Abstract
Roma are a socially and culturally distinct isolated population with genetically divergent subisolates, residing mainly across Central, Southern, and Eastern Europe. We evaluated the genetic etiology of hearing impairment (HI) in 15 Hungarian Roma families through exome sequencing. A family with autosomal dominant non-syndromic HI segregating a rare variant in the Calponin-homology 2 domain of PLS1, or Plastin 1 [p.(Leu363Phe)] was identified. Young adult Pls1 knockout mice have progressive HI and show morphological defects to their inner hair cells. There is evidence that PLS1 is important in the preservation of adult stereocilia and normal hearing. Four families segregated the European ancestral variant c.35delG [p.(Gly12fs)] in GJB2, and one family was homozygous for p.(Trp24*), an Indian subcontinent ancestral variant which is common amongst Roma from Slovakia, Czech Republic, and Spain. We also observed variants in known HI genes USH1G, USH2A, MYH9, MYO7A, and a splice site variant in MANBA (c.2158-2A>G) in a family with HI, intellectual disability, behavioral problems, and respiratory inflammation, which was previously reported in a Czech Roma family with similar features. Lastly, using multidimensional scaling and ADMIXTURE analyses, we delineate the degree of Asian/European admixture in the HI families understudy, and show that Roma individuals carrying the GJB2 p.(Trp24*) and MANBA c.2158-2A>G variants have a more pronounced South Asian background, whereas the other hearing-impaired Roma display an ancestral background similar to Europeans. We demonstrate a diverse genetic HI etiology in the Hungarian Roma and identify a new gene PLS1, for autosomal dominant human non-syndromic HI.
Subject terms: Genetics, Next-generation sequencing, Clinical genetics
Introduction
The Roma are a transnational founder population consisting of various socially and genetically divergent groups [1]. It is believed that the Roma migrated from Punjab, Haryana and Rajasthan regions of India to Europe between the 6th and 11th century. Migration led to a population bottleneck, followed by admixture with Eurasian populations. Founder effects, genetic drift, and differential admixture have shaped the current genetic landscape of the Roma. Their social traditions and limited gene flow between Romani groups have resulted in a noticeable genetic substructure [1]. The diverse Roma populations within Europe consist of genetically isolated founder subpopulations, with high rates of consanguinity [2]. The Roma entered Hungary in the 13th to 14th century. They currently compose ~ 3% of the Hungarian population (Hungarian Central Statistical Office), and are the largest minority.
Roma health is generally poor compared with other European populations [2], resulting from low socio-economic status and limited access to health care. Medical genetic research in the Roma has identified a number of private founder mutations causing rare Mendelian conditions that had not been previously described [3]. Genetic studies on hearing impairment (HI) in Roma subpopulations are scarce, and have mainly focused on GJB2 (connexin 26) variants, the major cause of autosomal recessive (AR) non-syndromic (NS) (HI) worldwide. In Spanish and Slovakian Roma, GJB2 accounts for up to 50% of all ARNSHI, mainly driven by the p.(Trp24*) variant, which by itself explains 39.5% and 23.2% of ARNSHI, respectively, in these populations [4, 5]. The p.(Trp24*) variant was also found to be prevalent in Czech HI patients of Roma heritage (9.7%) [6]. This variant is prevalent among Indian and Pakistani populations, and was probably brought by Roma to Europe from their Indian homeland [5]. Carrier rates of the p.(Trp24*) variant in Roma controls is 4–5% in the major linguistic and migrational categories of Balkan, Vlax, and western European Roma [7], however, we previously found that p.(Trp24*) is present in lower frequencies in Hungarian Roma controls (0.8%) [8]. Other GJB2 alleles observed in Slovakian Roma HI patients are p.(R127H) and c.35delG [p.(Gly12fs)] (underlying 19.4% and 8.3% of ARNSHI, respectively) and c.35delG [p.(Gly12fs)] in Spanish Roma (8.5% of ARNSHI) [4, 5]. In addition to GJB2, the MARVELD2 c.1331 + 2T > C splice site variant has been previously reported to play an important role in HI in Roma subisolates [9, 10].
Despite these efforts, studies using massive parallel sequencing interrogating the exome or genome are missing for the Roma. Hence, we evaluated the genetic etiology of HI in 15 Roma families.
Materials and methods
Sample collection and clinical evaluation
Institutional review board approval and consent were obtained from the University of Pecs (Protocol 8581-7/2017/EUIG) and Baylor College of Medicine and Affiliated Hospitals (Protocol H-17566). Written informed consent was obtained from all participating family members and peripheral blood samples were collected. Genomic DNA was isolated from peripheral ethylenediaminetetraacetic acid-anticoagulated blood samples using a standard desalting method. A clinical history was recorded. Pure-tone audiometry was performed at 250–8000 Hz in a sound proof room using a GSI-61 PT audiometer. Other causes of HI, including infections, ototoxic medications, and trauma, were evaluated. All individuals affected with HI underwent vestibular function testing including posturography, videonystagmography, and a Halmagyi test. They also underwent ophthalmological examinations, including vision tests and physical eye examinations, i.e., the slit lamp test, a general ophthalmological examination (subjective vision tests, numbers, letters, etc), optical coherence tomography, color vision tests, and microscopic examinations were performed.
Exome sequencing
Prior to exome sequencing, the MARVELD2 c.1331+2T>C splice site and the coding region of GJB2 were screened in affected individuals from all 15 families. Next, 19 affected individuals from all 15 families underwent exome sequencing using NimbleGen SeqCap EZv.2.0 followed by 70 bp paired-end sequencing on a HiSeq instrument (Illumina Inc., San Diego, CA), with a median read depth of 77×. We also exome sequenced DNA samples from affected family members with GJB2 variants that had been identified using Sanger sequencing, so the sequence data could be included in the multidimensional scaling (MDS) and ADMIXTURE analyses. Reads were aligned to the Human genome (Hg19/GRC37) using the Burrows-Wheeler transform software [11], polymerase chain reaction (PCR) duplicates were removed with Picard, and indel realignment was performed (GATK IndelRealigner). Recalibration and joint calling of single nucleotide variants and small insertions/deletions (indels) were performed with HaplotypeCaller (GATK) [12]. Last, variants were annotated using ANNOVAR [13], which includes dbSNFPv3.5 and dbscSNV1.1 [14, 15].
