Abstract
Clinical exome sequencing (CES) is increasingly being utilized; however, a large proportion of patients remain undiagnosed, creating a need for a systematic approach to increase the diagnostic yield. We have reanalyzed CES data for a clinically heterogeneous cohort of 102 probands with likely Mendelian conditions, including 74 negative cases and 28 cases with candidate variants, but reanalysis requested by clinicians. Reanalysis was performed by an interdisciplinary team using a validated custom-built pipeline, “Variant Explorer Pipeline” (VExP). This reanalysis approach and results were compared with existing literature. Reanalysis of candidate variants from CES in 28 cases revealed 1 interpretation that needed to be reclassified. A confirmed or potential genetic diagnosis was identified in 24 of 75 CES-negative/reclassified cases (32.0%), including variants in known disease-causing genes (n = 6) or candidate genes (n = 18). This yield was higher compared with similar studies demonstrating the utility of this approach. In summary, reanalysis of negative CES in a research setting enhances diagnostic yield by about a third. This study suggests the need for comprehensive, continued reanalysis of exome data when molecular diagnosis is elusive.
Subject terms: Genetic testing, Genetics research
Introduction
Whole-exome sequencing (WES) is becoming increasingly important in enabling tailored interventions in “personalized” or “precision” medicine. Recent studies suggest that in patients with suspected genetic disorders, clinical WES (CES) should be applied as a first-tier molecular test, in parallel with standard diagnostic care, to improve the cost-effectiveness of the diagnostic process [1, 2]. However, limitations of the current analytical methods [3], genetic and phenotypic heterogeneity [4], knowledge gaps in the gene–disease association [5], and interpretation/reporting differences [6] often hinder the identification of disease-causing variants. Ultimately, only 25–30% of families receive a diagnosis after CES [7, 8], leaving the genetic basis of the remaining families’ diseases unknown.
Recent studies indicate that periodic reanalysis of negative CES data using improved bioinformatic tools [9] and up-to-date gene–disease databases [5] can identify additional candidate variants, further enhanced by complementary methods including copy number variation (CNV) analysis [10], whole genome sequencing (WGS), and other targeted methods [11]. Despite the use of these approaches, many cases remain negative after reanalysis. Thus, there is a need to develop a comprehensive strategy for reanalyzing negative CES that will improve the analytical process, thereby providing better options for those who do not initially receive a molecular diagnosis.
In this study, using a CES cohort enrolled in the Manton Center for Orphan Disease Research at Boston Children’s Hospital, we present our strategy for retrospective reanalysis of negative CES studies. We describe a unique approach featuring a custom-built pipeline (“Variant Explorer Pipeline,” VExP), in conjunction with updated phenotypic information, literature, and databases, and in collaboration with an interdisciplinary team to enhance diagnostic yield while minimizing cost. As no methodological consensus guidelines for CES reanalysis currently exist, we have drawn from methods described in the literature [10–12], as well as our own reanalysis experience, to develop a standard strategy for negative CES reanalysis. This proposed strategy addresses each possible outcome after negative CES reanalysis, including cases that remain negative. Supported by the current literature, we have incorporated a novel bioinformatic strategy into an interdisciplinary research setting to introduce an innovative approach that addresses the need for comprehensive reanalysis of exome data.
Materials and methods
Recruitment of CES cohort
The Gene Discovery Core of the Manton Center for Orphan Disease Research (Manton GDC) utilizes an Institutional Review Board-approved research protocol at our institution established to enroll and study patients and families with a wide range of rare and undiagnosed disorders. The Manton GDC often obtains raw CES data from enrolled participants who have undergone prior clinical testing and information about the CES cohort can be found in Supplementary Table S1. These data can be negative, having demonstrated no candidate variant, or contain a candidate variant and are submitted by a referring physician to confirm the existing candidate or identify additional variants. The CES datasets utilized in this study were all performed by external Clinical Laboratory Improvement Amendments-certified genetic testing laboratories. Two hundred and seventy-five sequencing files (162 fastq, 63 bam, and 50 cram files) were included in our study. The clinical labs performed exome capture using different exome capture kits (e.g., Agilent SureSelectXT Human All Exon Kit V4 and V5) and sequenced captured libraries using Illumina HiSeq at varying levels of lab-specific coverage (>50 ×).
