Abstract
In clinical exome sequencing (cES), the American College of Medical Genetics and Genomics recommends limiting variant interpretation to established human-disease genes. The diagnostic yield of cES in intellectual disability and/or multiple congenital anomalies (ID/MCA) is currently about 30%. Though the results may seem acceptable for rare diseases, they mean that 70% of affected individuals remain genetically undiagnosed. Further analysis extended to all mutated genes in a research environment is a valuable strategy for improving diagnostic yields. This study presents the results of systematic research reanalysis of negative cES in a cohort of 313 individuals with ID/MCA. We identified 17 new genes not related to human disease, implicated 22 non-OMIM disease-causing genes recently or previously rarely related to disease, and described 1 new phenotype associated with a known gene. Twenty-six candidate genes were identified and are waiting for future recurrence. Overall, we diagnose 15% of the individuals with initial negative cES, increasing the diagnostic yield from 30% to more than 40% (or 46% if strong candidate genes are considered). This study demonstrates the power of such extended research reanalysis to increase scientific knowledge of rare diseases. These novel findings can then be applied in the field of diagnostics.
Subject terms: Next-generation sequencing, DNA sequencing
Introduction
Over the last decade, next-generation sequencing has revolutionized the world of rare diseases. After an extensive effort by researchers to identify new genes responsible for human diseases, clinical whole-exome sequencing (cES) is now currently used in the clinical setting for heterogeneous and rare genetic disorders, it leads to a positive diagnosis for about 30% of individuals with intellectual disability (ID) and/or multiple congenital anomalies (MCA) [1–5]. Two-thirds of these patients therefore remain without a molecular diagnosis after cES. This diagnostic yield is limited by the stringent criteria of the ACMG (American College of Medical Genetics and Genomics), which recommends restricting variant interpretation to the genes responsible for human diseases [6] mostly reported in the OMIM database, using a routine practice of wide diagnostic laboratories.
The exponential increase of scientific and genomic knowledge means that new genes are regularly linked to rare diseases with ID and/or MCA, which has resulted in ~30–60 new entries and 300–900 updated entries per month in the OMIM database (https://www.omim.org/statistics/update, 20th February 2019) [7]. The need for periodic reanalysis of ES data in undiagnosed patients therefore appears obvious. A recent reanalysis study, which diagnosed 10–15% more cases, endorsed systematic diagnostic reanalysis or reanalysis upon patient request to increase diagnostic yield [8]. However, in the vast majority of cases, successive cES analyses of OMIM disease-causing genes are inadequate. Such analyses could be enriched by the significant number of disease-causing genes published in the scientific literature and not referenced in the OMIM database. Indeed, with more than 250 genes associated with human diseases and more than 9200 novel pathogenic/likely pathogenic variants reported annually, knowledge moving faster than the data can be updated in the OMIM database, and a number of diagnoses are subsequently lacking [3, 8–12]. Moreover, information about variants has been updated in databases, such as OMIM or ClinVAR, because the reanalysis of ES data results in the reclassification of previously described variants. These changes are mainly variants of uncertain significance reclassified as affect functions or benign variants [3].
The reanalysis and reinterpretation of ES data in a research setting are made possible by a wide variety of tools and databases, and the abundant knowledge available in the scientific literature. Some resources can easily be implemented to the data workflow to help with the interpretation of ES data, including in silico predictive scores (PolyPhen, CADD, Grantham, GERP, http://mendel.stanford.edu/SidowLab/downloads/gerp/, SIFT, http://sift.jcvi.org/, and pLI), allele frequency in the population (EVS, http://evs.gs.washington.edu/EVS/, ExAC, and GnomAD), or in silico tools for splicing defects (HSF, http://www.umd.be/HSF/) [13–15].
A few studies that have used a range of currently available tools to reanalyze the cES data in the research environment have successfully increased diagnostic yield [3, 8, 16]. The diagnostic yield was dependent on the strategy and varied widely from study to study. Nambot et al. performed a systematic annual reanalysis of singleton ES and obtained 15% more diagnoses (24 individuals) from new and recently published genes. The combined results of these studies indicated that the OMIM database is slow to be updated and the drawbacks of restricting analyses to OMIM disease-causing genes were clearly demonstrated. Another effective strategy for accelerating the identification of new disease-causing genes is the use of trio-based ES. Eldomery et al. had a significantly higher diagnostic yield using trio-ES data in research analysis, with more than 50% of likely contributory genes, including candidate genes without recurrence [16]. These studies evidence the clear advantage of extending exploration to non-OMIM disease-causing genes, though further analyses will be essential to confirm the preliminary results [17].
In this study, singleton-ES data from 313 individuals with ID/MCA and negative cES were reanalyzed in a research setting. Our findings supplement a previously published study in the diagnostic setting [8]. We discuss the potential gains and consequences of reanalyzing ES data in individual care and for scientific knowledge.
Patients and methods
Patients
Singleton-ES data were obtained from a cohort of 313 unrelated patients, from 313 families, referred to the Reference Center for Congenital Anomalies and Malformative Syndromes in Dijon (France), or the Orphanomix units for genetics testing located in several hospitals in France. These data were reanalyzed in a research laboratory between July 2013 and December 2017 (Fig. 1a). The local ethics committee approved this study.
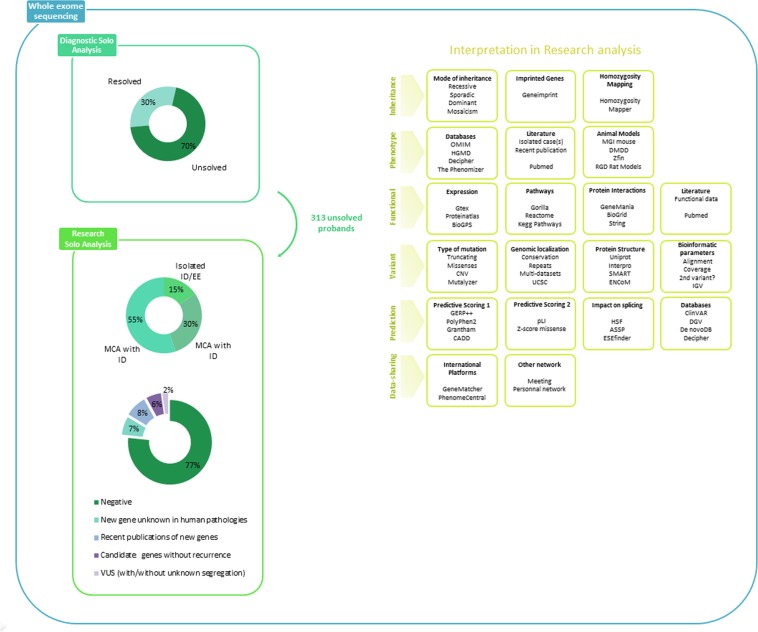
Fig. 1.
