Abstract
Intellectual disability (ID), megalencephaly, frontal predominant pachygyria, and seizures, previously called “thin” lissencephaly, are reported to be caused by recessive variants in CRADD. Among five families of different ethnicities identified, one homozygous missense variant, c.509G>A p.(Arg170His), was of Finnish ancestry. Here we report on the phenotypic variability associated for this potential CRADD founder variant in 22 Finnish individuals. Exome sequencing was used to identify candidate genes in Finnish patients presenting with ID. Targeted Sanger sequencing and restriction enzyme analysis were applied to screen for the c.509G>A CRADD variant in cohorts from Finland. Detailed phenotyping and genealogical studies were performed. Twenty two patients were identified with the c.509G>A p.(Arg170His) homozygous variant in CRADD. The majority of the ancestors originated from Northeastern Finland indicating a founder effect. The hallmark of the disease is frontotemporal predominant pachygyria with mild cortical thickening. All patients show ID of variable severity. Aggressive behavior was found in nearly half of the patients, EEG abnormalities in five patients and megalencephaly in three patients. This study provides detailed data about the phenotypic spectrum of patients with lissencephaly due to a CRADD variant that affects function. High inter- and intrafamilial phenotypic heterogeneity was identified in patients with pachygyria caused by the homozygous CRADD founder variant. The phenotype variability suggests that additional genetic and/or environmental factors play a role in the clinical presentation. Since frontotemporal pachygyria is the hallmark of the disease, brain imaging studies are essential to support the molecular diagnosis for individuals with ID and a CRADD variant.
Subject terms: Neurodevelopmental disorders, Clinical genetics
Introduction
The human brain is a highly complex structure and its normal development and function is critically dependent on the proper and tightly regulated activity of a large number of genes. Consequently, it is estimated that variants that affect function in more than 2000 genes can give rise to intellectual disability (ID) [1]. Of these genes, about half are still unknown [1–3]. ID is clinically and genetically heterogeneous and comprises several groups of neurodevelopmental disorders that in total affect 2–3% of the general population [4, 5]. One of these groups is characterized by rare congenital anomalies of the cerebral cortex caused by neuronal migration defects, called malformations of cortical development (MCDs) [6]. MCDs are classified into three major groups: malformations caused by abnormal cell proliferation, misdirected neuronal migration, or aberrant post-migrational cortical organization and connectivity [6, 7] and are usually diagnosed by brain imaging such as MRI (magnetic resonance imaging) [7]. To date, genetic variants that affect function in more than 100 genes are reported to cause MCDs [7].
Lissencephaly (LIS) spectrum of diseases is associated with deficient neuronal migration and abnormal formation of cerebral convolutions or gyri [7–9]. The LIS spectrum of diseases encompasses agyria, pachygyria, and subcortical band heterotopia (SBH). In addition, LIS is found in syndromic MCDs, such as Miller-Dieker syndrome [10]. Clinical manifestations of LIS include developmental delay, ID, early hypotonia with subsequent spastic quadriplegia, and seizures [11, 12]. To date, a total of 19 genes have been identified to underlie the LIS spectrum of brain malformations of which the most common mutated genes are LIS1 and DCX [11]. Based on the combination of brain imaging and molecular genetics data from several patients, a classification of LIS and SBH described 21 different patterns of the disease, including common patterns such as partial agyria-pachygyria and posterior mild pachygyria [9]. Among the rare types of LIS is frontal predominant pachygyria with mild cortical thickening, also called thin pachygyria or thin lissencephaly (TLIS) [9].
Recently, four homozygous variants that affect function of the CRADD gene have been reported to cause TLIS (OMIM #614499) [13–15]. The phenotype was characterized by TLIS, relative or absolute megalencephaly, and mild to moderate intellectual disability with no other congenital anomalies. Three out of 15 (20%) patients presented with seizures [13, 14]. One of the variants, c.509G>A p.(Arg170His) (GenBank: NM_003805.3), was identified in a homozygous state in a female of Finnish ancestry [13].
In search for genes underlying intellectual disability in Finnish families, the homozygous c.509G>A p.(Arg170His) variant that affects function in CRADD in 22 patients out of 15 Finnish families with ID was found. This suggests a founder variant in the isolated Finnish population. The presence of an identical variant in a large number of individuals presents a unique opportunity to analyze the phenotypic variation that can be observed for a single monogenic condition. We present detailed clinical phenotype and brain imaging findings of the families, showing that pachygyria is the hallmark of the disease in patients with the Finnish founder variant.
Methods
Patients were referred by physicians working in disability services and clinical geneticists. In total, 211 patients with ID of unknown cause were collected from Northeastern Finland for the study. In addition, six cases homozygous for c.509G>A p.(Arg170His) variant from Northern Finland Intellectual Disability (NFID) project [16] and two cases identified directly in the clinic by one of the authors (ER) were included into the detailed phenotype analysis. Finally, two Finnish siblings with pachygyria [17] belong to a collection of cases with prenatal ventriculomegaly and/or congenital, primary hydrocephalus whose genetic background was studied by the group of MA.
