Abstract
Hearing impairment (HI) is characterized by extensive genetic heterogeneity. To determine the population-specific contribution of known autosomal recessive nonsyndromic (ARNS)HI genes and variants to HI etiology; pathogenic and likely pathogenic (PLP) ARNSHI variants were selected from ClinVar and the Deafness Variation Database and their frequencies were obtained from gnomAD for seven populations. ARNSHI prevalence due to PLP variants varies greatly by population ranging from 96.9 affected per 100,000 individuals for Ashkenazi Jews to 5.2 affected per 100,000 individuals for Africans/African Americans. For Europeans, Finns have the lowest prevalence due to ARNSHI PLP variants with 9.5 affected per 100,000 individuals. For East Asians, Latinos, non-Finish Europeans, and South Asians, ARNSHI prevalence due to PLP variants ranges from 17.1 to 33.7 affected per 100,000 individuals. ARNSHI variants that were previously reported in a single ancestry or family were observed in additional populations, e.g., USH1C p.(Q723*) reported in a Chinese family was the most prevalent pathogenic variant observed in gnomAD for African/African Americans. Variability between populations is due to how extensively ARNSHI has been studied, ARNSHI prevalence and ancestry specific ARNSHI variant architecture which is impacted by population history. Our study demonstrates that additional gene and variant discovery studies are necessary for all populations and particularly for individuals of African ancestry.
Subject terms: Genetics, Mutation
Introduction
Of all congenital diseases that occur worldwide, hearing impairment (HI) has the highest rate for age-standardized disability life years [1]. Congenital HI affects 200-300 infants per 100,000 live births [2]. Fifty to sixty percent of congenital HI cases have a genetic etiology, of which 80% are non-syndromic (NS). Of these, ~80% are autosomal recessive (AR), 18% autosomal dominant (AD), 1–3% X-linked, and <1% mitochondrial [3]. Over 100 NSHI genes have been identified of which 68 underlie ARNSHI [4]. Some ARNSHI genes also underlie syndromic HI and/or ADNSHI.
There has been intensive work performed on identifying novel ARNSHI genes since GJB2 was identified in 1997 [5]. Screening known ARNSHI genes for either clinical or research purposes has led to the identification of novel variants in known genes and for pathogenic variants has led to more precise estimates of population-specific frequencies. Understudied populations can show a severe deficit of known ARNSHI pathogenic variants and genes. This deficit in knowledge hampers our understanding of the mechanism of hearing, development of therapeutic strategies, genetic diagnoses, prognosis and genetic counseling.
The goal of this study is to explore the population-specific allelic spectra of pathogenic variants in known ARNSHI genes using data from the Genome Aggregation Database (gnomAD) [6], which includes exome data on 123,136 individuals of African/African American, Ashkenazi Jewish, East Asian, Finnish, Latino, Non-Finnish European, and South Asian ancestries. We found that that the contribution of known pathogenic ARNSHI variants to HI etiology is highly variable between populations. Higher frequency rare variants make the largest contribution to ARNSHI prevalence. Even for populations with a high diversity of ARNSHI variants, HI prevalence due to very rare variants is low. Additionally, some ARNSHI variants that were not previously reported to be involved in ARNSHI for a specific population can play a considerable role in HI etiology for this ancestry group.
Methods
Variant annotation
A list of all ARNSHI related genes (N = 68) were obtained from ClinVar [7], the Deafness Variation Database (DVD) [8] and the Hereditary Hearing Loss Homepage [9]. Classification of variants within these 68 genes [i.e., pathogenic, likely pathogenic, variants of uncertain significance (VUS), likely benign, and benign] were acquired from ClinVar (FullRelease_2018-12) and DVD (v8_2017-19-09). Variants that were classified as pathogenic or likely pathogenic (PLP) in these databases were further studied. Variants with ambiguous pathogenicity e.g., were listed as being both pathogenic and benign or likely benign were further reviewed based on ClinVar submitter assertion criteria and the literature, and it was determined if these variants should truly be classified as PLP. Variants within ARNSHI genes that have been also reported to cause syndromic HI (six genes) or ADNSHI (eight genes) were not included in the analysis. The PLP variants were annotated with gnomAD exome (version 2.0.2_2017-10) population-specific frequencies [6].
GnomAD includes 7,652 Africans/African-Americans (AFR) of which 541 are Sub-Saharan Africans and the remainder are African-Americans; 4,925 Ashkenazi Jews (ASJ); 16,791 Latino Americans (LAT), of which ~11,700 are Mexican-American/Mexican, ~4,500 US-based Latinos, and ~500 of unknown origin; 8,624 East Asians (EAS), amongst there are ~2,000 Taiwanese, ~2,000 Korean, ~900 individuals from Hong Kong, ~100 Japanese, and the remaining ~3,500 individuals of undetermined East Asian ancestry; 11,150 Finns (FIN); 55,860 Non-Finnish Europeans (NFE) of which ~23,750 are North-western Europeans, ~13,100 Swedes, ~7,800 Southern Europeans, ~2,250 Bulgarians, 400 Estonians, and ~14,000 of unknown non-Finnish European ancestry; 15,391 South Asians (SAS), of which ~11,400 are of Pakistani origin and the remaining ~4,000 individuals are of undetermined South Asian ancestry; and 2,743 individuals with population unassigned (OTH). OTH was not further analyzed as an independent population, since their origin is unknown, but they were included in the analysis of the complete gnomAD data set. PLP variants with frequency >5.0 × 10−3 were further evaluated to determine if they were correctly classified by reviewing the literature. Copy number variants (CNVs) for ARNSHI were unavailable in gnomAD and therefore were not included in the analysis.
Population-specific cumulative PLP variant frequencies
Population cumulative PLP variant frequencies were obtained by summing the frequency of every PLP variant regardless of the ARNSHI gene in which it occurred:
Population-specific frequencies and prevalence of ARNSHI
A Χ2 test was used to evaluate each variant site within every population for deviations from Hardy-Weinberg equilibrium (HWE). Using reported gnomAD overall and population-specific frequencies for PLP ARNSHI variants, assuming HWE and linkage equilibrium, we estimated gene-specific cumulative frequency Gfgene for homozygous and compound heterozygous variants as follows:
where n is the number of PLP variant sites for each ARNSHI gene and p is the allele frequency for each variant site within a specific population. We also obtained the population-specific cumulative frequency for homozygous and compound heterozygous variants for all genes by summing the gene-specific estimates:
where g is the total number of genes with PLP variants within a specific population. It should be noted that unlike the cumulative PLP variant frequency, GfTotal is influenced by the number and frequency of variants within a specific gene, since compound heterozygous variants contribute to its frequency. Therefore GfTotal, unlike the cumulative PLP variant frequency, provides information on known PLP variants contribution to ARNSHI etiology.