Strategy of ES data analysis, databases, and tools used for variant interpretation in a research environment and the global results of this study. First, we filtered on the suspected mode of inheritance. Then, a large research, including literature and public databases associated with in silico scores, helping at the interpretation or indicates a possible link with the disease, allowed to select candidate genes. Finally, accordingly with the parental segregation, the variant can be shared in international platforms for the purpose of the identification of additional affected cases to confirm or rule out the candidate gene
Whole-exome sequencing research analysis and interpretation
Each research analysis was done immediately after the cES results were obtained. Rapid deployment is facilitated by our local translational integrative organization that combines a unit focused on diagnostic innovation and a research team. ES data are typically analyzed in two steps: (1) diagnostic interpretation restricted to disease-causing genes reported in the OMIM database, and (2) reanalysis in a research environment, including all detected variants. In each stage, a multidisciplinary team is implicated in the interpretation of ES data and some experts are involved in the two steps.
We used all of the ES data initially produced for the singleton cES analysis [8]. BAM files had been aligned to a human genome reference sequence (GRCh37/hg19) using BWA (Burrows–Wheeler Aligner; v0.7.15). All aligned reads underwent the same procedure: (i) duplicate paired-end reads were removed by Picard 2.4.1, (ii) indel realignment, and (iii) base quality score recalibration were done on the Genome Analysis Toolkit (GATK; v3.7). Variants with a quality score >30 and an alignment quality score >20 were annotated with SeattleSeq SNP Annotation (see Web resources). The annotation includes data of several public databases, such as HGMD (http://www.hgmd.cf.ac.uk/), ClinVAR (https://www.ncbi.nlm.nih.gov/clinvar/), and COSMIC (https://cancer.sanger.ac.uk/cosmic). CNV was detected with XHMM software (https://www.atgu.mgh.harvard.edu/) and annotated, using chromosomal coordinates of coding exonic sequences on the human genome (https://www.ncbi.nlm.nih.gov/refseq/). Rare variants present at a frequency above 1% in DGV, GnomAD Browser, ExAC Browser, and the NHLBI GO Exome Sequencing Project or present in 100 local exomes of unaffected individuals were excluded (see URL).
In the reanalysis for research purposes, we extended variant interpretation to genes not associated with human disease in the OMIM database. We also extended to atypical phenotypes unrelated to well-known genes in the OMIM database in prioritizing (i) biallelic variants, (ii) truncating and splice site variants, (iii) homozygous missense variants and in-frame deletion/insertion, and (iv) heterozygous missense variants and in-frame deletion/insertion. We first looked at a gene recently recognized in the literature as disease-causing but not listed in OMIM, and then we turned our attention to genes unknown to cause human diseases yet. To help with the interpretation, we used public databases listing variants or genes previously reported in human diseases, animal models (such as mouse, zebrafish, or rat), and gene expression data, and the impact of the variant in protein structure or function, protein interactions, or signaling pathways. We also used bioinformatics tools, including predictive scores of pathogenicity, conservation, or impact of missense and truncating variants (Fig. 1). We performed a systematic review of the literature to identify isolated cases, recent description of new genes, or functional data. In this study, cES data have been analyzed by two experts, and all of the results and candidate genes were presented and discussed in multidisciplinary assembly.
Variant validation and parental segregation
Candidate variants and parental segregation were confirmed by Sanger sequencing for SNV and quantitative PCR for CNV.
Genomic DNA was amplified by polymerase chain reaction (PCR) for Sanger sequencing, using HotStarTaq PCR kit (Qiagen) according to the manufacturer's protocol. PCR products were purified with the Agencourt CleanSEQ system (Beckman Coulter) and sequenced with the BigDye Terminator Cycle Sequencing kit, v3.1 (Applied Biosystems) in an ABI 3730 sequencer (Applied Biosystems). Sequence data were analyzed with Mutation Surveyor v4.0.9 (Softgenomics).
Genomic DNA was amplified for quantitative PCR with SYBR Green Master Mix kit according to the manufacturer's protocol.
Data sharing
For each candidate variant in a candidate gene, unknown in rare diseases or known but with a new phenotypic presentation, we actively searched for additional similar patients to confirm the genotype–phenotype correlation (Fig. 1). Multiple strategies were used to improve data sharing: exchanging information by e-mail, presenting case-report results in international congresses, scrutinizing a large series of ES studies in the literature or in public databases (i.e., DECIPHER, https://decipher.sanger.ac.uk/, de novo-db, http://denovo-db.gs.washington.edu/denovo-db/, and ClinVAR, https://www-ncbi-nlm-nih-gov.gate2.inist.fr/clinvar/), and international data-sharing platforms like GeneMatcher (https://www.genematcher.org/). All variants identified in new disease- causing genes have submitted in ClinVAR databases (SUB2871008, SUB2871014, SUB3604471, and SUB3731210).
Results
The cohort included 241/313 children (77%) with an average age of 9 years, and 72/313 adults (23%), including 185/313 males (59%) and 128/313 females (41%). Eleven patients were born to reported consanguineous parents. The ethnic origin was European (93.2%), African (6.5%), or Asian (0.3%). The individuals presented with isolated ID/epileptic encephalopathy (EE) (49/313 individuals), ID with MCA (93/313 individuals), or multiple congenital anomalies (MCA) without ID (171/313 patients) (Fig. 1). MCA included abnormality of brain morphology (HP: 0012443) (11%, 29/264 individuals), abnormality of calvarial morphology (HP:0002648) and abnormality of the face (HP:0000271) (57%, 150/264 individuals), abnormality of skeletal muscles (HP:0040290) (49%, 129/264 individuals), abnormality of the skin (HP:0000951) (20%, 53/264 individuals), abnormality of the gastrointestinal tract (HP:0011024) (17%, 45/264 individuals), abnormality of the genitourinary system (HP:0000119) (15%, 40/264 individuals), abnormality of the cardiovascular system (HP:0001626) (19%, 50/264 individuals), and abnormality of the respiratory system (HP:0002086) (13%, 34/264 individuals). Neurological features (142/313 individuals) included seizures (HP:0001250) (33%), global developmental delay (HP:0001263) (59%), ID (HP:0001249) (72%), and autism spectrum disorder (HP:0000729) (13%). Patients also had some abnormality of the endocrine system (HP:0000818) (6%), immune system (HP:0002715) (3%), or unspecific metabolic anomalies (3%). Detailed anonymized clinical data were available in the PhenomeCentral database using the standardized HPO (Human Phenotype Ontology) terms (https://www.phenomecentral.org/).
All individuals had received a negative or non-conclusive result after array-CGH and singleton cES analysis in the diagnostic setting. Of the 313 individuals, 133 were cases without a molecular diagnosis, who had been the focus of a previous study and 14 were individuals who have a candidate gene previously reported in the case of additional research strategy [8].
