Abstract
Height is a heritable and highly heterogeneous trait. Short stature affects 3% of the population and in most cases is genetic in origin. After excluding known causes, 67% of affected individuals remain without diagnosis. To identify novel candidate genes for short stature, we performed exome sequencing in 254 unrelated families with short stature of unknown cause and identified variants in 63 candidate genes in 92 (36%) independent families. Based on systematic characterization of variants and functional analysis including expression in chondrocytes, we classified 13 genes as strong candidates. Whereas variants in at least two families were detected for all 13 candidates, two genes had variants in 6 (UBR4) and 8 (LAMA5) families, respectively. To facilitate their characterization, we established a clustered network of 1025 known growth and short stature genes, which yielded 29 significantly enriched clusters, including skeletal system development, appendage development, metabolic processes, and ciliopathy. Eleven of the candidate genes mapped to 21 of these clusters, including CPZ, EDEM3, FBRS, IFT81, KCND1, PLXNA3, RASA3, SLC7A8, UBR4, USP45, and ZFHX3. Fifty additional growth-related candidates we identified await confirmation in other affected families. Our study identifies Mendelian forms of growth retardation as an important component of idiopathic short stature.
Subject terms: DNA sequencing, Disease genetics, Genetic counselling
Introduction
Human height is a heritable and highly heterogeneous trait [1]. Efforts to understand the genetic basis of growth have employed genome-wide association studies (GWAS) to systematically assess the effect on human height variation of common variants with a minor allele frequency > 5% [2]. 697 variants, mainly located in 423 noncoding loci, have been implicated in height variance in the population [2, 3]. Subsequent studies on rare variants, both at the nucleotide and genomic levels, further expanded the number of associated loci [3, 4]. So far, rare and common height-associated variants together explain about 27.4% of height heritability [3]. In addition, it is known from Mendelian forms of growth retardation that rare, large effect-size variants can have extremely large effects on growth development [3].
Short stature, defined auxologically as a height two standard deviations below the mean height in the population, affects about 3% of individuals and is a common medical concern. In a recent study combining systematic phenotyping and exome-based sequencing, we were able to identify a genetic cause in up to 33% of individuals with idiopathic short stature (ISS) [5]. Consequently, 67% of the affected individuals remained undiagnosed. Most forms of short stature have been attributed to Mendelian causes, highlighting defects in a diverse range of functional pathways [6, 7]. The most common monogenic causes include defects of the SHOX gene (2.4%) [8], heterozygous variants in ACAN (1.4%) [9] and many genes for rare syndromic forms as well as skeletal dysplasias [8, 10–12]. At least 477 genes have been found to affect human growth [13], but as yet there are no reliable estimates of the number of growth-associated genes. For most of these genes, though, no association with short stature has been found in humans. Affected individuals and their families would thus benefit from the identification of further genes associated with growth retardation. In this study in 254 unrelated individuals with ISS and their families, we used exome sequencing to identify and characterize novel candidate genes based on evolutionarily conserved networks.
Materials and methods
Individuals
The study was approved by the ethics committee of the Friedrich-Alexander-Universität Erlangen-Nürnberg (FAU). 565 individuals and their families were referred by medical specialists for evaluation of growth retardation after endocrine defects of the growth hormone pathway and other organic causes of growth retardation were excluded. After previous targeted testing including array analysis in some and exome sequencing failed to identify a known cause, we further assessed the exomes of 254 well-characterized families with at least 1 offspring whose growth standard deviation score (SDS) was ≥2 below the mean population height and/or the estimated family (est. height in Table 1) for potential candidate genes (Table 1 and Supplementary Table 2). Participants’ mean age (±standard deviation) was 9.2 ± 0.43 years, and 155/254 (61%) participants were female. Most index individuals (53%) presented with a height between 3 and 2 SDS below the mean, and 32% were born small for gestational age (Supplementary Figure 1). In 68/254 (27%) participants, additional features such as microcephaly, syndactyly, nail dysplasia or any nonspecific facial gestalt resulted in a diagnosis of syndromic short stature.
Table 1.
Clinical characteristics of 254 individuals with idiopathic short stature after exclusion of known causes
| Characteristic | No. (%) |
|---|---|
| Age group | |
| <4 yrs | 43 (17) |
| >4 yrs | 211 (83) |
| Small for gestational age | 81 (32) |
| Short stature [SDS]a | |
| −2 to −3 | 135 (53) |
| −3 to −4 | 55 (22) |
| −4 to −5 | 12 (5) |
| <−5 | 4 (2) |
| Below est. heightb | 48 (19) |
| Short stature type | |
| Isolated | 186 (73) |
| Syndromic | 68 (27) |
| Head circumference [SDS] | |
| >−2 | 153 (60) |
| −2 to −3 | 38 (15) |
| −3 to −5 | 32 (13) |
| <−5 | 3 (1) |
| Not available | 28 (11) |
| IQ | |
| Normal | 203 (80) |
| 70–85 | 51 (20) |
| Sex | |
| Female | 155 (61) |
| Male | 99 (39) |
| Bone age | |
| Accelerated | 11 (4) |
| Normal | 21 (8) |
| Delayed | 66 (26) |
| Not available | 156 (61) |
aAll 254 affected individuals presented with a height below the est. final adult height (est. height)
bAffected individuals with a height above −2 SDS, but below the est. height
Exome sequencing and variant assessment
We performed whole-exome sequencing in 185 affected individuals and both of their respective parents (trio analysis) and in 69 affected individuals (affected-only analysis) after enrichment by SureSelect targeted capturing on HiSeq 2500 (94.3%) or SOLiD 5500xl (5.7%). Exomes were analyzed by semiautomatic selection and data quality inspection of variants, followed by the interpretation in relation to the reported phenotypic spectrum (Supplementary Figures 2 and 3). Variants and familial segregation were confirmed by Sanger sequencing.