Exome data analysis/reanalysis of negative CES
Our exome-sequencing reanalysis approach uses an innovative, validated, custom-built pipeline VExP [13, 14] to narrow down potential candidate variants. VExP is a comprehensive system that integrates existing methods, genetic information, and probabilistic models into an automated pipeline for the identification of disease genes (Supplementary Fig. S1). VExP features include enhanced calling methodologies, novel variant detection algorithms, updated variant annotations, and validated filtering strategies. Independent families (102 total: 5 quads, 79 trios, 18 singletons) were processed using the VExP (v2018May), using the following four steps:
Step 1 (fastq to vcf): VExP starts with fastq raw data to avoid processing errors or reference mismatch data. When only bam files were available, PICARD (v2.9.2) was used to revert to fastq files. Raw data were processed to obtain vcf files using the human reference assembly 19, BWA (alignment, v0.7.15), PICARD (mark/delete duplicates), SAMTOOLS (variant calling, v1.4.1), and GATK (multi-sample variant calling, v3.7). Further, ANNOVAR (2018Apr18) and custom VExP scripts were used to add annotations from 53 relevant genetic databases (Supplementary Table S2) into the output of the system.
Step 2 (Variant filtering): We observed, on average, a total of 201,929 variants per individual (31,837 in coding/splicing sites). These uniform vcf files were next subjected to pedigree/relationship tests of each family to eliminate or correct discordant samples. Variant analysis was performed in each family using different inheritance models (assuming full penetrance) based on three filtering criteria: first, include variants predicted by ANNOVAR to have a potential functional coding consequence, including stop gain or loss, splice site disruption, indel, and nonsynonymous. Second, variants are filtered based on allele frequency in control populations (gnomAD, ExAC, EVS, 1000GP, and internal data from 2114 unaffected individuals from BCH). Heterozygous/hemizygous variants for dominant, de novo, and X-linked models were included if minor allele frequency (MAF) was <0.0005 (0.05%) in any database. In comparison, homozygous variants were included only if MAF was ≤0.00005 (0.005%) and for compound heterozygous models the MAF cutoff was ≤0.01 (1%) with no homozygous variant reported in any database. Phasing was performed if parents were available. The variants were further prioritized to include those with read depth ≥10 ×, alternative depth ≥5 ×, allele balance ≥0.20, and deleterious prediction (2 or more of 23 softwares, including PolyPhen, SIFT, FATHMM, and CADD; Supplementary Table S2). Third, an inheritance filter was also applied to identify any variants that may be incompletely penetrant or mosaic in the parent and these were analyzed and evaluated outside this workflow. These filters were calibrated and validated using data from 150 independent families with genetic diagnoses from Boston Children’s Hospital and the Broad Institute (Supplementary Data and Table S3).
Step 3 (Candidate genes): On average, 14 variants per quad, 16 per trio, and 131 per singleton family (Supplementary Fig. S2) were identified following this common filtering strategy (Steps 1 and 2). To target the analysis toward making a primary diagnosis, we subdivided our candidate genes into three categories: (a) likely pathogenic (LP) and pathogenic (P) variants in known disease-associated genes, (b) variants of uncertain significance (VUS) in known disease-associated genes, and (c) variants in novel genes. For (a) and (b), VExP uses the Human Phenotype Ontology (HPO) terms that define the phenotype of the patient to filter for variants that lie within an appropriate candidate gene list, built from 12 different medical databases (Online Mendelian Inheritance in Man, ClinVar, Mouse Genome Informatics, Human Gene Mutation Database (HGMD), Genetic Association Database, and HPO; see Table S2) to identify variants potentially correlated to the phenotype. For c) we selected variants predicted to be deleterious in candidate genes that were not confirmed previously to be implicated in human disease. Our database is integrated with 1,579 independent families (3,776 independent samples) with 178 different phenotypes that were processed using the same pipeline (VExP). We utilized the database to identify novel genes using a unique approach wherein we were able to exclude 40% of additional candidate genes (Supplementary Material and Supplementary Fig. S3).
Step 4 (Diagnostic): Candidate gene variants were further evaluated by our clinical review team that consists of genetic counselors and both internal and referring clinicians. The genomic and phenotypic data were combined with additional information including gene expression, protein domains, functional studies, animal models, interactomes, gene families, and literature [10, 12]. Potentially diagnostic candidate variants were deposited into data sharing platforms such as “Matchmaker Exchange” [15] and “GeneMatcher” [16] to corroborate findings and/or to find future collaborators for functional analysis. Variants in disease-associated genes were interpreted using the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP) guidelines for variant classification [17]. Likely diagnostic variants selected by our clinical review team were subsequently confirmed using Sanger sequencing in our laboratory at BCH and these confirmed variants were submitted to ClinVar (#SUB5116581). When available, other family members were used for segregation testing.
