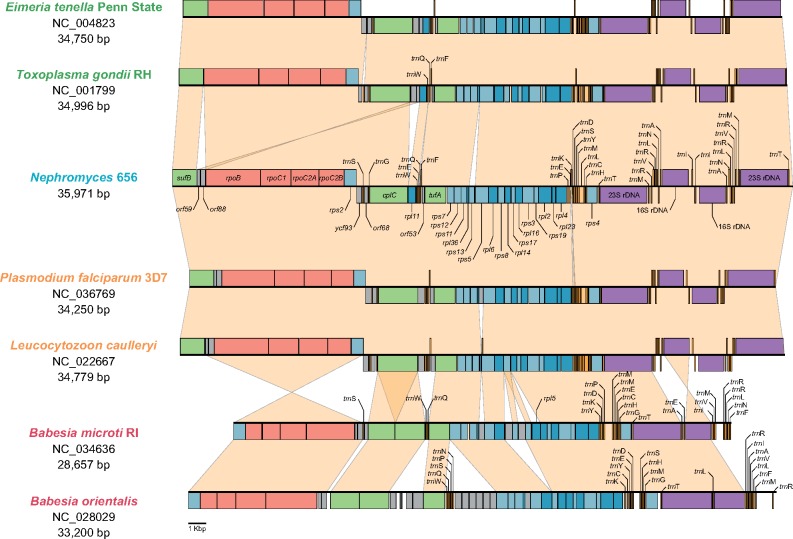
Our survey of the sequenced DNA library (i.e., metagenome) of the renal sac of the ascidian M. occidentalis revealed the presence of 13 contigs that resemble apicoplast genomes, each found at a different abundance (fig. 2, see also fig. 3 for a representative, and see Materials and Methods for further details). These contigs have sizes and G+C% contents that fall well within the range observed for apicoplast genomes (fig. 2; and see Muñoz-Gómez and Slamovits 2018). Within this range, however, the Nephromyces apicoplast genomes (named after their contig numbers) have slightly larger sizes and higher G+C% contents (averages of 35.62 kb and 22.16%, respectively; fig. 2; and see Muñoz-Gómez and Slamovits 2018 for comparative genomics of apicoplasts). These 13 apicoplast genomes are also very similar in terms of gene content and gene order (fig. 2 and supplementary fig. S6, Supplementary Material online) and do not differ considerably from the apicoplast genomes of haemosporidians and coccidians (fig. 4). A shared orf59-orf88 gene cluster (surrounded by sufB and rpoB) between Nephromyces and hematozoans support their monophyly as suggested by nuclues-gene trees. Moreover, all 13 of these genomes rely on a noncanonical genetic code where the UGA codon (the stop “opal” codon in the standard genetic code) invariably encodes tryptophan (see supplementary table S4, Supplementary Material online)—a feature also found in coccidians and in the apicomonad Chromera velia (Oborník and Lukeš 2015). Despite these similarities, these apicoplast genomes show differences among themselves, and also show greater diversity than apicoplast genomes within coccidians and haemosporidians. For example, the orf88 (rpl19) gene is absent from Nephromyces 705; rpl16 is absent from Nephromyces 461; the rps3-rps19-rpl2-rpl23-rpl4-rps4 gene cluster is absent from Nephromyces 638, and the rps13 gene is present only in Nephromyces 638, 654, and 656 (fig. 2 and supplementary fig. S6, Supplementary Material online). A phylogenetic tree inferred from 35 apicoplast protein-coding genes (6,403 amino acid sites) also shows that the apicoplast genomes are considerably divergent relative to each other (according to the estimated average number of amino acid replacements per site; see branch lengths in fig. 5); there is higher divergence among these 13 apicoplast genomes than that seen among species of Plasmodium or Toxoplasma (compare branch lengths in fig. 5). In summary, some features like 1) larger sizes, 2) higher G+C% contents, 3) a noncanonical genetic code, and 4) slower evolutionary rates (compare branch lengths in fig. 5), suggest some degree of stasis in the apicoplast genomes of Nephromyces relative to all other major sporozoan lineages.
Fig. 2.