After Sanger validation and parental segregation, the singleton-ES research analysis identified 84 candidate variants in 66 different genes in 73/313 individuals (23%) (Supplementary Table 1 and Table 1). These 84 variants included 55 missense variants (66%), 22 truncating variants (27%), 5 variants predicted to affect a splice site (6%), and 1 CNV (1%). The 84 variants were mainly de novo autosomal dominant (40%) or recessive (29%), and included 16 homozygous and 9 compound heterozygous variants. Of the 7.3% of X-linked variants, three occurred de novo and three were inherited from an unaffected mother. Three autosomal- dominant variants were confirmed by Sanger sequencing, but the parental segregation is unknown, because parental DNAs are not available (Fig. 2, Supplementary Table 1 and Table 1).
Table 1.
Summary of the 26 candidate genes identified by research ES reanalysis in this study
| Cases | Candidate gene | Mode of inheritance | Variant (hg19) | PolyPhen2 | Mis-Z score | pLI | HSF | Cohort | Phenotype | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| 49 | HOXB9 | De novo |
chr17:g.46700492 G > A NM_024017.4:c.523 C > T NM_024017.4: p.(Pro175Ser) |
Deleterious | 1.59 | 0.10 | MCA without ID | Spondylocostal dysostosis | ||
| 50 | PCSK7 | De novo | chr11:g.117079626 G > C NM_004716.2:c.1678C > G NM_004716.2: p.(Arg560Gly) | Tolerated | 2.75 | 0.19 | MCA without ID | Pai syndrome | ||
| 51 | SMARCB1 | De novo |
chr22:g.24175842 C > G NM_003073.3:c.1070 C > G NM_003073.3: p.(Thr357Arg) |
Deleterious | 4.51 | 1.00 | MCA without ID |
Facial dysmorphism Deafness Cardiac defects Pectus excavatum Hypoplasia of the corpus callosum |
Retterer et al. [54] | |
| 52 | SLC25A24 | De novo |
chr1:g.108700103 C > T NM_013386.3:c.650 G > A NM_013386.3: p.(Arg217His) |
Deleterious | 0.21 | 0.00 | MCA without ID |
Cutis laxa Progeroid facial appearance |
||
| 53 | SBNO2 | De novo |
chr19:g.1122996 A > G NM_014963.2:c.677 T > C NM_014963.2: p.(Leu226Pro) |
Deleterious | 2.07 | 0.02 | MCA with ID | Syndromic poikiloderma | ||
| 54 | NUP50 | AR |
chr22:g.45564079_45564082delGAAG NM_007172.3:c.21_24delGAAG NM_007172.3: p.(Lys8Asnfs*2) chr22:g.45579309 A > G NM_007172.3:c.1112 A > G NM_007172.3: p.(Asn371Ser) |
NA tolerated |
2.11 | 0.24 | Isolated ID/EE | ID | ||
| 55 | CASZ1 | AR |
chr1:g.10707901 C > T NM_001079843.2:c.3454 G > A NM_001079843.2: p.(Ala1152Thr) chr1:g.10714261 G > A NM_001079843.2:c.1853C > T NM_001079843.2: p.(Thr618Ile) |
Deleterious | 4.04 | 1.00 | MCA without ID |
Hyperextensible skin Thoracolumbar scoliosis Myopathy |
||
| 56 | SCUBE2 | AR |
chr11:g.9051581 G > A NM_020974.1:c.2266 C > T NM_020974.1: p.(Pro756Ser) chr11:g.9068923 C > T NM_020974.1:c.1895G > A NM_020974.1: p.(Gly632Asp) |
Deleterious | 0.46 | 0.00 | MCA without ID | Holoprosencephaly | ||
| 57 | ACO2 | AR |
chr22:g.41895778 C > T NM_001098.2:c.85 C > T NM_001098.2: p.(Arg29Trp) chr22:g.41903904 C > T NM_001098.2:c.283 C > T NM_001098.2: p.(Arg95Cys) |
Deleterious | 3.27 | 0.45 | Isolated ID/EE | ID | ||
| 58 | GBP3 | AR | chr1:g.89476647_89476648delAA NM_018284.2:c.1301_1302delTT NM_018284.2: p.(Phe434Tyrfs*13) | NA | −1.33 | 0.00 | MCA without ID |
Ataxia Facial dysmorphism |
||
| 59 | ABCA2 | AR |
chr9:g.139907733_139907739delCGAGCTG NM_001606.4:c.4582_4588delCAGCTCG NM_001606.4: p.(Gln1528*) |
NA | 5.37 | 1.00 | Isolated ID/EE | ID | Maddirevula et al. [11] | |
| 60 | CYFIP2 | AR | chr5:g.156746779 A > G NM_001037333.1:c.1366 A > G NM_001037332.2: p.(Met456Val) | Deleterious | 6.15 | 1.00 | MCA with ID |
ID Facial dysmorphism |
||
| 60 | TMEM71 | AR | chr8:g.133764099 G > T NM_001145153.1:c.246 C > A NM_001145153.1: p.Cys82* | NA | −0.98 | 0.00 | MCA without ID | Syndromic neutropenia | ||
| 61 | GPATCH11 | AR | chr2:g.37317866 G > T NM_174931.2:c.316 + 1 G > T | NA | −1.19 | 0.00 | Splice defect | MCA without ID | Syndromic neutropenia | |
| 62 | FAT4 | AR |
chr4:g.126238090 G > T NM_024582.4:c.524 G > T NM_024582.4: p.(Arg175Leu) chr4:g.126239320 C > A NM_024582.4:c.1754C > A NM_024582.4: p.(Pro585Gln) |
Deleterious Tolerated | −0.32 | 1.00 | MCA without ID | Severe arthrogryposis | ||
| 63 | SARS | AR |
chr1:g.109774299 G > T NM_006513.3:c.638 G > T NM_006513.3: p.(Arg213Leu) |
Deleterious | 3.13 | 1.00 | Isolated ID/EE | EE | ||
| 64 | CADPS | AR | chr3:g.62543079 C > A NM_003716.3:c.1753 + 1 G > T | NA | 3.41 | 0.05 | Splice defect | MCA with ID |