Analysis of exome Sequence data for HI variant identification
Variants were filtered, and exonic (missense, nonsense, stop/start altering, frameshift and in-frame variants) and splice and splice region (± 12 bp) sites with a minor allele frequency (MAF) < 0.02 in the exomes and genomes of the Genome Aggregation Database (gnomAD); Greater Middle East (GME) variome database; and the National Heart, Lung and Blood Institute (NHLBI) Exome Sequencing Project (ESP6500) database were retained [16–19]. Several inheritance models were considered and evaluated depending on the pedigree structure, i.e. de novo, autosomal dominant (AD), AR (homozygous and compound heterozygous), X-linked and digenic. Conservation and protein-altering effects of the variants were evaluated in silico, based on a variety of bioinformatic predictions included in dbSNFPv3.5 and dbscSNV1.1 [14, 15].
Variant validation and evaluation of segregation
Sanger sequencing was used to validate variants of interest and to evaluate familial segregation. Primers were generated using Primer3 [20], and regions surrounding the variants were amplified using PCR. ExoSAP-IT (USB Corp., Cleveland, OH, USA) was used to purify PCR-amplified products. Sequencing was performed using the BigDye Terminator v3.1 Cycle Sequencing Kit followed by capillary electrophoresis on an ABI 3730 DNA Analyzer (Applied Biosystems Inc, Foster City, CA USA). DNA sequences were aligned and analyzed using Codoncode Aligner v7.1.2 (CodonCode Corp., Centerville, MA, USA) or the Sequencher software v4.9 (GeneCodes Corp., Ann Arbor, MI, USA). All variants of interest have been deposited in ClinVar (www.ncbi.nlm.nih.gov/clinvar) under accession numbers SCV000853290–SCV000853304.
Multidimensional scaling analysis
Data sets included in the MDS analysis consisted of genomes from the phase three 1000 Genome project (N = 2504) and exome data from unrelated hearing-impaired Roma individuals (N = 16 one individual from each family with the exception for family 6003 where both affected parents II:1 and II:2 were included (Fig. 1); NimbleGen SeqCap EZv.2.0 exome kit). Analysis was performed using plinkv1.9 [21]. Each sample’s gender was verified and its relationship to other samples was evaluated to ensure that only unrelated individuals were included in the analysis. The data were then filtered by removing the following from each data set (i.e., 1000 Genomes and Roma): indels and variant sites with a MAF < 0.01, < 97% genotyping success rate or a significant deviation from Hardy–Weinberg equilibrium (calculated for each 1000 Genomes subpopulation and for the Roma; p < 1 × 10−7). The intersect between the genome and exome data sets were obtained and genomic locations with non-matching variant alleles between data sets were removed. A final subset of variants was selected by applying linkage disequilibrium (LD)-based pruning using a variance inflation factor (VIF = 2) and recursively removing SNPs within a sliding window. A final subset of 20,545 LD pruned variants for the 2520 individuals were obtained. MDS component (C) values were generated and imported into R for visualization.
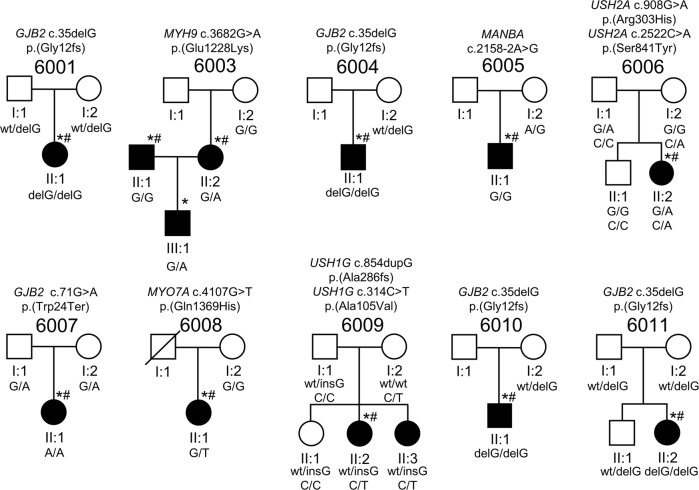
Fig. 1.
Pedigrees and variants identified in known HI genes in Hungarian Roma families. Drawings of the 10 Roma pedigrees that are segregating variants in known HI genes. Circles indicate females and squares males. Filled symbols represent individuals with HI and clear symbols denote individuals who are unaffected. The genotypes fare indicated below every individual for which a DNA sample was available. An asterisk (*) indicates family members whose DNA samples underwent exome sequencing and a pound sign (#) denotes exome sequence data that were included in the MDS and ADMIXTURE analysis. The HI status and genotype for (I:1) in pedigree 6003 and (I:1) in 6008 are unknown
Admixture analysis
Maximum likelihood estimations of individual ancestries were performed with ADMIXTURE v1.3 [22], using the same pruned data set as for MDS analysis. Estimates were obtained using K = 1–10 hypothetical ancestral populations. Cross-validation (CV) errors were evaluated to choose the optimal K-value.
Three-dimensional protein modeling
Homology modeling techniques were used to model the three-dimensional structure of wild-type and mutant PSL1 using the Protein Data Bank 1PXY crystal structure as a template [23]. The wild and mutant model were generated using the MODELLER8v1 software [24]. Figures were prepared using the UCSF-Chimera software [25].