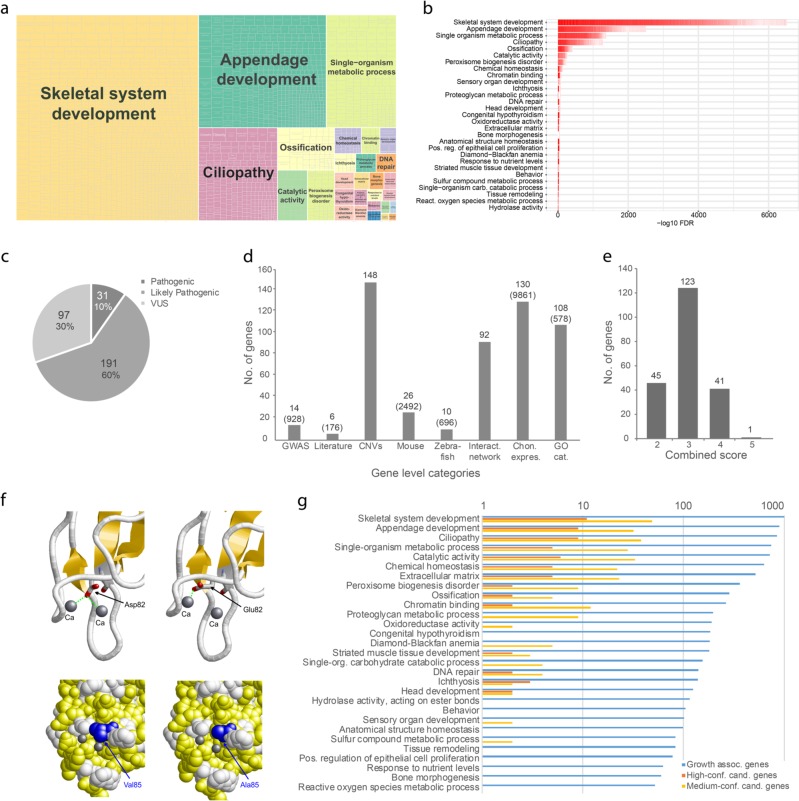
All 185 trios were analyzed for variants with de novo, compound heterozygous, homozygous, and X-linked recessive inheritance. We classified variants according to their population frequency and potential effect on gene function (Supplementary methods, Fig. 1c and Supplementary Tables 1–3). Population data from gnomAD was considered as most likely appropriate controls. After excluding benign and likely benign variants, we evaluated affected genes for their relevance to growth phenotypes. For this gene-level assessment, we included information from association studies, copy number variants, gene ontology (GO) terms, protein–protein interaction data, suitable mouse and zebrafish models, and a previous exome study [14] (Fig. 1d and Supplementary Table 4). Additional data on expression in chondrocytes was obtained by RNA sequencing (RNASeq) of cartilage tissue for all genes studied (Fig. 1d, Supplementary Methods and Figure 4). The combined results of the variant-level and gene-level assessments were then used to identify genes, which were further investigated with respect to the observed mode of inheritance in the 69 affected-only exomes (Fig. 1e, Supplementary Methods, Supplementary Figure 2 and Supplementary Table 5). Genes were finally classified as high-confidence and medium-confidence candidates based on the number of affected individuals and the combined variant-level and gene-level scores (Supplementary Table 6 and Supplementary Methods). Variants were uploaded to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/, Submission ID SUB5032330).
Fig. 1.
Enrichment analysis and candidate gene characterization. a Functional clustering of 1,025 known growth and short stature genes (b) into 29 biological clusters for growth. c Variant level assessment. Number of variants affecting function or likely affecting function and variants of unknown significance identified (Supplementary Tables 1, 2). d Categories of gene level assessment. Numbers represent the genes to which each category applies (see Supplementary Table 4). Numbers in brackets represent the genes among all selected known growth and short-stature genes. e The results of the variant and gene level evaluation were merged to a combined score (Shown is the highest score for each gene, Supplementary Table 5). f Based on structure analysis of variants of unknown significance (VUS), 5 variants in 4 of the high-confidence candidate genes were reclassified to likely pathogenic. Model of the RASA3 C2-domain showing the site of the Asp82Glu and Val85Ala variants. Both residues are located in a pocket of the C2 domains that contains two Ca2+ ions (Ca). Asp82 forms interactions with both Ca2+ ions (green dotted lines), whereas the longer glutamate side chain of the Asp82Glu variant can only interact with one of the Ca2+ ions, probably leading to a loss of the second Ca2+ ion from the binding pocket. Val85 (blue) is located on the lateral wall of this pocket, and the shorter alanine side chain in the Val85Ala variant affects the width of the pocket. g Distribution of the 63 high- and medium-confidence candidate genes in the growth-associated clusters
Functional clustering analysis
Functional enrichment analysis was performed using the Database for Annotation, Visualization, and Integrated Discovery, which comprises 1025 Genes using the keywords “growth delay” from the Human Phenotype Ontology (HPO) database and “short stature” from Online Mendelian Inheritance in Man (OMIM) and MedGen (Supplementary methods). Proteins were submitted to DAVID using human gene Ensembl identifiers. Significantly overrepresented annotation terms were retrieved with the options GOTERM_MF_ALL, GOTERM_CC_ALL, GOTERM_BP_ALL, KEGG_PATHWAY, UP_KEYWORDS, and OMIM_DISEASE. A false discovery rate (FDR) of 0.05 by the Benjamini and Hochberg approach was used to determine significant enrichment using all human gene Ensembl identifiers in the BioMart database as the background (Supplementary methods). Functional Annotation Clustering was performed using the DAVID functional annotation clustering tool with the following parameters: overlap = 5, initialSeed = 5, finalSeed = 5, linkage = 0.5, kappa = 20. This tool implements a fuzzy clustering algorithm to cluster functional annotation terms based on the degree of the overlap between associated genes. Raw p-values were used to compute the initial clusters. Annotation terms with an FDR > 0.05 were subsequently pruned. Given a list of candidate genes and an annotation term, we calculated (i) the odds of a human gene (Ensembl gene identifier) in the list being associated with the annotation term; and (ii) the odds of a human gene (Ensembl gene identifier) not in the list being associated with the term. The odds ratio was calculated by dividing the odds from (i) by the odds from (ii) using the fisher.test () function implemented in the R environment for statistical computing. Results were presented as a treemap based on the functional annotation clustering using R’s treemap package.
