Abstract
We identified a 14q21.2 microdeletion in a 16-year-old boy with autism spectrum disorder (ASD), IQ in the lower part of normal range but high-functioning memory skills. The deletion affects a gene desert, and the non-deleted gene closest to the microdeletion boundaries is LRFN5, which encodes a protein involved in synaptic plasticity and implicated in neuro-psychiatric disorders. LRFN5 expression was significantly decreased in the proband’s skin fibroblasts. The deleted region includes the pseudogene chr14.232.a, which is transcribed into a long non-coding RNA (lncLRFN5-10), whose levels were also significantly reduced in the proband’s fibroblasts compared to controls. Transfection of the patient’s fibroblasts with a plasmid expressing chr14.232.a significantly increased LRFN5 expression, while siRNA targeting chr14.232.a-derived lncLRFN5-10 reduced LRFN5 levels. In summary, we report on an individual with ASD carrying a microdeletion encompassing the pseudogene chr14.232.a encoding for lncLRFN5-10, which was found to affect the expression levels of the nearby, non-deleted LRFN5. This case illustrates the potential role of long non-coding RNAs in regulating expression of neighbouring genes with a functional role in ASD pathogenesis.
Subject terms: Development, Autism spectrum disorders
Introduction
Autism spectrum disorders (ASD) display impaired social interaction and communication, and restricted and repetitive behaviour/interest. ASD is a major health burden with an incidence of up to 1 in 68 children [1]. Affected children frequently show intellectual disability (ID) whereas only a minority of individuals have high-functioning ASD with IQ >70. Specific molecular defects can be detected in ~20% of ASD children who undergo both array-CGH and whole exome sequencing (WES) [2]. According to the AutDB autism database, 1036 genes (last update September 2018) have been implicated in ASD [3]. Regulatory elements in the genome are increasingly recognized as important players in ASD by affecting the expression of autism genes [4].
We report on an individual with high-functioning ASD who harbours a chromosomal microdeletion at 14q21.2 encompassing a pseudogene-derived long non-coding RNA (lncRNA). The microdeletion affects the expression of the closest non-deleted gene, namely LRFN5, which encodes a synaptic adhesion-like molecule [5, 6].
Case presentation
The patient is the first child of non-consanguineous parents and was born by caesarean section after 37 weeks of uneventful pregnancy with a birth weight of 3,000 g (50th centile) and unremarkable perinatal events. He was able to sit without support at 10 months and walked independently at 23 months. At 2 years of age, his language was delayed and he was noted to have poor social interactions. He did not have any facial dysmorphisms or congenital anomalies.
When he was 16 years old, his weight was 91.5 kg (99th centile), head circumference 56 cm (34th centile), height 168 cm (21st centile), and body mass index 32.4 kg/m2 ( > 99th centile, z-score = +2.8). Brain MRI, performed at the same age, was normal. Ophthalmological evaluations were unremarkable and both cardiac and abdominal ultrasounds did not reveal any abnormalities.
The proband’s cognitive skills, evaluated when he was 16-year-old, fell within the lower part of normal range (IQ score 75 at the Wechsler Intelligence Scale for Children-III). He met the criteria for autism defined by the Autism Diagnostic Observation Schedule and Autism Diagnostic Interview-Revised. His memory skills were enhanced, and he showed brilliant non-verbal index scores for both working memory and long-term memory upon the Italian version of test of memory and learning.
