Abstract
Multi-gene testing is useful in genetically heterogeneous conditions, including inherited cardiac pathologies. Increasing the number of genes analysed increases diagnostic yield of variants of certain, likely, or uncertain pathogenicity. Concerns exist regarding management of variants of uncertain/likely pathogenicity in conditions of oligogenic inheritance or variable expressivity. We surveyed a sample of colleagues across different specialties and departments internationally to compare management of patients with class 3 or class 4 variants in genes associated with non-syndromic cardiomyopathy or arrhythmia. An electronic survey regarding clinical management of variants (www.surveymonkey.com/r/cardiacvariants) was designed and distributed to colleagues internationally via professional bodies and direct email. 150 respondents (88 centres, 27 countries) completed the survey, most of whom were Clinical Geneticists or Genetic Counsellors. Most respondents offer pre-symptomatic testing to asymptomatic relatives of an individual with class 4 or class 5 variants. A minority of respondents offer pre-symptomatic testing for class 3 variants. Considering class 4 variants, 22 (15%) are fully reassuring that the patient with a negative predictive test would not develop the familial phenotype, while 123 (82%) counselled patients about the possibility of variant reclassification. Variability existed between and within centres and specialties. Multiple “free text” comments were provided. Recurring themes including need for multidisciplinary input, technical concerns, and concern regarding duty to review variants of uncertain significance. This study demonstrates that variability in management of likely pathogenic/uncertain variants exists. Close multi-disciplinary input is essential. The development of disorder or gene-specific evidence-based guidelines might ameliorate uncertainty in management.
Subject terms: Genetic testing, Cardiac hypertrophy, Arrhythmias, Genetic counselling
Introduction
Diagnostic testing using massively parallel genetic sequencing is the standard approach for individuals with phenotypes associated with heterogeneous genetic aetiology. Heritable cardiac pathologies such as channelopathies or cardiomyopathies can be phenotypically and genotypically diverse. Cardiac phenotypes associated with variants in different genes may be overlapping or indistinct. Multi-gene panels incorporating increased numbers of genes, whole exome or whole genome analyses are readily available for investigation of individuals with such phenotypes. These broad approaches have increased the diagnostic yield in such individuals in a time-efficient and cost-efficient manner, and are particularly useful in post-mortem analysis when the amount of DNA available for interrogation is finite [1]. Such approaches also increase the yield of variants of uncertain significance, of variants in genes with weak or no association with the specific phenotype in question, and of multiple variants in multiple genes [2–4].
Variants in genes associated with cardiac pathologies often demonstrate reduced penetrance and variable expressivity, and affected families often have private variants, adding to the challenge of variant classification [5, 6]. Genetic testing may provide essential information to guide therapy and inform prognostication; but may also provide results that cause confusion and anxiety to clinician and patient alike.
The average human genome contains between 4 and 5 million variants. Determining which variants are clinically significant depend on a number of factors, including the clinical context (phenotype and genetic locus in which the variant has been identified), population and disease frequency, computational and functional evidence, and segregation of the variant with disease. Plon et al. defined five classes of variants depending on probability of pathogenicity. Class 5 (pathogenic) variants are those variants with >99% probability of pathogenicity, and Class 4 (likely pathogenic) variants those with 95–99% probability of pathogenicity. Class 1 (benign) variants have <0.1%, and Class 2 (likely benign) variants 0.1–5% probability of pathogenicity. Those variants with 5–95% probability of pathogenicity are categorised as Class 3 variants (variants of uncertain significance) [7].
The American College of Medical Genetics (ACMG) have provided a framework to standardise classification of variants [8], the underlying principles of which largely relate to rare monogenic disorders. For common, genetically heterogeneous disorders, such as inherited cardiac pathologies, the various lines of evidence referenced in the guidelines are not always helpful. Most variants in genes associated with cardiac arrhythmias and cardiomyopathies are missense, and inherited, and a high frequency in population databases does not preclude pathogenicity of a variant with reduced penetrance. The recognition that these guidelines require adaptation for particular genes or genetic disorders have prompted amendments to the guidelines for MYH7, and genes in the RAS-MAPK pathway [9] and it is likely that similar amendments for other genes associated with common or genetically heterogenous disorders will follow [10].