Protein structure analysis
For all variants of unknown significance in high-confidence candidate genes and for missense variants in RASA3, we performed in silico structural analyses and reclassified variants based on predicted functional consequences (Fig. 1f and Supplementary Figure 8). Models for all wild-type protein domains were either obtained from Modbase [15] or modelled using HHpred [16] and Modeller [17]. The variant amino acid was exchanged using Swiss-Model [18] and visualized with RasMol [19]. Detailed information on individual modelling is provided in the Methods section in the supplementary information.
Results
Identification of novel candidate genes for ISS
Characterization of involved variants and genes and consideration of independent affected individuals carrying these variants revealed 63 candidate genes in 92 (36%) of the 254 affected individuals included in the analysis. We classified 13 genes as high-confidence genes and the remaining 50 genes as medium-confidence candidates (Tables 2 and 4, Supplementary Tables 7 and 11, and Supplementary Figures 6–18). The mode of inheritance was mainly autosomal dominant (71%), followed by X-linked recessive (17%), and autosomal recessive (11%). The most abundant variants were missense variants (78%), followed by nonsense (15%), splice site (6%), and non-frameshift insertion / deletion variants (1%). Nonsense variants were identified in the high-confidence candidate genes RASA3 and USP45. Based on in silico structural analysis, we reclassified 5 variants in 4 of the high-confidence candidate genes from “variants of unknown significance” (VUS) to “likely pathogenic” (Fig. 1f and Supplementary Table 7).
Table 2.
Categories, function and phenotype overview of identified high-confidence candidate genes
| Gene | No. pats. | Variant level classificatione | Gene level classification [No. of cat.]c | Combined score [1–4] | Main pathwayd | Phenotype | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| V | IV | III | Heigth [SDS ± s] | OFC [SDS ± s] | Syndromic | Common features (no. of individuals) | |||||
| CPZ | 3a | 3 | 1 | 3 | −3.6 ± 1.4 | −2.3 ± 1 | 0/3 | Short neck (2/3), hypertelorism (2/3), low set ears (3/3), wide intermammillary distance (2/3) | |||
| EDEM3 | 2 | 2 | 3 | 3 | Protein processing | −2 ± 0.1 | −1.7 ± 0.7 | 0/3 | High arched eye brows (2/2), thin upper lip (2/2) | ||
| FBRS | 3a | 2 | 1 | 3 | 3 | −2.2 ± −0.2 | −2 ± 0.4 | 0/3 | Thin hair (2/3), prominent ears (2/3), thin lips (2/3), brachydactyly (2/3) | ||
| IFT81 | 2 | 2 | 2 | 3 | Cilium Assembly | −2.9 ± 0 | −3.1 ± 1 | 0/3 | wide nasal bridge (2/2), high arched eye brows (2/2) | ||
| KCND1 | 2 | 2 | 1 | 3 | Cardiac conduction | −2.9 ± 0.4 | −2.4 ± 1.1 | 0/2 | Fair hair (2/2), sparse eye brows (2/2), thin lips (2/2), brachydactyly (2/2) | ||
| LAMA5 | 8b | 4 | 4 | 4 | 3 | Human papillomavirus infection | −2.9 ± 0.6 | −1.4 ± 1.1 | 2/8 | Thin lips (4/8), barrel chest (5/8), sandal gap (6/8) | |
| MED24 | 2 | 1 | 1 | 5 | 4 | Thyroid hormone signaling | −2.5 ± 0.4 | −1.5 ± 0.3 | 0/2 | Thin lips (2/2), brachydactyly (2/2) | |
| PLXNA3 | 2 | 2 | 5 | 3 | Axon guidance | −2 ± 0.3 | −0.8 ± 1 | 0/2 | Lateral hypoplasia of brows (2/2), broad thumbs (2/2) | ||
| RASA3 | 2 | 1 | 1 | 4 | 3–4 | Ras signaling | −2.7 ± 0.5 | −2 ± 0 | 0/2 | Broad nasal tip (2/2), thin lips (2/2), barrel chest (2/2) | |
| SLC7A8 | 2 | 2 | 3 | 3 | Protein degradation | −2.9 ± 0.7 | −2 ± 0.5 | 0/2 | Brachydactyly (2/2) | ||
| UBR4 | 6b | 5 | 1 | 4 | 2–3 | Human papillomavirus infection | −2.9 ± 0.5 | −1 ± 1.2 | 3/6 | Lateral sparse brows (4/6), barrel chest (4/6), brachydactyly (5/6) | |
| USP45 | 4 | 1 | 2 | 1 | 2 | 2–4 | DNA Repair | −2.9 ± 0.7 | −1.6 ± 1.1 | 0/4 | Brachydactyly (4/4) |
| ZFHX3 | 2 | 1 | 1 | 5 | 3–4 | Regulat.pluripotency of stem cells | −2.5 ± 0.7 | −2 ± 0 | 0/2 | Brachydactyly (2/2), thin lips (2/2), barrel chest (2/2) | |
aSegregation not available in 1 individual
bSegregation not available in 4 individuals
cmaximum of 8 gene level categories
dPathway from KEGG/Reactome with the highest frequency of selected genes with growth phenotype (see also Supplementary Table 10
eVariant level classification (Supplementary Tables 1-4): III—Variant of unknown significance, IV—likely pathogenic, V—pathogenic
Table 4.