—The Nephromyces clade (Nephromycida) is diverse and here comprises at least 13 distinct apicoplast genomes found in a metagenome derived from the renal sac of a single individual of the molgulid tunicate host, Molgula occidentalis. Each apicoplast genome is found at different abundances in the metagenome. The genomes show differences in gene content, G+C% content, and sequence divergence (see branch lengths in supplementary fig. S4A, Supplementary Material online). The guide cladogram is derived from the phylogram in supplementary figure S5, Supplementary Material online. The predicted genes orf59 and orf88 might be rps18 and rpl19, respectively, based on remote similarities to these genes in the GenBank nr database. Genome features were calculated in Anvi’o v4. The apicoplast genomes of Nephromyces 654 (26.24 kb) and 656 (26.78 kb) could not be circularized and appear as smaller than all others because they lack one or both rRNA-gene operons, respectively. Mean coverage: average depth of coverage across contig. Coverage STD: the standard deviation of coverage values for a given contig. Abundance: mean coverage of a contig divided by that sample’s overall mean coverage. Variability: number of reported single-nucleotide variants per kilo base pair.
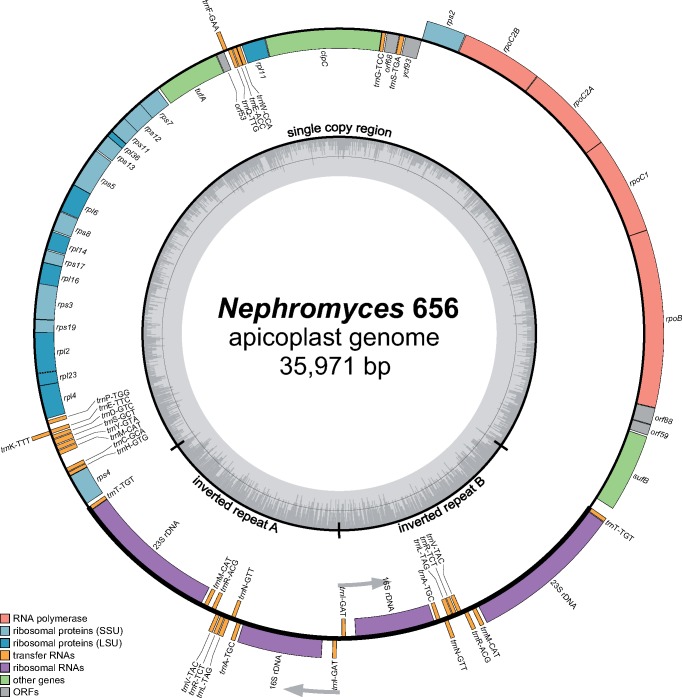
Fig. 3.
—The apicoplast genome of Nephromyces 656 as a representative of the Nephromicida. Nephromyces 656’s apicoplast genome has a size of 35,971 kb, encodes 31 protein-coding genes, two rRNA genes, and 24 tRNA genes, and is representative of all other Nephromyces lineages found in the renal sac of a single Molgula occidentalis individual (see fig. 2). The apicoplast genome map of Nephromyces 656 was drawn with OGDraw v1.1.1.
Fig. 4.
—The apicoplast genome of Nephromyces closely resembles those of other sporozoans such as hematozoans and coccidians in terms of general organization (i.e., one single copy region and two inverted repeats that contain the rRNA-gene operons), and gene content and order. However, a shared orf59-orf88 gene cluster (surrounded by sufB and rpoB) between Nephromyces and hematozoans support their monophyly as suggested by nuclues-gene trees. Two divergent representatives are shown for the Haemosporidia, Piroplasmida, and Coccidia. Piroplasmids have slightly more divergent apicoplast genomes. Gene coloring scheme follows that in Fig. 3. Genes colored in gray represent ORFs or highly divergent genes that are otherwise syntenic with those in other apicoplast genomes.
Fig. 5.
—(A) A maximum-likelihood tree for the Apicomplexa inferred by IQ-TREE using the site-heterogeneous LG+PMSF(C60)+F+G4 model on 35 plastid-encoded genes (6,403 amino acid sites). Branch support values are 100% SH-aLRT, 100% UFBoot2+NNI, 100% SBS, and 1.0 PP unless annotated. (B) Progressive removal of the fastest evolving sites in increments of 5% and variation of SBS, SH-aLRT, and UFBoot2+NNI branch support values under the site-heterogeneous LG+PMSF(C60)+F+G4 model for the branch that united the Hematozoa and the Coccidia.