Assuming the PLP variants in its homozygous or compound heterozygous state are fully penetrant, the prevalence of ARNSHI due to PLP variants was calculated for each population per 100,000 individuals by multiplying Gfgene and GfTotal by 100,000.
Results
ARNSHI variant data set
After evaluation of the ClinVar and DVD data a total of 1183 ARNSHI PLP variants, i.e., synonymous, missense, nonsense, frameshift, insertion/deletion (indel), and splice site were obtained. Of the 1183 PLP ARNSHI variants, 515 (including 71 indels) variants were observed in 53 ARNSHI genes in gnomAD and 668 (including 201 indels) were not observed (Table 2 and Supplementary Tables S1 and S2). Of the 515 variants that were observed in gnomAD none deviated from HWE (p > 5.0 × 10−8).
Table 2.
PLP SNVs and indels
| Number of variant sites observed in gnomAD | Number of variant sites not observed | |||||
|---|---|---|---|---|---|---|
| Functional Classification | SNVs | Indels | Total | SNVs | Indels | Total |
| nonsynonymous SNV | 293 | – | 293 | 308a | – | 308 |
| frameshift deletion | – | 52 | 52 | – | 99 | 99 |
| frameshift insertion | – | 8 | 8 | – | 59 | 59 |
| frameshift substitution | – | – | – | – | 10 | 10 |
| stop-loss | 1 | – | 1 | – | – | – |
| stop-gain | 87 | 3 | 90 | 92 | 3 | 95 |
| non-frameshift deletion | – | 8 | 8 | – | 21 | 21 |
| non-frameshift insertion | – | – | – | – | 4 | 4 |
| non-frameshift substitution | – | – | – | – | 1 | 1 |
| Synonymous | 14 | – | 14 | 1 | – | 1 |
| Splicing | 46 | – | 46 | 63 | 4 | 67 |
| Intronic | 2 | – | 2 | 3 | – | 3 |
| UTR5 | 1 | – | 1 | – | – | – |
| Total number of variant sites | 444 | 71 | 515 | 467 | 201 | 668 |
aIncludes four double nucleotide variants
Forty-three ARNSHI genes were reported to have PLP indels, including GJB2 with the most indels followed by OTOF and MYO15A (Supplementary Tables S1 and S2). In gnomAD a total of 28 genes were observed to harbor 71 indels. The lengths of these indels [3.32 base pairs (bp), 5.50 bp standard deviation (stdev)] were not statistically different (Welch two sample T test p = 0.55) than those not observed (3.78 bp, stdev 5.51).
For 15 ARNSHI genes no PLP variants were found in gnomAD. These genes have a total of 22 known PLP variants, with none of them being reported in more than two families and 11 were reported only in a single family (Supplementary Table S3).
Cumulative PLP variants frequencies
The highest cumulative PLP variant frequency of 6.57% was observed for ASJ, which is 1.7x greater than the frequency observed for NFE (3.76%) and 3.6x greater than the frequency for FIN (1.82%), the lowest cumulative PLP variant frequency observed. AFR had a cumulative variant frequency of 1.96%, the second lowest cumulative PLP variant frequency observed. SAS had a slightly higher cumulative PLP variant frequency (3.99%) than EAS (3.13%). LAT (2.77%) had a higher cumulative PLP variant frequency than FIN or AFR but lower than other populations (Table 1).
Table 1.
Overview of PLP ARNSHI variants by gnomAD population
| Pop | Description | Number of PLP Variantsa | Number of Genesb | Number of Individualsc | Vcumf d | GfTotale | ARNSHI prevalencef |
|---|---|---|---|---|---|---|---|
| AFR | African/African American | 104 | 28 | 7,151 | 1.96 × 10−2 | 5.21 × 10−5 | 5.2 |
| ASJ | Ashkenazi Jewish | 44 | 18 | 4,733 | 6.57 × 10−2 | 9.69 × 10−4 | 96.9 |
| EAS | East Asian | 90 | 29 | 8,205 | 3.13 × 10−2 | 1.71 × 10−4 | 17.1 |
| FIN | Finnish | 42 | 18 | 10,504 | 1.82 × 10−2 | 9.52 × 10−5 | 9.5 |
| LAT | Latino | 158 | 36 | 16,064 | 2.77 × 10−2 | 2.61 × 10−4 | 26.1 |
| NFE | Non-Finnish European | 317 | 44 | 52,253 | 3.76 × 10−2 | 2.67 × 10−4 | 26.7 |
| SAS | South Asian | 162 | 40 | 14,694 | 3.99 × 10−2 | 3.37 × 10−4 | 33.7 |
| ALLg | Total | 515 | 53 | 116,218 | 3.45 × 10−2 | 2.14 × 10−4 | 21.4 |
aThe number of PLP variants cannot be compared between populations due to differences in sample sizes
bInformation on each gene’s contribution to ARNSHI can be found in Supplementary Table S5 and S6
cBased on the average exome read depth i.e., ≥10 × and genotype quality score i.e., ≥20 for PLP variants
dCummulative variant frequency for each population
eEstimated frequency of individuals with ARNSHI due to PLP variants
fARNSHI prevalence due to known PLP variants represented as the number affected per 100,000 individuals
gIncludes all gnomAD populations (including “Other”). Pop: population code; PLP: Pathogenic and Likely pathogenic variants
Comparison of frequency (GfTotal) of individuals with ARNSHI by population
The estimated frequency of individuals with ARNSHI (GfTotal) for each population varies greatly, with AFR (5.21 × 10−5) having the lowest GfTotal and ASJ the highest (9.69 × 10−4), with an 18.4-fold greater GfTotal for ASJ than AFR. SAS has the second highest GfTotal (3.37 × 10−4) and 6.5 fold greater GfTotal than AFR. The ASJ founder population also has a GfTotal which is 10.0x greater than for the FIN founder population (Table 1).