Results of gene cluster analysis and functional distribution growth associated genes and candidate genes
| Cluster name | Growth associated genes | High-confidence candidate genes | Medium-confidence candidate genes | ||||
|---|---|---|---|---|---|---|---|
| No. | Mean fold-enrichment | No. | Mean fold-enrichment | Name | No. | Mean fold-enrichment | |
| Skeletal system development | 996 | 3.2 | 11 | 0.7 | CPZ, EDEM3, FBRS, IFT81, KCND1, PLXNA3, RASA3, SLC7A8, UBR4, USP45, ZFHX3 | 49 | 0.9 |
| Appendage development | 894 | 2.8 | 9 | 0.5 | CPZ, EDEM3, FBRS, IFT81, KCND1, PLXNA3, RASA3, SLC7A8, ZFHX3 | 32 | 1.4 |
| Ciliopathy | 845 | 3.6 | 9 | 1.5 | EDEM3, IFT81, KCND1, PLXNA3, RASA3, SLC7A8, UBR4, USP45, ZFHX3 | 38 | 1.2 |
| Single-organism metabolic process | 740 | 3.7 | 5 | 0.1 | CPZ, EDEM3, KCND1, RASA3, SLC7A8 | 28 | 0.6 |
| Catalytic activity | 721 | 1.7 | 6 | 0.2 | CPZ, EDEM3, RASA3, UBR4, USP45, ZFHX3 | 33 | 1.8 |
| Chemical homeostasis | 629 | 1.9 | 5 | 1.0 | EDEM3, IFT81, KCND1, RASA3, SLC7A8 | 22 | 0.8 |
| Extracellular matrix | 521 | 1.4 | 5 | 1.1 | CPZ, EDEM3, FBRS, PLXNA3, SLC7A8 | 23 | 0.9 |
| Peroxisome biogenesis disorder | 361 | 6.0 | 2 | 0.1 | IFT81, SLC7A8 | 9 | 0.3 |
| Ossification | 284 | 5.6 | 2 | 0.4 | CPZ, EDEM3 | 5 | 1.6 |
| Chromatin binding | 265 | 2.9 | 2 | 0.2 | PLXNA3, ZFHX3 | 12 | 0.8 |
| Proteoglycan metabolic process | 196 | 3.0 | 1 | 0.5 | EDEM3 | 9 | 0.8 |
| Oxidoreductase activity | 190 | 2.7 | 2 | 0.1 | |||
| Congenital hypothyroidism | 184 | 4.2 | |||||
| Diamond-Blackfan anemia | 182 | 3.5 | 5 | 0.4 | |||
| Striated muscle tissue development | 180 | 2.5 | 2 | 0.9 | KCND1, RASA3 | 3 | 0.4 |
| Single-organism carbohydrate catabolic process | 155 | 2.4 | 4 | 1.8 | |||
| DNA repair | 141 | 4.4 | 2 | 0.6 | 4 | 2.5 | |
| Ichthyosis | 139 | 3.9 | 3 | 0.5 | EDEM3 | 2 | 0.6 |
| Head development | 125 | 2.8 | 2 | 2.8 | PLXNA3, ZFHX3 | 2 | 0.3 |
| Hydrolase activity. acting on ester bonds | 116 | 1.9 | 1 | 1.6 | EDEM3 | 1 | 0.1 |
| Behavior | 105 | 2.3 | 1 | 0.3 | ZFHX3 | 1 | 0.2 |
| Sensory organ development | 101 | 4.0 | 2 | 4.3 | |||
| Anatomical structure homeostasis | 99 | 3.1 | |||||
| Sulfur compound metabolic process | 83 | 3.5 | 2 | 0.3 | |||
| Tissue remodeling | 83 | 2.4 | 1 | 0.7 | RASA3 | ||
| Positive regulation of epithelial cell proliferation | 78 | 3.2 | 1 | 1.5 | PLXNA3 | 1 | 0.4 |
| Response to nutrient levels | 63 | 3.4 | 1 | 0.3 | |||
| Bone morphogenesis | 60 | 6.3 | 1 | 2.4 | |||
| Reactive oxygen species metabolic process | 52 | 2.6 | |||||
Previously reported candidate genes
Of the 63 candidate genes we identified, 6 (9.5%) were previously found to be associated with short stature or syndromes featuring short stature (Table 3 and Supplementary Table 12). A de novo loss of the start codon in ZBED4 was found in another, smaller exome study in individuals with ISS [14]. A missense variant in BRD4 was reported to segregate in one family with short stature [20]. Variants in FZD2 and LZTR1 were described in individuals with Robinow syndrome-like phenotype (FZD2) [21, 22] and Noonan syndrome (LZTR1), respectively [23, 24]. Recently, variants in AMMECR1 were observed in individuals with midface hypoplasia, hearing impairment, elliptocytosis, and nephrocalcinosis (OMIM 300990) [25, 26]. Here, short stature is a constant feature. Biallelic variants in IFT81 underlie a severe form of short stature (short-rib thoracic dysplasia 19; OMIM 617895). Interestingly, a heterozygous missense variant segregates with the growth deficit in one family, suggesting autosomal dominant inheritance.
Table 3.
Previously reported short stature associated candidate genes
| Gene | Candidate Gene confidence level | No. pats. | Variant level classificationa | Height | Phenotype | ||||
|---|---|---|---|---|---|---|---|---|---|
| Propoportionate | Syndromic | Main features | |||||||
| V | IV | III | |||||||
| IFT81 | high | 2 | 2 | −2.6 & −2.9 | 2/2 | 0/2 | Wide nasel bridge, high arched eye brows | ||
| AMMECR1 | medium | 1 | 1 | −3.2 | 1/1 | 0/1 | Lacrimal duct aplasia | ||
| BRD4 | medium | 1 | 1 | −2.9 | 1/1 | 1/1 | Short neck, low set reas, sparse eyebrows, frontal bossing | ||
| FZD2 | medium | 2 | 1 | 1 | −2.9 & −4.2 | 2/2 | 2/2 | Posteriorly rotated ears, abnormalities of the eye brows | |
| LZTR1 | medium | 2 | 1 | 1 | −3.2 & −3.0 | 1/2 | 1/2 | none | |
| ZBED4 | medium | 1 | 1 | −2.6 | 1/1 | 0/1 | Brachydactyly, broad philtrum, low set ears | ||
Enrichment analysis and clustering of genes known to be related to growth retardation
1025 genes potentially involved in growth delay or growth regulation were selected using the keywords “growth delay” from the Human Phenotype Ontology (HPO) database and “short stature” from OMIM and MedGen (Supplementary methods). Based on their significantly enriched functions, pathways and other biological features, we identified 29 clusters, including skeletal system development (GO:0001501), appendage development (GO:0048736), and ciliopathy and metabolic processes (GO:0044710) (Fig. 1b). Moreover, clusters were often related to central processes like chromatin binding (GO:0003682) or extracellular matrix (GO:0310122).