Approximately 5–16% of heritable cardiac pathologies have an underlying di-genic or oligo-genic inheritance [4, 11], which may result in atypical phenotypes or inheritance patterns. Accurate risk estimation may therefore be complex. Likely pathogenic variants in most genes associated with monogenic disorders are considered clinically actionable, for which cascade testing of the specific variant identified may be offered to affected relatives (confirmatory testing) and unaffected relatives (pre-symptomatic testing). Colloquially, the term “predictive testing” usually refers to specific variant testing offered to relatives of a proband in whom a variant is identified, although some of these relatives may already demonstrate a phenotype.
Considering inherited cardiac pathologies, the lack of definitive evidence for pathogenicity, and possibility of unidentified di-genic or oligo-genic contribution, may preclude full reassurance of relatives in whom a negative pre-symptomatic result is obtained. Pre-symptomatic testing of variants of uncertain significance is not recommended; but testing of other affected individuals for such variants may be helpful for the purposes of segregation analysis [7].
Based on our experience, in the Republic of Ireland, a proportion of diagnostic genetic testing for inherited cardiac pathologies is performed by cardiologists in a small number of tertiary cardiac centres, while most subsequent pre-symptomatic testing and all remaining diagnostic tests are requested by genetic healthcare professionals in a single Clinical Genetics unit. Monthly multi-disciplinary meetings are attended by cardiologists, geneticists and clinical scientists, generating discussion regarding management of patients with variants of uncertain or likely pathogenicity, and with respect to testing and surveillance of their unaffected relatives. Prompted by these discussions, we aimed to investigate the practices of international colleagues in managing symptomatic and asymptomatic carriers of variants of uncertain or likely pathogenicity in genes associated with cardiac pathology.
Materials and methods
The authors designed a survey (Supplementary Figure 1) to determine how symptomatic and asymptomatic patients with, or at risk of inheriting, variants of different classes of pathogenicity in genes associated with cardiac pathologies, are managed by colleagues internationally. Data was also collected with respect to the occupation and subspecialty interest of the respondent, and characteristics of the centre in which they practiced. The link to the survey (www.surveymonkey.ie/cardiacvariants) was distributed to colleagues internationally via professional bodies (European Society of Human Genetics, Association of Genetic Counsellors and Nurses) and by direct email to colleagues in clinical laboratories and in Departments of Cardiology and Clinical Genetics. All data was tabulated and analysed using SPSS v.23 (IBM). Relationships between two categorical variables were analysed using Chi-squared tests or Fisher’s exact test as appropriate. Distribution of continuous data was assessed by Shapiro–Wilk test. Normal data are described as mean ± standard deviation, and data that are not normally distributed as median (range).
Thematic analysis of free text comments was performed by a single author.
Results
Respondent demographics
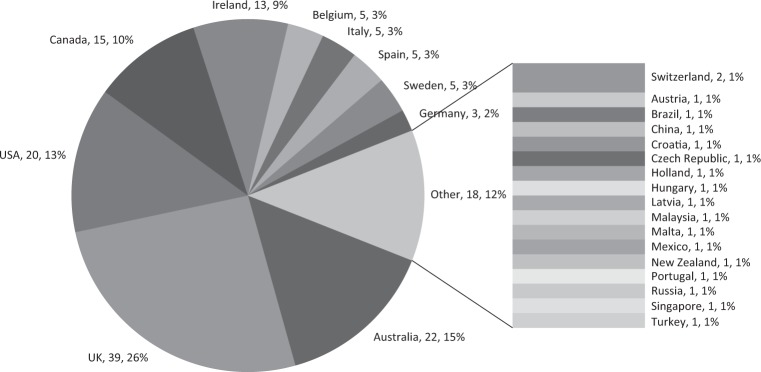
In total, 150 respondents from 88 centres across 27 countries completed the survey. Most respondents (73%) were from five countries; including UK (n = 39, 26%), Australia (n = 22, 15%), USA (n = 20, 13%), Canada (n = 15, 10%) or Ireland (n = 13, 9%) (Fig. 1). The number of responses per centre is outlined in Supplementary Table 1. The median number of “cardiac patients” attending each service per year was 200 (3–10,000). The majority of respondents were clinicians working in Clinical Genetics, including consultant (n = 61, 41%) and trainee clinical geneticists (n = 10, 7%), genetic counsellors (n = 48, 32%) and clinical scientists (n = 19, 13%), with a minority of responses from cardiologists (n = 12, 8%). One hundred and one (70%) respondents had a special interest and/or protected time in their working contract to practice cardiac genetics (Table 1). Respondents from a total of 58 (66%) centres stated that at least one consultant geneticist practicing in their centre had a sub-specialty interest in Cardiac Genetics; although there were conflicting answers about this from different respondents in seven centres. Next generation sequencing analysis for diagnostic cardiac genetic testing was reportedly performed in house in 44 centres (19 countries), while otherwise testing was reportedly outsourced. There were conflicting responses regarding availability of in-house Next-Generation Sequencing (NGS)-based testing for inherited cardiac pathologies from respondents in three centres.