Results
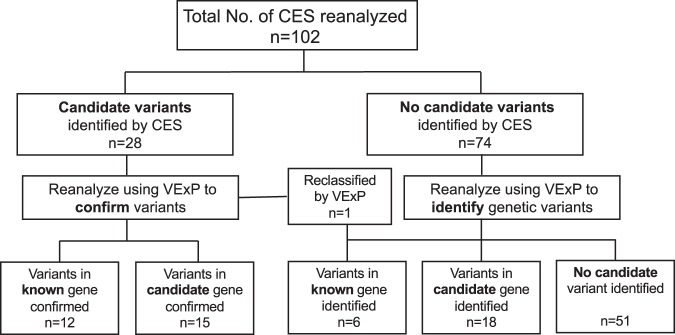
These 102 CES cases were sequenced over a period of 5 years from 2013 to 2018 (2013: 5 cases; 2014: 7 cases; 2015: 14 cases; 2016: 27 cases; 2017: 37 cases; and 2018: 12 cases). The average and median times between original analysis and reanalysis in this study were 1.9 ± 1.4 years and 2 years, respectively. Data from these cases were reanalyzed and classified into one of the five following groups: (a) variants in known genes (that match phenotype) identified by CES and confirmed by reanalysis; (b) variants in candidate (novel and phenotypic expansion) genes identified by CES and confirmed by reanalysis; (c) variants in known genes missed by CES and identified by reanalysis; (d) variants in candidate genes missed by CES and identified by reanalysis; and (e) no likely diagnostic variant identified by CES or reanalysis (Fig. 1).
Fig. 1.
Flowchart of CES reanalysis and outcomes. All 102 CES cases with fastq data were reanalyzed by VExP and classified into one of the five following groups: (a) confirmed variants in known disease-causing genes previously identified by CES; (b) confirmed variants in candidate genes previously identified by CES; (c) variants in known disease-causing genes identified on reanalysis; (d) variants in candidate genes identified on reanalysis; and (e) no candidate variant identified on reanalysis. CES, clinical exome sequencing; VExP, Variant Explorer Pipeline
Of the 102 reanalyzed cases, 28 had previously identified variants in known (n = 12) and candidate genes (n = 15), and one needed to be reclassified. Reanalysis of those 28 cases was requested by the referring physicians in order to confirm existing findings and/or potentially identify additional disease-causing variants. Meanwhile, of the 74 negative and 1 reclassified case (n = 75), our reanalysis determined six instances of missed diagnoses. In addition, we identified 18 probands with variants in candidate genes. The remaining 51 cases stayed unsolved despite our reanalysis.
The known disease-causing genes and candidate genes in which variants were identified or confirmed by reanalysis are listed in Table 1. The majority of the variants were de novo (29/51), followed by recessive (22/51), which included compound heterozygous (9/51), homozygous (7/51), and hemizygous (6/51).
Table 1.
List of known and candidate disease-causing genes in which variants were identified or confirmed by reanalysis
| Inheritance | Known genes | Candidate genes (novel/phenotypic expansion) |
|---|---|---|
| De novo | PPM1D, FBXO11, CAMK2B, PRKAR1A, *CLTC, *MAP1B, ATP1A3, ADCY5, CBL, SLC6A1, PTCH1, UBE3A, FAR1 | EIF2AK2, GABRB2, MORC2, EZH1, TMEM104, *PTBP1, *NBEA, HNRNPU, HNRNPDL, *RIN3, KDM4B, SEMA6B, TFE3, CELF2, WDR37, MPP5 |
| Hemizygous | BCORL1, L1CAM, PGK1, *PIR, TSPAN6, TAF1 | |
| Homozygous | ASNS, EXOSC9 | CIT, SLC25A26, *EEFSEC, *TMX2, WSB2 |
| Compound heterozygous | *PAH, *RBFOX1, ECHS1 | WDFY4, LDB3, KIF14, HCN1, KAT14, HIBCH |
*Singleton
Phenotypic details associated with known disease-causing genes identified by reanalysis are listed in Table 2. These variants were either recently published, such as CAMK2B (NM_001220.4:c.416C > T, p.(Pro139Leu)) [18], CLTC (NM_004859.3:c.2669C > T, p.(Pro890Leu)) [19], MAP1B (NM_005909.4:c.5368C > T, p.(Arg1790*)) [20], or high impact variants (nonsense, frameshift, hotspot mutation or mutation in functional domains with high conservation) found in disease-causing genes with a clinical presentation that fit the reported patient phenotype, including PPM1D (NM_003620.3:c.1258dupA, p.(Lys420fs)), FBXO11 (NM_025133.4:c.2145G > C, p.(Lys715Asn)), and PRKAR1A (NM_001278433.1: c.620A > G, p.(Tyr207Cys)).