Enrichment analysis revealed that 95% of the candidate genes were mapped to 26 of the 29 clusters previously implicated in growth or short stature (Fig. 1g and Table 4). LAMA5 and MED24 were not mapped to any of these 29 clusters (Table 4 and Supplementary Tables 7, 10, and 11). The main involved KEGG pathways for the 13 high-confidence candidate genes were: metabolic pathways (hsa:01100), PI3K-Akt signaling pathway (hsa:04151), signaling pathways regulating pluripotency of stem cells (hsa:04550), thyroid hormone signaling pathway (hsa:04919), and pathways in cancer (hsa:05200) (Table 2 and Supplementary Table 10).
Functional overview of 13 high-confidence candidate genes
Intensive review of the known function of the 13 high-confidence genes led to the localization in 7 functional groups: Wnt signaling (CPZ, IFT81, LAMA5), cellular growth regulation (EDEM3, KCND1, SLC7A8, UBR4), thyroid hormone signaling (CPZ, KCND1, MED24), zebrafish phenotype (PLXNA3), Ras MAPK signaling (RASA3), growth hormone signaling interaction (ZFHX3), and ubiquitination (UBR4, USP45) (Supplementary Table 9). 3 genes are known to be involved in Wnt regulation. CPZ is induced by thyroid hormones and modulates Wnt signaling pathways by modification of the activity of Wnt-4 and thereby regulates the terminal differentiation of growth plate chondrocytes. IFT81 encodes a member of the IFT complex B core. Together with IFT74, IFT81 is required for ciliogenesis. LAMA5 encodes the laminin subunit alpha 5 and regulates Wnt- and PI3K signaling. Besides CPZ, KCND1 and MED24 have a reported function in thyroid hormone signaling. KCND1 is expressed in the thyroid gland which might imply a function in the hormonal regulation of growth. MED24 encodes one of the thyroid hormone receptor-associated proteins, that forms a complex with the thyroid receptor via TRAP220 [27]. 5 high-confidence genes, EDEM3, KCND1, SLC7A8, UBR4, USP45, are involved in the regulation of cellular growth, either by protein degradation (EDEM3), thyroid hormone regulation (KCND1), the mTOR pathway (SLC7A8) or ubiquitination (UBR4, USP45). Furthermore, ZFHX3 interacts with POU1F1, a member of the growth hormone pathway. RASA3 encodes a Ras-GTPase activating protein and is thus part of the RAS MAPK pathway.
Discussion
Growth-related disorders constitute a very heterogeneous group of disorders. Based on the results from large GWAS and gene-expression studies, we have estimated that at least 1000 genes are involved [5, 28]. These studies highlighted the observation that rare, large effect-size variants of growth retardation are inherited according to the three classical patterns of Mendelian inheritance [28]. Nevertheless, 67% of individuals with ISS remain without a diagnosis [5]. Using exome sequencing, we therefore aimed to identify novel genes associated with short stature in 254 affected families in whom known causes of short stature had previously been excluded (Table 2, Supplementary Tables 7 and 11, and Supplementary Figure 1) [5]. We identified variants in 63 candidate genes in a total of 92 independent families. Based on a classification scheme using variant and gene information as well as the number of independent affected individuals, we classified 13 genes as high-confidence genes and 50 other genes as medium-confidence candidates (Fig. 1c–e and Supplementary Figure 2).
To facilitate their characterization, we compiled a list of 1025 genes from HPO, OMIM, and MedGen based on the keywords “growth delay” and “short stature” and clustered their significantly enriched functions, pathways and other biological features. This resulted in 29 clusters, the largest of which were skeletal system development, appendage development, metabolic processes, and ciliopathy. These clusters reflect not only processes of skeletal development but also genes involved in basic processes like cellular growth (Fig. 1a, b, Table 4, and Supplementary Figures 5–7). Interestingly, the clusters with the highest mean enrichment were bone morphogenesis (6.3-fold), peroxisome biogenesis disorder (6.0-fold), and ossification (5.6-fold). These results confirmed a broad functional range of potentially growth-related genes. In addition, we generated a functional map of growth-associated genes that provides general information on functions, pathways and other biological features overrepresented in this gene set. We were able to map 95% of the 63 candidate genes onto at least one of the 29 clusters, supporting their relevance to growth (Fig. 1g and Table 4). These include the aforementioned clusters with a high mean enrichment, but also more specific clusters such as extracellular matrix, ossification, bone morphogenesis, and several catalytic processes.
The sensitivity of our approach to identify candidate genes for short stature was further supported by the identification of six genes (IFT81, AMMECR1, BRD4, FZD2, LZTR1, ZBED4) recently found to be associated with this phenotype (Table 3, Supplementary Table 12). Both the previously reported mode of inheritance and the type of variant were observed for AMMECR1 (X-linked recessive loss of function variants) [25, 26], BRD4 (autosomal dominant missense variants) [20] and ZBED4 (de novo nonsense variants) [14]. In addition, the clinical phenotype of the affected individuals was part of the phenotypic spectrum reported, providing additional evidence for their implication (Table 3 and Supplementary Table 12). Interestingly, while nonsense variants in FZD2 cause severe skeletal dysplasia phenotypes [21, 22], we identified missense variants in this gene in individuals with ISS, suggesting that missense variants are associated with a milder phenotype. IFT81 has previously been shown to be associated with an autosomal recessive syndromic form of short stature [24, 29, 30]. We propose that heterozygous variants affecting only one allele cause only short stature, a mechanism similar to that recently demonstrated for variants in ACAN [9]. For LZTR1, which is involved in the RAS-Map kinase pathway, the situation is more complex, since variant type-dependent recessive and dominant inheritance modes were observed to cause a Noonan-spectrum disorder with short stature as a consistent phenotype [23, 24]. We hypothesize that specific missense variants may be associated with isolated short stature.