Results
Clinical findings
All affected individuals displayed non-progressive HI, with the majority presenting with non-syndromic hearing impairment (NSHI). HI was most likely congenital in all affected individuals and the diagnosis of HI varied between 2 and 7 years of age. Pure-tone audiograms are displayed in Fig. 2 and Supplementary Figure S1. Some of the affected family members presented with syndromic HI or had an additional phenotype unlikely to be part of the HI etiology. Hearing-impaired member of family 6005 (II:1) also displayed intellectual disability and respiratory inflammations treated as cystic fibrosis. This child was also diagnosed with attention-deficit/hyperactivity disorder. Hearing-impaired members (II:2 and II:3) of pedigree 6009 also suffer from polycystic ovarium syndrome and hypothyreosis, although it is unclear whether this is related to their HI, as these are common medical conditions. The hearing-impaired member of family 6014 (II:1) has hypertelorism and mild micrognathia. No vestibular dysfunction nor vision problems were identified in any of the hearing-impaired family members, however both the unaffected and affected siblings of family 6006 have hypermetropia, which is likely unassociated with HI. No features of Usher syndrome or retinitis pigmentosa (RP) were found in any of the affected individuals (Table 1, Fig. 1, and Supplementary Figure S2).
Fig. 2.
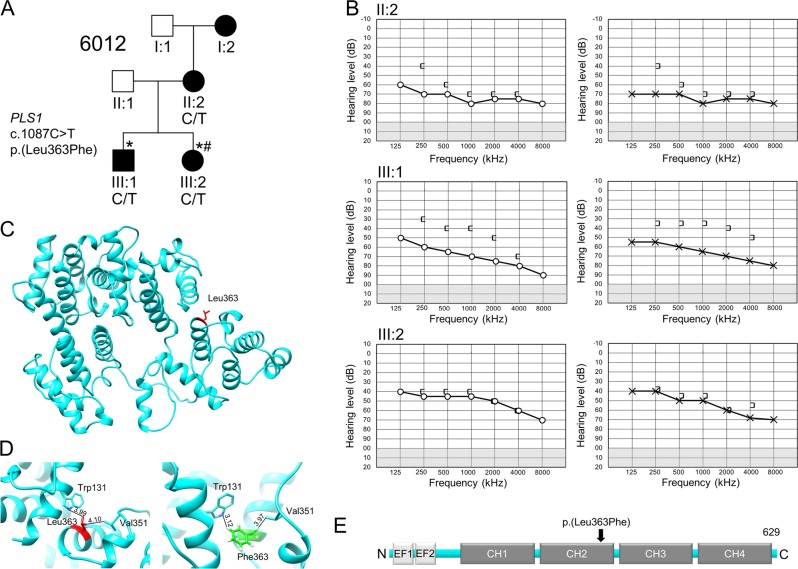
Family 6012 segregates an autosomal dominant variant in PLS1. a Pedigree drawing of family 6012 with genotypes of the PLS1 variant indicated for those family members with available DNA samples. Circles denote females and squares males. Filled symbols represent individuals with HI and clear symbols individuals who are unaffected. An asterisk (*) indicates family members whose DNA sample that underwent exome sequencing and a pound sign (#) denotes exome sequence data that were included in the MDS analysis. There was no family history of HI on the paternal side. b Right air (O) and bone ([) conductions, and left ear air (x) and bone (]) conductions for 6012 affected family members. II:2: age 34 years; III:1: age 8 years; III:2: age 11 years c. Three-dimensional representation of PLS1. The residues are represented by a stick model. The variant position of p.(Leu363) is highlighted in red. d A zoomed in view of hydrophobic interactions at position 363 of the wild-type (left) and mutated (right) PLS1 protein. e Domain structure of the PLS1 protein with the variant p.(Leu363Phe) indicated
Table 1.
Rare HI variants identified in the Roma families
| Family | Gene | Phenotype | Known/novel Gene | Genotype | Segr info | Transcript | cDNA | Protein | Known/novel HI variant | GERP + + RS | CADD | rs_ID | gnomAD All MAF | gnomAD MAF NFE | gnomAD MAF SAS | ACMGa |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6001 | GJB2 | HI | Known | Hom | Recessive | NM_004004.5 | c.35delG | p.(Gly12fs) | Known | NA | NA | rs80338939 | 6.19 × 10−3 | 9.58 × 10−3 | 8.17 × 10−4 | Pathogenic |
| 6003 | MYH9 | HI | Known | Het | Dominant | NM_002473.5 | c.3682G>A | p.(Glu1228Lys) | Novel | 4.98 | 31 | rs746956415 | 1.19 × 10−5 | 1.76 × 10−5 | 0 | VUS |
| 6004 | GJB2 | HI | Known | Hom | Recessive | NM_004004.5 | c.35delG | p.(Gly12fs) | Known | NA | NA | rs80338939 | 6.19 × 10−3 | 9.58 × 10−3 | 8.17 × 10−4 | Pathogenic |
| 6005 | MANBA | HI, ID, RI, ADHD | Known syndromic | Hom | Recessive | NM_005908.3 | c.2158-2A>G | Splicingb | Known | 5.17 | 23.4 | rs772852668 | 2.39 × 10−5 | 1.76 × 10−5 | 0 | Pathogenic |
| 6006 | USH2A | HIc | Known | Comp Het | Recessive | NM_206933.2 | c.908G>A | p.(Arg303His) | Known | 4.93 | 34 | rs371777049 | 3.90 × 10−5 | 6.23 × 10−5 | 0 | Pathogenic |
| 6006 | USH2A | HIc | Known | Comp Het | Recessive | NM_206933.2 | c.2522C>A | p.(Ser841Tyr) | Known | 6.03 | 24.5 | rs111033282 | 6.06 × 10−3 | 8.92 × 10−3 | 1.11 × 10−3 | VUS |
| 6007 | GJB2 | HI | Known | Hom | Recessive | NM_004004.5 | c.71G>A | p.(Trp24*) | Known | 5.21 | 36 | rs104894396 | 5.22 × 10−4 | 6.26 × 10−5 | 4.38 × 10−3 | Pathogenic |
| 6008 | MYO7A | HI | Known | Het | Dominantd | NM_000260.3 | c.4107G>T | p.(Gln1369His) | Novel | 4.69 | 24.6 | . | 0 | 0 | 0 | VUS |
| 6009 | USH1G | HIe | Known | Comp Het | Recessive | NM_173477.4 | c.854dupG | p.(Ala286fs) | Novel | NA | NA | . | 0 | 0 | 0 | Pathogenic |
| 6009 | USH1G | HIe | Known | Comp Het | Recessive | NM_173477.4 | c.314C>T | p.(Ala105Val) | Novel | 2.8 | 24.9 | . | 0 | 0 | 0 | Likely pathogenic |
| 6010 | GJB2 | HI | Known | Hom | Recessive | NM_004004.5 | c.35delG | p.(Gly12fs) | Known | NA | NA | rs80338939 | 6.19 × 10−3 | 9.58 × 10−3 | 8.17 × 10−4 | Pathogenic |
| 6011 | GJB2 | HI | Known | Hom | Recessive | NM_004004.5 | c.35delG | p.(Gly12fs) | Known | NA | NA | rs80338939 | 6.19 × 10−3 | 9.58 × 10−3 | 8.17 × 10−4 | Pathogenic |