Plastid-Gene Trees Conflict with Nucleus-Gene Trees and Place the Nephromycida as Sister to Both the Hematozoa and Coccidia
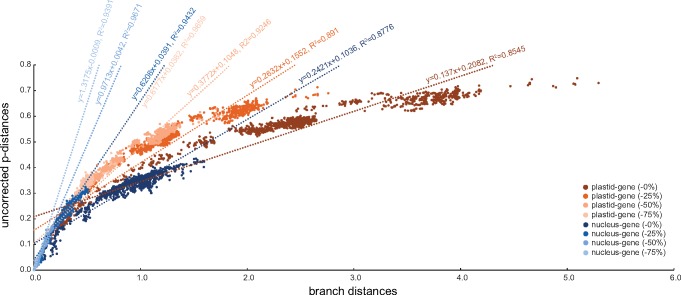
Phylogenetic trees inferred from plastid-encoded genes place the Nephromycida in a more “basal” position as sister to both the Hematozoa and Coccidia (fig. 5A and supplementary fig. S7, Supplementary Material online). This topology conflicts with those inferred from nucleus-encoded genes (fig. 1A and supplementary figs. S1–S3, Supplementary Material online). We progressively removed fast-evolving sites from our much smaller plastid-gene data set (35 genes; 6,403 amino acid sites) but found that support for the topology where the Nephromycida is sister to all other sporozoans (the Hematozoa and Coccidia) remains stable (fig. 5B). We hypothesize that the source of the incongruence between plastid- and nucleus-gene trees most probably lies in the fast-evolving nature of the extremely A+T%-rich apicoplast genomes. Indeed, the plastid-gene data set is much more saturated than the nucleus-gene data set. For example, plastid-gene trees show higher rates of estimated amino acid replacements per site than nucleus-gene trees (e.g., compare branch lengths and units in figs. 1A and 5A). Furthermore, taxon–taxon branch lengths (corrected distances) deviate much more from the observed uncorrected taxon–taxon p-distances in the plastid-gene data set than in the nucleus-gene data set (fig. 6). The plastid-gene data set also “desaturates” much more slowly than the nucleus-gene data set as fast-evolving sites are progressively removed (compare slopes for regression lines in fig. 6). Most recently, Kwong et al. (2019) also found conflicts between nucleus-gene and mitochondrion-gene trees on the one hand, and plastid-gene trees on the other hand, when attempting to place the novel corallicoid lineage, which further casts doubts on the evolutionary signal and usefulness of plastid-encoded genes for resolving deep relationships in the Apicomplexa. The combination of saturation, fewer genes and amino acid sites, and a less diverse set of species (e.g., Cryptosporidium, gregarines and colpodellids have lost their plastid genomes) makes the plastid-gene data set less reliable than the nucleus-gene data set to resolve deep relationships, and therefore the place of the Nephromycida, in the Apicomplexa tree.
Fig. 6.
—Saturation plot that shows the relationship between taxon–taxon uncorrected p-distances and (corrected) branch distances for both plastid-gene and nucleus-gene data sets after 0%, 25%, 50%, and 75% of the fastest-evolving sites have been removed.