RASA3 and FGF18 are two novel candidates implicated in known signaling pathways. In RASA3, we identified a de novo frameshift variant leading to a pre-terminal stop codon in one individual, and a de novo missense variants located in the C2 domain in a second individual, suggesting a loss-of-function effect. Through collaboration, we identified one additional individual with ISS and a de novo missense variant in this domain. Molecular modeling revealed that these missense variants potentially interfere with proper function of the C2 domain, which is reported to be relevant in targeting the protein to the cell membrane (Fig. 1f) [31]. Both the disruption of GTPase, mediated by a pre-terminal stop codon, and mislocation in the cell may potentially interfere with its proper function and thus lead to reduced inactivation of Ras-signaling and further hyperactivation of the pathway [32]. Likewise, FGF18 plays an important role in chondrogenesis and osteogenesis by binding to FGFR2 and FGFR3 [33]. Gain-of-function variants in both receptors were reported to cause mainly syndromic forms of short stature [34–36]. We propose that, by analogy with other members of this group of growth factors, a disease causing variant in the interacting protein leads to constitutional activation of the receptors in the complex [37].
Our classification, in addition to variant and gene information, is based on the number of independent affected individuals. All 13 high-confidence candidate genes were identified in at least 2 families, and 5 candidates even exhibited variants in up to 8 families. The most promising candidates were LAMA5 with 4 variants likely affecting the function and 4 variants of unknown significance in 8 families, and UBR4 with 5 variants likely affecting the function and 1 variant of unknown significance in 6 families (Table 2). All affected individuals carrying variants in either of these genes presented with proportionate short stature and a mean height that was 2.9 SDS below average. LAMA5 encodes the laminin subunit alpha 5, which plays a crucial role in development by regulating Wnt and PI3K signaling during osteoblast differentiation [38–40]. Recent reports demonstrated an involvement in syndromes including kidney disease, osteoarthritis and hypothyroidism, and an association with reduced height in elderly individuals from Southern Italy [41–44]. Studies in Lama5 mice with a hypomorphic allele indicated an essential role for LAMA5 in growth development [45]. Correspondingly, UBR4, the ubiquitin protein ligase E3 component n-recognin 4, is involved in cancer cell growth [46]. In Ubr4 knockout mice, growth retardation was reported in fetuses [47]. A 3.46 Mb deletion encompassing UBR4 was reported in an affected individual with short stature [48]. Intolerance to missense variants as indicated by a z-score of 5.98 (ExAC) suggested that the variants identified in the 6 affected individuals are highly likely to be associated with the phenotype [48, 49]. For another 50 genes, we found strong functional evidence for their involvement, albeit some were identified only in a single family. Thus, we cannot exclude that some genes may be unrelated to short stature (Supplementary Table 8-10).
If we were to consider only the 13 high-confidence candidates, this would yield an exome sequencing detection rate of 15% in highly selected families in whom known causes were previously excluded. This probably constitutes an underestimation as we assume that some of the other 50 candidate genes may be confirmed in subsequent studies. We may have missed smaller structural variants, variants in noncoding or insufficiently covered regions, or epigenetic changes, which have also been reported in connection with short stature [4, 50]. Furthermore, polygenic inheritance, mating selection and familial height variability may have hampered the clinical characterization of, and hence the identification of, the underlying variant.
In conclusion, we identified and characterized 13 high-confidence candidate genes with variants in two or more independent families and another 50 medium-confidence candidate genes. Of these candidate genes, 95% are annotated with functions, pathways and other biological features that are significantly enriched among 1,025 growth-associated genes. These results illustrate that in entities with extremely high heterogeneity and complexity, such as growth defects, clinical characterization and variant-level and gene-level information need to be combined to identify candidate genes. Our study also suggests that single gene defects are an important contributor to the extreme lower end of the growth distribution.
Supplementary information
Acknowledgements
We thank all participants and their families for taking part in this study. We acknowledge the excellent technical support of Farah Radwan, Evelyn Galsterer, Angelika Diem, and Heike Friebel. We also thank the Exome Aggregation Consortium and the groups that provided exome variant data for comparison. A full list of contributing groups can be found at http://exac.broadinstitute.org/about. This study utilized data generated by the DECIPHER community. A full list of centers that contributed to data generation is available online from http://decipher.sanger.ac.uk and via email from decipher@sanger.ac.uk. This study was supported by grants from the German Research Foundation (DFG; grants TH 896/3-3 and TH 896/3-4), the Interdisciplinary Centre for Clinical Research (IZKF) of the Friedrich-Alexander-Universität Erlangen-Nürnberg (Project F4) and by the ELAN Fonds (14-08-06-1) of the Faculty of Medicine of the Friedrich-Alexander Universität Erlangen-Nürnberg (FAU) to CT.
Author contributions
CTT designed the study. MZ, CZ, AW, RAJ, EK, AMJ, TRR, DW, ARa, and CTT contributed data from affected individuals. All affected individuals were clinically evaluated by HGD and CTT. Statistical analysis was done by LT, BP, SU, FF, CB, ABE, and HS. PK and FF contributed the chondrocyte expression data. CK and UT performed clinical diagnostic testing. Splice site validation was performed by NNH and ES. Data was analyzed by NNH, BP, SS, FF, SU, CB, HS, AR, and CTT. NNH and CTT interpreted the results. NNH, AR and CTT wrote the draft manuscript. All co-authors provided feedback on the estimates and contributed to the subsequent versions of the manuscript. All authors read and approved the final version of the manuscript.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41431-019-0362-0) contains supplementary material, which is available to authorized users.