Prevalence of ARNSHI due to PLP variants by population
We estimated the prevalence of ARNSHI etiology due to PLP variants in seven gnomAD populations (Table 1). Prevalence estimates due to known PLP variants display considerable variability between populations, i.e., Africans/African-Americans (5.2 affected per 100,000 individuals), Ashkenazi Jews (96.9 affected per 100,000 individuals), East Asians (17.1 affected per 100,000 individuals), Finns (9.5 affected per 100,000 individuals), Latinos (26.1 affected per 100,000 individuals), non-Finnish Europeans (26.7 affected per 100,000 individuals), and South Asians (33.7 affected per 100,000 individuals) (Table 1). For Ashkenazi Jews, the high ARNSHI prevalence due to PLP variants is largely driven by several high frequency GJB2 variants (Fig. 1; Supplementary Table S4).
Fig. 1.
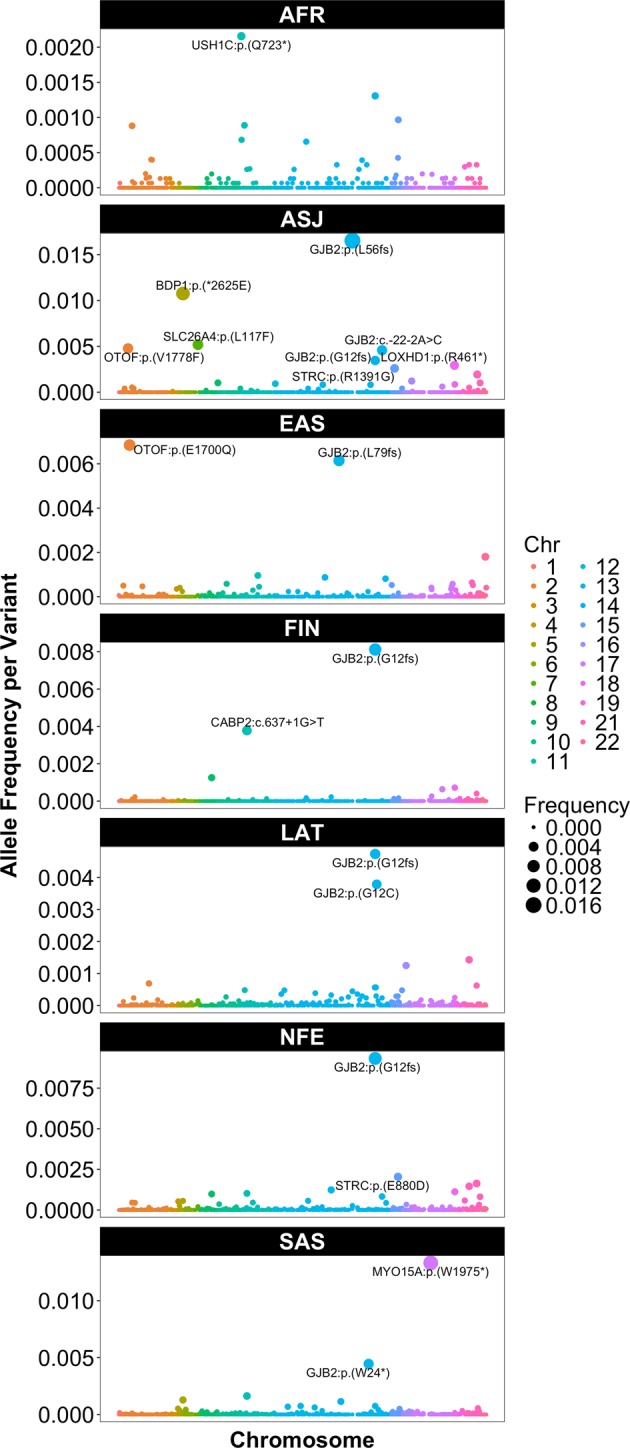
Frequency per PLP ARNSHI variant in each gnomAD population. PLP variants are ordered by chromosome and position. Those with a frequency ≥2.0 × 10−3 in at least one population are labeled (see Supplementary Table S4 for additional information). Note that the Y-axis was adapted for each population due to the large variability in frequencies
Population-specific allelic spectra of variants
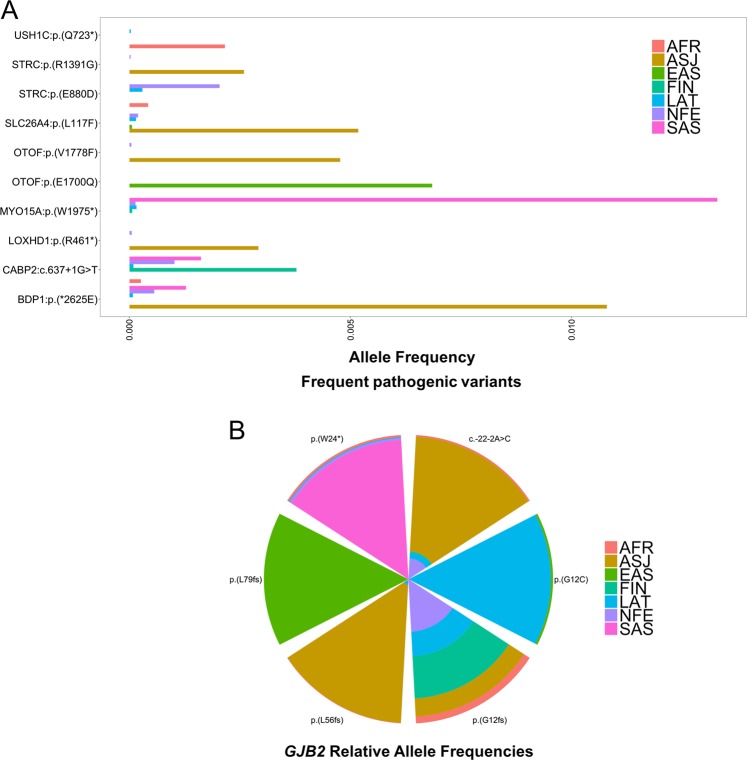
The allelic spectra of the seven gnomAD populations revealed 16 variants with higher frequencies, i.e., frequencies ≥2.0 × 10−3, some exclusive to specific populations (Fig. 1; Fig. 2; Supplementary Table S4; Supplementary Fig. S1). The AFR population had one higher frequency PLP ARNSHI variant, USH1C p.(Q723*) (frequency = 2.16 × 10−3) and the remaining variants all have very low frequencies < 1.4 × 10−3. The only other population where the USH1C p.(Q723*) variant is observed is LAT, an admixed population which would include individuals with African ancestry. In contrast the ASJ population displays eight rare PLP variants with frequencies ≥ 2.0 × 10−3 [BDP1 (1), GJB2 (3), LOXHD1 (1), OTOF (1), SLC26A4 (1) and STRC (1)]. The Asian populations each had two higher frequency rare PLP variants [EAS: OTOF p.(E1700Q) (frequency = 6.85 × 10−3) and GJB2 p.(L79fs) (frequency = 6.15 × 10−3) and SAS; MY015A p.(W1975*) (frequency = 1.33 × 10−2), and GJB2 p.(W24*) (frequency = 4.45 × 10−3)]. Likewise, for FIN and NFE a higher frequency variant is observed in GJB2 p.(G12fs) (c.35delG) [FIN (frequency = 8.12 × 10−3) and NFE (frequency = 9.33 × 10−3)]. Also, FIN has a higher frequency variant in CABP2 c.637 + 1G > T (frequency = 3.78 × 10−3) and NFE in STRC p.(E880D) (frequency = 2.04 × 10−3). For the LAT population both higher frequency variants are within GJB2, i.e., p.(G12fs) (c.35delG) (frequency = 4.74 × 10−3) and p.(G12C) (frequency = 3.79 × 10−3).