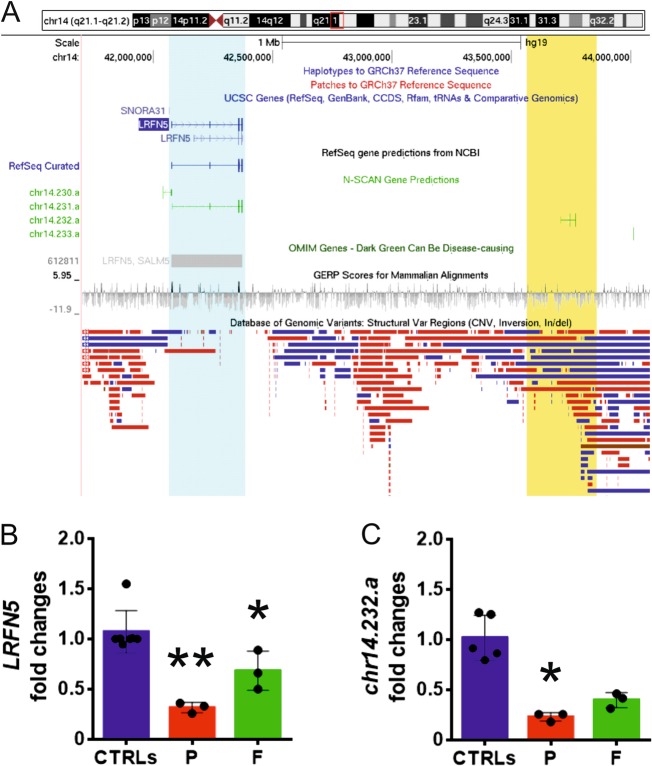
Metabolic screening tests and analysis for Fragile-X syndrome were all negative. Array-CGH, at a resolution of 40 kb in the backbone and 20 kb in the targeted regions [Sureprint G3, 4 × 180 Agilent, (Santa Clara)], showed a 291 kb microdeletion on chromosome 14q21.2, NC_000014.8:g.(43516648_43545282)_(43837068_43856485)del (ISCN nomenclature 14q21.2(43545282_43837068) × 1) (Fig. 1a) that was also detected in his father but not in his mother. The 14q21.2 deletion was confirmed by another array-CGH (CGX Hd, PerkinElmer, Waltham) with similar resolution. At the Wechsler Adult Intelligence Scale, his 52-year-old father showed an IQ of 78, consistent with a lower part of normal range cognition. The microdeletion is not entirely mapped in any control individuals from the Database of Genomic Variants (Fig. 1a). The microdeletion is devoid of coding genes and LRFN5 (chr14:42076764-42373752) is the closest coding gene, being located 1.12 Mb from the centromeric breakpoint (Fig. 1a). Direct sequencing of LRFN5 (NM_152447.4) exons and intron-exon boundaries (Supporting Methods) did not show any variants, ruling out pathogenic variants in the non-deleted allele. Whole-exome sequencing (WES) libraries were prepared using the SureSelectQXT Clinical Research Exome V1 kit (Agilent) and run on a NextSeq500 system (Illumina). A total of 77,657 high-quality variants were identified by WES in the proband’s genome and, among these variants, 862 were rare (frequency < 0.01). The prioritization process based on trio segregation, selection of variants with potential loss-of-function effect and higher Combined Annotation Dependent Depletion (CADD) score showed three de novo, one homozygous, and one X-linked variants as top candidates (Table 1 and Supplementary Material).
Fig. 1.
a Microdeletion at 14q21.2 (chr14:43545282-43837068 according to hg19), detected in the proband, is highlighted in yellow, whereas LRFN5 (chr14:42076764-42373752), at 1.12 Mb distance, is highlighted in the light blue area. b Expression levels of LRFN5 in the proband (P), his father (F) and controls (CTRLs) (n = 7) were significantly reduced in the proband and his father (**p < 0.001, *p < 0.005). c Expression levels of chr14.232.a in the proband (P), his father (F) and controls (CTRLs) (n = 5) were significantly reduced in the proband (*p < 0.05)
Table 1.
Candidate gene variants detected by whole-exome sequencing
| Chr | Position | Gene | Transcript accession# | Exon | Nucleotide change | Amino acid change | ExAc | Gnomad | CADD |
|---|---|---|---|---|---|---|---|---|---|
| De novo | |||||||||
| 9 | 39103792 | CNTNAP3 | NM_033655.3 | 16 | c.2485G > C | p.(Val829Leu) | −1 | −1 | 23.4 |
| 2 | 202251090 | TRAK2 | NM_015049.2 | 14 | c.1814A > C | p.(Tyr605Ser) | −1 | −1 | 29.4 |
| 11 | 86947710 | TMEM135 | NM_022918.3 | 6 | c.494_498del | p.(Tyr165fs) | −1 | −1 | |
| Homozygous | |||||||||
| 3 | 127323612 | MCM2 | NM_004526.2 | 3 | c.398G > A | p.(Arg133His) | 0.003 | 0.0018 | 27 |
| X-linked | |||||||||
| X | 47060300 | UBA1 | NM_003334.3 | 6 | c.488T > C | p.(Val163Ala) | −1 | −1 | 25.5 |
WES was performed on the trio by SureSelect Clinical Research Exome (Agilent, Technologies) and NextSeq500 sequencing system (Illumina, hg 19 human reference genome). A customized pipeline based on Burrows-Wheeler Alignment tool, Genome Analysis Toolkit and ANNOVAR were used to call, annotate, filter, and prioritize variants. Variants are submitted in LOVD repository (as Lab-ID ULOele-15-22; https://databases.lovd.nl/shared/individuals/00228899).