Fig. 1.
Country of practice of respondents
Table 1.
Occupation and sub-specialty interest of respondents
| Occupation | Number of respondents (N) | Specialist interest in cardiac genetics (N) |
|---|---|---|
| Consultant clinical geneticist | 61 | 36 (59%) |
| Trainee in clinical genetics | 10 | 3 (30%) |
| Genetic counsellor | 48 | 35 (73%) |
| Clinical scientist | 19 | 17 (89%) |
| Cardiologist | 12 | 10 (83%) |
| Total | 150 | 101 (67%) |
Cascade testing
Most respondents would offer pre-symptomatic testing to the relatives of a patient in whom a class 5 variant (n = 146, 97%) or class 4 variant (123, 82%) (Table 2) had been identified (Fig. 2). A proportion of respondents would only offer specific variant testing to affected individuals for familial class 5 (1, 1%) or class 4 (23, 15%) variants. Two (2%) individuals (both genetic health professionals) would not offer pre-symptomatic testing for either class 4 or class 5 variants.
Table 2.
Predictive testing for variants of class 3/4/5
| Do you offer predictive testing for the following variants? | Yes | No | Only to affected individuals | No response |
|---|---|---|---|---|
| Class 5 variant | 146 (97%) | 2 (1%) | 1 (1%) | 1 (1%) |
| Class 4 variant | 123 (82%) | 2 (1%) | 23 (15%) | 2 (1%) |
| Class 3 variant | 18 (12%) | 35 (23%) | 95 (63%) | 2 (1%) |
| Considering predictive testing for class 4 variants | Yes | No | Defer to Cardiologist to decide | No response |
|---|---|---|---|---|
| Are you entirely reassuring that non-carriers have low risk of developing the family phenotype? | 22 (15%) | 101 (67%) | 21 (14%) | 6 (4%) |
| Do you recommend cardiac assessment of non-carrier if one has not been performed? | 95 (63%) | 37 (25%) | 10 (7%) | 8 (5%) |
| Do you recommend discontinuation of cardiac follow-up of asymptomatic individuals with negative test? | 40 (27%) | 51 (34%) | 52 (35%) | 7 (5%) |
| Do you counsel patients about the possibility of reclassification of variants? | 123 (82%) | 14 (9%) | 13 (9%) | 0 |
Fig. 2.
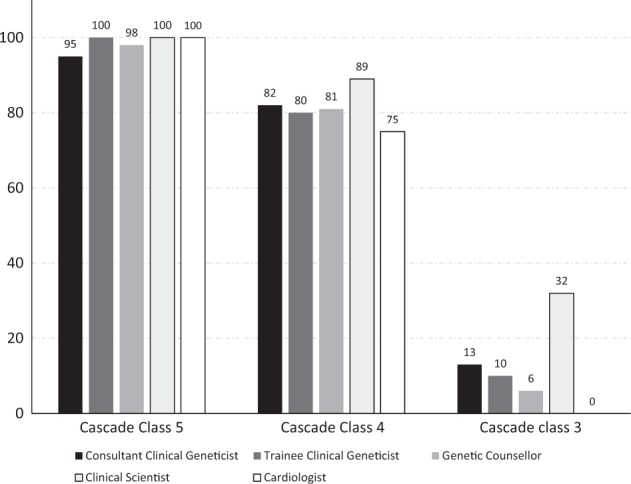
Proportion of respondents who would offer pre-symptomatic testing of familial variants of different classes to unaffected relatives
Eighteen respondents (12%) would offer pre-symptomatic testing to relatives for a familial class 3 variant, including eight consultant clinical geneticists, three genetic counsellors, six clinical scientists, and one clinical genetics trainee; while no cardiologist reported offering pre-symptomatic testing for such variants (p = 0.053). Of the remaining respondents, 35 (23%) would not offer pre-symptomatic testing for such variants, while 95 (63%) would offer confirmatory-specific variant testing of such variants to affected family members to facilitate segregation analysis.