The apicoplast of Nephromyces has a conserved set of biosynthetic capabilities
In apicomplexans, like in many other plastid-bearing eukaryotes, the metabolic pathways for the biosynthesis of isoprenoids, fatty acids, and heme are localized to the plastid or apicoplast (Ralph et al. 2004; Lim and McFadden 2010; van Dooren and Hapuarachchi 2017); the heme biosynthetic pathway is only partly localized to the apicoplast. The dependency of the cytoplasm on these apicoplast biosynthetic pathways explains the persistence of these nonphotosynthetic plastids (i.e., leucoplasts) in parasitic cells and other heterotrophic eukaryotes with a photosynthetic ancestry (Janouškovec et al. 2015; Kamikawa et al. 2017; Graupner et al. 2018; Muñoz-Gómez and Slamovits 2018; Dorrell et al. 2019). These three biosynthetic capabilities are all conserved in coccidians and haemosporidians, but piroplasmids have lost the capacity to biosynthesize heme and fatty acids in their apicoplasts (e.g., Janouškovec et al. 2015). We searched our Nephromyces-enriched metatranscriptome, which was estimated to be 92.7% complete (see Materials and Methods), for the main enzymes (many of which are plastid-derived or of plastid origin) involved in these three biosynthetic pathways. These enzymes most likely localize to the apicoplast, as they do in most sporozoans and other plastid-bearing eukaryotes (Janouškovec et al. 2015), although there are a few exceptions for heme biosynthetic enzymes (Lim and McFadden 2010; Seeber and Soldati-Favre 2010). We found that Nephromyces has all of the main enzymes involved in the biosynthesis of heme (i.e., ALAS, ALAD, PBGD, UROS, UROD, CPOX, PPOX, and FECH), isoprenoids (i.e., DXS, IspC, IspD, IspE, IspF, IspG, and IspH), and fatty acids (i.e., FabD, FabG, FabH, FabZ, FabI, FabB/F, and ACP) (see supplementary table S5, Supplementary Material online). Many of these are plastid-derived enzymes (see supplementary fig. S8, Supplementary Material online) and especially those involved in fatty acid and isoprenoid biosynthesis appear to have signal and transit peptides, as predicted by SignalP v4.1 and ChloroP v1.1, that might direct them to the apicoplast (supplementary table S5, Supplementary Material online). Similar to coccidians and Cryptosporidium (Janouškovec et al. 2015), but unlike hematozoans, Nephromyces also appears to have a cytosolic multi-domain type I fatty acid synthase (FASI); there are, however, several FASI-like enzymes, like polyketide synthases or PKSs, that have functions other than de novo fatty acid synthesis (Zhu et al. 2004; Kohli et al. 2016). We infer the presence of a type I FASI in Nephromyces based on transcripts that are most similar to the type I FASI of other apicomplexans in the NCBI GenBank nr database (supplementary table S6, Supplementary Material online) and that have all or most of the type I FASI domains in a conserved order (i.e., KS, AT, DH, ER, KR, ACP, and SDR). Even though FASI-like enzymes cannot be easily distinguished among themselves without a systematic and rigorous study of their evolutionary history and domain composition, Nephromyces contrasts with its divergent sister group, the Hematozoa, by retaining FASI-like enzymes. These findings are consistent with the phylogenetic placement of Nephromyces as sister to the Hematozoa, and also suggest that Nephromyces has retained the ancestral metabolic state of the sporozoan apicoplast.
A Diverse Community of Nephromyces Lineages Co-Exist Inside a Single Renal Sac
Our finding of 13 distinct apicoplast genomes that are most closely related to each other (see figs. 2 and 5A and supplementary fig. S7, Supplementary Material online) in a single renal sac metagenome suggests that divergent Nephromyces lineages can infect not only the same host species but a single host individual. Preliminary evidence for this diversity had been seen in PCR surveys that revealed distinct 18S rRNA genes from total DNA isolated from a renal sac of a single M. occidentalis individual (Saffo et al. 2010). Indeed, a similar pattern could be observed when identifying orthologous sequences for phylogenetic inference; several full-length orthologs for Nephromyces endosymbionts of M. occidentalis formed clades in our single gene trees (see Mendeley Data and supplementary fig. S7, Supplementary Material online for examples). In support of these observations, experiments in the laboratory have shown that Nephromyces cells isolated from one Molgula species or individual can infect a laboratory-raised symbiont-free Molgula individual (Saffo and Davis 1982; Saffo and Nelson 1983) and that molgulid tunicates remain susceptible to infection by Nephromyces throughout most of their mature development (Saffo and Nelson 1983). Multiple infections and the co-existence of several and related endosymbionts (parasitic or otherwise) have been documented for other apicomplexans (see Votýpka et al. 2017 for a discussion). For example, 1) the malaria parasites P.falciparum and Plasmodium vivax can infect the same human host (Mayxay et al. 2004; see King et al. 2015 for a general review), 2) Plasmodium berghei and Plasmodium yoelii can co-exist and even hybridize in the same rodent host (Ramiro et al. 2015), 3) several different species of Eimeria can be found infecting the same chicken or rabbit host (Duszynski and Couch 2013; Clark et al. 2016; Votýpka et al. 2017), 4) the most pathogenic and widespread type strains of T.gondii are hybrids of two ancestral lines (Grigg et al. 2001), and 5) some invertebrate hosts are also known to harbor several and distinct, usually closely related, species of gregarines in their guts (e.g., Clopton et al. 1992; Leander et al. 2003). Taken together, these independent observations converge on the “multiple infection” hypothesis which states that several most closely related, but genetically divergent enough, lineages of “Nephromyces” infect the same host throughout its development.