−1: indicate absence in control dataset. Additional interpretation of these variants is provided in Supplementary material
Chr chromosome, ExAC Exome Aggregation Consortium, Gnomad Genome Aggregation Database, CADD Combined Annotation Dependent Depletion
Methods
The presented case has been submitted to Decipher with ID#381198 (https://decipher.sanger.ac.uk/patient/381198?consent=1#overview/patient-general). Skin fibroblasts of the proband, his father and age- and gender-matched normal controls were cultured at 37 °C with 5% CO2 in DMEM medium supplemented with 20% v/v fetal bovine serum and penicillin/streptomycin. One microgram of extracted RNA was reverse transcribed (High Capacity cDNA Kit, Applied Biosystems) and real-time PCR was performed using 2 µg of cDNA (LRFN5 assay ID Hs00290466_m1; housekeeping RNF111 assay ID Hs00405638_m1 and B2M assay ID Hs99999907_m1, ThermoFisher) and TaqMan Universal PCR MasterMix 2X on the Applied Biosystems 7300 according to the manufacturer’s protocols.
Short interfering RNA (siRNA) and scramble control sequences have been designed by the Invivogen design online tool. Specificity for the chr14.232.a-derived lncLRFN5-10 has been evaluated by using Basic Local Alignment Search Tool (BLAST) (Supporting Materials). Fibroblast cells were cultured in DMEM plus 10% fetal bovine serum (FBS) and 5% penicillin/streptomycin. Cells were transfected with siRNA for the chr14.232.a-derived lncLRFN5-10 or scramble control. Lipofectamine RNAiMAX Transfection Reagent (ThermoFisher Scientific) was used for transfections according to the manufacturer’s instructions. Cells were harvested 72 h after transfection for RNA extraction. For transfections, cells up to 80% confluency were trypsinized, washed in phosphate buffer solution and resuspended in buffer R at a concentration of 100,000 cells per 10 µL with plasmids expressing lncLRFN5-10 synthetized by GeneArt or green fluorescent protein both under the control of a CMV promoter. Cells were microporated with Neon Transfection System (ThermoFisher) with pulse of 1700 volts, pulse width 20 ms, pulse number 1. Cells were seeded and harvested after 48 h for RNA extraction.
Real-time PCR for the chr14.232.a-derived lncLRFN5-10 and for LRFN5 after siRNA transfection were performed with the primers reported in Supporting Methods using iQ Supermix SYBR Green 2X on the Bio-Rad iCycler (Biorad) according to the manufacturer’s protocols. Real-time PCR reactions were performed in triplicate.
Statistical analysis
Two-tailed Student t-tests were used for mean comparisons using the R package for statistical computing version 3.3.1, Prism7. P-values below 0.05 were considered statistically significant.
Results
Expression of LRFN5 in the patient’s skin fibroblasts was significantly reduced compared to control fibroblasts (n = 7) (p < 0.0005) (Fig. 1b). LRFN5 expression was also reduced in skin fibroblasts from the patient’s father, who was also a carrier of the 14q21.2 microdeletion (p < 0.005) (Fig. 1b). No evidence of regulatory elements such as enhancers and insulators were detected in the microdeleted region (Supporting Methods). The three-dimensional structure of the genome is emerging as a modulator of long-range gene expression regulation and most enhancer–promoter interactions occur within domains of a self-interacting chromatin, called topologically associating domains (TADs) [7]. However, no evidence of TAD boundaries and insulators was detected in the microdeletion region (Supporting Methods). The 14q21.2 microdeleted region instead was found to encompass the 886-nucleotide pseudogene chr14.232.a consisting of 2 exons (Fig. 1a). Within the microdeleted region, analysis of LNCipedia [8] revealed the presence of a 333-nucleotide lncRNA, named lncLRFN5-10, sharing 100% identity over a 280-nucleotide segment with exon two of chr14.232.a (Supplementary Results). Based on this observation, we assumed that lncLRFN5-10 is encoded by chr14.232.a. The mRNA levels of chr14.232.a were significantly reduced in the proband’s fibroblasts compared to controls (n = 5) (p < 0.05), whereas the reduction in the father’s fibroblasts was not significant (p = 0.12) (Fig. 1c).