Management of carriers and non-carriers of likely pathogenic variants
Considering Class 4 variants (Table 2), most respondents demonstrated caution in reassuring individuals undergoing pre-symptomatic testing for such variants, with 110 (81%) counselling about residual uncertainty with respect to pathogenicity of a class 4 variant, and a similar proportion (111, 82%) counselling about the possibility of reclassification of the variant in the future.
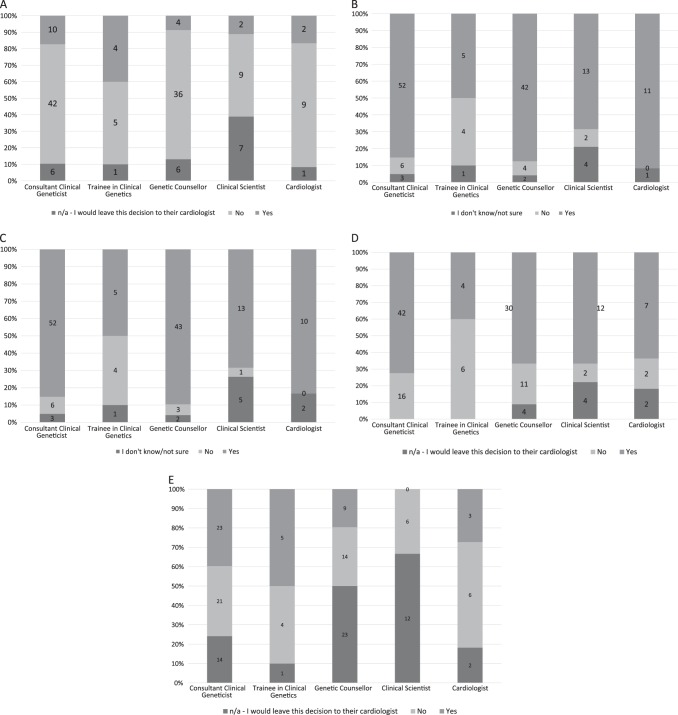
For individuals with negative pre-symptomatic tests for familial class 4 variants, a minority of respondents (22, 15%) would be entirely reassuring that such individuals were free from risk of developing the familial phenotype. Trainees in Clinical Genetics were more likely than any other group of professionals to be reassuring in this circumstance (p = 0.064, Fig. 3a). Trainees were also more likely to omit discussion regarding uncertainty of pathogenicity of a class 4 variant (Fig. 3b, p = 0.037), or possibility of reclassification of such variants (Fig. 3c, p = 0.007).
Fig. 3.
Respondent answers by specialty. a Participant response regarding whether they would be fully reassuring that the asymptomatic individuals having a negative predictive test for familial class 4 variant would not develop the familial phenotype. b Participant response regarding discussing uncertainty about pathogenicity of a class 4 variant with asymptomatic individual having negative predictive test. c Participant response regarding whether they mention potential reclassification of class 4 variant to asymptomatic individuals having negative predictive test. d Participant response regarding referral of asymptomatic individuals having negative predictive test for familial class 4 variant for cardiac assessment. e Participant response regarding whether they recommend discharge of asymptomatic individuals having negative predictive tests for familial class 4 variant to asymptomatic individuals having negative predictive test
Most respondents (95, 63%) would recommend that asymptomatic individuals would have a cardiac assessment if one had not yet been undertaken, even in the setting of a negative pre-symptomatic test for a familial class 4 variant. Clinical Genetics Trainees were less likely to do so than other groups (Fig. 3d, p = 0.009, X2), and were more likely to recommend discharge of asymptomatic individuals with negative pre-symptomatic tests for familial class 4 variants (Fig. 3e, p = 0.002, X2). Variability was noted between and within centres (Supplementary Figures 1–5).
Variant reinterpretation and reclassification
Most respondents would recommend that routine reinterpretation and reclassification of variants be attempted at least every year (82, 55%) (Table 3). Most respondents (N = 101, 67%)) felt that attempts at variant reclassification should be part of standard practice. Most respondents felt that variant reclassification should be attempted when another relative in a known family developed a phenotype (N = 114, 76%), or with development of new conflicting data regarding the familial phenotype (N = 124, 83%), through population databases (N = 124, 83%), or in the published literature (N = 121, 81%). A proportion felt variant reclassification should be mandated by professional bodies (N = 79, 53%).
Table 3.