The growth and occipitofrontal head circumference (OFC) of the Finnish patients were compared with the age and sex matched Finnish standard reference curves [18]. The new Finnish reference for OFC was used for patients between 0–7 years [19]. As the new reference was not standardized for patients older than 7 years, the old Finnish reference was used for patients older than 7 years [20]. OFC was compared to height and relative megalencephaly was defined as OFC more than 2 standard deviations (SD) above height centile for age.
The CRADD variant data were submitted to LOVD database (www.LOVD.nl/CRADD) hosted at Leiden University Medical Center, the Netherlands. The parents or legal guardians of all patients in this study provided written informed consent. The study was approved by the ethics committees at the Helsinki University Central Hospital, Northern Ostrobothnia Hospital District, Hospital District of Helsinki and Uusimaa, and the Radboud University Medical Center.
Exome sequencing (ES)
High-throughput sequencing was performed for sibling pairs in FIN-1 and FIN-2 families (Fig. 1) at BGI (BGI-Shenzhen, Shenzhen, China). Exome capturing was carried out by using Agilent SureSelect Target Enrichment V5 (Agilent Technologies, Santa Clara, CA, USA). Sequencing was performed by using the Illumina HiSeq 2000 platform (Illumina, Inc. San Diego, CA). Illumina base calling software v1.7 was employed to analyze the raw image files with default parameters. Data analysis were performed using the Roche Newbler software (v.2.3) using human genome build hg19/GRCh37.
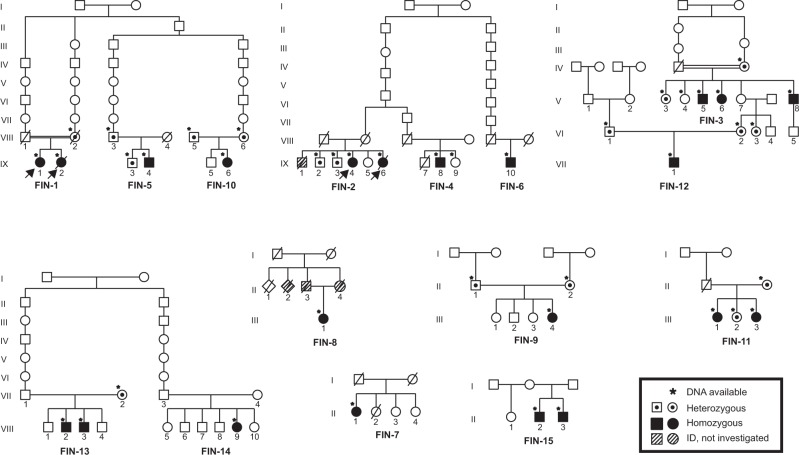
Fig. 1.
Pedigrees of the families. Pedigree of the family indicating genotype of the individuals who were available for testing. The arrow indicates the proband in each pedigree where ES was performed
Exome sequencing and targeted Sanger sequencing were also used to analyze the data of 757 patients with ID of unknown etiology belonging to the NFID cohort to identify the CRADD founder variant in six additional cases with pachygyria, see detailed description of the project in Kurki et al. [16].
Filtering and annotation
Selection of the variants for sibling pairs in FIN-1 and FIN-2 was performed by using a seven tier filtering strategy: (i) inclusion of variants present in ≥2 reads and present in ≥70% of all reads (ii) exclusion of variants present with a frequency ≥1% in unaffected controls (Dept of Human Genetics, Nijmegen, 12,000 controls), (iii) exclusion of variants within intergenic, intronic, and UTR regions, (iv) exclusion of variants present in dbSNP142, 1000 Genome, NHLBI Exome Variant Server (EVS) database or the Genome Aggregation Database (gnomAD) with a frequency ≥1% and number of homozygotes >2, (v) inclusion of missense variants with a Combined Annotation Dependent Depletion (CADD) score of ≥15, (vi) selected variants in genes that are expressed in the brain based on their EST profile in the Unigene database (transcripts per million – TPM ≥5), and (vii) inclusion of variants that segregate with the disease in the respective pedigree as determined by using Sanger sequencing (Supplementary information).
Variant screening by PCR restriction fragment length polymorphism
Restriction fragment length polymorphism (RFLP) analysis was performed on the PCR fragment amplified following the protocol used for Sanger sequencing validation. Enzymatic digestion was performed using AfeI (New England BioLabs, Massachusetts, USA) following the manufacturer’s protocol.
Brain imaging
Brain MRI data apart from the previously published FIN-15 siblings [17] was analyzed by an experienced neuroradiologist (MKB). Due to geographic and temporal differences, brain magnetic resonance imaging of these new patients was performed with various 1.5T equipment and different imaging protocols. One patient’s computer tomography images were also re-analyzed due to suboptimal MR-images.