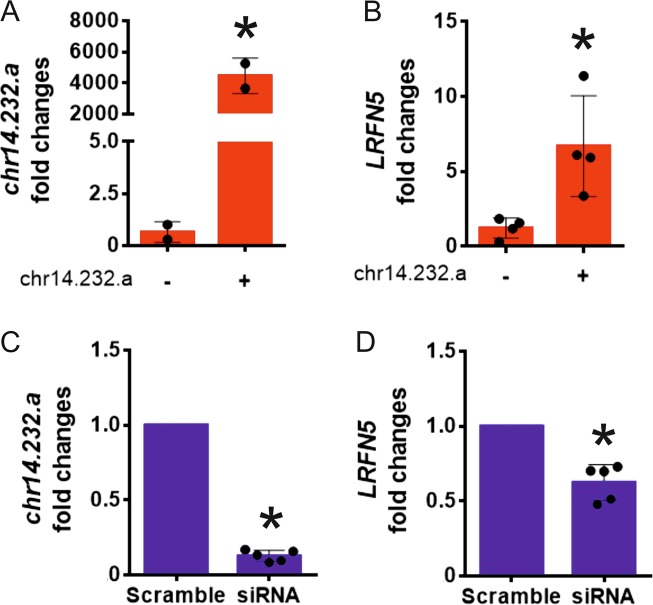
To investigate whether chr14.232.a affects LRFN5 expression, we transfected a plasmid expressing the chr14.232.a in the proband’s fibroblasts. A significant increase in chr14.232.a expression in both the patient’s and control transfected cells was detected (Fig. 2a and Supplementary Fig. S1), and the transfected proband’s cells showed a significant increase in LRFN5 expression compared to cells transfected with a control plasmid (p < 0.01) (Fig. 2b), whereas no changes were detected in control fibroblasts (Supplementary Fig. S2). Transfection with siRNA knocking down chr14.232.a in fibroblasts from five different wild-type controls resulted in significant reduction of LRFN5 mRNA levels (Fig. 2c, d), further supporting the chr14.232.a-dependent regulation of LRFN5 expression.
Fig. 2.
a Expression levels of chr14.232.a were increased following transfection of a plasmid expressing chr14.232.a in the proband’s skin fibroblasts. (*p < 0.05) b LRFN5 expression levels were significantly increased after chr14.232.a transfection in the proband’s skin fibroblasts. (*p < 0.05). c SiRNA knock-down of chr14.232.a decreased chr14.232.a expression in control fibroblasts (n = 5) (*p < 0.05). d LRFN5 expression levels were significantly reduced after transfection of siRNA targeting chr14.232.a in control fibroblasts (n = 5) (*p < 0.05)
Discussion
We describe an individual with ASD harbouring a 14q21.2 microdeletion devoid of protein-coding genes, which affects LRFN5 expression levels in skin fibroblasts by position effect. LRFN5 is a remarkably isolated gene located within a 5-Mb interval lacking other genes. It is mainly expressed in the brain, contributes to synaptic plasticity [6, 9], and has been implicated in schizophrenia [10].
One individual with developmental delay, seizures, and microcephaly was found to harbour a deletion affecting the last five exons of LRFN5 and to have a 47,XXX karyotype [11], the latter potentially contributing to the phenotype [12]. Although there have been no other published cases with a microdeletion overlapping the one reported here, a patient with ASD was previously reported with a balanced de novo translocation between chromosomes 14 and 21 leading to reduced LRFN5 expression in fibroblasts, along with a 2.6 Mb 2q31.1 deletion [13]. However, although supposed to have a pathogenic role, the 2q31.1 deletion was not considered the only cause of the patient’s phenotype [13]. The same paper also reported two other subjects carrying large chromosomal deletions including LRFN5, who presented with cognitive impairments but not ASD [13]. LRFN5 was also identified by WES as a candidate gene in a large cohort of undiagnosed patients with neurodevelopmental disorder [14], and is listed in the Simons Foundation Autism Research Initiative (SFARI) database with evidence of causality [15] and among the de novo variants in subjects with ASD of the denovo-db database [16].
We observed reduced LRFN5 expression in the fibroblasts of the ASD individual with 14q21.2 microdeletion presented herein. We found no evidence of either putative regulatory elements (enhancers, promoters, locus control regions) or TAD boundaries and insulators mapping within the deleted region. Therefore, we focused on the pseudogene chr14.232.a localized within the microdeletion interval. As expected, expression of chr14.232.a was significantly reduced in the proband’s fibroblasts. However, we also found that transfection of chr14.232.a significantly increased LRFN5 expression in the proband’s fibroblasts and knock-down of chr14.232.a in control fibroblasts decreased LRFN5 expression.