Musculoskeletal defects Facial dysmorphism Seizure |
|
| 65 | RFT1 | AR |
chr3:g.53126558 C > T NM_052859.3:c.1285 G > A NM_052859.3: p.(Val429Met) chr3:g.53156410 G > A NM_052859.3:c.436 C > T NM_052859.3: p.(His146Tyr) |
Deleterious | −0.35 | 0.00 | MCA with ID |
ID Facial dysmorphism Hyperextensible skin Hypotonia |
||
| 66a | ARHGAP6 | X-linked |
chrX:g.11160407 A > G NM_013427.2:c.2203 T > C NM_013427.2: p.(Trp735Arg) |
Deleterious | 1.99 | 0.99 | Isolated ID/EE | ID | ||
| 66a | CHIC1 | X-linked | chrX:g.72783341 G > C NM_001039840.2:c.221 G > C NM_001039840.2: p.(Arg74Pro) | Deleterious | 1.71 | 0.68 | Isolated ID/EE | ID | ||
| 67 | MAGED1 | X-linked | chrX:g.51638620 G > A NM_001005332.1:c.517 G > A NM_001005332.1: p.(Ala173Thr) | Deleterious | 1.42 | 0.98 | MCA with ID |
ID Facial dysmorphism Autism |
||
| 68 | PLXNA1 | AD | chr3:g.126733621 C > T NM_032242.3:c.2824 C > T NM_032242.3:p.(Arg942Trp) | Deleterious | 5.15 | 1.00 | MCA with ID |
Facial dysmorphism Eye abnormality Seizure |
||
| 69 | PLCB4 | AD |
chr20:g.9389801 A > T NM_000933.2:c.1936A > T NM_000933.3: p.(Met646Leu) |
Deleterious | 2.78 | 0.27 | MCA without ID |
Anodontia Facial dysmorphism |
||
| 70 | ACTN2 | NA |
chr1:g.236881186 T > G NM_001103.3:c.155 T > G NM_001103.3: p.(Leu52Arg) |
Tolerated | 1.76 | 1.00 | MCA without ID | Myopathy | ||
| 71 | FEZF2 | NA |
chr3:g.62355766 G > A NM_018008.3:c.1372 C > T NM_018008.3: p.(Gln458*) |
NA | 3.73 | 0.90 | MCA without ID |
Facial dysmorphism Deafness |
Anazi et al. 2017 | |
| 72 | MTOR | NA | chr1:g.11227521_11227522delinsAA NM_004958.3:c.4306_4307delinsTT NM_004958.3: p.(Ala1436Phe) | Deleterious | 7.89 | 1.00 | MCA with ID |
Macrocephaly Facial dysmorphism Cardiac defects Hypotonia EE Splenomegaly Hepatomegaly |
OMIM 616638 |
NA not available, AR autosomal recessive, AD autosomal dominant, HSF human splice finding, PP2 PolyPhen2 score, Mis-Zscore Z-score for missense (constraint metrics from GnomAD), pLI probability of loss-of-function intolerance (constraint metrics from GnomAD)
aTwo candidate genes in the same case. A detailed phenotype is available upon request. Candidate genes included unknown disease-causing genes and known disease-causing genes inconclusive in our patients because of the absence of parental segregation or atypical phenotype. One year or more has passed since the initial cES analysis and this reanalysis
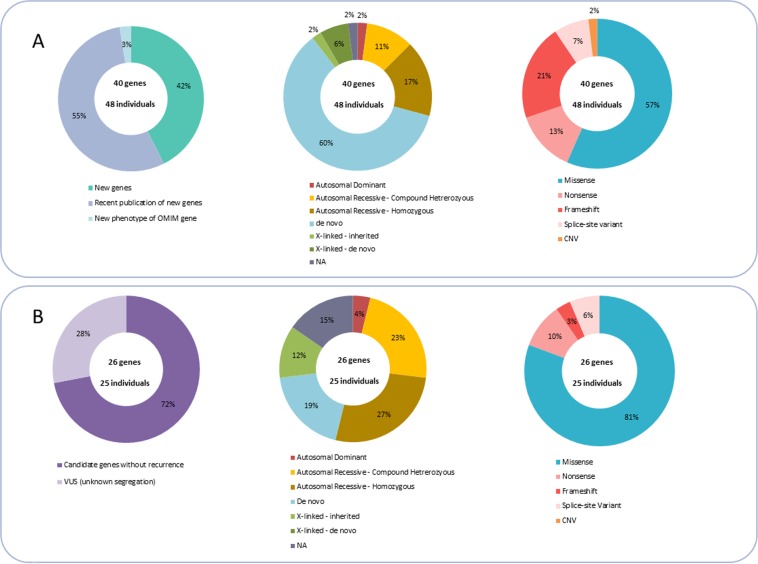
Fig. 2.
Repartition of the mode of inheritance and type of variant in validated disease-causing genes (a) or in candidate genes (b)
All 66 candidate genes were shared with the national and/or international scientific and medical communities. We submitted the individual genotypes, mainly new candidate genes or genes with low recurrence and/or associated with an atypical phenotype to the GeneMatcher platform. A match was found for 16 submitted genes. Our data-sharing strategies confirmed that 40/66 genes as responsible for disease (Supplementary Table 1) and the remaining 26/66 are currently considered candidate genes (Table 1), because there was no or insufficient recurrence (Fig. 2). The number of recurrence to confirm the involvement of a gene in a disease is estimated to three unrelated cases with homozygous or compound heterozygous variants in autosomal recessive phenotypes, and five affected cases with heterozygous variants for autosomal-dominant phenotype [18]. In the absence of insufficient recurrence, the genes remain a candidate with a variant of uncertain significance. Many affected function variants were missense variants (57%), most frequently identified in the DI/EE cohort (75%) (Figs. 2a and 3a). The autosomal or X-linked sporadic variants appear to affect function, while half of autosomal-recessive or X-linked inherited variants remain variants of uncertain significance (Figs. 2a and 3d). The vast majority of nonsense and truncating variants (about 75%) were confirmed responsible for diseases, and around 40% of missense and splice site variants remained candidates (Fig. 3e). Data sharing resulted in national and international collaborations for 21 genes (27 patients), 15 of which were used for functional studies. Our results have led to 15 scientific publications, and 12 papers are currently in progress, 6 of which are being led by our team [19–33].
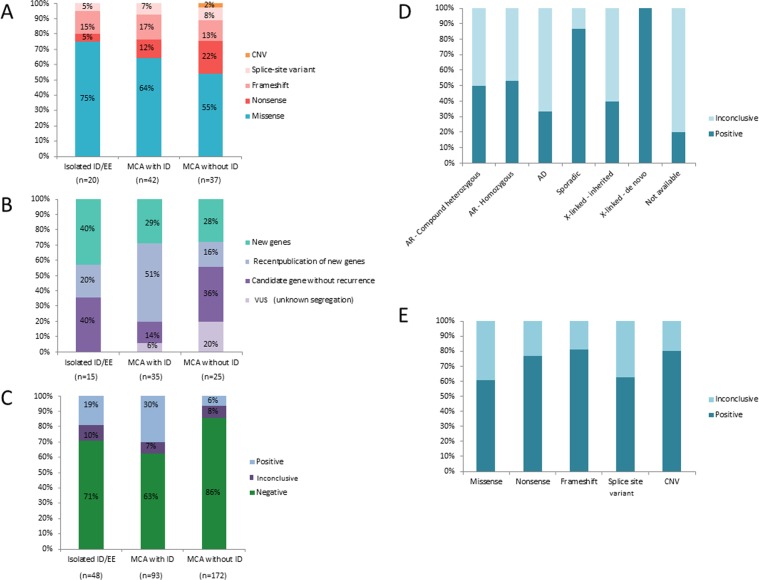
Fig. 3.