Results
Following filtering of ES data, we identified and confirmed by Sanger sequencing a previously reported homozygous c.509G>A (p.(Arg170His)) variant that affects function [13] in the CRADD gene in the two affected sibling pairs of families FIN-1 and FIN-2 (Fig. 1). This variant was found heterozygous in all unaffected tested family members. Subsequently, by using ES, PCR-RFLP, and Sanger sequencing, 18 additional patients carrying the same homozygous CRADD variant were identified in different cohorts (Supplementary information). Altogether, a total of 23 patients of Finnish origin homozygous for the c.509G>A (p.(Arg170His)) variant in CRADD has been found to date ([13], Table 1). The allele frequency of c.509G>A in the European non-Finnish population is 0.01%, in contrast to the Finnish population frequency of 0.59% [21] (Fig. 3), showing that this allele is enriched in the latter. As reported in the SISu database [22] there are 125 heterozygous and no homozygous healthy individuals carrying the variant in 10,389 genotypes. Furthermore, the variant frequency reaches up to 1.25% in Eastern and Northern parts of Finland [22]. Based on previously published haplotype analysis data of 9363 Finns with both GWAS and exome sequencing data [23], the estimated haplotype length in heterozygote carriers of c.509G>A in CRADD is 1,684,758 bp.
Table 1.
Clinical details of the patients
| Family ID, patient ID | Learning disability | Occipitofrontal circumference | Relative OFCa | Frontotemporal pachygyria on brain MRI | Seizures | Abnormality in the EEG | Strabismus | Aggressive bursts | Distractability/hyperactivity | LOVD IDb |
|---|---|---|---|---|---|---|---|---|---|---|
| FIN-1, IX/1 | Moderate ID | 56.5 cm (+1.3 SD) | +1.9 SD | + | – | – | – | + | – | 00207614 |
| FIN-1, IX/2 | Moderate ID | 56 cm (+0.8 SD) | +1.2 SD | N/A | – | Photosensitivity | – | – | – | 00207617 |
| FIN-2, IX/4 | Moderate ID | 53.5 cm (−1.2 SD) | +2.0 SD | N/A | – | – | + | – | – | 00207623 |
| FIN-2, IX/6 | Moderate ID | 56 cm (+0.8 SD) | +2.2 SD | + | – | – | – | – | – | 00207624 |
| FIN-3, V/5 | Mild ID | N/A | N/A | + | – | – | + | – | + | 00207625 |
| FIN-3, V/6 | Mild ID | 56.7 cm (+1.4 SD) | +1.8 SD | + | + | Photosensitivity | – | – | – | 00207626 |
| FIN-3, V/8 | N/A | N/A | N/A | N/A | – | – | – | – | – | 00207627 |
| FIN-4, IX/8 | N/A | 56.5 cm (−0.7 SD) | N/A | + | + | Photosensitivity | + | + | + | 00207630 |
| FIN-5, IX/4 | Mild ID | 60 cm (+1.9 SD) | +2.7 SD | + | – | N/A | – | + | – | 00207631 |
| FIN-6, IX/10 | Moderate ID | 56.6 cm (−0.7 SD) | N/A | + | – | N/A | – | – | – | 00207632 |
| FIN-7, II/1 | Moderate ID | 58 cm (+2.4 SD) | N/A | + | – | – | + | + | – | 00207633 |
| FIN-8, III/1 | Moderate ID | 54.8 cm (0 SD) | +1.7 SD | + | – | N/A | + | + | + | 00207634 |
| FIN-9, III/4 | Moderate ID | 52.3 cm (−2.0 SD) | −1.9 SD | + | – | – | – | + | – | 00207635 |
| FIN-10, IX/6 | Mild ID | 55 cm (0 SD) | +1.3 SD | + | – | – | + | – | + | 00207636 |
| FIN-11, III/1 | Moderate ID | 55 cm (0 SD) | +0.8 SD | + | – | – | – | + | + | 00207638 |
| FIN-11, III/3 | Moderate ID | 56 cm (+0.8 SD) | +1.3 SD | + | – | N/A | + | + | + | 00207639 |
| FIN-12,VII/1 | Moderate ID | 50 cm (−1.7 SD) | −1.5 SD | + | – | – | + | – | + | 00207640 |
| FIN-13, VIII/2 | Mild ID | 54.7 cm (+0.1 SD) | +0.5 SD | + | – | + | + | + | – | 00207641 |
| FIN-13, VIII/3 | Moderate ID | 56.5 cm (−0.2 SD) | 0 SD | + | – | N/A | – | – | – | 00207642 |
| FIN-14,VIII/9 | Global developmental delay | 50.5 cm (+0.1 SD) | +1 SD | + | – | + | – | – | – | 00207643 |
| FIN-15, II/2c | Moderate ID | 53.5 cm (+0.2 SD) | +0.4 SD | + | – | – | – | + | – | 00207644 |
| FIN-15, II/3c | Mild ID | 55 cm (+1.5 SD) | +1.9 SD | + | – | – | – | – | – | 00207646 |
N/A not available, ID intellectual disability, EEG electroencephalography.
aOFC compared to height
bLeiden Open Variation Database Individual ID
cAvela et al. [17]
Fig. 3.