Table 2.
Phenotypic details associated with known disease-causing genes identified by reanalysis
| ID | Phenotype | Variants (hg19) | MIM no. | ACMG/AMP |
|---|---|---|---|---|
| 1 | Microcephaly, failure to thrive, dysmorphic features, severe periodic limb movements, GDD, gastroesophageal reflux | PPM1D chr17:58734200, NM_003620.3:c.1258dupA, p.(Lys420fs) (de novo) | 617450 | P |
| 2 | GDD, speech and motor apraxia, dysmorphic facial features, relative microcephaly, bifid uvula, submucosal cleft palate | FBXO11 chr2:48040455, NM_001190274.1:c.2145G > C, p.(Lys715Asn) (de novo) | 618089 | LP |
| 3 | Intellectual disability, GDD, hypotonia, abnormal movements | CAMK2B chr7:44283125, NM_001220.4:c.416C > T, p.(Pro139Leu) (de novo) | 617799 | LP |
| 4 | Skeletal dysplasia, GDD, hypotonia, facial dysmorphism and absent speech | PRKAR1A, chr17:66521965, NM_001276290.1: c.620A > G, p.(Tyr207Cys) (de novo) | 101800 | LP |
| 5 | Congenital hypotonia, GDD, short stature, lack of coordination, prominent fetal pads on fingers/toes | *CLTC chr17:57754422, NM_004859.3:c.2669C > T, p.(Pro890Leu) | 617854 | VUS |
| 6 | Metopic craniosynostosis, GDD, attention deficit hyperactivity disorder, hypotonia, mild dysmorphic features, pyloric stenosis | *MAP1B chr5:71494550, NM_005909.4:c.5368C > T, p.(Arg1790*) | 157129 | P |
*Singleton
GDD global developmental delay, LP likely pathogenic, P pathogenic, VUS variant of unknown significance.
Overall, the diagnostic yield for reanalyzed negative plus one reclassified CES cases was 32.0% (24/75). The diagnostic rate was slightly higher (no significant difference) in trios than in singletons (32.8% and 28.6%, respectively; Supplementary Table S4). Reanalysis of older CES cases had a higher chance to be diagnostic, although not statistically significant (38.1% for 2013–15 vs. 29.6% for 2016–18, P-value > 0.05). Diagnosis rate for each year is depicted in Supplementary Fig. S4. Variant assessments, literature searches, and collaboration with other laboratories contributed to the validation of candidate genes in these patients. Using the reanalysis workflow described, we have achieved a confirmed or likely genetic diagnosis for 24 probands in the 75 negative or reclassified rare disease families after reanalysis, improving the diagnostic yield.
Discussion
Here we describe a comprehensive and collaborative approach for reanalyzing negative CES cases with heterogeneous phenotypes that maximizes diagnostic yield. Our computational approach employs an innovative and validated custom-built pipeline (VExP) [13, 14] that integrates enhanced calling, variant detection and annotations, filtering strategies, and probabilistic models using 53 different databases. Out of 75 negative/reclassified cases, the confirmed and proposed genetic diagnosis account for 8.0% and 24%, respectively, resulting in an overall diagnostic yield of 32.0%. This is an improvement from prior studies that report diagnostic yields ranging from 7.7% to 27.2% [5, 9, 12, 21, 22], supporting our comprehensive approach. Further, compared with our study, higher diagnostic yields have been reported, in studies of consanguineous families [23, 24], or with utilization of additional methods such as CNV analysis [10], WGS, and others [11]. Overall, the yield of all reported studies including ours is 21% (472/2250), 14.9% for known and 6.1% for candidate genes (Table 3). It should be noted that the comparison in Table 3 is limited in precision by variable confounding factors including family type (trios, singletons, etc.), ethnicity, phenotypic variability, and methodology.
Table 3.