| 6012 | PLS1 | HI | Novel, causes HI in mice | Het | Dominant | NM_002670.2 | c.1087C>T | p.(Leu363Phe) | Novel | 5.73 | 29.9 | . | 0 | 0 | 0 | VUS |
Abbreviations are as follows:
Genotypes: Hom: Homozygous; Comp Het: Compound heterozygous; Het: Heterozygous
Phenotype codes: ADHD: attention-deficit/hyperactivity disorder; HI: hearing impairment; ID: intellectual disability; RI: respiratory inflammation
Population codes: NFE: Non-Finnish European; SAS: South Asian
Scores and frequencies: ACMG: American College of Medical Genetics and Genomics classification of variants; CADD, Combined Annotation Dependent Depletion v1.3; gnomAD, Genome Aggregation Database v.2.1; NA, not available; MAF, minor allele frequency; VUS: variant of unknown significance
aMore details can be found in Supplementary Table S2
bNG_012804.1:g.130948 A > G. The dbscSNV ADA and RF scores are 1 and 0.932, respectively
cAffected and unaffected siblings both are affected with hypermetropia
dPotentially autosomal recessive
eAffected siblings also suffer from polycystic ovarium syndrome and hypothyreosis
Genetic screening
Prior to exome sequencing, we screened the coding region of GJB2 and the Roma ancestral MARVELD2 splice site variant (c.1331+2T>C) in all families via Sanger sequencing. Five families were carriers of homozygous GJB2 variants, four segregated GJB2 c.35delG[p.(Gly12fs)], and one segregated the Indian subcontinent ancestral GJB2 variant p.(Trp24*) (Fig. 1; Table 1). The MARVELD2 c.1331+2T>C splice site variant was not observed for any of the affected members of the 15 pedigrees.
Exome sequencing was performed, and we identified a missense variant, p.(Leu363Phe), in PLS1 that segregated with HI with an AD mode of inheritance in family 6012 (Table 1; Fig. 2). This gene was previously shown to be associated with HI in mice, but not humans [26]. The identified p.(Leu363Phe) variant affects the Calponin-homology 2 (CH2) domain, is highly conserved, deemed deleterious by a number of bioinformatics tools, and is absent from exome and genomes data from GME, NHLBI-ESP, and gnomAD (Table 1; Fig. 2).
In the remaining families, exome sequencing revealed rare and predicted damaging variants in known HI genes. Novel NSHI variants in MYO7A [p.(Gln1369His); family 6008] and MYH9 [p.(Glu1228Lys) family 6003] that segregate with an AD mode of inheritance were observed. Family 6003 is bilineal and the p.(Glu1228Lys) variant in MYH9 was only observed to segregate from the affected mother II:2 to her affected son III:3 and was not observed in the affected father II:1 (Fig. 1). Family 6009 segregates novel compound AR heterozygous variants in USH1G [p.(Ala286fs) & p.(Ala105Val)], a gene that is involved either NSHI or Usher Syndrome 1G [27]. This family does not have any features of Usher syndrome. Family 6006 segregates known Usher Syndrome 2A gene USH2A [p.(Arg303His) & p.(Ser841Tyr)] variants with an AR compound heterozygous mode of inheritance (Fig. 1; Table 1). Both children, unaffected (II:1) and affected (II:2), from family 6006 suffer from hypermetropia but do not show any signs of RP. As RP can be variable and have a slow or late onset, it is possible that the children of families 6006 (II:2, age at examination was 16 years) and 6009 (II:2, age at examination was 16 years and II:3, 13 years) could still develop RP [28]. The affected member of family 6005 displays a phenotype that includes intellectual disability and respiratory inflammation (beta-mannosidosis) and segregates a previously reported splice site variant (c.2158-2A>G) in MANBA. This variant is predicted to alter splicing, which was experimentally confirmed in a previous study (Table 1) [14].
HI etiology remains unresolved in four Roma families and for the affected father (II:1) in bilineal family 6003. For three of these families, variants of unknown significance (VUS) were identified. The affected father (II:1) in family 6003 (is heterozygous for a rare variant in MAP3K1 p.(Arg183Gln). Variants in Map3k1 cause HI in the mouse and this gene is crucial for the survival of hair cells [29, 30]. In addition, in family 6014 we found suggestive evidence of digenic inheritance with heterozygous variants observed in BMP2 p.(Met60Leu) and CHSY1 p.(Arg260Gln), genes which interact with each other to facilitate inner ear development [31]. At last, in family 6015 AR compound heterozygous PTPRS [p.(Ala1115Thr) & p.(Arg851Leu)] variants were observed. PTPRS is involved in neurogenesis and is expressed in the inner ear, including the hair cells [32]. In two families (6002 and 6013), we did not identify any causal variants or variants of interest that segregate with HI. Additional information is provided in Supplementary Figure S2 and Supplementary Table S1.