Fig. 2.
Frequent PLP ARNSHI variants for each gnomAD population. a Cumulative frequency of all PLP variants with a frequency of ≥2.0 × 10−3 in at least one gnomAD population (with exception of GJB2). Some variants are highly prevalent in a specific population, e.g., USH1C p.(Q723*) in AFR and LOXHD1 p.(R461*) in ASJ. b The relative (%) frequency for all PLP GJB2 variants with a frequency of ≥2.0 × 10−3 in at least one population
GJB2 is established as the most common cause of ARNSHI [10]. The spectrum and prevalence of some GJB2 variants are dependent on population origin [11]. Six GJB2 PLP variants were found to have frequencies ≥2.0 × 10−3 in at least one population (Fig. 2b). The p.(G12fs) (c.35delG) variant is observed in all populations, except for EAS (Supplementary Fig. S2), which is consistent with previous reports [11]. Four of these GJB2 variants are present in higher frequencies in one specific population, i.e., p.(L79fs) [EAS (frequency = 6.15 × 10−3)]; p.(G12C) [LAT (frequency = 3.79 × 10−3)]; c.-22-2A >C [ASJ (frequency = 4.58 × 10−3)]; and p.(L56fs) (c.167delT) [ASJ (frequency = 1.65 × 10−2)] (Fig. 2b), but are observed in low frequencies in other populations, with c.167delT being an established ASJ founder variant [12]. Lastly, p.(W24*) (frequency = 4.45 × 10−3) is highly prevalent in SAS and is an established founder variant in this population [13].
Genes that contribute highly to the prevalence of ARNSHI
Over all gnomAD populations, GJB2 PLP variants are the greatest contributor to ARNSHI prevalence (17.7 affected per 100,000 individuals) compared to PLP variants in other ARNSHI genes. Other genes with a substantial contribution to the prevalence of ARNSHI over all gnomAD populations include TMPRSS3 (1.1 affected per 100,000 individuals), MYO15A (1.0 affected per 100,000 individuals), OTOF (0.5 affected per 100,000 individuals), and STRC (0.4 affected per 100,000 individuals) with variants in these genes being observed in each population. Gene-specific contributions to ARNSHI prevalence are often driven by higher frequency rare PLP variants of which some are ancient founder variants, e.g., for GJB2 [p.(G12fs);c.35delG, p.(W24*), and p.(L56fs);c.167delT] (Fig. 1; Supplementary Tables S4–S7). GJB2 is the primary contributor to ARNSHI prevalence in all gnomAD populations; with the exception of the SAS population with MYO15A being the highest contributors with a prevalence of 20.5 affected per 100,000 individuals and GJB2 having a prevalence of 11.8 affected per 100,000 individuals (Supplementary Table S6) [4]. The contribution of MYO15A to ARNSHI etiology in SAS is driven by p.(W1975*) (frequency = 1.33 × 10−2).
In the AFR population, USH1C plays the second greatest role in ARNSHI etiology with a prevalence of 0.8 affected per 100,000 individuals after GJB2 which has a prevalence of 3.0 affected per 100,000 individuals. The contribution of USH1C in AFR is driven by variant p.(Q723*) (frequency = 2.16 × 10−3). For both the NFE and LAT populations the second highest contributing gene to ARNSHI etiology after GJB2 is TMPRSS3 with two different variants p.(A426T) [LAT (frequency = 1.43 × 10−3)] and p.A138E [NFE (frequency = 1.63 × 10−3]. For the ASJ, EAS, and FIN the second highest contributing genes are BSND1, OTOF, and CABP2, respectively whose frequencies are driven by a single variant (Supplementary Tables S4–S7).
Discussion
The availability of massively parallel sequencing (MPS) has significantly accelerated the identification of genes involved in Mendelian phenotypes. For AR traits with extreme locus heterogeneity the study of multi-generational consanguineous families has been extremely beneficial to identify novel genes. However, the availability of consanguineous pedigrees is limited to specific populations e.g., South Asian, Middle Eastern, and North African. The majority of ARNSHI genes have been identified through the study of consanguineous families. Sequencing of known ARNSHI genes for research or clinical purposes has increased the identification of pathogenic variants in known ARNSHI genes, but has not allowed for the identification of novel genes. Although MPS has accelerated gene identification, many genes and pathogenic variants remain to be identified even in well-studied populations such as South Asians.