Comparison of this study with other publications related to negative CES reanalysis
| Study no. | No. of patients (singleton/trio) | Diagnosis | Diagnostic yield (known + candidate genes) | Methods | Reference | |
|---|---|---|---|---|---|---|
| Known genes | Candidate genes | |||||
| 1 | 23 (NA) | 4 | 0 | 17.4% | (1) BTI | Bone et al. [9] |
| 2 | 934 (NA) | 37 | 35 | 7.7% | (1) Unique candidate gene criteria | Hagman et al. [12] |
| 3 | 40 (40/0) | 4 | 0 | 10.0% | (1) L/DU | Wenger et al. [5] |
| 4 | 74 (6/68) | 27 | 11 | 51.4% | (1) BTI and (2) employed WES from additional family members | Eldomery et al. [10] |
| 5 | 33 (33 Consanguineous) | 24 | 5 | 87.9% | (1) Improved filtration strategy: positional mapping | Shamseldin et al. [24] |
| 6 | 50 (41 Consanguineous) | 6 | 20 | 52.0% | (1) BTI, (2) L/DU, and (3) phenotype update | Al-Nabhani et al. [23] |
| 7 | 156 (NA) | 24 | 0 | 15.4% | (1) Collaborations, (2) L/DU, (3) functional studies, and (4) targeted molecular testing | Nambot et al. [21] |
| 8 | 827 (0/827) | 182 | 43 | 27.2% | (1) BTI and (2) L/DU | Wright et al. [22] |
| 9 | 38 (1/37) | 21 | 5 | 68.4% | (1) BTI, (2) phenotype update, (3) targeted molecular testing, (4) WGS, and (5) collaborations | Shashi et al. [11] |
| 10 | 75 (15/60) | 6 | 18 | 32.0% | (1) BTI, (2) L/DU, (3) phenotype update, and (4) collaborations | This study |
| Total | 2250 | 335 (14.9%) | 137 (6.1%) | 21.0% | ||
BTI bioinformatics tools improvement, L/DU literature and database update, NA not available, WES whole exome sequencing, WGS whole genome sequencing
The continued accumulation of genetic knowledge during the time that elapsed between the CES and reanalysis may have affected our diagnostic yield [5, 21]. Every year, hundreds of new genes and thousands of new variants are linked to various phenotypes, making it necessary to reanalyze the genomic data [10, 11, 25]. Indeed, of the six cases where reanalysis identified diagnostic variants in established genes, five of these were in genes whose disease associations were published after the initial testing, the rest one (absent in the original VCF file) was detected by variant calling improvement. In addition, for 18 families we have a candidate gene (missing in the original analysis) and additional workup is ongoing. The diagnostic rate for early years (2013–15) was higher (although not statistically significant) compared with later years (2016–18); therefore, it is important to reanalyze the negative CES data periodically, preferably annually, with new methodologies, updated genomic datasets, improved pipelines/variants callings, and clinical experts. In addition, the families that do not receive a genetic diagnosis should have access to resources including research studies and patient registries to benefit from new disease discoveries over time.
Proposed strategy for negative CES reanalysis
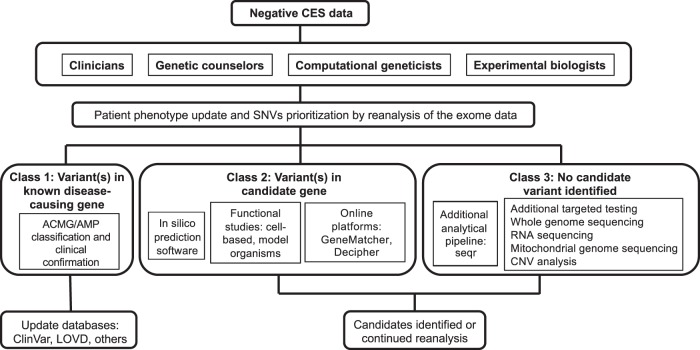
Negative CES reanalysis is governed by an intricate process with many collaborative components that is constantly subject to change based on the latest developments [11, 26]. Lack of standardization of this process leads to high variability in diagnostic yield, as well as improper care for patients who do not receive a diagnosis after reanalysis. In order to improve the outcomes for these patients, we proposed a comprehensive strategy for reanalyzing negative exomes as illustrated in Fig. 2. This strategy encompasses the range of methods described in the current literature and integrates novel technological advancements and information from our own database. In addition to updating initial reanalysis, our proposed strategy addresses each possible outcome after negative CES reanalysis. These outcomes can be divided into three classes: (1) variant(s) in known disease-causing gene; (2) variant(s) in novel candidate gene; and (3) no candidate variant identified.
Fig. 2.