Conclusions
Nephromyces stands out as an unusual apicomplexan because of its unique mosaic combination of ancestral (e.g., apicoplasts, peroxisomes, urate oxidase, and a single marine invertebrate host) and derived (e.g., bacterial endosymbionts, and probable mutualism and extracellular gigantism) features. Here, we sequenced a Nephromyces-enriched renal sac metagenome and metatranscriptome with the specific goals of deciphering the phylogenetic placement of Nephromyces, searching for and describing its apicoplast genome, and reconstructing its apicoplast metabolism. By performing sophisticated phylogenetic analyses that relied on complex site-heterogeneous models, we found that Nephromyces is sister to both the Haemosporidia and the Piroplasmida (the Hematozoa). These results suggest that the Nephromycida is a novel major lineage in the Apicomplexa. Future taxon sampling, especially for the diverse gregarines, is required to better understand the evolutionary history of the Nephromycida as well as the diversification of the Apicomplexa more generally. We have also shown that Nephromyces possess apicoplasts with a reduced genome and a set of biosynthetic pathways that resemble those of other parasitic apicomplexans such as coccidians (e.g., Toxoplasma) and haemosporidians (e.g., Plasmodium). We furthermore compile evidence for the co-existence of several genetically divergent Nephromyces lineages inside the renal sac of a single molgulid tunicate host. The data and insights provided by this study and others (e.g., Paight et al. 2019) pave the way for a renewed understanding of Nephromyces’ biology.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by a Killam Predoctoral Scholarship and a Nova Scotia Graduate Scholarship to S.A.M.-G., the Natural Sciences and Engineering Research Council of Canada (Discovery Grant RGPIN/05754-2015 to C.H.S.), and the National Institutes of Health (AI124092 to C.E.L. and C.P.) and the National Science Foundation (DEB 1541510 to C.E.L. and C.P.). We thank Kassandra Kennedy for help in manually annotating apicoplast genomes, Marlena Dlutek for technical advice regarding Illumina MiSeq sequencing, Tommy Harding for advice in Illumina library preparation, and Bruce Curtis for sharing his bioinformatic expertise with us.
Data deposition: Sequencing data were deposited in NCBI GenBank under the BioProject PRJNA524113. The Nephromyces-enriched renal sac metagenome and metatranscriptome of Molgula occidentalis were deposited in NCBI GenBank under the accessions SOZB00000000.1 and GHIL00000000.1. Raw sequencing reads were deposited on the NCBI SRA archive under the accessions SRR8618777, SRR8618778, SRR8618779, and SRR8618780. Multi-gene data sets as well as phylogenetic trees inferred in this study were deposited at Mendeley Data under the doi:10.17632/2xgz7vm5f3.1.