The prevalence of ARNSHI due to known PLP variants is influenced by the diversity of genetic variation and their frequency. Additionally, genetic similarity or admixture with well-studied populations can also impact the prevalence of ARNSHI due to known PLP variants. This prevalence is also impacted by the true prevalence of ARNSHI within a population. Within the literature, prevalence estimates for HI are highly variable even for the same population, because of how they are calculated, e.g., different age and decibel level cut-offs, as well as data being obtained from different sources, e.g., audiometry, otoacoustic emissions. The World Health Organization in 2018 reported the prevalence of HI in children (>30 dB in the better hearing ear) with South Asians having the highest prevalence, followed by sub-Saharan Africans, Latin Americans, and Europeans [14]. Additional information which is required to estimate the prevalence of ARNSHI is knowledge of the genetic proportion of HI, that can vary by population due to rates of infectious diseases which impact hearing. For genetic cases of HI, it is necessary to know what proportion is nonsyndromic and AR which can be influenced by consanguinity rates within a population.
We demonstrated that there is great variability in population-specific ARNSHI prevalence due to known PLP variants. Of all the gnomAD populations, with perhaps the exception of the Ashkenazi Jews, there are likely deficits in the estimated prevalence of ARNSHI due to known PLP variants, since pathogenic ARNSHI variants remain to be identified. Of the seven gnomAD populations, the one with the lowest ARNSHI prevalence due to known PLP variants, which mostly have very low frequencies, is the African/African Americans. This deficit is most likely impacted by the lack of genetic HI studies and clinical resequencing than there being a lower prevalence of ARNSHI in African/African-Americans compared to other ancestry groups. The Ashkenazi Jewish population has an ARNSI prevalence due to known PLP variants which is almost 3x higher than that estimated for South Asians, which is the gnomAD population with the second highest ARNSH prevalence due to known PLP variants. The prevalence of HI for Ashkenazi Jews is similar to other European populations [15], therefore the higher prevalence of known PLP variants is most likely due to reduced locus and allelic heterogeneity compared to other populations, the extensive study and clinical sequencing of known ARNSHI genes in this population.
Although a large number of variants were observed for African-Americans, due to their rarity they do not contribute highly to the prevalence of ARNSHI (5.2 affected per 100,000 individuals). Despite extensive genetic heterogeneity for ARNSHI, only a few genes [16] have been studied in sub-Saharan African populations, with the majority of studies investigating GJB2 [17]. There have also been limited studies on known ARNSHI genes in African Americans [18]. No novel ARNSHI genes have been identified in any African ancestry populations. The AFR population in gnomAD mainly consists of African Americans (93%). Pathogenic ARNSHI variants identified in African Americans can originate from Europeans, Sub-Saharan Africans or indigenous American populations. The GJB2 p.(G12fs) (c.35delG) variant, which is frequent in many populations is the second most common PLP variant within the AFR population (Fig. 2). This variant has been observed in Sudanese [19] and African Americans [20] and was likely inherited through European or Middle Eastern admixture. We also found several variants which were previously unknown to be highly prevalent in individuals of African ancestry. For example, USH1C variant p.(Q723*) is prevalent in AFR (frequency = 2.16 × 10−3), but was only previously reported in a Chinese family with ARNSHI [21] and by diagnostic laboratories in ClinVar [21].
The estimated prevalence of ARNSHI in the Finnish population due to known PLP variants is 9.5 affected per 100,000 individuals. The FIN population also has the lowest population-specific cumulative PLP variant frequency (1.82%) observed in gnomAD. Two major factors that might explain these observations: the scarcity of genetic HI studies in the Finnish population [22] and their unique genetic background which include multiple bottlenecks [23], that can cause rare variants to increase in frequency and decrease variant diversity. Therefore, isolated populations, e.g., Finnish and Ashkenazi Jewish often have a depletion of variants in the rare frequency spectrum and can even have damaging variants present at relatively high frequencies [24]. Founder populations can also have increased or decreased frequencies of specific diseases compared to other populations. Bottlenecks could be the reason why the CABP2 c.637 + 1G>T variant, has a high frequency (3.78 × 10−3) in FIN (Fig. 1). This variant was originally identified in three consanguineous Iranian families [25], and is also present in four other gnomAD populations at lower frequencies. This variant is 3.7x and 2.3x more frequent in FIN compared to NFE and SAS, respectively. Possibly, it was introduced into Finland in one of the early waves of immigration, and enriched due to increased drift and reduced selective pressure [24].
The prevalence of HI for Ashkenazi Jews is similar to other European populations [15], but Ashkenazi Jews have an estimated ARNSHI prevalence due to known PLP variants that is almost 3x higher than that for South Asians, which is the gnomAD population with the second highest ARNSHI prevalence due to known PLP variants. The high prevalence of ARNHI due to know PLP variants (96.9 affected per 100,000 individuals) for Ashkenazi Jews is likely because of its genetically isolated background, low diversity including highly prevalent PLP founder variants, and extensive study. The Ashkenazi Jews who have a Middle Eastern origin have undergone population bottlenecks, admixture with other European populations, and positive selection [26]. Interestingly, we did identify variants that are highly present in ASJ [OTOF: p.(V1778F) (frequency = 4.77 × 10−3) and GJB2: c.-22-2A>C (frequency = 4.58 × 10−3)] which were not previously reported to underlie HI in the Ashkenazi Jewish population, with the c.-22-2A>C being highly prevalent in ASJ (Fig. 2b). The c.-22-2A>C variant leads to mild post-lingual HI when homozygous, which may go undiagnosed, and was previously only described in three siblings in Spain [27] and one proband in Italy [28], but has, to our knowledge, never been reported in an Ashkenazi Jewish HI patient.
In the South Asian population, ARNSHI prevalence due to known PLP variants is 33.7 affected per 100,000 individuals. In gnomAD 74% of SAS are Pakistani and the majority of ARNSHI genes have been identified in consanguineous Pakistani pedigrees. Population diversity in South Asia could explain why the prevalence due to known PLP variants is low. The South Asians consist of >4,000 well-defined population groups, shaped by unique cultural, geographical, linguistic, and religious patterns [29].
In contrast to South Asians, for East Asians the ASNSHI due to known PLP variants is 17.1 affected per 100,000 individuals. Novel ARNSHI gene discovery studies in East Asia are less frequent than for South Asians, although, studies examining the allelic spectra of known HI genes in the Han Chinese are numerous [30]. The allelic spectrum of PLP ARNSHI variants in EAS is unique, the GJB2 p.(G12fs) (c.35delG) variant, which is highly prevalent in all other populations is absent, while GJB2 founder variant p.(L79fs) (c.235delC) [31] and OTOF variant p.(E1700Q) are highly present (Fig. 2) [32].