Repartition of the type of variants (a), classification of identified genes (b), and conclusion (c) in MCA and ID/EE cohorts. Percentage of inconclusive with variants of uncertain significance or positive diagnosis by the mode of inheritance (d) or type of variants (e)
Overall, we confirmed the involvement of 40 different genes in human disease in 48/313 individuals (15%). These 48 cases provided a molecular diagnosis for 30% of our MCA and ID cases, 19% of our isolated ID/EE cases, and 6% of our MCA without ID cases. The 40 disease-causing genes can be classified as (1) 17 new genes unknown in human disease; (2) 22 previously published genes with low recurrence and not yet referenced in the OMIM database (https://www.omim.org/); (3) 1 established OMIM gene with a new previously unrelated phenotype (Fig. 2a).
The results were of uncertain significance in 26/313 individuals (8%) who made up 7% of our MCA and ID cases, 10% of isolated ID/EE cases, and 8% of the MCA without ID cohort (Fig. 3c). Indeed, the 26 genes are now candidates because of lacking recurrence (Table 1), and were mostly missense variants (81%), illustrating the difficulties to interpret the functional impact of missense variants in the gene only based on parental segregation and in silico scores in the absence of recurrence after data sharing (Fig. 2b).
Discussion
Research reanalysis after negative singleton cES has demonstrated its ability to rapidly improve diagnostic yield and scientific knowledge. Using this approach, we confirmed the involvement of 40 different disease-causing genes in 15% of individuals with negative cES. We were thus able to increase the positive yield from 30% with diagnostic cES analysis alone to 40% with extended singleton-ES reanalysis in a research setting.
Identification of new disease-causing genes improves scientific knowledge
Analyzing singleton-ES data for research purposes provides a valuable opportunity to identify new disease-causing genes. We identified 17 new genes in 21 individuals (Supplementary Table 1). Among these 17 genes, 9 genes presented autosomal-recessive truncating and/or missense variants and 7 presented de novo missense variants (Fig. 2a). Five genes were selected because of de novo truncated variants in a gene with a highly loss-of-function intolerance (pLI = 1 in ExAc database; http://exac.broadinstitute.org/; http://gnomad.broadinstitute.org/). Of the 17 new genes, 11 were confirmed as involved after significant recurrence through data sharing, via GeneMatcher (6/11) or other data-sharing strategies (5/11), including congresses and national or personal networks. Strategies combining research reanalysis and international data sharing foster national and international collaborations (21 collaborative projects) and have improved scientific knowledge of rare disorders. Our results have resulted in 15 papers in scientific publications [19, 22–33] and 12 papers are currently in progress.
Confirmation of previously published disease-causing gene
cES analysis is often restricted to disease-causing genes reported in the OMIM database, in which 5.102 disease-causing genes are associated with a genetic disease in OMIM database (updated 19th February 19, 2019). When next-generation sequencing-based technologies appeared, the number of known genes responsible for human diseases increased exponentially, and new genes are published every day in the scientific literature, in particular for ID/MCA [7]. Despite regular updates, the OMIM database remains incomplete and provides only limited resources for the diagnosis of rare diseases.
Access to all available scientific literature is essential, because it allows researchers to identify genes previously published but not yet recognized in the OMIM database [11]. Nineteen of our 40 involved genes from 7% of the 313 reanalyzed individuals, had previously been published but were not referenced in the OMIM database. TBR1 (MIM 604616) variants were identified in two patients with ID and autism. The TBR1 gene was first reported in patients with autism in 2012, and more than ten unrelated patients were later reported in different large cohorts of autistic individuals. Functional data and animal models also underlined the involvement of TBR1 in autism/ID, though it was not recognized in the OMIM database [34–39]. Our data sharing identified 20 additional individuals with ID and TBR1 variants, definitively establishing causality (Nambot et al., in press).
Regular updates to the OMIM database encourage prospective diagnostic reanalysis. This can lead to new diagnoses from recently identified genes that are progressively reported in the OMIM database, but the molecular diagnosis is generally delayed by at least 1 year [8]. Three of our 40 disease-causing genes (1% of the 313 reanalyzed individuals) had recently been described in the literature (Fig. 2, Supplementary Table 1). Frequent literature reviews and prospective updates of bioinformatics pipelines would ensure the diagnosis of rare diseases linked to recently identified genes.
Extension of the phenotype–genotype spectrum of well-known genes
Another fundamental challenge for variant interpretation is to provide a positive diagnosis for well-known genes, when the phenotype or genotype is atypical. Clinical heterogeneity is common in rare diseases, ranging from vast, overlapping clinical spectrums to completely different phenotypes [40–42]. In the OMIM database, 729/2664 disease-causing genes are associated with two distinct or overlapping phenotypes, and 235/2664 genes have more than four phenotypes, highlighting the complexity of correlating genotypes–phenotypes (updated 7th April, 2018). Phenotypic variability can be explained by the impact of variants, hypomorphic, or null alleles [41], gain or loss of function [40], and the genetic and/or environmental background [43, 44]. In ultrarare diseases, knowledge is limited by the lack of individuals with causal variants in the same gene. Research analysis could therefore identify new phenotypes linked to well-known genes, but only recognized for only one phenotype with different clinical presentations in the OMIM database. If there is no correlation with the OMIM phenotypic reference, reverse phenotyping limits the chances of delivering an initial diagnosis. In an individual affected with a newly identified severe Bohring–Opitz-like syndrome, ES revealed a homozygous variant in the KLHL7 gene (MIM 611119). KLHL7 was previously reported in cold-induced sweating syndrome 3, which is characterized by clinical features that partly overlap with our patient’s presentation (MIM 617055) [23]. We submitted the case to the GeneMatcher platform and at an international congress in the hope of identifying additional patients, and thus recruited five additional patients with a Bohring–Opitz-like presentation. We finally expanded the clinical spectrum of KLHL7 autosomal-recessive variants by describing a syndrome with features overlapping cold-induced sweating syndrome 3 and Bohring–Opitz syndrome [23].
Ongoing challenges in variant interpretation and candidate genes
Variant interpretation in ES research analysis is mainly based on suspected inheritance, phenotypic, variant and functional databases, variant prediction scores, animal models, accessible literature, and data sharing (Fig. 1). Alas, even with access to multiple data sources and the many tools designed to exploit them, many candidates remain (Fig. 2b).
The use of a singleton strategy for diagnosis has an economic advantage [8], but shows its limits when the analysis is extended to research. The absence of parental segregation reduces the ability to select candidate variants on the mode of inheritance, since de novo variants are confirmed to affect function in almost 90% of sporadic individuals. However, autosomal dominant or X-linked inherited variants remain variants of uncertain significance in 60–70% of cases, and in 50% of cases for recessive variants. Most inherited variants are variants of uncertain significance, because it is difficult to distinguish a causal variant from among all the inherited variants (~50% of variants), as well as incomplete penetrance and intra-familial variability, which have been described in some rare genetic diseases. Access to mutational parental segregation and detailed phenotyping would certainly be an advantage in this highly complex context.