All identified SNV variants in this study were predicted damaging by at least three bioinformatic tools, had a Combined Annotation Dependent Depletion C-score > 20 (indicating that a variant is at least among the top 1% of deleterious variants in the genome), and showed conservation across species. Detailed bioinformatic scores, evidence available for the American College of Medical Genetics classification and a short explanation of the interpretation per variant is listed in Supplementary Table S2.
Three-dimensional modeling of PLS1
We used homology modeling techniques to model the three-dimensional structure of wild-type and mutant PSL1 p.(Leu363Phe), located in the CH2 domain (Fig. 2). We demonstrate a difference in distances and small local perturbation in the side chains at nearby residues in the CH1 [p.(Trp131)] and CH2 [p.(Val351)] domains between wild-type and mutant, owing to the substitution of small aliphatic Leu residue to the large aromatic and hydrophobic Phe residue at position 363.
Ancestry analysis
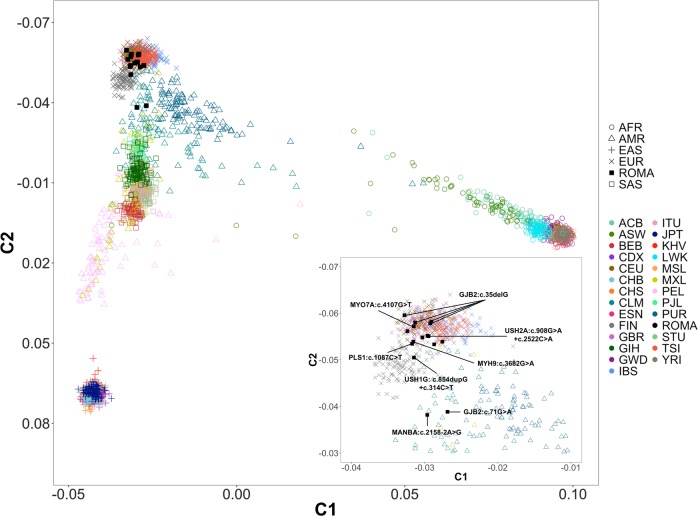
We evaluated the genetic background of the Roma Hungarian families with HI, because of their known diverse genetic substructure and the genetic distinctiveness of each Roma settlement within Europe. First, MDS components analysis was performed. We included exome sequence data from 16 unrelated affected individuals from the 15 Roma families and compared it to sequence data from the 1000 genomes project (Phase3), which includes data on individuals from 26 different ancestries. The MDS analysis demonstrate that most of the Hungarian Romani display a similar genetic background to individuals of European ancestry (Fig. 3; Supplementary Figure S3), demonstrating likely a high level of European admixture, which is comparable to what has been observed in previous larger population-based studies of Hungarian Roma [33]. Individuals carrying [GJB2:c.71 G > A;p.(Trp24*)] and [MANBA:c.2158-2A>G] variants showing a divergent background compared with other families clustering between Asians and Europeans.
Fig. 3.
Multidimensional scaling analysis. This plot shows the first two MDS components. Samples with variants in HI genes present in Table 1 are labeled in the right zoom insert. Populations codes: ROMA: Romani hearing-impaired family members included in this study, AFR: African; AMR: Admixed American (Latinos); EAS: East Asian; EUR: European; SAS: South Asian. Subpopulations: ACB: African Caribbean in Barbados; ASW: Americans of African Ancestry in SW USA; BEB: Bengali from Bangladesh; CDX: Chinese Dai in Xishuangbanna, China; CEU: Utah Residents (CEPH) with Northern and Western European Ancestry; CHB: Han Chinese in Beijing, China; CHS: Southern Han Chinese; CLM: Colombians from Medellin, Colombia; ESN: Esan in Nigeria; FIN: Finnish in Finland; GBR: British in England and Scotland; GIH: Gujarati Indian from Houston; GWD: Gambian in Western Divisions in the Gambia; Texas; IBS: Iberian Population in Spain; ITU: Indian Telugu from the UK; JPT: Japanese in Tokyo, Japan; KHV: Kinh in Ho Chi Minh City, Vietnam; LWK: Luhya in Webuye, Kenya; MSL: Mende in Sierra Leone; MXL: Mexican Ancestry from Los Angeles USA; PEL: Peruvians from Lima, Peru; PJL: Punjabi from Lahore, Pakistan; PUR: Puerto Ricans from Puerto Rico; STU: Sri Lankan Tamil from the UK; TSI: Toscani in Italy; YRI: Yoruba in Ibadan, Nigeria
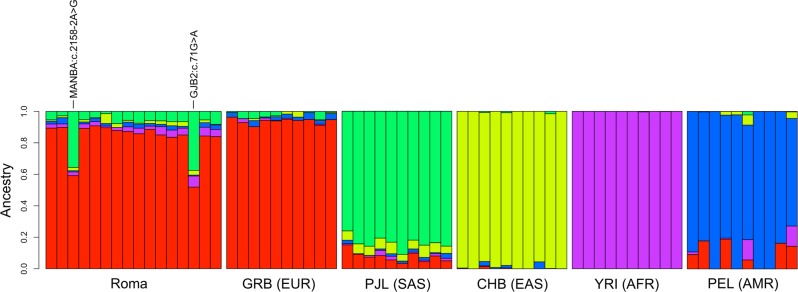
To support the MDS findings, we also performed ancestry estimates on the same sample set implementing the ADMIXTURE [22] method, using K = 1–10 hypothetical ancestries. We chose a hypothetical ancestry of K = 5 as optimal, based on CV values and the number of populations (Fig. 4; Supplementary Figure S4). Our data confirm that Roma share ancestral backgrounds with mainly Europeans (EUR) and South Asians (SAS), previously both shown to be an important source in the origin of Roma ancestry [33]. The results from the ADMIXTURE analyses are also comparable with the MDS analyses, with two individuals [GJB2:c.71G>A;p.(Trp24*)] and [MANBA:c.2158-2A>G] showing a more pronounced South Asian background. Additional SAS subpopulations and K-value estimates can be found in Supplementary Figures S4-6.
Fig. 4.