References
- 1.Visscher PM, Medland SE, Ferreira MA, et al. Assumption-free estimation of heritability from genome-wide identity-by-descent sharing between full siblings. PLoS Genet. 2006;2:e41. doi: 10.1371/journal.pgen.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wood AR, Esko T, Yang J, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet. 2014;46:1173–86. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marouli E, Graff M, Medina-Gomez C, et al. Rare and low-frequency coding variants alter human adult height. Nature. 2017;542:186–90. doi: 10.1038/nature21039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zahnleiter D, Uebe S, Ekici AB, et al. Rare copy number variants are a common cause of short stature. PLoS Genet. 2013;9:e1003365. doi: 10.1371/journal.pgen.1003365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hauer Nadine N, Popp Bernt, Schoeller Eva, Schuhmann Sarah, Heath Karen E, Hisado-Oliva Alfonso, Klinger Patricia, Kraus Cornelia, Trautmann Udo, Zenker Martin, Zweier Christiane, Wiesener Antje, Abou Jamra Rami, Kunstmann Erdmute, Wieczorek Dagmar, Uebe Steffen, Ferrazzi Fulvia, Büttner Christian, Ekici Arif B, Rauch Anita, Sticht Heinrich, Dörr Helmuth-Günther, Reis André, Thiel Christian T. Clinical relevance of systematic phenotyping and exome sequencing in patients with short stature. Genetics in Medicine. 2017;20(6):630–638. doi: 10.1038/gim.2017.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Warman ML, Cormier-Daire V, Hall C, et al. Nosology and classification of genetic skeletal disorders: 2010 revision. Am J Med Genet A. 2011;155A:943–68. doi: 10.1002/ajmg.a.33909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amin N, Mushtaq T, Alvi S. Fifteen-minute consultation: the child with short stature. Arch Dis Child Educ Pract Ed. 2015;100:180–4. doi: 10.1136/archdischild-2014-306488. [DOI] [PubMed] [Google Scholar]
- 8.Rappold GA, Fukami M, Niesler B, et al. Deletions of the homeobox gene SHOX (short stature homeobox) are an important cause of growth failure in children with short stature. J Clin Endocrinol Metab. 2002;87:1402–6. doi: 10.1210/jcem.87.3.8328. [DOI] [PubMed] [Google Scholar]
- 9.Hauer NN, Sticht H, Boppudi S, et al. Genetic screening confirms heterozygous mutations in ACAN as a major cause of idiopathic short stature. Sci Rep. 2017;7:12225. doi: 10.1038/s41598-017-12465-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonafe L, Cormier-Daire V, Hall C, et al. Nosology and classification of genetic skeletal disorders: 2015 revision. Am J Med Genet A. 2015;167A:2869–92. doi: 10.1002/ajmg.a.37365. [DOI] [PubMed] [Google Scholar]
- 11.Seaver LH, Irons M. American College of Medical Genetics Professional P, Guidelines C: ACMG practice guideline: genetic evaluation of short stature. Genet Med. 2009;11:465–70. doi: 10.1097/GIM.0b013e3181a7e8f8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allen DB, Cuttler L. Clinical practice. Short stature in childhood—challenges and choices. N Engl J Med. 2013;368:1220–8. doi: 10.1056/NEJMcp1213178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stevens A, Hanson D, Whatmore A, Destenaves B, Chatelain P, Clayton P. Human growth is associated with distinct patterns of gene expression in evolutionarily conserved networks. BMC Genom. 2013;14:547. doi: 10.1186/1471-2164-14-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo MH, Shen Y, Walvoord EC, et al. Whole exome sequencing to identify genetic causes of short stature. Horm Res Paediatr. 2014;82:44–52. doi: 10.1159/000360857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pieper U, Webb BM, Dong GQ, et al. ModBase, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2014;42:D336–346. doi: 10.1093/nar/gkt1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zimmermann Lukas, Stephens Andrew, Nam Seung-Zin, Rau David, Kübler Jonas, Lozajic Marko, Gabler Felix, Söding Johannes, Lupas Andrei N., Alva Vikram. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. Journal of Molecular Biology. 2018;430(15):2237–2243. doi: 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 17.Webb B, Sali A. Protein structure modeling with MODELLER. Methods Mol Biol. 2017;1654:39–54. doi: 10.1007/978-1-4939-7231-9_4. [DOI] [PubMed] [Google Scholar]
- 18.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–23. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 19.Sayle RA, Milner-White EJ. RASMOL: biomolecular graphics for all. Trends Biochem Sci. 1995;20:374. doi: 10.1016/S0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 20.Jin HS, Kim J, Kwak W, Jeong H, Lim GB, Lee CG. Identification of a novel mutation in BRD4 that causes autosomal dominant syndromic congenital cataracts associated with other neuro-skeletal anomalies. PLoS One. 2017;12:e0169226. doi: 10.1371/journal.pone.0169226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saal HM, Prows CA, Guerreiro I, et al. A mutation in FRIZZLED2 impairs Wnt signaling and causes autosomal dominant omodysplasia. Hum Mol Genet. 2015;24:3399–409. doi: 10.1093/hmg/ddv088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagasaki K, Nishimura G, Kikuchi T, et al. Nonsense mutations in FZD2 cause autosomal-dominant omodysplasia: Robinow syndrome-like phenotypes. Am J Med Genet A. 2018;176:739–42. doi: 10.1002/ajmg.a.38623. [DOI] [PubMed] [Google Scholar]
- 23.Yamamoto GL, Aguena M, Gos M, et al. Rare variants in SOS2 and LZTR1 are associated with Noonan syndrome. J Med Genet. 2015;52:413–21. doi: 10.1136/jmedgenet-2015-103018. [DOI] [PubMed] [Google Scholar]
- 24.Johnston JJ, van der Smagt JJ, Rosenfeld JA, et al. Autosomal recessive Noonan syndrome associated with biallelic LZTR1 variants. Genet Med. 2018;20:1175–85. [DOI] [PMC free article] [PubMed]
- 25.Basel-Vanagaite Lina, Pillar Nir, Isakov Ofer, Smirin-Yosef Pola, Lagovsky Irina, Orenstein Naama, Salmon-Divon Mali, Tamary Hannah, Zaft Tami, Bazak Lily, Meyerovitch Joseph, Pelli Tal, Botchan Shay, Farberov Luba, Weissglas-Volkov Daphna, Shomron Noam. X-linked elliptocytosis with impaired growth is related to mutated AMMECR1. Gene. 2017;606:47–52. doi: 10.1016/j.gene.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 26.Moyses-Oliveira M, Giannuzzi G, Fish RJ, et al. Inactivation of AMMECR1 is associated with growth, bone, and heart alterations. Hum Mutat. 2018;39:281–91. doi: 10.1002/humu.23373. [DOI] [PubMed] [Google Scholar]