Proposed strategy for reanalyzing negative exomes. Collaborations between multidisciplinary clinical and research teams facilitate reanalysis of negative CES to reach a diagnosis. These negative CES cases can be divided into three classes after reanalysis: (1) candidate variants linked to the phenotype but of limited pathogenic evidence; (2) candidate variants in new genes not linked to the phenotype; (3) no candidate variants detected by CES after reanalysis. CES, clinical exome sequencing; CNV, copy number variation; VExP, Variant Explorer Pipeline
Outcomes categorized as class 1 in Fig. 2 may include one or more VUS that can only partially explain patients’ phenotypes at the time of diagnosis. In addition, multiple variants in two or more genes can be responsible for rare diseases, leading to a mixed phenotype without a clear diagnosis [27]. Once class 1 variants are categorized as P or LP variants according to the ACMG/AMP guidelines and clinically confirmed by the referring physicians, they should be deposited into databases including ClinVar, HGMD, and LOVD (Leiden Open Variation Database).
Outcomes categorized as class 2 should first be evaluated for damaging effects using in-silico predictions tools and variants predicted to be deleterious should be subjected to further functional studies in vitro and in vivo, in order to assess the pathogenicity of the candidate variants. Novel potential disease genes of interest or undergoing functional evaluation should be uploaded onto online data sharing platforms (e.g., GeneMatcher [16], Matchmaker Exchange) [15], in order to facilitate the collection of pathogenic evidence through collaborative efforts. Confirmed or published novel gene discoveries can then be added to the gene–disease databases to improve the power of the analytical pipeline. Class 2 outcomes should also be reanalyzed regularly to account for updates in analytical methods and database information.
Class 3 consists of outcomes in which no potentially diagnostic variant was identified by reanalysis. Possible factors contributing to this result include disease-causing variants with allele balance <20%, which are prone to misclassification as false-negative due to sequencing and mapping errors from CES [28]. These variants are often missed in cases of somatic mosaicism (e.g., GNAQ in Sturge–Weber syndrome: allele balance = 1–18%) [29]. Trinucleotide repeat diseases such as Fragile X syndrome, Huntington’s disease, and myotonic dystrophy are also frequently missed by CES [30]. Additional target testing can be used to improve the detection rate of trinucleotide repeat diseases [31] and low-allele balance variants, as has been demonstrated in cases of mosaicism [32]. Negative outcomes may also result from variants that lie in non-coding regions, unmappable reads arising from repeated regions, or structural variants that CES cannot detect. In these cases, WGS and long read sequencing can be applied to improve the detection rate, as these methods allow additional detection of both structural and non-exonic variants, and insertion of mobile elements such as transposons [33].
RNA-sequencing and mitochondrial genome sequencing can aid in the diagnosis when CES fails. By detecting both coding and non-coding variants at gene expression levels and alternative splicing, transcriptome sequencing holds considerable diagnostic potential for rare disorders [34]. Further, although CES data are increasingly being employed for CNV analysis, technical limitations due to inherent heterogeneity in exome capture may result in false negatives [35]. In such cases, CNV analysis techniques such as microarray-based CNV profiling or reanalyzing data from WGS for CNVs can be employed [36].
In conclusion, we report our cumulative experience from reanalysis of a CES cohort enrolled in our institution using a custom-built comprehensive variant detection and analysis pipeline in conjunction with updated phenotypic information, literature, and databases in collaboration with an interdisciplinary team. These advances allowed us to reach a confirmed or potential diagnosis for up to a third of previously negative CES cases, demonstrating a meaningful improvement in diagnostic yield. We also compare our findings with other studies, review literature, and propose an overall strategy for reanalyzing negative exome data that will standardize the analytical process and provide appropriate medical care for patients with no diagnosis by CES.
Supplementary information
Acknowledgements
We thank all the referring physicians and caregivers, and especially the patients and families for their participation in this research. We also thank Heather Paterson, Tina Truong, Grace VanNoy, and Clair McHugh for assistance with patient enrollment and sample and data collection. The Manton Center Gene Discovery Core is supported by a generous gift from The Manton Foundation. This work was also supported by grants NIAMS R01AR068429, NICHD/NHGRI U19HD077671, NICHD R01HD075802, and NICHD K12HD052896, and by the resources of the IDDRC Molecular Genetics Core funded by U54HD090255, from the US National Institutes of Health.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Klaus Schmitz-Abe and Qifei Li
Supplementary information
The online version of this article (10.1038/s41431-019-0401-x) contains supplementary material, which is available to authorized users.