ADMIXTURE analysis. Results with K = 5 hypothetical ancestries are shown. Each column represents one individual. Individuals with [GJB2:c.71G>A;p.(Trp24*)] and [MANBA:c.2158-2A>G] variants are labeled. Ten random individuals from each of the five main 1000 Genome ancestries are also plotted for comparison purposes. PJL (Punjabi) were chosen to represent SAS as North-Western Indian populations play an important role in the South Asian ancestry component of Roma [33]. Populations codes: ROMA: Romani hearing-impaired family members included in this study, AFR: African; AMR: Admixed American (Latinos); EAS: East Asian; EUR: European; SAS: South Asian. Subpopulations: CHB: Han Chinese in Beijing, China; GBR: British in England and Scotland; PEL: Peruvians from Lima, Peru; PJL: Punjabi from Lahore, Pakistan; YRI: Yoruba in Ibadan, Nigeria
Discussion
This study aimed to investigate the genetic background of HI in the Hungarian Roma, a genetic sub-isolate of the genetically diverse Roma founder population. We illustrate the presence of European admixture, as was observed by previous population studies [33], show the presence of HI variants of European origin, and that the genetics of HI in Hungarian Roma is diverse with some HI variants being unique to this population. We also highlight a family that segregates a rare and predicted damaging variant in PLS1, which was previously found to be implicated in HI in mice [26].
In family 6012, we identified a predicted damaging novel missense variant in PLS1 (Plastin 1) in a conserved amino acid that segregates with HI with an AD mode of inheritance. The introduction of p.(Leu363Phe) affects the CH2, or Calponin-homology 2 domain, within PLS1 that is important for its actin binding properties, and leads to disturbances in residue interactions within the CH2 domain of the protein and between CH1 and CH2 (Fig. 2). The PLS1 gene also shows low number of observed loss-of-function (gnomAD o/e metric = 0.42 [CI95:0.28–0.66]) and missense variants (gnomAD o/e metric = 0.76 [CI95:0.69–0.85]), suggesting that there is some intolerance in this gene toward these types of variants.
The HI of affected family members in family 6012 ranges from mild-to-profound, with the higher frequencies more severely affected. Affected members II:2 and III:1 display mixed hearing loss while individual III:2 displays sensorineural HI (Fig. 2). Affected individual III:1 was also diagnosed with mild otitis media with effusion, explaining the presence of a conductive hearing loss in addition to sensorineural HI. Pls1 is expressed the murine developing and adult inner ear, including in the inner and outer hair cells [34]. In the human inner ear, PLS1 is expressed in both the vestibule and cochlea [35]. Plastin 1 is an abundant actin-bundling protein of the stereocilia, and young adult knockout mice have a moderate form of HI across all frequencies, which progresses to severe HI with age [26]. Conversely to other actin-bundling proteins, PLS1 is unessential in the initial formation of stereocilia, however, the stereocilia of inner hair cells KO mice showed morphological defects. This suggests a specific role for plastin 1 in the preservation of adult stereocilia and normal hearing. Variant p.(Leu363Phe) in family 6012 affects the CD2 domain within PLS1, which is the major determinant for the distinct functions of plastins, and crucial for its interaction with F-actin [36]. It was proposed that variants in the human PLS1 gene may be associated with HI [26]. Our finding supports this hypothesis, and this is the first description in humans of a predicted function altering variant in PLS1. Additional PLS1 HI families would aid in supporting that PLS1 is involved in the etiology of ADNSHI.
In 9 out of 15 families, we identified rare and predicted damaging variants in five known HI genes, GJB2, USH1G, USH2A (AR), and MYH9, MYO7A (AD; Fig. 1; Table 1). Variants in GJB2 were the most common and were identified five families. Four families segregate GJB2 c.35delG [p.(Gly12fs)], which has a European origin [37], and one family segregates GJB2 p.(Trp24*), an ancestral Indian subcontinent variant, which is a common GJB2 variant amongst Roma from Slovakia, Czech Republic, and Spain [4–6]. Previous genetic studies focused on ancestral analyses have shown that the Roma people show a high West Eurasian ancestry (~ 80%), likely originating from Central/East Europe, in addition to a South Asian ancestry, likely originating from North–West India and possibly also Pakistan [33]. Using MDS and ADMIXTURE analyses, we show that the individual with the Indian ancestral c.71 G > A:p.(Trp24*) variant shows a more pronounced South Asian ancestry, reflecting the Indian origin of the variant, whereas the individuals with c.35delG [p.(Gly12fs)] variants are very similar to individuals from European ancestry (Figs. 3 and 4), reflecting their European admixture. In addition, three HI variants [MYH9: p.(Glu1228Lys); MANBA: c.2158-2A>G; USH2A: p.(Arg303His)] we identified are present in low frequency in European populations (Table 1), but not in individuals of South Asian ancestry (Table 1), suggesting these HI variants may have been introduced to the Hungarian Roma by European admixture, similar to GJB2 c.35delG [p.(Gly12fs)].
The variants in MYO7A and USH1G, to our knowledge, have never been reported in hearing-impaired individuals or databases (Table 1). Variants in MYO7A can cause ADNSHI and ARNSHI, and Usher syndrome Type 1B. The observed MYO7A variant p.(Gln1369His) with a likely AD mode of inheritance in family 6008 is located in the first FERM domain. MYO7A variants underlying ADNSHI, are rarer than ARNSHI MYO7A variants, and the majority are reported in the motor domain with some in the neck region [38], but not in the FERM domain. It is possible that the affected child II:1 is the carrier of a compound heterozygous variant and we did not observe the second allele, although we carefully checked each exon using the Integrative Genomics Viewer and no other rare variants in MYO7A were found.