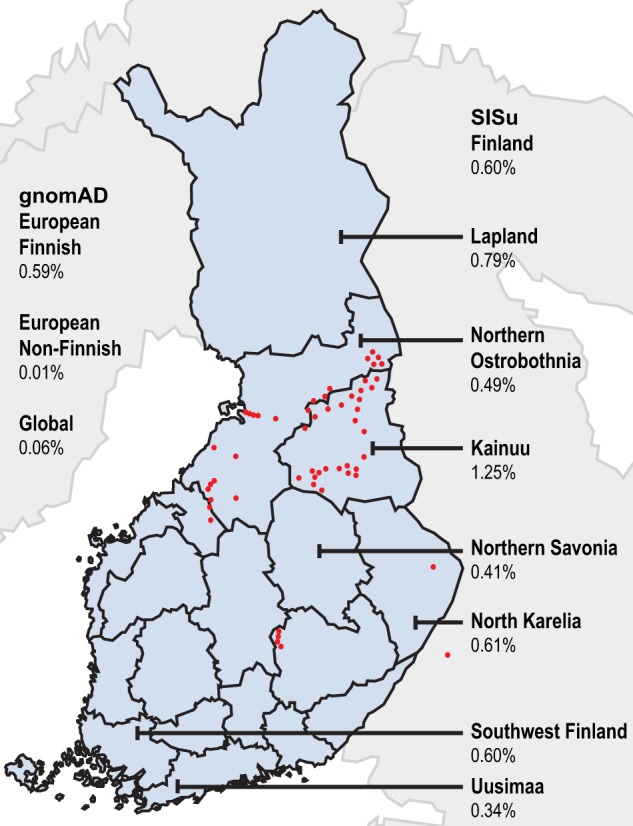
Allele frequency distribution of the CRADD c.509G>A variant in Finland and birthplaces of the grandparents of the patients. Allele frequencies according to the gnomAD browser and SISu databases. Red dots represent the birthplaces of the grandparents of the patients. Blue areas on the map represent the regions of Finland. (color figure online).
Genealogical studies
To further demonstrate that the c.509G>A (p.(Arg170His)) variant in CRADD is a founder variant, we performed a genealogical study of the Finnish families in accordance with previously published criteria [24]. We traced ancestors from Finnish Population Registries and scrutinized microfilm copies available in the National Archives of Finland. Several core families show inter- and intra-familial consanguinity (Fig. 1) confirming the founder effect of the c.509G>A variant in CRADD in the Finnish population. Tracing back to the 1790s forefathers revealed that 50% of the grandparents originate from Northeastern Finland (Fig. 3).
Clinical delineation of patients
Early psychomotor development was normal in all patients. The patients were characterized by either mild to moderate ID (N = 21/22) or global developmental delay (N = 1/22, VIII/9, FIN-14, at the age of 4 years) ([17], Table 1). Language development was markedly delayed in young patients. The patients in the younger generations received speech therapy. The patients in the older generations did not receive speech therapy, and their speech improved spontaneously by school age. Five out of the 22 patients showed EEG abnormalities, such as photosensitivity (N = 3) and general background slowing (N = 2), and two individuals were diagnosed with epilepsy (N = 2/22, 9%). Strabismus and visual problems were observed in nine out of the 22 patients (N = 9/22, 41%). Distractability or hyperactive behavior at school-going age was present in seven patients (N = 7/22, 32%). Neuropsychiatric problems characterized by aggressive bursts started usually at school age but also much later in adulthood requiring neuroleptic medication in 10 patients (N = 10/22, 45%). Characteristically, adult patients have preferred working in assisting everyday tasks and some have educated themselves. Majority of adult patients have moved to assisted living facilities where they can get help in everyday tasks. Some of them can live relatively independently. No striking growth abnormalities nor common characteristic dysmorphic features were present. Patient II/1, FIN-7, had megalencephaly (OFC 58 cm;+2.4 SD). In the rest of the patients studied the head circumference was in normal range compared to Finns [18, 19] and varied between −2.0 SD and + 1.9 SD from the mean reference values matched for age, sex, and ethnicity. We also compared the OFC-to-height and patients IX/4, FIN-5, and IX/4, FIN-2, had relative megalencephaly (OFC compared to height, +2.2 SD and +2.7 SD respectively) and in rest of the patients OFC-to-height was between −1.9 SD and +2.0 SD from the mean reference values matched for age, sex, and ethnicity (Table 1). Detailed clinical phenotypes of all patients are provided in the clinical descriptions of patients’ section in the Supplementary information and in ref. [17]. The clinical phenotype of the heterozygous carriers was normal.