The interpretation of missense variants also continues to be a challenge, even more so than truncating variants (40% of missense variants remained variant of uncertain significance). Missense variants can be located in functional or structural protein domains (CSNK2A1) or not (SAMD9, CLTC), clustered (PACS2, NACC1), or dispersed around the gene (NR2F1) with gain or loss-of- function impacts (Supplementary Table 1 and Table1). The absence of functional studies makes predicting the impact of missense variants difficult. Since missense variants make up 75–80% of all rare variants in the exome, the risk of identifying a missense variant as candidate variants is statistically increased. Algorithms have been developed to help with variant interpretation, but are not sufficient without biological validation (Fig. 1). This highlights the importance of the international effort to complete public variant databases, such as ClinVAR or DECIPHER [45, 46]. In addition to the missense variants, a large portion of splice site variants remained candidates (Fig. 3e).
Our work shows that data sharing is essential for establishing human genotype–phenotype relationships and conclusively classifying variants. The Matchmaker Exchange Initiative offers a data-sharing platform that can be used to match patients all over the world, according to their phenotypic or genotypic features [47]. Data-sharing tools increase diagnostic yield and should be actively used by the scientific and medical communities. In this study, data sharing was used to determine the involvement of 40/66 genes in rare disease, mainly for de novo missense variants. Low or no recurrence, however, did not allow us to establish the implication of 26/66 genes in human disease. Because these genes are candidates for ultrarare phenotypes and the number of clinicians or scientists sharing genotypic data is relatively small, additional cases have not yet been identified. In time, the increasing amount of data shared on international platforms should provide additional chances to conclude [25, 30].
Numerous cES remains negative because the cohort comprised various and heterogeneous disorders with the unknown etiology; thus, they can be included in non-Mendelian diseases or environmental/external causes of diseases [48]. In addition, the detection of variants remains, mainly depending of the sequencing technique, the exome capture kit, the exome coverage, and the bioinformatics pipeline used [49–51]. A reanalysis of cES data with an updated pipeline that reflects the lastest knowledge and the progression of bioinformatics software appears to be a determining factor in the identification of new variants during reanalysis, notably in low-covered genes, exon/intron boundaries, and noncoding sequences [48].
An effective strategy for increasing positive yield
This study demonstrates how singleton-ES research reanalysis can efficiently and rapidly increase scientific knowledge in rare diseases by identifying new disease-causing genes, implicating recent known genes not reported in the OMIM database, or extending the phenotype or genotype spectrum of well-known genes. This strategy is also a rapid means of obtaining a diagnosis, with positive results in 15% of individuals with negative cES, leading to nearly 11% of additional diagnoses in the initial cohort. The limits of the singleton strategy could be overcome with a second-step strategy based on trio ES. Complementary ES strategies increase the diagnostic ability of ES, and should be explored further and would be interesting to be quickly integrated after an initial diagnostic analysis in changing diagnostic laboratory practices in the strategy of exome analysis in the future.
Supplementary information
Acknowledgements
We thank all collaborators, clinicians, patients, and their families.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ange-Line Bruel, Sophie Nambot
These authors jointly supervised this work: Laurence Faivre, Christel Thauvin-Robinet
Contributor Information
Ange-Line Bruel, Phone: +33(0)380393238, Email: ange-line.bruel@u-bourgogne.fr.
Orphanomix Physician’s Group:
Alain Verloès, Alexandra Karsenti, Alice Goldenberg, Aurélia Jacquette, Béatrice Jouret, Béatrice Laudier, Christine Coubes, Christine Francannet, Daphné Lehalle, David Geneviève, Delphine Heron, Didier Lacombe, Elise Schaefer, Elodie Lacaze, Emmanuel Jacquemin, Fabienne Prieur, Fanny Laffarge, Florence Petit, François Feillet, Gilles Morin, Gwenaëlle Diene, James Lespinasse, Jeanne Amiel, Judith Melki, Laëtitia Lambert, Laurence Perrin, Lucile Pinson, Marie-Line Jacquemont, Marie-Pierre Cordier-Alex, Marine Lebrun, Marion Gérard-Blanluet, Marjolaine Willems, Massimiliano Rossi, Nicolas Chassaing, Nicole Philip, Renaud Touraine, Salima El-Chehadeh, Séverine Audebert-Bellanger, Sophie Blesson, and Yline Capri
Supplementary information
The online version of this article (10.1038/s41431-019-0442-1) contains supplementary material, which is available to authorized users.
References
- 1.Farwell KD, Shahmirzadi L, El-Khechen D, Powis Z, Chao EC, Tippin Davis B, et al. Enhanced utility of family-centered diagnostic exome sequencing with inheritance model-based analysis: results from 500 unselected families with undiagnosed genetic conditions. Genet Med. 2015;17:578–86. doi: 10.1038/gim.2014.154. [DOI] [PubMed] [Google Scholar]
- 2.Lee H, Deignan JL, Dorrani N, Strom SP, Kantarci S, Quintero-Rivera F, et al. Clinical Exome Sequencing for Genetic Identification of Rare Mendelian Disorders. JAMA. 2014;312:1880–7. doi: 10.1001/jama.2014.14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wright CF, Fitzgerald TW, Jones WD, Clayton S, McRae JF, van Kogelenberg M, et al. Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet. 2015;385:1305–14. doi: 10.1016/S0140-6736(14)61705-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med. 2013;369:1502–11. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Y, Muzny DM, Xia F, Niu Z, Person R, Ding Y, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–9. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–23. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vissers LELM, Gilissen C, Veltman JA. Genetic studies in intellectual disability and related disorders. Nat Rev Genet. 2016;17:9. doi: 10.1038/nrg3999. [DOI] [PubMed] [Google Scholar]
- 8.Nambot S, Thevenon J, Kuentz P, Duffourd Y, Tisserant E, Bruel A-L, et al. Clinical whole-exome sequencing for the diagnosis of rare disorders with congenital anomalies and/or intellectual disability: substantial interest of prospective annual reanalysis. Genet Med. 2017;20:645–654. doi: 10.1038/gim.2017.162. [DOI] [PubMed] [Google Scholar]