Transcribed pseudogenes are increasingly recognized as regulators of gene expression and encode lncRNAs, defined as RNA-transcribed molecules of at least 200 nucleotides with no protein-coding capacity [17]. LncRNAs with high levels of expression in the brain are emerging as major architects of cognitive evolution [18] and are co-expressed with several genes implicated in ASD [19, 20]. In addition, they have been proposed as therapeutic candidates to restore expression of specific genes in ASD [21].
The functions of the vast majority of lncRNA transcripts are unknown, although various modes of action have been proposed, including epigenetic pre- and post-transcriptional regulations, and enhancer-like functions [21]. Moreover, they can act as sponge/decoy for miRNA [22]. Interestingly, the 3′UTR of LFRN5 and lncLRFN5-10 shared six microRNA recognition elements (MRE) (Supplementary Table S1), and thus it is conceivable that lncLRFN5-10 might function as a sponge and compete with miRNA-targeting LRFN5 to relieve miRNA-mediated repression. Alternatively, lncLRFN5-10 might exert an enhancer-like function on LRFN5 because expression of several lncRNA positively correlates with those of their neighbouring coding genes [21].
The father of the index case carried the same deletion but showed milder neurocognitive impairment. However, the proband was also found to carry a missense de novo variant in the CNTNAP3 gene that encodes for a member of family of proteins implicated in ASD [23]. Although functional evidence is not available, the CNTNAP3 variant might explain the phenotypic difference between the proband and his father, because co-inheritance of two or more rare genetic variants causing the same disease has been proposed in neurodevelopmental disorders [24].
In summary, we describe an individual with ASD and an unusual and heterogeneous cognitive profile, characterized by a talent with peaks and savant features in working memory who harboured a 14q21.2 microdeletion containing the chr14.232.a located at ∼1.12 Mb from the LRFN5 gene. Interestingly, we found that chr14.232.a increases expression of LRFN5 in his fibroblasts. Reduced expression of LRFN5 in fibroblasts should be interpreted with caution as these cells might not be representative of expression in brain cells, which are involved in the neurodevelopmental phenotype. Nevertheless, this study provides a potentially novel mechanism involved in ASD pathogenesis based on the involvement of pseudogene-encoded lncRNA regulating the expression of neighbouring protein-coding genes.
Supplementary information
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41431-019-0430-5) contains supplementary material, which is available to authorized users.
References
- 1.Christensen DL, Baio J, Van Naarden Braun K, Bilder D, Charles J, Constantino JN, et al. Prevalence and characteristics of autism spectrum disorder among children aged 8 years—autism and developmental disabilities monitoring network, 11 sites, United States, 2012. MMWR Surveill Summ. 2016;65:1–23. doi: 10.15585/mmwr.ss6503a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tammimies K, Marshall CR, Walker S, Kaur G, Thiruvahindrapuram B, Lionel AC, et al. Molecular diagnostic yield of chromosomal microarray analysis and whole-exome sequencing in children with autism spectrum disorder. JAMA. 2015;314:895–903. doi: 10.1001/jama.2015.10078. [DOI] [PubMed] [Google Scholar]
- 3.Pereanu W, Larsen EC, Das I, Estevez MA, Sarkar AA, Spring-Pearson S, et al. AutDB: a platform to decode the genetic architecture of autism. Nucleic Acids Res. 2018;46:D1049–D1054. doi: 10.1093/nar/gkx1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Short PJ, McRae JF, Gallone G, Sifrim A, Won H, Geschwind DH, et al. De novo mutations in regulatory elements in neurodevelopmental disorders. Nature. 2018;555:611–6. doi: 10.1038/nature25983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi Y, Nam J, Whitcomb DJ, Song YS, Kim D, Jeon S, et al. SALM5 trans-synaptically interacts with LAR-RPTPs in a splicing-dependent manner to regulate synapse development. Sci Rep. 2016;6:26676. doi: 10.1038/srep26676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goto-Ito S, Yamagata A, Sato Y, Uemura T, Shiroshima T, Maeda A, et al. Structural basis of trans-synaptic interactions between PTPdelta and SALMs for inducing synapse formation. Nat Commun. 2018;9:269. doi: 10.1038/s41467-017-02417-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spielmann M, Lupianez DG, Mundlos S. Structural variation in the 3D genome. Nat Rev Genet. 2018;19:453–67. doi: 10.1038/s41576-018-0007-0. [DOI] [PubMed] [Google Scholar]