How frequently should variant re-classification be undertaken?
| Consultant clinical geneticist | Trainee in linical genetics | Genetic counsellor | Clinical scientist | Cardiologist | |
|---|---|---|---|---|---|
| Six-monthly | 4 (7%) | 0 | 3 (6%) | 0 | 1 (8%) |
| Annually | 24 (39%) | 4 (40%) | 25 (52%) | 16 (84%) | 5 (42%) |
| 3-yearly | 5 (8%) | 2 (20%) | 3 (6%) | 0 | 0 |
| 5-yearly | 19 (31%) | 4 (40%) | 16 (33%) | 2 (11%) | 6 (50%) |
| Not at all | 8 (13%) | 0 | 0 | 1 (5%) | 0 |
| Not specified | 1 (2%) | 0 | 1 (2%) | 0 | 0 |
| Total | 61 | 10 | 48 | 19 | 12 |
Concerns regarding the use of NGS for investigation of inherited cardiac pathology
A free-text comment box captured concerns of the respondents with many common themes including (Table 4); lack of consensus between or within departments in management of carriers and non-carriers of class 3 or class 4 variants; concerns about technical issues, including coverage, and sensitivity of panels in detecting large genomic rearrangements; continuous expansion of the number of genes on specific disease panels; the increased potential of detecting variants of uncertain significance, or variants in genes not typically associated with the phenotype in question; variability in genes included on panels between different laboratories—a common issue in genetically heterogeneous conditions [12].
Table 4.
Concerns of respondents expressed in free-text box
| Comments | N |
|---|---|
| Multidisciplinary Input required for accurate phenotyping and classification | 22 |
| Lack of consistency within and between units/labs | 8 |
| Technical limitations | 8 |
| Management and Interpretation of Incidental/uncertain results | 15 |
| Reduced penetrance | 8 |
Incomplete penetrance, variable expressivity and oligogenic causes of cardiac pathology were also cited as factors complicating the interpretation of cardiac genetic variants. One respondent expressed concern about a lack of clarification as to whom should bear responsibility to investigate and reclassify variants (laboratory or clinical staff). The nature of working relationship between cardiologists and clinical geneticists was an area that a proportion of respondents felt could be improved by closer co-operation and regular multi-disciplinary team meetings. Some respondents expressed concern about counselling and consenting for genetic testing being undertaken without the input of a genetics team.
Discussion
These results show clear variability in the management of carriers and non-carriers of class 4 variants, and in pre-symptomatic testing practices internationally. Variability was evident between and within centres, and between and within specialties. Surprisingly, a proportion of respondents, all professionals working in genetics, would offer pre-symptomatic testing to unaffected family members for a familial class 3 variant, while a minority of individuals would not offer such testing for class 4 or class 5 variants. A number of clinicians emailed the authors or utilised the free text box to clarify their responses, suggesting that differences in insurance implications, and funding of genetic testing impacted their practice; while other clinicians suggested that they did not consider these to be modifying factors in decision-making. Clinicians in some centres treated class 4 variants with caution; and would not offer pre-symptomatic testing to unaffected relatives until variant could be definitively reclassified as a class 5 variant.
There was considerable variability in the proposed management of asymptomatic relatives of a patient in whom a class 4 variant had been identified, especially with respect to ongoing management of those individuals with a negative pre-symptomatic test result. Standardisation of management of such variants may be difficult, as a significant proportion of respondents suggested that management of carriers and non-carriers of familial variants of different classes was dependent on the individual and familial phenotype, implying that decisions may be made on a case-by-case, ad hoc basis. Certain respondents suggested that management decisions may be altered depending on the specific probability of pathogenicity of a particular variant. Tavtigian et al. have published a Bayesian adaptation of the ACMG classification guidelines to develop a quantitative approach to variant classification [13], and it remains to be seen if a continuous rather than an ordinal scale for classification will be considered more informative for patient management.
The results of this study suggest that most clinicians in most centres consider class 4 and class 5 variants to be clinically actionable, for which pre-symptomatic testing should be offered. However, most respondents demonstrate caution with respect to management of asymptomatic non-carriers of class 4 variants, and most suggest reversion to phenotype information to determine ongoing management; which, arguably, calls into question the utility of offering testing for such variants to asymptomatic individuals. Trainees in Clinical Genetics were more likely to be reassuring to family members testing negative for familial class 4 variants, and were also more likely to omit discussing uncertainty in variant interpretation. This may reflect a lack of awareness, or lack of confidence among trainees, reflecting potential gaps in training that should be addressed.