- 27.Zhang J, Fondell JD. Identification of mouse TRAP100: a transcriptional coregulatory factor for thyroid hormone and vitamin D receptors. Mol Endocrinol. 1999;13:1130–40. doi: 10.1210/mend.13.7.0295. [DOI] [PubMed] [Google Scholar]
- 28.Marouli E, Graff M, Medina-Gomez C, et al. Rare and low-frequency coding variants alter human adult height. Nature. 2017;542:186–90. [DOI] [PMC free article] [PubMed]
- 29.Perrault I, Halbritter J, Porath JD, et al. IFT81, encoding an IFT-B core protein, as a very rare cause of a ciliopathy phenotype. J Med Genet. 2015;52:657–65. doi: 10.1136/jmedgenet-2014-102838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duran I, Taylor SP, Zhang W, et al. Destabilization of the IFT-B cilia core complex due to mutations in IFT81 causes a Spectrum of Short-Rib Polydactyly Syndrome. Sci Rep. 2016;6:34232. doi: 10.1038/srep34232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang D, Aravind L. Identification of novel families and classification of the C2 domain superfamily elucidate the origin and evolution of membrane targeting activities in eukaryotes. Gene. 2010;469:18–30. doi: 10.1016/j.gene.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aoki Y, Niihori T, Inoue S, Matsubara Y. Recent advances in RASopathies. J Hum Genet. 2016;61:33–39. doi: 10.1038/jhg.2015.114. [DOI] [PubMed] [Google Scholar]
- 33.Ohbayashi N, Shibayama M, Kurotaki Y, et al. FGF18 is required for normal cell proliferation and differentiation during osteogenesis and chondrogenesis. Genes & Dev. 2002;16:870–9. doi: 10.1101/gad.965702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shiang R, Thompson LM, Zhu YZ, et al. Mutations in the transmembrane domain of FGFR3 cause the most common genetic form of dwarfism, achondroplasia. Cell. 1994;78:335–42. doi: 10.1016/0092-8674(94)90302-6. [DOI] [PubMed] [Google Scholar]
- 35.Ornitz DM, Legeai-Mallet L. Achondroplasia: development, pathogenesis, and therapy. Dev Dyn. 2017;246:291–309. doi: 10.1002/dvdy.24479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li ZL, Chen X, Zhuang WJ, et al. FGFR2 mutation in a Chinese family with unusual Crouzon syndrome. Int J Ophthalmol. 2016;9:1403–8. doi: 10.18240/ijo.2016.10.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naski MC, Wang Q, Xu J, Ornitz DM. Graded activation of fibroblast growth factor receptor 3 by mutations causing achondroplasia and thanatophoric dysplasia. Nat Genet. 1996;13:233–7. doi: 10.1038/ng0696-233. [DOI] [PubMed] [Google Scholar]
- 38.Spenle C, Simon-Assmann P, Orend G, Miner JH. Laminin alpha5 guides tissue patterning and organogenesis. Cell Adhes Migr. 2013;7:90–100. doi: 10.4161/cam.22236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ritie L, Spenle C, Lacroute J, et al. Abnormal Wnt and PI3Kinase signaling in the malformed intestine of lama5 deficient mice. PLoS One. 2012;7:e37710. doi: 10.1371/journal.pone.0037710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hong D, Chen HX, Yu HQ, et al. Morphological and proteomic analysis of early stage of osteoblast differentiation in osteoblastic progenitor cells. Exp Cell Res. 2010;316:2291–2300. doi: 10.1016/j.yexcr.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sampaolo S, Napolitano F, Tirozzi A, et al. Identification of the first dominant mutation of LAMA5 gene causing a complex multisystem syndrome due to dysfunction of the extracellular matrix. J Med Genet. 2017;54:710–20. doi: 10.1136/jmedgenet-2017-104555. [DOI] [PubMed] [Google Scholar]
- 42.Braun Daniela A, Warejko Jillian K, Ashraf Shazia, Tan Weizhen, Daga Ankana, Schneider Ronen, Hermle Tobias, Jobst-Schwan Tilman, Widmeier Eugen, Majmundar Amar J, Nakayama Makiko, Schapiro David, Rao Jia, Schmidt Johanna Magdalena, Hoogstraten Charlotte A, Hugo Hannah, Bakkaloglu Sevcan A, Kari Jameela A, El Desoky Sherif, Daouk Ghaleb, Mane Shrikant, Lifton Richard P, Shril Shirlee, Hildebrandt Friedhelm. Genetic variants in the LAMA5 gene in pediatric nephrotic syndrome. Nephrology Dialysis Transplantation. 2018;34(3):485–493. doi: 10.1093/ndt/gfy028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Luca M, Crocco P, De Rango F, Passarino G, Rose G. Association of the Laminin, Alpha 5 (LAMA5) r4925386 with height and longevity in an elderly population from Southern Italy. Mech Ageing Dev. 2016;155:55–59. doi: 10.1016/j.mad.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 44.De Luca M, Crocco P, Wiener H, Tiwari HK, Passarino G, Rose G. Association of a common LAMA5 variant with anthropometric and metabolic traits in an Italian cohort of healthy elderly subjects. Exp Gerontol. 2011;46:60–64. doi: 10.1016/j.exger.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shannon MB, Patton BL, Harvey SJ, Miner JH. A hypomorphic mutation in the mouse laminin alpha5 gene causes polycystic kidney disease. J Am Soc Nephrol. 2006;17:1913–22. doi: 10.1681/ASN.2005121298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huh KW, DeMasi J, Ogawa H, Nakatani Y, Howley PM, Munger K. Association of the human papillomavirus type 16 E7 oncoprotein with the 600-kDa retinoblastoma protein-associated factor, p600. Proc Natl Acad Sci USA. 2005;102:11492–7. doi: 10.1073/pnas.0505337102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakaya T, Ishiguro K, Belzil C, et al. p600 plays essential roles in fetal development. PLoS One. 2013;8:e66269. doi: 10.1371/journal.pone.0066269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Firth HV, Richards SM, Bevan AP, et al. DECIPHER: database of chromosomal imbalance and phenotype in humans using Ensembl resources. Am J Hum Genet. 2009;84:524–33. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waldman LA, Chia DJ. Towards identification of molecular mechanisms of short stature. Int J Pediatr Endocrinol. 2013;2013:19. doi: 10.1186/1687-9856-2013-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.