ARNSHI genes and variants explain 26.7 affected per 100,000 individuals in non-Finnish Europeans, although they are very well-studied through research and clinical sequencing of known ARNSHI genes. The extensive study of known ARNSHI genes perhaps explains why of the 53 ARNSHI genes observed in gnomAD, 44 were observed in NFE. However, since ARNSHI gene discovery studies in Europeans are scarce a significant proportion of the prevalence that is not explained may be due to genes that are yet to be discovered.
For Latinos, the prevalence is 26.1 affected per 100,000 individuals for known PLP ARNSHI variants. The LAT population in gnomAD mainly includes samples of Mexican ancestry (70%) and Latinos residing in the US (27%). The low prevalence due to known ARNSHI variants might be attributable to the fact that most HI studies include few Latino participants [33]. Based on reports from single families and small studies, variants such as GJB2 p.(G12fs) (c.35delG) and p.(R216fs) (c.645delTAGA) [34] are most commonly associated with HI in Hispanic and Latino populations [33]. We found that GJB2, TMPRSS3, and OTOF are the most prevalent ARNSHI genes in the LAT gnomAD population. We also detected a GJB2 variant that is highly prevalent in LAT p.(G12C) (c.34G>T) (frequency = 3.79 × 10−3); Fig. 2b, which was reported previously in North American studies in individuals with unreported ethnic background [35, 36], and also individuals of Mexican origin [34]. It was suggested that this variant occurs in the US due to Mexican immigration [34]. Based on the large number of Mexicans in the LAT gnomAD cohort and its frequency here (3.79 × 10−3), p.(G12C) might be an indigenous Mexican founder variant.
It has been recommended to use a population-specific frequency threshold of ≤5.0 × 10−3 for AR traits to elucidate pathogenic variants [37]. We have included a few variants that exceeded this threshold for one population, i.e., three variants in GJB2, and one variant each in MYO15A, OTOF, BDP1, and SLC26A4 (Supplementary Table S1). Some ARNSHI variants have an exceptionally high frequency compared to those for other Mendelian phenotypes. For example, several GJB2 variants are founder variants that are highly prevalent in specific populations which may be due to a selective advantage for heterozygote carriers (Fig. 2) [38]. The MYO15A p.(W1975*) variant was originally identified in two Iranian families [39]. This variant, a suspected founder allele in the South Asian population, affects an exon of MYO15A which shows alternate splicing in the inner ear [40], and was suggested as exempt to the frequency cut-off rule [37]. The BDP1 p.(*2625E) variant causes an elongation of 11 residues of the BDP1 protein [41], and the only reported ARNSHI variant in this gene so far. Individuals that are homozygous for this variant usually have high frequency HI and therefore many affected individuals may go undiagnosed. SLC26A4 [p.(L117F)] [42] and OTOF [p.(E1700Q)] [32] variants were also included in this study since they are considered pathogenic by a large number of sources, including recent evaluations by clinical laboratories. It should be noted that SLC26A4 [p.(L117F)] has been reported for both non-syndromic HI and Pendred syndrome in ClinVar.
Some of the missing ARNSHI prevalence in all populations could be due to larger CNVs and genomic re-arrangements, which are, at the time of this study, unavailable in gnomAD. It is well known that CNVs contribute to ARNSHI etiology. One ARNSHI gene with a large number of CNVs is STRC, which contributes predominantly to individuals of European ancestry, but is rare in other populations [42]. The del(GJB6-D13S1830) deletion is common amongst Ashkenazi Jews and Western Europeans [43].
There are six PLP regulatory and intronic variants of which three are not observed in gnomAD exomes (Table 2). These three intronic variants, HGF (c.482 + 1986_1988delTGA and c.482 + 1991_2000delGATGATGAAA) and MARVELD2 (c.1331 + 1_4delGTGA) were all previously identified in Pakistani families [44], are not present in gnomAD genomes which could be due to there being no SAS genomes in gnomAD or their extremely low frequencies or absents in other populations. This deficit of ARNSHI PLP intronic and regulatory variants could be due to the understudy of these genomic regions, as previous commonly performed Sanger sequencing-based gene discovery studies generally only focused on exons. Although currently custom, exome or whole genome sequencing is usually performed, even with WGS providing appropriate data for their discovery, noncoding variants remain difficult to interpret.
Most PLP variants which were not observed in gnomAD are extremely rare. For indels there is no difference in the length between those observed in gnomAD and those which were not, so the ability to identify indels likely does not impact their lack of inclusion in gnomAD. In addition to the rarity of variants impacting frequency in gnomAD, population ascertainment can also affect whether a variant is detected in gnomAD, e.g., poor representation of individuals of Arabic ancestry in gnomAD.
In general, the missing genetic contributions to ARNSHI prevalence can also be attributed to our limited knowledge on pathogenic ARNSHI variants. It has been hypothesized that there may be >1,000 genes contributing to HI etiology [45] and therefore likely many ARNSHI genes have not yet been identified, particularly in understudied and diverse populations such as Sub-Saharan Africans.
The results of this study highlight the need to further study ARNSHI even though many genes have been discovered to date. Although it is clear there is still a necessity to study consanguineous families to identify novel ARNSHI genes, it would be of great benefit to also study non-consanguineous families from European, East Asian, Latino, and African populations to discover novel ARNSHI genes, as some might be ancestry specific. A better understanding of noncoding variants will aid in elucidating their contribution to ARNSHI etiology. In addition, the allelic spectrum of pathogenic variants of the known ARNSHI genes has not been fully elucidated, and the further examination of these genes in understudied populations will highly improve our knowledge on pathogenic ARNSHI variants.
Web Resources
Centers for Disease Control and Prevention (CDC), https://www.cdc.gov/
Deafness Variation Database, http://deafnessvariationdatabase.org/
Genome Aggregation Database (gnomAD), http://gnomad.broadinstitute.org/about
Hereditary Hearing Loss Homepage, http://hereditaryhearingloss.org/
Supplementary information
Acknowledgements
This work was supported by the National Institute on Deafness and Other Communication Disorders grants R01 DC011651, R01 DC003594, and R01 DC016593, and the National Institute of Genome Research Grant UM1 HG006493 to SML. LCF was supported by the Swiss National Science Foundation (Advanced Postdoc Mobility 177853).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41431-019-0417-2) contains supplementary material, which is available to authorized users.