The ACMG guidelines [8] have provided an excellent framework for variant classification; which should help standardisation of variant interpretation across laboratories. The inclusion of high allelic frequency (>5%) in exome or genome sequencing projects as standalone evidence against pathogenicity has triggered reclassification of numerous variants [14]. Some lines of evidence; such as segregation data, inheritance information or functional evidence; which might help definitively classify variants as pathogenic or benign, may not be readily available to every laboratory, and it is still possible that identical variants may be differentially classified between different laboratories where different lines of evidence in the framework have been applied. Furthermore, the ACMG guidelines have not been universally adopted in laboratories internationally.
The reduced penetrance, variable expressivity and genetic heterogeneity of inherited cardiac pathologies are well reported [5, 15–17]. Individuals found to carry a pathogenic variant in a gene associated with an arrhythmia are deemed to have a diagnosis of the condition, even in the absence of ECG findings [18]; where previously such a finding in an asymptomatic individual was considered simply a marker of risk rather than a diagnostic criterion. This may have serious psychological implications for the patient and may have economic consequences, impacting medical insurance premia in certain jurisdictions. Decisions with respect to offering pre-symptomatic testing may be made by clinical teams based on variant classification, familial phenotype, or other factors, but uptake of that offer may be based on multiple interacting biopsychosocial factors, as well as clinical recommendations [19, 20].
Identification of a highly penetrant pathogenic variant associated with an inherited cardiac pathology in an asymptomatic individual facilitates intervention by surveillance, medication, or device implantation to prevent, or enable early identification and management of a phenotype, and, hopefully, to prevent occurrence of a sudden cardiac death in a family. Misinterpretation of a variant in a family with a cardiac phenotype can have negative consequences, with potential mortality or serious morbidity if an actionable mutation is missed, or with potential over-investigation, inappropriate medicalisation, and increased anxiety in patients where pathogenicity is incorrectly ascribed to a benign variant. Misclassification, and reclassification, of variants may be a particular issue in patients from populations underrepresented in population databases [21]. The workload associated with cascade testing and variant classification and reclassification is substantial; particularly in large families [22] or families with di-genic or oligo-genic contributions, which make up a proportion of our cases. Currently, laboratories are not expected to perform systematic reanalysis of old data, unless specific information or request from laboratory or clinical community prompts a review of a particular variant [23]. Most respondents in this study were of the opinion that reinterpretation of variants should be systematically undertaken.
It is interesting to note that a third of respondents do not recommend discontinuation of cardiac follow-up in unaffected individuals with negative pre-symptomatic tests for class 4 familial variants, suggesting heightened awareness of possibility of future re-classification and downgrading of such variants, as well as awareness of potentially unidentified di-genic or oligogenic contributions.
In Ireland, our limited resources already struggle to keep up with demand for testing. Increased use of testing using expanded panels or whole exome or whole genome approaches is going to stretch these resources even further. Increased education and training of clinicians offering genetic testing for and management of inherited cardiac pathologies is critical, to ensure that testing is only performed in appropriate circumstances, that interpretation of variants is performed with accuracy, and to ensure that patients are counselled appropriately with respect to any uncertainty or potential outstanding risk. It is also crucial that robust systems are in place to facilitate identification and re-contacting of families where reinterpretation and re-classification of a familial variant occurs.
Limitations of the study
This is a survey-based study with self-reported opinions from a heterogeneous group of clinicians providing care to patients with or at risk of inherited cardiac issues. In this study, we considered heritable channelopathies and cardiomyopathies, but did not consider congenital heart diseases or vasculopathies. There were differences between respondents in terms of specialty, sub-specialty experience, and practice volume. The survey was disseminated by direct email to relevant clinicians, and through professional bodies. The authors cannot comment on what proportion of individuals who were contacted responded to the survey. The vast majority of responses were from five countries, and there is an over-representation of individuals with a special sub-specialty interest in cardiac genetics. Therefore, the results of the study may not be reflective of the opinion or practice of generalists, or in clinicians practicing in under-represented countries where practice may be influenced by local guidelines for surveillance, or implications for insurance. Data was not specifically undertaken regarding criteria for testing, or technology used in testing.