Brain imaging findings
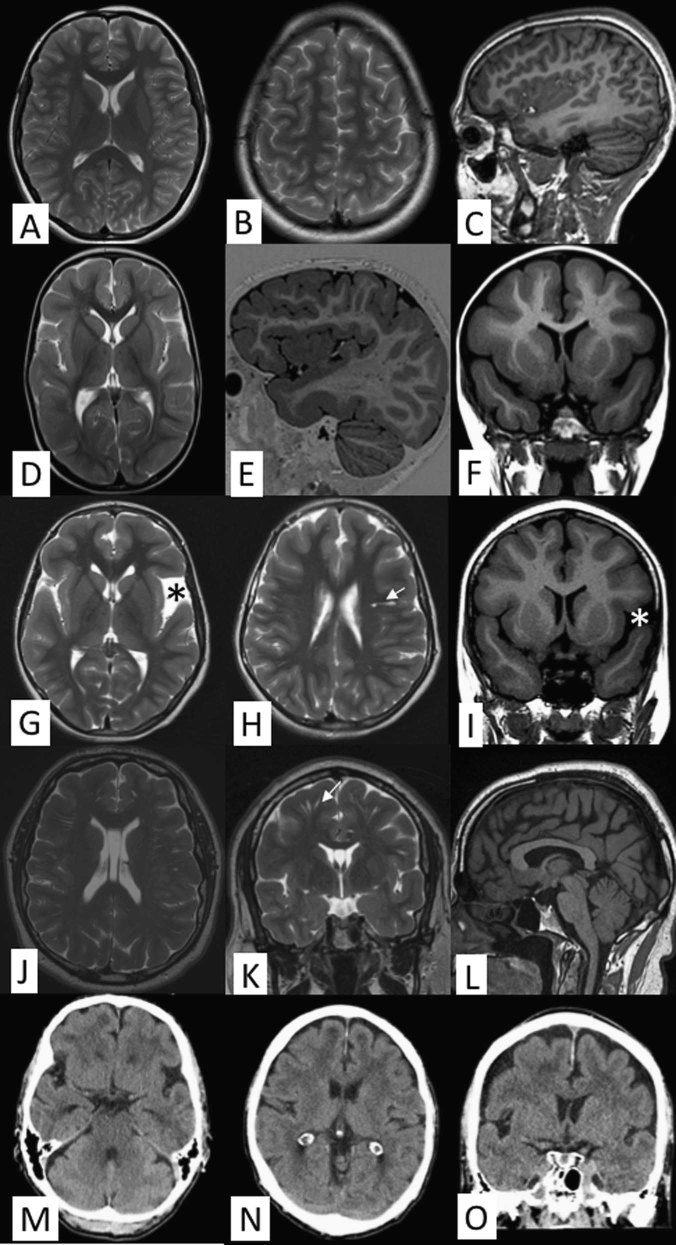
Brain MRI data available for 17 affected individuals revealed pachygyria and a mild cortical thickening up to 8 mm predominantly ranging from 5 to 7 mm with a frontotemporal predilection. Similar MRI-findings were reported by Avela et al. (2012). High-quality CT was able to confirm the main finding adequately in one patient. Some of the most representative images of patients in various ages are presented in Fig. 2. Data on other MRI findings is available in Supplementary Table 3. White matter was normally myelinized. No obvious midline anomalies were detected. Basal ganglia and thalami were considered normal. In some of the elderly patients, brain atrophy and white matter hyperintensities probably unrelated to the genetic condition was detected. In patients VII/1, FIN-12, and VIII/2, FIN-13 the lateral ventricles appeared slightly larger than usual and patients III/4, FIN-9, and II/1, FIN-7 had small arachnoid cysts. Prominent perivascular spaces were seen in all the patients. Insignificant findings included cavum septum pellucidum (cum vergae) and enlarged cisterna magna.
Fig. 2.
Brain imaging findings. a–c Axial T2-weighted images at different levels (a, b) and sagittal volumetric acquisition T1-weighted 3D-MPR thin slice image (c) of a normal 25-year-old individual. Note normal dense gyral pattern and normal cortical thickness (around 3 mm) in comparison with the following images. d–f Axial T2-weighted image (d), sagittal volumetric acquisition T1-weighted 3D-IR thin slice image (e), and coronal T1 FLAIR (fluid attenuated inversion recovery) image (f) in patient VIII/9, FIN-14 at the age of 2 years 8 months demonstrates symmetric pachygyria (broad and shallow sulci) with mild cortical thickening up to 8 mm. The frontotemporal predilection of both pachygyria and cortical thickening is nicely depicted in figure e, whereas the occipitoparietal regions show much milder changes. g–i Axial T2-weighted images at different levels (g, h) and coronal T1-FLAIR image (i) in patient III/4, FIN-9 at the age 9 years 6 months show similar findings as in figures d–f. A small temporal arachnoid cyst is marked by an asterisk and prominent perivascular space by an arrow. j–l Axial (j) and coronal (k) T2-weighted images in patient IX/4, FIN-5 at the age of 43 years demonstrate a bit less pronounced although evident pachygyria and mild cortical thickening of 6–7 mm. Prominent perivascular spaces (arrow) are also evident on both sides (k). T1-weighted sagittal image shows normal midline structures (l). m–o Good-quality thin-slice computer tomography (CT) scan shows pachygyria and mild cortical thickening almost as good as MRI in an elderly male (IX/10, FIN-6). Multiplanar reconstruction (MPR) CT-images in axial (m, n) and coronal (o) planes
Discussion
The first variants that affect function in CRADD were identified by Puffenberger et al. [15] and Di Donato et al. [13] in patients of different ethnicities presenting generally frontal predominant pachygyria, seizures, megalencephaly, and mild to moderate ID. A possible homozygous founder variant (c.382G>C; (p.(Gly128Arg)) was identified in two Mennonite families with seven affected patients presenting with mild to moderate ID [13, 15]. Brain MRI performed in three of seven affected individuals revealed TLIS with normal cerebellum, as reported with the MRI features in all other CRADD affected subjects. In addition, four other homozygous variants in the CRADD gene have been reported to cause TLIS [13, 14]. One of the missense variants that affect function, c.509G>A p.(Arg170His), was found in an American patient of Finnish ancestry who also presented megalencephaly (OFC 60.9 cm, +3.6 SD), aggressive bursts, and seizures [13].