References
- 1.Stark Z, Schofield D, Alam K, Wilson W, Mupfeki N, Macciocca I, et al. Prospective comparison of the cost-effectiveness of clinical whole-exome sequencing with that of usual care overwhelmingly supports early use and reimbursement. Genet Med. 2017;19:867–74. doi: 10.1038/gim.2016.221. [DOI] [PubMed] [Google Scholar]
- 2.Stark Z, Tan TY, Chong B, Brett GR, Yap P, Walsh M, et al. A prospective evaluation of whole-exome sequencing as a first-tier molecular test in infants with suspected monogenic disorders. Genet Med. 2016;18:1090–6. doi: 10.1038/gim.2016.1. [DOI] [PubMed] [Google Scholar]
- 3.Xue Y, Ankala A, Wilcox WR, Hegde MR. Solving the molecular diagnostic testing conundrum for Mendelian disorders in the era of next-generation sequencing: single-gene, gene panel, or exome/genome sequencing. Genet Med. 2015;17:444–51. doi: 10.1038/gim.2014.122. [DOI] [PubMed] [Google Scholar]
- 4.Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12:745–55. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 5.Wenger AM, Guturu H, Bernstein JA, Bejerano G. Systematic reanalysis of clinical exome data yields additional diagnoses: implications for providers. Genet Med. 2017;19:209–14. doi: 10.1038/gim.2016.88. [DOI] [PubMed] [Google Scholar]
- 6.Pena LD, Jiang YH, Schoch K, Spillmann RC, Walley N, Stong N, et al. Looking beyond the exome: a phenotype-first approach to molecular diagnostic resolution in rare and undiagnosed diseases. Genet Med. 2018;20:464–9. doi: 10.1038/gim.2017.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee H, Deignan JL, Dorrani N, Strom SP, Kantarci S, Quintero-Rivera F, et al. Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA. 2014;312:1880–7. doi: 10.1001/jama.2014.14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farwell KD, Shahmirzadi L, El-Khechen D, Powis Z, Chao EC, Davis BT, et al. Enhanced utility of family-centered diagnostic exome sequencing with inheritance model–based analysis: results from 500 unselected families with undiagnosed genetic conditions. Genet Med. 2015;17:578–86. doi: 10.1038/gim.2014.154. [DOI] [PubMed] [Google Scholar]
- 9.Bone WP, Washington NL, Buske OJ, Adams DR, Davis J, Draper D, et al. Computational evaluation of exome sequence data using human and model organism phenotypes improves diagnostic efficiency. Genet Med. 2016;18:608–17. doi: 10.1038/gim.2015.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eldomery MK, Coban-Akdemir Z, Harel T, Rosenfeld JA, Gambin T, Stray-Pedersen A, et al. Lessons learned from additional research analyses of unsolved clinical exome cases. Genome Med. 2017;9:26. doi: 10.1186/s13073-017-0412-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shashi V, Schoch K, Spillmann R, Cope H, Tan QKG, Walley N, et al. A comprehensive iterative approach is highly effective in diagnosing individuals who are exome negative. Genet Med. 2019;21:161–72. doi: 10.1038/s41436-018-0044-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagman KDF, Shinde DN, Mroske C, Smith E, Radtke K, Shahmirzadi L, et al. Candidate-gene criteria for clinical reporting: diagnostic exome sequencing identifies altered candidate genes among 8% of patients with undiagnosed diseases. Genet Med. 2017;19:224–35. doi: 10.1038/gim.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmitz-Abe K, Ciesielski SJ, Schmidt PJ, Campagna DR, Rahimov F, Schilke BA, et al. Congenital sideroblastic anemia due to mutations in the mitochondrial HSP70 homologue HSPA9. Blood. 2015;126:2734–8. doi: 10.1182/blood-2015-09-659854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jamuar SS, Schmitz-Abe K, D’Gama AM, Drottar M, Chan WM, Peeva M, et al. Biallelic mutations in human DCC cause developmental split-brain syndrome. Nat Genet. 2017;49:606–12. doi: 10.1038/ng.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Philippakis AA, Azzariti DR, Beltran S, Brookes AJ, Brownstein CA, Brudno M, et al. The Matchmaker Exchange: a platform for rare disease gene discovery. Hum Mutat. 2015;36:915–21. doi: 10.1002/humu.22858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat. 2015;36:928–30. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Küry S, van Woerden GM, Besnard T, Onori MP, Latypova X, Towne MC, et al. De novo mutations in protein kinase genes CAMK2A and CAMK2B cause intellectual disability. Am J Hum Genet. 2017;101:768–88. doi: 10.1016/j.ajhg.