Conclusion
As more broad genetic testing such as whole exome or genome sequencing becomes more readily available, management of variants of uncertain significance, or secondary incidental detection of variants in genes unrelated to the clinical phenotype will become increasingly challenging. With respect to variants in genes associated with cardiac pathologies, it is critical that interpretation be undertaken with multidisciplinary input. It is important that clinicians are mindful of the genetic heterogeneity of the disorders, and that missing heritability is considered when providing screening recommendations to the relatives of a proband with an uninformative test result. It is also important to consider expressivity and age-related penetrance when discharging apparently unaffected individuals based on phenotype only in families where the genetic basis has not been fully established. It is also important that clinicians consider the possibility of reinterpretation and reclassification of variants when returning results to patients, so that patients can be counselled appropriately [24]. Standardisation of practice, and on-going intradepartmental and interdepartmental audits of reporting and management of affected families would be beneficial to both patients and clinicians alike. It should improve confidence in testing process and reduce inconsistency.
Supplementary information
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41431-019-0391-8) contains supplementary material, which is available to authorised users.
References
- 1.Neubauer J, Haas C, Bartsch C, Medeiros-Domingo A, Berger W. Post-mortem whole-exome sequencing (WES) with a focus on cardiac disease-associated genes in five young sudden unexplained death (SUD) cases. Int J Legal Med. 2016;130:1011–21. doi: 10.1007/s00414-016-1317-4. [DOI] [PubMed] [Google Scholar]
- 2.Ouellette AC, Mathew J, Manickaraj AK, Manase G, Zahavich L, Wilson J, et al. Clinical genetic testing in pediatric cardiomyopathy: is bigger better? Clin Genet. 2018;93:33–40. doi: 10.1111/cge.13024. [DOI] [PubMed] [Google Scholar]
- 3.Cirino AL, Lakdawala NK, McDonough B, Conner L, Adler D, Weinfeld M, et al. A comparison of whole genome sequencing to multigene panel testing in hypertrophic cardiomyopathy patients. Circul Cardiovasc Genet. 2017;10:e001768. doi: 10.1161/CIRCGENETICS.117.001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gómez J, Reguero JR, Coto E. The ups and downs of genetic diagnosis of hypertrophic cardiomyopathy. Rev Esp Cardiol. 2016;69:61–8. doi: 10.1016/j.recesp.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 5.Michels M, Soliman OI, Phefferkorn J, Hoedemaekers YM, Kofflard MJ, Dooijes D, et al. Disease penetrance and risk stratification for sudden cardiac death in asymptomatic hypertrophic cardiomyopathy mutation carriers. Eur Heart J. 2009;30:2593–8. doi: 10.1093/eurheartj/ehp306. [DOI] [PubMed] [Google Scholar]
- 6.Christiaans I, Birnie E, van Langen IM, van Spaendonck-Zwarts KY, van Tintelen JP, van den Berg MP, et al. The yield of risk stratification for sudden cardiac death in hypertrophic cardiomyopathy myosin-binding protein C gene mutation carriers: focus on predictive screening. Eur Heart J. 2010;31:842–8. doi: 10.1093/eurheartj/ehp539. [DOI] [PubMed] [Google Scholar]
- 7.Plon SE, Eccles DM, Easton D, Foulkes WD, Genuardi M, Greenblatt MS, et al. Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum Mutat. 2008;29:1282–91. doi: 10.1002/humu.20880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gelb BD, Cave H, Dillon MW, Gripp KW, Lee JA, Mason-Suares H, et al. ClinGen’s RASopathy Expert Panel consensus methods for variant interpretation. Genet Med. 2018;20:1334–45. doi: 10.1038/gim.2018.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelly Melissa A, Caleshu Colleen, Morales Ana, Buchan Jillian, Wolf Zena, Harrison Steven M, Cook Stuart, Dillon Mitchell W, Garcia John, Haverfield Eden, Jongbloed Jan D H, Macaya Daniela, Manrai Arjun, Orland Kate, Richard Gabriele, Spoonamore Katherine, Thomas Matthew, Thomson Kate, Vincent Lisa M, Walsh Roddy, Watkins Hugh, Whiffin Nicola, Ingles Jodie, van Tintelen J Peter, Semsarian Christopher, Ware James S, Hershberger Ray, Funke Birgit. Adaptation and validation of the ACMG/AMP variant classification framework for MYH7-associated inherited cardiomyopathies: recommendations by ClinGen’s Inherited Cardiomyopathy Expert Panel. Genetics in Medicine. 