Here we report the detailed clinical phenotype of an additional 22 new cases from 15 Finnish families with mild to moderate intellectual disability who carry the c.509G>A p.(Arg170His) variant in CRADD homozygously, suggesting a founder effect in the isolated Finnish population. All patients whose brain imaging data was available (N = 19; 86%) showed frontotemporal predominant pachygyria. The presence of such founder variant allowed an in-depth analysis of the possible phenotypic variation that can be observed for an identical CRADD variant. Neurobehavioural problems were present in nearly half of the cases and EEG abnormalities, including photosensitivity and background slowing, were present in five (N = 5/17, 29%) patients.
Detailed analysis of the phenotype showed inter- and intrafamilial variability in disease severity. The hallmark of the disease was pachygyria with frontotemporal predilection and mild cortical thickening in brain MRI (Fig. 2). These findings are compatible with TLIS phenotype described by Di Donato et al. [13]. However, in contrast to the earlier study [13], absolute megalencephaly was found only in one patient and relative megalencephaly was found in two additional patients in our cohort. The phenotype was characterized by normal early development, followed by delayed speech and language development. Some patients developed neurobehavioural problems including distractibility and short attention span in childhood, and in less than half of the patients aggressive bursts started to appear during adolescence. In the end, the disease manifests mainly as mild to moderate ID diagnosed during childhood. Special education was given for the younger generations of the patients. Phenotypic variability suggests that additional genetic and/or environmental factors play a role in the clinical presentation. Modifying genes are likely, as even in the same family patients can display a variable phenotype. In addition, patients with a mild phenotype may remain undiagnosed as exemplified in a previous study [14, 15], where a family with the CRADD founder variant had been reported initially with non-syndromic ID. However, brain MRI revealed TLIS. Likewise in this study, two young patients presenting with global developmental delay and predominantly frontotemporal pachygyria in brain MRI were further identified to carry the homozygous founder CRADD variant by using targeted Sanger sequencing. In fact, the vast majority of the patients presented with the non-syndromic ID and showed no clinical features suggesting a possible CRADD variant that affects function. In addition, two males (V/5 and V/8, FIN-3; Fig. 2) were found to carry the homozygous founder CRADD variant after they participated in targeted genetic screening due to their sister’s diagnosis. In most cases MRIs were performed retrospectively after the identification of the homozygous CRADD variant or the scans were done previously but the neuroradiological interpretation of pachygyria remained unnoticed. Finally, these results are compatible with clinical and brain imaging data of two patients described by Avela et al. [17]. This indicates that brain imaging studies are essential to support the molecular diagnosis for individuals with intellectual disability.
The CRADD gene encodes an adapter protein containing a caspase-recruitment domain (CARD) and death domain (DD) that interacts with PIDD (p53-induced protein with death domain) and caspase-2 (CASP2) to initiate apoptosis [25–28]. CRADD/caspase-2 signaling plays an essential role in the human cortical architecture, synaptic plasticity, and cognitive function during brain development [13, 28]. Disruption of this pathway would accordingly result in cortical malformations, such as megalencephaly and lissencephaly. Indeed, the phenotype of Cradd−/− mice included megalencephaly and seizures, although without changes in cortical thickness or lamination [13]. In contrast, studies with Casp2−/− mice reported no brain phenotype [29, 30]. The megalencephaly described for Cradd−/− mice and the lack of brain abnormalities in Casp2−/− mice are not consistent to the TLIS and ID observed in patients with CRADD variants that affect protein function. Thus, the CRADD/caspase-2 signaling pathway may have a slightly different function in mice and human, resulting in overgrowth without MCD in mice and MCD without overgrowth in humans.
The allele frequency of c.509G>A in the Finnish population is 0.59% [21] (Fig. 3). In comparison, the European non-Finnish frequency is 0.01%, showing that this allele is enriched in the Finnish population. As reported in the SISu database [22], there are 125 heterozygous and no homozygous healthy individuals carrying the variant in 10,389 genotypes. In addition, the variant frequency is even higher in Eastern and Northern parts of Finland, and reaches up to 1.25% [22]. According to our genealogical analysis, 50% of our patients’ ancestors originate from Northeastern sub-isolate (Fig. 3). Here, several rare autosomal recessive diseases, such as Northern epilepsy [31], Salla disease [32, 33], and nonketotic hyperglycinemia [34], and their founder variants have previously been reported displaying strong geographical clustering. This is due to the Finnish internal migrations during the 16th century, when the regional sub-isolates were formed and grew without further immigration until the 20th century, resulting in reduced genetic diversity [35, 36]. The enriched variant distribution in Eastern and Northern parts of Finland is in agreement with the genealogical analysis of the identified patients in this study, and with the haplotype analysis [23] and carrier frequencies previously estimated in the Finnish disease heritage [37]. Martin et al. [23] have studied haplotype sharing of heterozygote carriers of recessive Finnish Disease Heritage variants. Based on data in Martin et al. [23], the allele frequency of c.509 G > A in Finnish population (0.59%) is on par with Cornea Plana disease variant (0.52%, rs121917858) and they both have similar high haplotype enrichment (22.3 for CRADD c.509G>A and 23.5 for rs121917858) (see Table 1 in Martin et al. [23]). This supports the hypothesis of a founder effect of the CRADD variant in the isolated Northeastern Finnish population.