For the affected member (II:1) of family 6005, we identified an AR variant in MANBA (c.2158-2A>G; Table 1, Fig. 1). This same MANBA splice site variant was previously reported in a beta-mannosidosis Czech Roma family displaying an AR mode of inheritance [39]. Human beta-mannosidosis a lysosomal storage disease caused by a deficiency of the enzyme beta-mannosidase. The phenotypic manifestation of human beta-mannosidosis amongst patients is variable, even among family members there are wide ranges of symptoms and age of onset. Our patient presents with a milder phenotype including moderate-to-profound HI (World Health Organization classification; Supplementary Figure S2), intellectual disability, behavioral problems and respiratory inflammation, which are recurrent features of beta-mannosidosis. The c.2158-2A>G splice site variant was previously shown to lead to two mutant mRNAs through activation of a cryptic splice site in the exon downstream and exon skipping [39]. This variant is observed in very low frequencies in gnomADv2.1 Latino (MAF = 1.16 × 10-4) and non-Finnish European (MAF = 1.76 × 10−5) populations, but not in South Asian or any other gnomAD populations. MDS and ADMIXTURE analysis of this family demonstrate they have a more pronounced South Asian ancestry than other Roma individuals in this study (Figs. 3 and 4). Therefore, the origin of this variant is unclear, although it is now reported to have been observed in two Roma families.
We also identified several VUS in genes of interest, including BMP2, CHSY, PTPRS, and MAP3K1 (Supplementary Table S1; Supplementary Figure S2). This includes family 6014 with possible digenic heterozygous variants in both BMP2 and CHSY1. Variants in CHSY1 cause a syndrome including brachydactyly and hearing loss [31], and microdeletions that include BMP2 lead to a syndrome with variable expressivity including cleft palate, facial dysmorphism, Pierre-Robin sequence, and HI [40]. Bmp2 and Chsy1 are both expressed in the inner ear and hair cells during development [34], and interestingly, Bmp2b-chsy1 interaction modulates inner ear development in zebrafish [31]. The hearing-impaired individual (II:1) of family 6014 also presents with hypertelorism and mild micrognathia.
In conclusion, we illustrate a diverse genetic HI etiology in the Hungarian Roma, which might reflect their genetic substructure and Asian and European admixture. In addition, we identified a rare variant in PLS1 which is likely a novel cause of ADSNHI in humans. PLS1 has previously been shown to be important in the preservation of adult stereocilia and was also implicated in hearing loss in mice.
Supplementary information
Acknowledgements
We thank the families for their participation in this study. This work was supported by the National Institute on Deafness and Other Communication Disorders grants R01 DC011651 and R01 DC003594 (to S.M.L), and the National Scientific Research Program (NKFI) K 119540, GINOP-2.3.2-15-2016-00039 and EFOP 3.6.1-16-2016-00004 (to J.B and B.M).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zsolt Szabo is deceased
Supplementary information
The online version of this article (10.1038/s41431-019-0372-y) contains supplementary material, which is available to authorized users.
References
- 1.Morar B, Azmanov DN, Kalaydjieva L. Roma (Gypsies): Genetic Studies. In: eLS. John Wiley & Sons, Ltd: Chichester, UK, 2013 10.1002/9780470015902.a0006239.pub3.
- 2.Hajioff S, McKee M. The health of the Roma people: a review of the published literature. J Epidemiol Community Health. 2000;54:864–9. doi: 10.1136/jech.54.11.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalaydjieva L, Gresham D, Calafell F. Genetic studies of the Roma (Gypsies): a review. BMC Med Genet. 2001;2:5. doi: 10.1186/1471-2350-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Álvarez A, del Castillo I, Villamar M, Aguirre LA, González-Neira A, López-Nevot A, et al. High prevalence of theW24X mutation in the gene encoding connexin-26 (GJB2) in Spanish Romani (gypsies) with autosomal recessive non-syndromic hearing loss. Am J Med Genet Part A. 2005;137A:255–8. doi: 10.1002/ajmg.a.30884. [DOI] [PubMed] [Google Scholar]
- 5.minárik G, Ferák V, Feráková E, Ficek A, Poláková H, Kádasi L. High frequency of GJB2 mutation W24X among Slovak Romany (Gypsy) patients with non-syndromic hearing loss (NSHL) Gen Physiol Biophys. 2003;22:549–56. [PubMed] [Google Scholar]
- 6.Seeman P, Malíková M, Rašková D, Bendová O, Groh D, Kubálková M, et al. Spectrum and frequencies of mutations in the GJB2 (Cx26) gene among 156 Czech patients with pre-lingual deafness. Clin Genet. 2004;66:152–7. doi: 10.1111/j.1399-0004.2004.00283.x. [DOI] [PubMed] [Google Scholar]
- 7.Bouwer S, Angelicheva D, Chandler D, Seeman P, Tournev I, Kalaydjieva L. Carrier rates of the ancestral Indian W24X mutation in GJB2 in the general gypsy population and individual subisolates. Genet Test. 2007;11:455–8. doi: 10.1089/gte.2007.0048. [DOI] [PubMed] [Google Scholar]
- 8.Sipeky C, Matyas P, Melegh M, Janicsek I, Szalai R, Szabo I, et al. Lower carrier rate of GJB2 W24X ancestral Indian mutation in Roma samples from Hungary: implication for public health intervention. Mol Biol Rep. 2014;41:6105–10. doi: 10.1007/s11033-014-3488-8. [DOI] [PubMed] [Google Scholar]
- 9.Mašindová I, Šoltýsová A, Varga L, Mátyás P, Ficek A, Hučková M, et al. MARVELD2 (DFNB49) mutations in the hearing impaired Central European Roma population - prevalence, clinical impact and the common origin. PLoS ONE. 2015;10:e0124232. doi: 10.1371/journal.pone.0124232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Šafka Brožková D, Laštůvková J, Štěpánková H, Krůtová M, Trková M, Myška P, et al. DFNB49 is an important cause of non-syndromic deafness in Czech Roma patients but not in the general Czech population. Clin Genet. 2012;82:579–82. doi: 10.1111/j.1399-0004.2011.01817.x. [DOI] [PubMed] [Google Scholar]