Literature Cited
- Aldeyarbi HM, Karanis P.. 2016. The ultra-structural similarities between Cryptosporidium parvum and the gregarines. J Eukaryot Microbiol. 63(1):79–85. [DOI] [PubMed] [Google Scholar]
- Bankevich A, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner P. 1930. Tier und Pflanze in symbiose. Second edition Berlin (Germnay: ): Borntraeger [Google Scholar]
- Buchner P. 1965. Endosymbiosis of animals with plant micro-organisms (trans. B. Mueller, F.H. Foeckler). New York: Wiley-Interscience [Google Scholar]
- Burki F, Okamoto N, Pombert J-F, Keeling PJ.. 2012. The evolutionary history of haptophytes and cryptophytes: phylogenomic evidence for separate origins. Proc Biol Sci. 279(1736):2246–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T.. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25(15):1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. 1993a. Kingdom protozoa and its 18 phyla. Microbiol Rev. 57(4):953–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier‐Smith T. 1993b. The protozoan phylum opalozoa. J Eukaryot Microbiol. 40:609–615. [Google Scholar]
- Cavalier-Smith T. 2003. Protist phylogeny and the high-level classification of Protozoa. Eur J Protistol. 39(4):338–348. [Google Scholar]
- Cavalier-Smith T. 2014. Gregarine site-heterogeneous 18S rDNA trees, revision of gregarine higher classification, and the evolutionary diversification of Sporozoa. Eur J Protistol. 50(5):472–495. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. 2018. Kingdom Chromista and its eight phyla: a new synthesis emphasising periplastid protein targeting, cytoskeletal and periplastid evolution, and ancient divergences. Protoplasma 255(1):297–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T, Chao EE.. 2004. Protalveolate phylogeny and systematics and the origins of Sporozoa and dinoflagellates (phylum Myzozoa nom. nov.). Eur J Protistol. 40(3):185–212. [Google Scholar]
- Ciancio A, et al. 2008. Redescription of Cardiosporidium cionae (Van Gaver and Stephan, 1907) (Apicomplexa: Piroplasmida), a plasmodial parasite of ascidian haemocytes. Eur J Protistol. 44(3):181–196. [DOI] [PubMed] [Google Scholar]
- Clark EL, et al. 2016. Cryptic Eimeria genotypes are common across the southern but not northern hemisphere. Int J Parasitol. 46(9):537–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clopton RE, Janovy J, Percival TJ.. 1992. Host stadium specificity in the gregarine assemblage parasitizing Tenebrio molitor. J Parasitol. 78(2):334–337. [PubMed] [Google Scholar]
- de Lacaze-Duthiers H. 1874. Histoire des ascidies simples des cotes de France. I. Archives de Zoologie Expérimentale et Générale . Histoire Naturelle—Morphologie—Histologie – Évolution Des Animaux. 3:304–313. [Google Scholar]
- de Vienne DM, Ollier S, Aguileta G.. 2012. Phylo-MCOA: a fast and efficient method to detect outlier genes and species in phylogenomics using multiple co-inertia analysis. Mol Biol Evol. 29(6):1587–1598. [DOI] [PubMed] [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G.. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrell RG, et al. 2019. Principles of plastid reductive evolution illuminated by nonphotosynthetic chrysophytes. Proc Natl Acad Sci USA. 116(14):6914–6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duszynski DW, Couch L.. 2013. The biology and identification of the Coccidia (Apicomplexa) of rabbits of the world. London, UK:Elsevier. [Google Scholar]
- Eren AM, et al. 2015. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ 3:e1319.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giard MA. 1888. On Nephromyces, a new genus of Fungi parasitic in the kidney of the Molgulidæ. J Nat Hist Ser. 61: 386–388. [Google Scholar]
- Graupner N, et al. 2018. Evolution of heterotrophy in chrysophytes as reflected by comparative transcriptomics. FEMS Microbiol Ecol. 94:fiy039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigg ME, Bonnefoy S, Hehl AB, Suzuki Y, Boothroyd JC.. 2001. Success and virulence in Toxoplasma as the result of sexual recombination between two distinct ancestries. Science 294(5540):161–165. [DOI] [PubMed] [Google Scholar]