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamdan FF, Myers CT, Cossette P, Lemay P, Spiegelman D, Laporte AD, et al. High rate of recurrent de novo mutations in developmental and epileptic encephalopathies. Am J Hum Genet. 2017;101:664–85. doi: 10.1016/j.ajhg.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walters GB, Gustafsson O, Sveinbjornsson G, Eiriksdottir VK, Agustsdottir AB, Jonsdottir GA, et al. MAP1B mutations cause intellectual disability and extensive white matter deficit. Nat Commun. 2018;9:3456. doi: 10.1038/s41467-018-05595-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nambot S, Thevenon J, Kuentz P, Duffourd Y, Tisserant E, Bruel AL, et al. Clinical whole-exome sequencing for the diagnosis of rare disorders with congenital anomalies and/or intellectual disability: substantial interest of prospective annual reanalysis. Genet Med. 2018;20:645–54. doi: 10.1038/gim.2017.162. [DOI] [PubMed] [Google Scholar]
- 22.Wright CF, McRae JF, Clayton S, Gallone G, Aitken S, FitzGerald TW, et al. Making new genetic diagnoses with old data: iterative reanalysis and reporting from genome-wide data in 1,133 families with developmental disorders. Genet Med. 2018;20:1216–23. doi: 10.1038/gim.2017.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al‐Nabhani M, Al‐Rashdi S, Al‐Murshedi F, Al‐Kindi A, Al‐Thihli K, Al‐Saegh A, et al. Re‐analysis of exome sequencing data of intellectual disability samples: yields and benefits. Clin Genet. 2018;94:495–501. doi: 10.1111/cge.13438. [DOI] [PubMed] [Google Scholar]
- 24.Shamseldin HE, Maddirevula S, Faqeih E, Ibrahim N, Hashem M, Shaheen R, et al. Increasing the sensitivity of clinical exome sequencing through improved filtration strategy. Genet Med. 2017;19:593–8. doi: 10.1038/gim.2016.155. [DOI] [PubMed] [Google Scholar]
- 25.Mersch J, Brown N, Pirzadeh-Miller S, Mundt E, Cox HC, Brown K, et al. Prevalence of variant reclassification following hereditary cancer genetic testing. JAMA. 2018;320:1266–74. doi: 10.1001/jama.2018.13152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bergant G, Maver A, Lovrecic L, Čuturilo G, Hodzic A, Peterlin B. Comprehensive use of extended exome analysis improves diagnostic yield in rare disease: a retrospective survey in 1,059 cases. Genet Med. 2018;20:303–12. doi: 10.1038/gim.2017.142. [DOI] [PubMed] [Google Scholar]
- 27.Deltas C. Digenic inheritance and genetic modifiers. Clin Genet. 2018;93:429–38. doi: 10.1111/cge.13150. [DOI] [PubMed] [Google Scholar]
- 28.Li H. Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics. 2014;30:2843–51. doi: 10.1093/bioinformatics/btu356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shirley MD, Tang H, Gallione CJ, Baugher JD, Frelin LP, Cohen B, et al. Sturge–Weber syndrome and port-wine stains caused by somatic mutation in GNAQ. N Engl J Med. 2013;368:1971–9. doi: 10.1056/NEJMoa1213507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tankard RM, Delatycki MB, Lockhart PJ, Bahlo M. Detecting known repeat expansions with standard protocol next generation sequencing, towards developing a single screening test for neurological repeat expansion disorders. Am J Hum Genet. 2018;103:858–73. doi: 10.1016/j.ajhg.2018.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wenstrom KD. Fragile X and other trinucleotide repeat diseases. Obstet Gynecol Clin North Am. 2002;29:367–88. doi: 10.1016/S0889-8545(01)00005-5. [DOI] [PubMed] [Google Scholar]
- 32.Gajecka M. Unrevealed mosaicism in the next-generation sequencing era. Mol Genet Genomics. 2016;291:513–30. doi: 10.1007/s00438-015-1130-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lionel AC, Costain G, Monfared N, Walker S, Reuter MS, Hosseini SM, et al. Improved diagnostic yield compared with targeted gene sequencing panels suggests a role for whole-genome sequencing as a first-tier genetic test. Genet Med. 2018;20:435–43. doi: 10.1038/gim.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Byron SA, Van Keuren-Jensen KR, Engelthaler DM, Carpten JD, Craig DW. Translating RNA sequencing into clinical diagnostics: opportunities and challenges. Nat Rev Genet. 2016;17:257–71. doi: 10.1038/nrg.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43:838–46. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tammimies K, Marshall CR, Walker S, Kaur G, Thiruvahindrapuram B, Lionel AC, et al. Molecular diagnostic yield of chromosomal microarray analysis and whole-exome sequencing in children with autism spectrum disorder. JAMA. 2015;314:895–903. doi: 10.1001/jama.2015.10078. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.