References
- 1.Reference GH. Nonsyndromic hearing loss. Genetics Home Reference.https://ghr.nlm.nih.gov/condition/nonsyndromic-hearing-loss. Accessed 23 Oct 2018.
- 2.Quick statistics about hearing. NIDCD. 2015. https://www.nidcd.nih.gov/health/statistics/quick-statistics-hearing. Accessed 23 Oct 2018.
- 3.Morton CC, Nance WE. Newborn Hearing screening - a silent revolution. New Engl J Med. 2006;354:2151–64. doi: 10.1056/NEJMra050700. [DOI] [PubMed] [Google Scholar]
- 4.Mahboubi H, Dwabe S, Fradkin M, Kimonis V, Djalilian HR. Genetics of hearing loss: where are we standing now? Eur Arch Otorhinolaryngol. 2012;269:1733–45. doi: 10.1007/s00405-011-1910-6. [DOI] [PubMed] [Google Scholar]
- 5.Shearer AE, Hildebrand MS, Smith RJ. Hereditary hearing loss and deafness overview. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJ, Stephens K, et al., editors. GeneReviews®. Seattle (WA): University of Washington; 1993.
- 6.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shearer AE, Eppsteiner RW, Booth KT, Ephraim SS, Gurrola J, Simpson A, et al. Utilizing ethnic-specific differences in minor allele frequency to recategorize reported pathogenic deafness variants. Am J Hum Genet. 2014;95:445–53. doi: 10.1016/j.ajhg.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hereditary Hearing Loss - Hereditary Hearing loss Homepage. http://hereditaryhearingloss.org/
- 10.Cohn ES, Kelley PM. Clinical phenotype and mutations in connexin 26 (DFNB1/GJB2), the most common cause of childhood hearing loss. Am J Med Genet. 1999;89:130–6. doi: 10.1002/(SICI)1096-8628(19990924)89:3<130::AID-AJMG3>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 11.Dzhemileva LU, Barashkov NA, Posukh OL, Khusainova RI, Akhmetova VL, Kutuev IA, et al. Carrier frequency of GJB2 gene mutations c.35delG, c.235delC and c.167delT among the populations of Eurasia. J Hum Genet. 2010;55:749–54. doi: 10.1038/jhg.2010.101. [DOI] [PubMed] [Google Scholar]
- 12.Morell RJ, Kim HJ, Hood LJ, Goforth L, Friderici K, Fisher R, et al. Mutations in the Connexin 26 Gene (GJB2) among Ashkenazi Jews with Nonsyndromic Recessive Deafness. New Engl J Med. 1998;339:1500–5. doi: 10.1056/NEJM199811193392103. [DOI] [PubMed] [Google Scholar]
- 13.Salman M, Bashir R, Imtiaz A, Maqsood A, Mujtaba G, Iqbal M, et al. Mutations of GJB2 encoding connexin 26 contribute to nonsyndromic moderate and severe hearing loss in Pakistan. Eur Arch Otorhinolaryngol. 2015;272:2071–5. doi: 10.1007/s00405-015-3523-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.WHO | Estimates. WHO. http://www.who.int/deafness/estimates/en/. Accessed 23 Oct 2018.
- 15.International-Hearing-Loss-Data-Table. https://www.cdc.gov/ncbddd/hearingloss/documents/International-Hearing-Loss-Data-Table.pdf. Accessed 23 Oct 2018.
- 16.Meyer CG, Gasmelseed NM, Mergani A, Magzoub MMA, Muntau B, Thye T, et al. Novel TMC1 structural and splice variants associated with congenital nonsyndromic deafness in a Sudanese pedigree. Hum Mutat. 2005;25:100. doi: 10.1002/humu.9302. [DOI] [PubMed] [Google Scholar]
- 17.Lasisi AO, Bademci G, Foster J, Blanton S, Tekin M. Common genes for non-syndromic deafness are uncommon in sub-Saharan Africa: a report from Nigeria. Int J Pedia Otorhinolaryngol. 2014;78:1870–3. doi: 10.1016/j.ijporl.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rudman JR, Kabahuma RI, Bressler SE, Feng Y, Blanton SH, Yan D, et al. The genetic basis of deafness in populations of African descent. J Genet Genom. 2017;44:285–94. doi: 10.1016/j.jgg.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 19.Gasmelseed NMA, Schmidt M, Magzoub MMA, Macharia M, Elmustafa OM, Ototo B, et al. Low frequency of deafness-associated GJB2 variants in Kenya and Sudan and novel GJB2 variants. Hum Mutat. 2004;23:206–7. doi: 10.1002/humu.9216. [DOI] [PubMed] [Google Scholar]
- 20.Pandya A, Arnos KS, Xia XJ, Welch KO, Blanton SH, Friedman TB, et al. Frequency and distribution of GJB2 (connexin 26) and GJB6 (connexin 30) mutations in a large North American repository of deaf probands. Genet Med. 2003;5:295–303. doi: 10.1097/01.GIM.0000078026.01140.68. [DOI] [PubMed] [Google Scholar]
- 21.Ouyang XM, Xia XJ, Verpy E, Du LL, Pandya A, Petit C, et al. Mutations in the alternatively spliced exons of USH1C cause non-syndromic recessive deafness. Hum Genet. 2002;111:26–30. doi: 10.1007/s00439-002-0736-0. [DOI] [PubMed] [Google Scholar]
- 22.Soini HK, Karjalainen MK, Hinttala R, Rautio A, Hallman M, Uusimaa J. Mitochondrial hearing loss mutations among Finnish preterm and term-born infants. Audio Res. 2017;7:189. doi: 10.4081/audiores.2017.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jakkula E, Rehnström K, Varilo T, Pietiläinen OPH, Paunio T, Pedersen NL, et al. The genome-wide patterns of variation expose significant substructure in a founder population. Am J Hum Genet. 2008;83:787–94. doi: 10.1016/j.ajhg.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chheda H, Palta P, Pirinen M, McCarthy S, Walter K, Koskinen S, et al. Whole-genome view of the consequences of a population bottleneck using 2926 genome sequences from Finland and United Kingdom. Eur J Hum Genet. 2017;25:477–84. doi: 10.1038/ejhg.2016.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schrauwen I, Helfmann S, Inagaki A, Predoehl F, Tabatabaiefar MA, Picher MM, et al. A mutation in CABP2, expressed in cochlear hair cells, causes autosomal-recessive hearing impairment. Am J Hum Genet. 2012;91:636–45. doi: 10.1016/j.ajhg.2012.