- Haas BJ, et al. 2013. De novo transcript sequence reconstruction from RNA-Seq: reference generation and analysis with Trinity. Nat Protoc. 8(8):1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harant H. 1931. Contribution a l’histoire naturelle des ascides et de leurs parasites. 2. Chytridinées. Monaco (Europe: ): Ann. Inst. Océanogr. [Google Scholar]
- Harmer J, Yurchenko V, Nenarokova A, Lukeš J, Ginger ML.. 2018. Farming, slaving and enslavement: histories of endosymbioses during kinetoplastid evolution. Parasitology 145(10):1311–1323. [DOI] [PubMed] [Google Scholar]
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS.. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janouškovec J, et al. 2015. Factors mediating plastid dependency and the origins of parasitism in apicomplexans and their close relatives. Proc Natl Acad Sci USA. 112(33):10200–10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamikawa R, et al. 2017. A non-photosynthetic diatom reveals early steps of reductive evolution in plastids. Mol Biol Evol. 34(9):2355–2366. [DOI] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T.. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33(2):511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King KC, Stelkens RB, Webster JP, Smith DF, Brockhurst MA.. 2015. Hybridization in parasites: consequences for adaptive evolution, pathogenesis, and public health in a changing world. PLoS Pathog. 11(9):e1005098.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli GS, John U, Van Dolah FM, Murray SA.. 2016. Evolutionary distinctiveness of fatty acid and polyketide synthesis in eukaryotes. ISME J. 10(8):1877–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong WK, del Campo J, Mathur V, Vermeij MJA, Keeling PJ.. 2019. A widespread coral-infecting apicomplexan with chlorophyll biosynthesis genes. Nature 568(7750):103.. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Philippe H.. 2004. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 21(6):1095–1109. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Rodrigue N, Stubbs D, Richer J.. 2013. PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst Biol. 62(4):611–615. [DOI] [PubMed] [Google Scholar]
- Leander BS. 2008. Marine gregarines: evolutionary prelude to the apicomplexan radiation? Trends Parasitol. 24(2):60–67. [DOI] [PubMed] [Google Scholar]
- Leander BS, Harper JT, Keeling PJ.. 2003. Molecular phylogeny and surface morphology of marine aseptate gregarines (Apicomplexa): Selenidium spp. and Lecudina spp. J Parasitol. 89(6):1191–1205. [DOI] [PubMed] [Google Scholar]
- Lim L, McFadden GI.. 2010. The evolution, metabolism and functions of the apicoplast. Philos Trans R Soc Lond B Biol Sci. 365(1541):749–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayxay M, Pukrittayakamee S, Newton PN, White NJ.. 2004. Mixed-species malaria infections in humans. Trends Parasitol. 20(5):233–240. [DOI] [PubMed] [Google Scholar]
- Muñoz-Gómez SA, Slamovits CH.. 2018. Chapter Three - Plastid Genomes in the Myzozoa In: Chaw S-M, Jansen RK, editors. Advances in botanical research. Vol. 85 Plastid Genome Evolution. London, UK:Academic Press; p. 55–94. [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oborník M, Lukeš J.. 2015. The organellar genomes of chromera and vitrella, the phototrophic relatives of apicomplexan parasites. Annu Rev Microbiol. 69:129–144. [DOI] [PubMed] [Google Scholar]
- Paight C, Slamovits CH, Saffo MB, Lane CE.. 2019. Nephromyces encodes a urate metabolism pathway and predicted peroxisomes, demonstrating that these are not ancient losses of apicomplexans. Genome Biol Evol. 11(1):41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph SA, et al. 2004. Tropical infectious diseases: metabolic maps and functions of the Plasmodium falciparum apicoplast. Nat Rev Microbiol. 2(3):203–216. [DOI] [PubMed] [Google Scholar]
- Ramiro RS, et al. 2015. Hybridization and pre-zygotic reproductive barriers in Plasmodium. Proc Biol Sci. 282(1806):20143027.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan U, Paparini A, Monis P, Hijjawi N.. 2016. It’s official – cryptosporidium is a gregarine: what are the implications for the water industry? Water Res. 105:305–313. [DOI] [PubMed] [Google Scholar]