- 9.Bowling KM, Thompson ML, Amaral MD, Finnila CR, Hiatt SM, Engel KL, et al. Genomic diagnosis for children with intellectual disability and/or developmental delay. Genome Med. 2017;9. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5448144/ [DOI] [PMC free article] [PubMed]
- 10.Costain G, Jobling R, Walker S, Reuter MS, Snell M, Bowdin S, et al. Periodic reanalysis of whole-genome sequencing data enhances the diagnostic advantage over standard clinical genetic testing. Eur J Hum Genet Ejhg. 2018;26:740–744. doi: 10.1038/s41431-018-0114-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maddirevula S, Alzahrani F, Al-Owain M, Al Muhaizea MA, Kayyali HR, AlHashem A, et al. Autozygome and high throughput confirmation of disease genes candidacy. Genet Med. 2018;21:736–742. doi: 10.1038/s41436-018-0138-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wenger AM, Guturu H, Bernstein JA, Bejerano G. Systematic reanalysis of clinical exome data yields additional diagnoses: implications for providers. Genet Med. 2017;19:209–14. doi: 10.1038/gim.2016.88. [DOI] [PubMed] [Google Scholar]
- 13.Dong C, Wei P, Jian X, Gibbs R, Boerwinkle E, Wang K, et al. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum Mol Genet. 2015;24:2125–37. doi: 10.1093/hmg/ddu733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jian X, Boerwinkle E, Liu X. In silico tools for splicing defect prediction—a survey from the viewpoint of end-users. Genet Med. 2014;16:497–503. doi: 10.1038/gim.2013.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi T, Tsang WY, Li J, Lane W, Dynlacht BD. Centriolar kinesin Kif24 interacts with CP110 to remodel microtubules and regulate ciliogenesis. Cell. 2011;145:914–25. doi: 10.1016/j.cell.2011.04.028. [DOI] [PubMed] [Google Scholar]
- 16.Eldomery MK, Coban-Akdemir Z, Harel T, Rosenfeld JA, Gambin T, Stray-Pedersen A, et al. Lessons learned from additional research analyses of unsolved clinical exome cases. Genome Med. 2017;9:26. doi: 10.1186/s13073-017-0412-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chong JX, Buckingham KJ, Jhangiani SN, Boehm C, Sobreira N, Smith JD, et al. The genetic basis of mendelian phenotypes: discoveries, challenges, and opportunities. Am J Hum Genet. 2015;97:199–215. doi: 10.1016/j.ajhg.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilissen C, Hoischen A, Brunner HG, Veltman JA. Disease gene identification strategies for exome sequencing. Eur J Hum Genet. 2012;20:490–7. doi: 10.1038/ejhg.2011.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Assoum M, Philippe C, Isidor B, Perrin L, Makrythanasis P, Sondheimer N, et al. Autosomal-recessive mutations in AP3B2, adaptor-related protein complex 3 beta 2 subunit, cause an early-onset epileptic encephalopathy with optic atrophy. Am J Hum Genet. 2016;99:1368–76. doi: 10.1016/j.ajhg.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Assoum M, Lines MA, Elpeleg O, Darmency V, Whiting S, Edvardson S, et al. Further delineation of the clinical spectrum of de novo TRIM8 truncating mutations. Am J Med Genet A. 2018;176:2470–2478. doi: 10.1002/ajmg.a.40357. [DOI] [PubMed] [Google Scholar]
- 21.Basilicata MF, Bruel A-L, Semplicio G, Valsecchi CIK, Aktaş T, Duffourd Y, et al. De novo mutations in MSL3 cause an X-linked syndrome marked by impaired histone H4 lysine 16 acetylation. Nat Genet. 2018;50:1442–51. doi: 10.1038/s41588-018-0220-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bruel A-L, Masurel-Paulet A, Rivière J-B, Duffourd Y, Lehalle D, Bensignor C, et al. Autosomal recessive truncating MAB21L1 mutation associated with a syndromic scrotal agenesis. Clin Genet. 2017;91:333–8. doi: 10.1111/cge.12794. [DOI] [PubMed] [Google Scholar]
- 23.Bruel A-L, Bigoni S, Kennedy J, Whiteford M, Buxton C, Parmeggiani G, et al. Expanding the clinical spectrum of recessive truncating mutations of KLHL7 to a Bohring-Opitz-like phenotype. J Med Genet. 2017;54:830–5. doi: 10.1136/jmedgenet-2017-104748. [DOI] [PubMed] [Google Scholar]
- 24.El Chehadeh S, Kerstjens-Frederikse WS, Thevenon J, Kuentz P, Bruel A-L, Thauvin-Robinet C, et al. Dominant variants in the splicing factor PUF60 cause a recognizable syndrome with intellectual disability, heart defects and short stature. Eur J Hum Genet Ejhg. 2016;25:43–51. doi: 10.1038/ejhg.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Esteve C, Francescatto L, Tan PL, Bourchany A, De Leusse C, Marinier E, et al. Loss-of-function mutations in UNC45A cause a syndrome associating cholestasis, diarrhea, impaired hearing, and bone fragility. Am J Hum Genet. 2018;102:364–74. doi: 10.1016/j.ajhg.2018.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamdan FF, Myers CT, Cossette P, Lemay P, Spiegelman D, Laporte AD, et al. High Rate of recurrent de novo mutations in developmental and epileptic encephalopathies. Am J Hum Genet. 2017;101:664–85. doi: 10.1016/j.ajhg.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lehalle D, Mosca-Boidron A-L, Begtrup A, Boute-Benejean O, Charles P, Cho MT, et al. STAG1 mutations cause a novel cohesinopathy characterised by unspecific syndromic intellectual disability. J Med Genet. 2017;54:479–88. doi: 10.1136/jmedgenet-2016-104468. [DOI] [PubMed] [Google Scholar]
- 28.Lennox AL, Jiang R, Suit L, Fregeau B, Sheehan CJ, Aldinger KA, et al. Pathogenic DDX3X mutations impair RNA metabolism and neurogenesis during fetal cortical development. bioRxiv. 2018;317974. 10.1101/317974. [DOI] [PMC free article] [PubMed]
- 29.Marsh APL, Heron D, Edwards TJ, Quartier A, Galea C, Nava C, et al. Mutations in DCC cause isolated agenesis of the corpus callosum with incomplete penetrance. Nat Genet. 2017;49:511–4. doi: 10.1038/ng.3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olson HE, Jean-Marçais N, Yang E, Heron D, Tatton-Brown K, van der Zwaag PA, et al. A recurrent de novo PACS2 heterozygous missense variant causes neonatal-onset developmental epileptic encephalopathy, facial dysmorphism, and cerebellar dysgenesis. Am J Hum Genet. 2018;102:995–1007. doi: 10.1016/j.ajhg.2018.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saunier C, Støve SI, Popp B, Gérard B, Blenski M, AhMew N, et al. Expanding the phenotype associated with NAA10-related n-terminal acetylation deficiency. Hum Mutat. 2016;37:755–64. doi: 10.1002/humu.23001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thauvin-Robinet C, Duplomb-Jego L, Limoge F, Picot D, Masurel A, Terriat B, et al. Homozygous FIBP nonsense variant responsible of syndromic overgrowth, with overgrowth, macrocephaly, retinal coloboma and learning disabilities. Clin Genet. 2016;89:e1–4. doi: 10.1111/cge.12704. [DOI] [PubMed] [Google Scholar]