- 11.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164–e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu X, Wu C, Li C, Boerwinkle E. dbNSFPv3.0: a one-stop database of functional predictions and annotations for human nonsynonymous and splice-site SNVs. Hum Mutat. 2016;37:235–41. doi: 10.1002/humu.22932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jian X, Boerwinkle E, Liu X. In silico prediction of splice-altering single nucleotide variants in the human genome. Nucleic Acids Res. 2014;42:13534–44. doi: 10.1093/nar/gku1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scott EM, Halees A, Itan Y, Spencer EG, He Y, Azab MA, et al. Characterization of Greater Middle Eastern genetic variation for enhanced disease gene discovery. Nat Genet. 2016;48:1071–6. doi: 10.1038/ng.3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu W, O’Connor TD, Jun G, Kang HM, Abecasis G, Leal SM, et al. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2012;493:216–20. doi: 10.1038/nature11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Auton A, Abecasis GR, Altshuler DM, Durbin RM, Abecasis GR, Bentley DR, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23:1289–91. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- 21.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–64. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhati M, Lee C, Nancarrow AL, Lee M, Craig VJ, Bach I, et al. Implementing the LIM code: the structural basis for cell type-specific assembly of LIM-homeodomain complexes. EMBO J. 2008;27:2018–29. doi: 10.1038/emboj.2008.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen M, et al. Current Protocols in Protein Science. Hoboken, NJ, USA: John Wiley & Sons, Inc; 2007. Comparative Protein Structure Modeling Using MODELLER; pp. 2.9.1–2.9.31. [DOI] [PubMed] [Google Scholar]
- 25.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF chimera? A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 26.Taylor R, Bullen A, Johnson SL, Grimm-Günter EM, Rivero F, Marcotti W, et al. Absence of plastin 1 causes abnormal maintenance of hair cell stereocilia and a moderate form of hearing loss in mice. Hum Mol Genet. 2015;24:37–49. doi: 10.1093/hmg/ddu417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maria Oonk AM, van Huet RAC, Leijendeckers JM, Oostrik J, Venselaar H, van Wijk E, et al. Nonsyndromic hearing loss caused by USH1G mutations. Ear Hear. 2015;36:205–11. doi: 10.1097/AUD.0000000000000095. [DOI] [PubMed] [Google Scholar]
- 28.Tsujikawa M, Wada Y, Sukegawa M, Sawa M, Gomi F, Nishida K, et al. Age at onset curves of retinitis pigmentosa. Arch Ophthalmol. 2008;126:337. doi: 10.1001/archopht.126.3.337. [DOI] [PubMed] [Google Scholar]
- 29.Parker A, Cross SH, Jackson IJ, Hardisty-Hughes R, Morse S, Nicholson G, et al. The goya mouse mutant reveals distinct newly identified roles for MAP3K1 in the development and survival of cochlear sensory hair cells. Dis Model Mech. 2015;8:1555–68. doi: 10.1242/dmm.023176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yousaf R, Meng Q, Hufnagel RB, Xia Y, Puligilla C, Ahmed ZM, et al. MAP3K1 function is essential for cytoarchitecture of the mouse organ of Corti and survival of auditory hair cells. Dis Model Mech. 2015;8:1543–53. doi: 10.1242/dmm.023077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, Laue K, Temtamy S, Aglan M, Kotan LD, Yigit G, et al. Temtamy preaxial brachydactyly syndrome is caused by loss-of-function mutations in chondroitin synthase 1, a potential target of BMP signaling. Am J Hum Genet. 2010;87:757–67. doi: 10.1016/j.ajhg.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meathrel K, Adamek T, Batt J, Rotin D, Doering LC. Protein tyrosine phosphatase?-deficient mice show aberrant cytoarchitecture and structural abnormalities in the central nervous system. J Neurosci Res. 2002;70:24–35. doi: 10.1002/jnr.10382. [DOI] [PubMed] [Google Scholar]
- 33.Melegh BI, Banfai Z, Hadzsiev K, Miseta A, Melegh B. Refining the South Asian origin of the Romani people. BMC Genet. 2017;18:82. doi: 10.1186/s12863-017-0547-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen J, Scheffer DI, Kwan KY, Corey DP. SHIELD: an integrative gene expression database for inner ear research. Database (Oxf) 2015;2015:bav071. doi: 10.1093/database/bav071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schrauwen I, Hasin-Brumshtein Y, Corneveaux JJ, Ohmen J, White C, Allen AN, et al. A comprehensive catalogue of the coding and non-coding transcripts of the human inner ear. Hear Res. 2016;333:266–74. doi: 10.1016/j.heares.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang R, Chang M, Zhang M, Wu Y, Qu X, Huang S. The structurally plastic CH2 domain is linked to distinct functions offimbrins/plastins. J Biol Chem. 2016;291:17881–96. doi: 10.1074/jbc.M116.730069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kokotas H, Grigoriadou M, Villamar M, Giannoulia-Karantana A, del Castillo I, Petersen MB. Hypothesizing an ancient Greek origin of the GJB2 35delG mutation: can science meet history? Genet Test Mol Biomark. 2010;14:183–7. doi: 10.1089/gtmb.2009.0146. [DOI] [PubMed] [Google Scholar]
- 38.Li L, Yuan H, Wang H, Guan J, Lan L, Wang D, et al. Identification of a MYO7A mutation in a large Chinese DFNA11 family and genotype–phenotype review for DFNA11. Acta Otolaryngol. 2018;138:463–70. doi: 10.1080/00016489.2017.1397743. [DOI] [PubMed] [Google Scholar]
- 39.Alkhayat AH, Kraemer SA, Leipprandt JR, Macek M, Kleijer WJ, Friderici KH. Human beta-mannosidase cDNA characterization and first identification of a mutation associated with human beta-mannosidosis. Hum Mol Genet. 1998;7:75–83. doi: 10.1093/hmg/7.1.75. [DOI] [PubMed] [Google Scholar]
- 40.Sahoo T, Theisen A, Sanchez-Lara PA, Marble M, Schweitzer DN, Torchia BS, et al. Microdeletion 20p12.3 involving BMP2 contributes to syndromic forms of cleft palate. Am J Med Genet Part A. 2011;155:1646–53. doi: 10.1002/ajmg.a.34063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.