- 8.Volders PJ, Verheggen K, Menschaert G, Vandepoele K, Martens L, Vandesompele J, et al. An update on LNCipedia: a database for annotated human lncRNA sequences. Nucleic Acids Res. 2015;43:4363–4. doi: 10.1093/nar/gkv295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin Z, Liu J, Ding H, Xu F, Liu H. Structural basis of SALM5-induced PTPdelta dimerization for synaptic differentiation. Nat Commun. 2018;9:268. doi: 10.1038/s41467-017-02414-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu B, Woodroffe A, Rodriguez-Murillo L, Roos JL, van Rensburg EJ, Abecasis GR, et al. Elucidating the genetic architecture of familial schizophrenia using rare copy number variant and linkage scans. Proc Natl Acad Sci USA. 2009;106:16746–51. doi: 10.1073/pnas.0908584106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mikhail FM, Lose EJ, Robin NH, Descartes MD, Rutledge KD, Rutledge SL, et al. Clinically relevant single gene or intragenic deletions encompassing critical neurodevelopmental genes in patients with developmental delay, mental retardation, and/or autism spectrum disorders. Am J Med Genet Part A. 2011;155:2386–96. doi: 10.1002/ajmg.a.34177. [DOI] [PubMed] [Google Scholar]
- 12.Otter M, Schrander-Stumpel CT, Curfs LM. Triple X syndrome: a review of the literature. Eur J Hum Genet. 2010;18:265–71. doi: 10.1038/ejhg.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Bruijn DRH, van Dijk AHA, Pfundt R, Hoischen A, Merkx GFM, Gradek GA, et al. Severe progressive autism associated with two de novo hanges: a 2.6-Mb 2q31.1 deletion and a balanced t(14;21)(q21.1; p11.2) translocation with long-range epigenetic silencing ofLRFN5 expression. Mol Syndromol. 2010;1:46–57. doi: 10.1159/000280290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farwell Hagman KD, Shinde DN, Mroske C, Smith E, Radtke K, Shahmirzadi L, et al. Candidate-gene criteria for clinical reporting: diagnostic exome sequencing identifies altered candidate genes among 8% of patients with undiagnosed diseases. Genet Med. 2017;19:224–35. doi: 10.1038/gim.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abrahams BS, Arking DE, Campbell DB, Mefford HC, Morrow EM, Weiss LA, et al. SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs) Mol Autism. 2013;4:36. doi: 10.1186/2040-2392-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turner TN, Yi Q, Krumm N, Huddleston J, Hoekzema K, FS HA, et al. Denovo-db: a compendium of human de novo variants. Nucleic Acids Res. 2017;45:D804–D811. doi: 10.1093/nar/gkw865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knauss JL, Sun T. Regulatory mechanisms of long noncoding RNAs in vertebrate central nervous system development and function. Neuroscience. 2013;235:200–14. doi: 10.1016/j.neuroscience.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barry G. Integrating the roles of long and small non-coding RNA in brain function and disease. Mol Psychiatry. 2014;19:410–6. doi: 10.1038/mp.2013.196. [DOI] [PubMed] [Google Scholar]
- 19.Parikshak NN, Swarup V, Belgard TG, Irimia M, Ramaswami G, Gandal MJ, et al. Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature. 2016;540:423–7. doi: 10.1038/nature20612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Briggs JA, Wolvetang EJ, Mattick JS, Rinn JL, Barry G. Mechanisms of long non-coding RNAs in mammalian nervous system development, plasticity, disease, and evolution. Neuron. 2015;88:861–77. doi: 10.1016/j.neuron.2015.09.045. [DOI] [PubMed] [Google Scholar]
- 21.Wahlestedt C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat Rev Drug Discov. 2013;12:433–46. doi: 10.1038/nrd4018. [DOI] [PubMed] [Google Scholar]
- 22.Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17:272–83. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 23.Poot M. A candidate gene association study further corroborates involvement of contactin genes in autism. Mol Syndromol. 2014;5:229–35. doi: 10.1159/000362891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pizzo L, Jensen M, Polyak A, Rosenfeld JA, Mannik K, Krishnan A, et al. Rare variants in the genetic background modulate cognitive and developmental phenotypes in individuals carrying disease-associated variants. Genet Med. 2019;21:816–25. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.