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bray SM, Mulle JG, Dodd AF, Pulver AE, Wooding S, Warren ST. Signatures of founder effects, admixture, and selection in the Ashkenazi Jewish population. PNAS. 2010;107:16222–7. doi: 10.1073/pnas.1004381107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gandía M, Del Castillo FJ, Rodríguez-Álvarez FJ, Garrido G, Villamar M, Calderón M, et al. A novel splice-site mutation in the GJB2 gene causing mild postlingual hearing impairment. PLoS ONE. 2013;8:e73566. doi: 10.1371/journal.pone.0073566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stanghellini I, Genovese E, Palma S, Ravani A, Falcinelli C, Guarnaccia MC, et al. New and rare GJB2 alleles in patients with nonsyndromic sensorineural hearing impairment: a genotype/auditory phenotype correlation. Genet Test Mol Biomark. 2014;18:839–44. doi: 10.1089/gtmb.2014.0185. [DOI] [PubMed] [Google Scholar]
- 29.Ayub Q, Tyler-Smith C. Genetic variation in South Asia: assessing the influences of geography, language and ethnicity for understanding history and disease risk. Brief Funct Genom. 2009;8:395–404. doi: 10.1093/bfgp/elp015. [DOI] [PubMed] [Google Scholar]
- 30.Yang T, Wei X, Chai Y, Li L, Wu H. Genetic etiology study of the non-syndromic deafness in Chinese Hans by targeted next-generation sequencing. Orphanet J Rare Dis. 2013;8:85. doi: 10.1186/1750-1172-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yao J, Lu Y, Wei Q, Cao X, Xing G. A systematic review and meta-analysis of 235delC mutation of GJB2 gene. J Transl Med. 2012;10:136. doi: 10.1186/1479-5876-10-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang F, Ma D, Wang Y, Qiu Y, Liu F, Wang Q, et al. Novel compound heterozygous mutations in the OTOF Gene identified by whole-exome sequencing in auditory neuropathy spectrum disorder. BMC Med Genet. 2017;18:35. doi: 10.1186/s12881-017-0400-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mittal R, Patel AP, Nguyen D, Pan DR, Jhaveri VM, Rudman JR, et al. Genetic basis of hearing loss in Spanish, Hispanic and Latino populations. Gene. 2018;647:297–305. doi: 10.1016/j.gene.2018.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hernández-Juárez AA, Lugo-Trampe J, de J, Campos-Acevedo LD, Lugo-Trampe A, Treviño-González JL, de-la-Cruz-Ávila I, et al. GJB2 and GJB6 mutations are an infrequent cause of autosomal-recessive nonsyndromic hearing loss in residents of Mexico. Int J Pedia Otorhinolaryngol. 2014;78:2107–12. doi: 10.1016/j.ijporl.2014.09.016. [DOI] [PubMed] [Google Scholar]
- 35.Putcha GV, Bejjani BA, Bleoo S, Booker JK, Carey JC, Carson N, et al. A multicenter study of the frequency and distribution of GJB2 and GJB6 mutations in a large North American cohort. Genet Med. 2007;9:413–26. doi: 10.1097/GIM.0b013e3180a03276. [DOI] [PubMed] [Google Scholar]
- 36.Tang HY, Fang P, Ward PA, Schmitt E, Darilek S, Manolidis S, et al. DNA Sequence Analysis of GJB2, Encoding Connexin 26. Am J Med Genet A. 2006;140:2401–15. doi: 10.1002/ajmg.a.31525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rehman AU, Bird JE, Faridi R, Shahzad M, Shah S, Lee K, et al. Mutational Spectrum of MYO15A and the Molecular Mechanisms of DFNB3 Human Deafness. Hum Mutat. 2016;37:991–1003. doi: 10.1002/humu.23042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.D’Adamo P, Guerci VI, Fabretto A, Faletra F, Grasso DL, Ronfani L, et al. Does epidermal thickening explain GJB2 high carrier frequency and heterozygote advantage? Eur J Hum Genet. 2009;17:284–6. doi: 10.1038/ejhg.2008.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fattahi Z, Shearer AE, Babanejad M, Bazazzadegan N, Almadani SN, Nikzat N, et al. Screening for MYO15A gene mutations in autosomal recessive nonsyndromic, GJB2 negative Iranian deaf population. Am J Med Genet A. 2012;158A:1857–64. doi: 10.1002/ajmg.a.34411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang Y, Wang A, Belyantseva IA, Anderson DW, Probst FJ, Barber TD, et al. Characterization of the human and mouse unconventional myosin XV genes responsible for hereditary deafness DFNB3 and shaker 2. Genomics. 1999;61:243–58. doi: 10.1006/geno.1999.5976. [DOI] [PubMed] [Google Scholar]
- 41.Girotto G, Mezzavilla M, Abdulhadi K, Vuckovic D, Vozzi D, Khalifa Alkowari M, et al. Consanguinity and hereditary hearing loss in Qatar. Hum Hered. 2014;77:175–82. doi: 10.1159/000360475. [DOI] [PubMed] [Google Scholar]
- 42.Sloan-Heggen CM, Bierer AO, Shearer AE, Kolbe DL, Nishimura CJ, Frees KL, et al. Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum Genet. 2016;135:441–50. doi: 10.1007/s00439-016-1648-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.del Castillo I, Moreno-Pelayo MA, del Castillo FJ, Brownstein Z, Marlin S, Adina Q, et al. Prevalence and Evolutionary Origins of the del(GJB6-D13S1830) Mutation in the DFNB1 Locus in Hearing-Impaired Subjects: a Multicenter Study. Am J Hum Genet. 2003;73:1452–8. doi: 10.1086/380205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shafique S, Siddiqi S, Schraders M, Oostrik J, Ayub H, Bilal A, et al. Genetic spectrum of autosomal recessive non-syndromic hearing loss in pakistani families. PLoS ONE. 2014;9:e100146. doi: 10.1371/journal.pone.0100146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holder S. Hereditary hearing loss and its syndromes Robert J. Gorlin, Helen V. Toriello, and M. Michael J. Cohen, Jr. New York: Oxford University Press, 1995, 457 pp. Am J Med Genet. 1996;61:101.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.