This study shows that CRADD c.509G>A variant is a Finnish founder variant. This study establishes that among the whole spectrum of lissencephaly [9, 38], the clinical phenotype caused by the founder variant in CRADD is considerably milder than other types of lissencephaly. It is of notice that some patients have educated themselves, live independently and have had families and children (this study) [14]. The mild phenotype highlights the importance of performing brain imaging studies when dissecting molecular diagnosis for individuals with intellectual disability.
Supplementary information
Acknowledgements
We are grateful to the families for participating in the study. We thank Shaffaq Raza, Minna Varhala, and Eija Hämäläinen for excellent technical help. We thank all the clinicians who have recruited patients to this study. This work was supported by the European Union’s Seventh Framework Program (Gencodys; grant 241995 to HvB). DLP is recipient of a CAPES Fellowship (99999.013311/2013–01).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Daniel L. Polla, Elisa Rahikkala, Michaela K. Bode
These authors jointly supervised this work: Hans van Bokhoven, Irma Järvelä
Change history
9/10/2019
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
Supplementary information
The online version of this article (10.1038/s41431-019-0383-8) contains supplementary material, which is available to authorized users.
References
- 1.van Bokhoven H. Genetic and epigenetic networks in intellectual disabilities. Annu Rev Genet. 2011;45:81–104. doi: 10.1146/annurev-genet-110410-132512. [DOI] [PubMed] [Google Scholar]
- 2.Willemsen MH, Kleefstra T. Making headway with genetic diagnostics of intellectual disabilities. Clin Genet. 2014;85:101–10. doi: 10.1111/cge.12244. [DOI] [PubMed] [Google Scholar]
- 3.Riazuddin S, Hussain M, Razzaq A, Iqbal Z, Shahzad M, Polla DL, et al. Exome sequencing of Pakistani consanguineous families identifies 30 novel candidate genes for recessive intellectual disability. Mol Psychiatry. 2017;22:1604–14. doi: 10.1038/mp.2016.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Institute of Medicine (U.S.). Committee to evaluate the supplemental security income disability program for children with mental disorders. In: Boat TF, Wu JT, editors. National Academies of Sciences Engineering and Medicine: mental disorders and disabilities among low-income children. Washington, D.C.: National Academies Press, 2015. . [PubMed]
- 5.Ropers HH. Genetics of early onset cognitive impairment. Annu Rev Genom Hum Genet. 2010;11:161–87. doi: 10.1146/annurev-genom-082509-141640. [DOI] [PubMed] [Google Scholar]
- 6.Barkovich AJ, Guerrini R, Kuzniecky RI, Jackson GD, Dobyns WB. A developmental and genetic classification for malformations of cortical development: update 2012. Brain. 2012;135:1348–69. doi: 10.1093/brain/aws019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guerrini R, Dobyns WB. Malformations of cortical development: clinical features and genetic causes. Lancet Neurol. 2014;13:710–26. doi: 10.1016/S1474-4422(14)70040-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guerrini R, Marini C. Genetic malformations of cortical development. Exp Brain Res. 2006;173:322–33. doi: 10.1007/s00221-006-0501-z. [DOI] [PubMed] [Google Scholar]
- 9.Di Donato N, Chiari S, Mirzaa GM, Aldinger K, Parrini E, Olds C, et al. Lissencephaly: expanded imaging and clinical classification. Am J Med Genet A. 2017;173:1473–88. doi: 10.1002/ajmg.a.38245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reiner O, Carrozzo R, Shen Y, Wehnert M, Faustinella F, Dobyns WB, et al. Isolation of a Miller-Dieker lissencephaly gene containing G protein beta-subunit-like repeats. Nature. 1993;364:717–21. doi: 10.1038/364717a0. [DOI] [PubMed] [Google Scholar]
- 11.Parrini E, Conti V, Dobyns WB, Guerrini R. Genetic basis of brain malformations. Mol Syndromol. 2016;7:220–33. doi: 10.1159/000448639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fry AE, Cushion TD, Pilz DT. The genetics of lissencephaly. Am J Med Genet C Semin Med Genet. 2014;166C:198–210. doi: 10.1002/ajmg.c.31402. [DOI] [PubMed] [Google Scholar]