- 33.Thevenon J, Milh M, Feillet F, St-Onge J, Duffourd Y, Jugé C, et al. Mutations in SLC13A5 cause autosomal-recessive epileptic encephalopathy with seizure onset in the first days of life. Am J Hum Genet. 2014;95:113–20. doi: 10.1016/j.ajhg.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deriziotis P, O’Roak BJ, Graham SA, Estruch SB, Dimitropoulou D, Bernier RA, et al. De novo TBR1 mutations in sporadic autism disrupt protein functions. Nat Commun. 2014;5:4954. doi: 10.1038/ncomms5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamdan FF, Srour M, Capo-Chichi J-M, Daoud H, Nassif C, Patry L, et al. De novo mutations in moderate or severe intellectual disability. PLoS Genet. 2014;10:e1004772. doi: 10.1371/journal.pgen.1004772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McDermott JH, Study DDD, Clayton-Smith J, Briggs TA. The TBR1-related autistic-spectrum-disorder phenotype and its clinical spectrum. Eur J Med Genet. 2018;61:253–6. doi: 10.1016/j.ejmg.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 37.Notwell JH, Heavner WE, Darbandi SF, Katzman S, McKenna WL, Ortiz-Londono CF, et al. TBR1 regulates autism risk genes in the developing neocortex. Genome Res. 2016;26:1013–22. doi: 10.1101/gr.203612.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–22. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palumbo O, Fichera M, Palumbo P, Rizzo R, Mazzolla E, Cocuzza DM, et al. TBR1 is the candidate gene for intellectual disability in patients with a 2q24.2 interstitial deletion. Am J Med Genet A. 2014;164A:828–33. doi: 10.1002/ajmg.a.36363. [DOI] [PubMed] [Google Scholar]
- 40.Isidor B, Lindenbaum P, Pichon O, Bézieau S, Dina C, Jacquemont S, et al. Truncating mutations in the last exon of NOTCH2 cause a rare skeletal disorder with osteoporosis. Nat Genet. 2011;43:306–8. doi: 10.1038/ng.778. [DOI] [PubMed] [Google Scholar]
- 41.Mougou-Zerelli S, Thomas S, Szenker E, Audollent S, Elkhartoufi N, Babarit C, et al. CC2D2A mutations in Meckel and Joubert syndromes indicate a genotype-phenotype correlation. Hum Mutat. 2009;30:1574–82. doi: 10.1002/humu.21116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thauvin-Robinet C, Thomas S, Sinico M, Aral B, Burglen L, Gigot N, et al. OFD1 mutations in males: phenotypic spectrum and ciliary basal body docking impairment. Clin Genet. 2013;84:86–90. doi: 10.1111/cge.12013. [DOI] [PubMed] [Google Scholar]
- 43.Chandler CH, Chari S, Tack D, Dworkin I. Causes and consequences of genetic background effects illuminated by integrative genomic analysis. Genetics. 2014;196:1321–36. doi: 10.1534/genetics.113.159426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hernández-Porras Isabel, Jiménez-Catalán Beatriz, Schuhmacher Alberto J, Guerra Carmen. The impact of the genetic background in the Noonan syndrome phenotype induced by K-RasV14I. Rare Diseases. 2015;3(1):e1045169. doi: 10.1080/21675511.2015.1045169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bragin E, Chatzimichali EA, Wright CF, Hurles ME, Firth HV, Bevan AP, et al. DECIPHER: database for the interpretation of phenotype-linked plausibly pathogenic sequence and copy-number variation. Nucleic Acids Res. 2014;42:D993–1000. doi: 10.1093/nar/gkt937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–8. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sobreira NLM, Arachchi H, Buske OJ, Chong JX, Hutton B, Foreman J, et al. Matchmaker exchange. Curr Protoc Hum Genet. 2017;95:9.31. 1–9.31.15. doi: 10.1002/cphg.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shamseldin HE, Maddirevula S, Faqeih E, Ibrahim N, Hashem M, Shaheen R, et al. Increasing the sensitivity of clinical exome sequencing through improved filtration strategy. Genet Med. 2017;19:593–8. doi: 10.1038/gim.2016.155. [DOI] [PubMed] [Google Scholar]
- 49.Chilamakuri CSR, Lorenz S, Madoui M-A, Vodák D, Sun J, Hovig E, et al. Performance comparison of four exome capture systems for deep sequencing. BMC Genom. 2014;15:449. doi: 10.1186/1471-2164-15-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hwang S, Kim E, Lee I, Marcotte EM. Systematic comparison of variant calling pipelines using gold standard personal exome variants. Sci Rep. 2015;5. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4671096/ [DOI] [PMC free article] [PubMed]
- 51.Meynert AM, Ansari M, FitzPatrick DR, Taylor MS. Variant detection sensitivity and biases in whole genome and exome sequencing. BMC Bioinforma. 2014;15:247. doi: 10.1186/1471-2105-15-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stessman Holly A.F., Willemsen Marjolein H., Fenckova Michaela, Penn Osnat, Hoischen Alexander, Xiong Bo, Wang Tianyun, Hoekzema Kendra, Vives Laura, Vogel Ida, Brunner Han G., van der Burgt Ineke, Ockeloen Charlotte W., Schuurs-Hoeijmakers Janneke H., Klein Wassink-Ruiter Jolien S., Stumpel Connie, Stevens Servi J.C., Vles Hans S., Marcelis Carlo M., van Bokhoven Hans, Cantagrel Vincent, Colleaux Laurence, Nicouleau Michael, Lyonnet Stanislas, Bernier Raphael A., Gerdts Jennifer, Coe Bradley P., Romano Corrado, Alberti Antonino, Grillo Lucia, Scuderi Carmela, Nordenskjöld Magnus, Kvarnung Malin, Guo Hui, Xia Kun, Piton Amélie, Gerard Bénédicte, Genevieve David, Delobel Bruno, Lehalle Daphne, Perrin Laurence, Prieur Fabienne, Thevenon Julien, Gecz Jozef, Shaw Marie, Pfundt Rolph, Keren Boris, Jacquette Aurelia, Schenck Annette, Eichler Evan E., Kleefstra Tjitske. Disruption of POGZ Is Associated with Intellectual Disability and Autism Spectrum Disorders. The American Journal of Human Genetics. 2016;98(3):541–552. doi: 10.1016/j.ajhg.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chiu A.T.G., Pei S.L.C., Mak C.C.Y., Leung G.K.C., Yu M.H.C., Lee S.L., Vreeburg M., Pfundt R., van der Burgt I., Kleefstra T., Frederic T.M.-T., Nambot S., Faivre L., Bruel A.-L., Rossi M., Isidor B., Küry S., Cogne B., Besnard T., Willems M., Reijnders M.R.F., Chung B.H.Y. Okur-Chung neurodevelopmental syndrome: Eight additional cases with implications on phenotype and genotype expansion. Clinical Genetics. 2018;93(4):880–890. doi: 10.1111/cge.13196. [DOI] [PubMed] [Google Scholar]
- 54.Kosho T, Okamoto N, Coffin-Siris Syndrome International Collaborators. Genotype-phenotype correlation of Coffin-Siris syndrome caused by mutations in SMARCB1, SMARCA4, SMARCE1, and ARID1A. Am J Med Genet C Semin Med Genet. 2014;166C:262–75. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.