- Saffo MB. 1978. Studies on the renal sac of the ascidian Molgula manhattensis. I. Development of the renal sac. J Morphol. 155(3):287–309. [DOI] [PubMed] [Google Scholar]
- Saffo MB. 1981. The enigmatic protist Nephromyces. Biosystems 14(3–4):487–490. [DOI] [PubMed] [Google Scholar]
- Saffo MB. 1982. Distribution of the endosymbiont nephromyces giard within the Ascidian family Molgulidae. Biol Bull. 162(1):95–104. [Google Scholar]
- Saffo MB. 1990. Symbiosis within a symbiosis: intracellular bacteria within the endosymbiotic protist Nephromyces. Mar Biol. 107(2):291–296. [Google Scholar]
- Saffo MB. 1991. Symbiogenesis and the evolution of mutualism: lessons from nephromyces-bacterial-molgulid symbiosis In: Symbiosis as a source of evolutionary innovation: speciation and morphogenesis. Cambridge, MA:MIT Press; p. 410–429. [Google Scholar]
- Saffo MB, Davis WL.. 1982. Modes of infection of the ascidian molgula manhattensis by its endosymbiont nephromyces giard. Biol Bull. 162(1):105–112. [Google Scholar]
- Saffo MB, Lowenstam HA.. 1978. Calcareous deposits in the renal sac of a molgulid tunicate. Science 200(4346):1166–1168. [DOI] [PubMed] [Google Scholar]
- Saffo MB, McCoy AM, Rieken C, Slamovits CH.. 2010. Nephromyces, a beneficial apicomplexan symbiont in marine animals. Proc Natl Acad Sci USA. 107(37):16190–16195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffo MB, Nelson R.. 1983. The cells of Nephromyces: developmental stages of a single life cycle. Can J Bot. 61(12):3230–3239. [Google Scholar]
- Seeber F, Soldati-Favre D.. 2010. Chapter 5 – Metabolic pathways in the apicoplast of apicomplexa In: Jeon KW, editor. International review of cell and molecular biology. Vol. 281 International review of cell and molecular biology. London, UK:Academic Press; p. 161–228. [DOI] [PubMed] [Google Scholar]
- Simdyanov TG, et al. 2017. A new view on the morphology and phylogeny of eugregarines suggested by the evidence from the gregarine Ancora sagittata (Leuckart, 1860) Labbé, 1899 (Apicomplexa: Eugregarinida). PeerJ 5:e3354.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susko E, Roger AJ.. 2007. On reduced amino acid alphabets for phylogenetic inference. Mol Biol Evol. 24(9):2139–2150. [DOI] [PubMed] [Google Scholar]
- Thompson RCA, Koh WH, Clode PL.. 2016. Cryptosporidium—what is it? Food Waterborne Parasitol. 4: 54–61. [Google Scholar]
- Toso MA, Omoto CK.. 2007. Gregarina niphandrodes may lack both a plastid genome and organelle. J Eukaryot Microbiol. 54(1):66–72. [DOI] [PubMed] [Google Scholar]
- van Dooren GG, Hapuarachchi SV.. 2017. Chapter five – the dark side of the chloroplast: biogenesis, metabolism and membrane biology of the apicoplast In: Hirakawa Y, editor. Advances in botanical research. Vol. 84 Secondary endosymbioses. London, UK:Academic Press; p. 145–185. [Google Scholar]
- Vaidya G, Lohman DJ, Meier R.. 2011. SequenceMatrix: concatenation software for the fast assembly of multi‐gene datasets with character set and codon information. Cladistics 27(2):171–180. [DOI] [PubMed] [Google Scholar]
- Verbruggen H. 2018. SiteStripper version 1.03. http://www.phycoweb.net/software; last accessed July 25, 2019
- Votýpka J, Modrý D, Oborník M, Šlapeta J, Lukeš J.. 2017. Apicomplexa In: Archibald JM, Simpson AGB, Slamovits CH, editors. Handbook of the protists. Cham (Switzerland: ): Springer International Publishing; p. 567–624. [Google Scholar]
- Wang H-C, Minh BQ, Susko E, Roger AJ.. 2018. Modeling site heterogeneity with posterior mean site frequency profiles accelerates accurate phylogenomic estimation. Syst Biol. 67(2):216–235. [DOI] [PubMed] [Google Scholar]
- Zhu G, et al. 2004. Expression and functional characterization of a giant Type I fatty acid synthase (CpFAS1) gene from Cryptosporidium parvum. Mol Biochem Parasitol. 134(1):127–135. [DOI] [PubMed] [Google Scholar]
- Zhu G, Marchewka MJ, Keithly JS.. 2000. Cryptosporidium parvum appears to lack a plastid genome. Microbiology 146(2):315–321. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.