2018;20(3):351–359. doi: 10.1038/gim.2017.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rigato I, Bauce B, Rampazzo A, Zorzi A, Pilichou K, Mazzotti E, et al. Compound and digenic heterozygosity predicts lifetime arrhythmic outcome and sudden cardiac death in desmosomal gene-related arrhythmogenic right ventricular cardiomyopathy. Circulation. 2013;6:533–42. doi: 10.1161/CIRCGENETICS.113.000288. [DOI] [PubMed] [Google Scholar]
- 12.Easton DF, Pharoah PDP, Antoniou AC, Tischkowitz M, Tavtigian SV, Nathanson KL, et al. Gene-panel sequencing and the prediction of breast-cancer risk. N Engl J Med. 2015;372:2243–57. doi: 10.1056/NEJMsr1501341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tavtigian SV, Greenblatt MS, Harrison SM, Nussbaum RL, Prabhu SA, Boucher KM, et al. Modeling the ACMG/AMP variant classification guidelines as a Bayesian classification framework. Genet Med. 2018;20:1054–60. doi: 10.1038/gim.2017.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andreasen C, Nielsen JB, Refsgaard L, Holst AG, Christensen AH, Andreasen L, et al. New population-based exome data are questioning the pathogenicity of previously cardiomyopathy-associated genetic variants. Eur J Hum Genet. 2013;21:918–28. doi: 10.1038/ejhg.2012.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Priori SG, Napolitano C, Schwartz PJ. Low penetrance in the long-QT syndrome: clinical impact. Circulation. 1999;99:529–33. doi: 10.1161/01.CIR.99.4.529. [DOI] [PubMed] [Google Scholar]
- 16.Goldenberg I, Horr S, Moss AJ, Lopes CM, Barsheshet A, McNitt S, et al. Risk for life-threatening cardiac events in patients with genotype-confirmed long-QT syndrome and normal-range corrected QT intervals. J Am Coll Cardiol. 2011;57:51–9. doi: 10.1016/j.jacc.2010.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amin AS, Wilde AAM. The future of sudden cardiac death research. Prog Pediatr Cardiol. 2017;45:49–54. doi: 10.1016/j.ppedcard.2017.02.008. [DOI] [Google Scholar]
- 18.Priori SG, Wilde AA, Horie M, Cho Y, Behr ER, Berul C, et al. HRS/EHRA/APHRS expert consensus statement on the diagnosis and management of patients with inherited primary arrhythmia syndromes: document endorsed by HRS, EHRA, and APHRS in May 2013 and by ACCF, AHA, PACES, and AEPC in June 2013. Heart Rhythm. 2013;10:1932–63. doi: 10.1016/j.hrthm.2013.05.014. [DOI] [PubMed] [Google Scholar]
- 19.Christian S, Atallah J, Clegg R, Giuffre M, Huculak C, Dzwiniel T, et al. Uptake of predictive genetic testing and cardiac evaluation for children at risk for an inherited arrhythmia or cardiomyopathy. J Genet Couns. 2018;27:124–30. doi: 10.1007/s10897-017-0129-0. [DOI] [PubMed] [Google Scholar]
- 20.Christiaans I, Birnie E, Bonsel GJ, Wilde AA, van Langen IM. Uptake of genetic counselling and predictive DNA testing in hypertrophic cardiomyopathy. Eur J Hum Genet. 2008;16:1201–7. doi: 10.1038/ejhg.2008.92. [DOI] [PubMed] [Google Scholar]
- 21.Manrai AK, Funke BH, Rehm HL, Olesen MS, Maron BA, Szolovits P, et al. Genetic misdiagnoses and the potential for health disparities. New Engl J Med. 2016;375:655–65. doi: 10.1056/NEJMsa1507092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McVeigh TP, Donnelly D, Al Shehhii M, Jones EA, Murray A, Wedderburn S, et al. Towards establishing consistency in triage in a tertiary specialty. Eur J Hum Genet. 2019;27:547–55. doi: 10.1038/s41431-018-0322-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matthijs G, Souche E, Alders M, Corveleyn A, Eck S, Feenstra I, et al. Guidelines for diagnostic next-generation sequencing. Eur J Hum Genet. 2016;24:2–5. doi: 10.1038/ejhg.2015.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van El CG, Cornel MC, Borry P, Hastings RJ, Fellmann F, Hodgson SV, et al. Whole-genome sequencing in health care. Recommendations of the European Society of Human Genetics. Eur J Hum Genet. 2013;21(Suppl. 1):S1–S5. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.