- 13.Di Donato N, Jean YY, Maga AM, Krewson BD, Shupp AB, Avrutsky MI, et al. Mutations in CRADD result in reduced caspase-2-mediated neuronal apoptosis and cause megalencephaly with a rare lissencephaly variant. Am J Hum Genet. 2016;99:1117–29. doi: 10.1016/j.ajhg.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harel T, Hacohen N, Shaag A, Gomori M, Singer A, Elpeleg O, et al. Homozygous null variant in CRADD, encoding an adaptor protein that mediates apoptosis, is associated with lissencephaly. Am J Med Genet A. 2017;173:2539–44. doi: 10.1002/ajmg.a.38347. [DOI] [PubMed] [Google Scholar]
- 15.Puffenberger EG, Jinks RN, Sougnez C, Cibulskis K, Willert RA, Achilly NP, et al. Genetic mapping and exome sequencing identify variants associated with five novel diseases. PLoS ONE. 2012;7:e28936. doi: 10.1371/journal.pone.0028936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurki MI, Saarentaus E, Pietilainen O, Gormley P, Lal D, Kerminen S et al. Contribution of rare and common variants to intellectual disability in a high-risk population sub-isolate of Northern Finland. bioRxiv. 2019;10:410. [DOI] [PMC free article] [PubMed]
- 17.Avela K, Toiviainen-Salo S, Karttunen-Lewandowski P, Kauria L, Valanne L, Salonen-Kajander R. Frontotemporal pachygyria-two new patients. Eur J Med Genet. 2012;55:753–7. doi: 10.1016/j.ejmg.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 18.Sorva R, Tolppanen EM, Lankinen S, Perheentupa J. [Evaluation of childhood growth] Duodecim. 1985;101:465–76. [PubMed] [Google Scholar]
- 19.Karvonen M, Hannila ML, Saari A, Dunkel L. New Finnish reference for head circumference from birth to 7 years. Ann Med. 2012;44:369–74. doi: 10.3109/07853890.2011.558519. [DOI] [PubMed] [Google Scholar]
- 20.Saari A, Sankilampi U, Hannila ML, Kiviniemi V, Kesseli K, Dunkel L. New Finnish growth references for children and adolescents aged 0 to 20 years: Length/height-for-age, weight-for-length/height, and body mass index-for-age. Ann Med. 2011;43:235–48. doi: 10.3109/07853890.2010.515603. [DOI] [PubMed] [Google Scholar]
- 21.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sequencing Initiative Suomi project (SISu), Institute for Molecular Medicine Finland (FIMM), University of Helsinki, Finland. http://sisuproject.fi. [SISu v4.1, 03/2018].
- 23.Martin AR, Karczewski KJ, Kerminen S, Kurki MI, Sarin AP, Artomov M, et al. Haplotype sharing provides insights into fine-scale population history and disease in Finland. Am J Hum Genet. 2018;102:760–75. doi: 10.1016/j.ajhg.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varilo T, Nikali K, Suomalainen A, Lonnqvist T, Peltonen L. Tracing an ancestral mutation: genealogical and haplotype analysis of the infantile onset spinocerebellar ataxia locus. Genome Res. 1996;6:870–5. doi: 10.1101/gr.6.9.870. [DOI] [PubMed] [Google Scholar]
- 25.Duan H, Dixit VM. RAIDD is a new ‘death’ adaptor molecule. Nature. 1997;385:86–89. doi: 10.1038/385086a0. [DOI] [PubMed] [Google Scholar]
- 26.Lin Y, Ma W, Benchimol S. Pidd, a new death-domain-containing protein, is induced by p53 and promotes apoptosis. Nat Genet. 2000;26:122–7. doi: 10.1038/79102. [DOI] [PubMed] [Google Scholar]
- 27.Berube C, Boucher LM, Ma W, Wakeham A, Salmena L, Hakem R, et al. Apoptosis caused by p53-induced protein with death domain (PIDD) depends on the death adapter protein RAIDD. Proc Natl Acad Sci USA. 2005;102:14314–20. doi: 10.1073/pnas.0506475102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ribe EM, Jean YY, Goldstein RL, Manzl C, Stefanis L, Villunger A, et al. Neuronal caspase 2 activity and function requires RAIDD, but not PIDD. Biochem J. 2012;444:591–9. doi: 10.1042/BJ20111588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shalini S, Dorstyn L, Wilson C, Puccini J, Ho L, Kumar S. Impaired antioxidant defence and accumulation of oxidative stress in caspase-2-deficient mice. Cell Death Differ. 2012;19:1370–80. doi: 10.1038/cdd.2012.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Padalecki SS, Chaudhuri AR, De Waal E, Goins BA, Grubbs B, et al. Caspase-2 deficiency enhances aging-related traits in mice. Mech Ageing Dev. 2007;128:213–21. doi: 10.1016/j.mad.2006.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ranta S, Zhang Y, Ross B, Lonka L, Takkunen E, Messer A, et al. The neuronal ceroid lipofuscinoses in human EPMR and mnd mutant mice are associated with mutations in CLN8. Nat Genet. 1999;23:233–6. doi: 10.1038/13868. [DOI] [PubMed] [Google Scholar]
- 32.Aula P, Autio S, Raivio KO, Rapola J, Thoden CJ, Koskela SL, et al. “Salla disease”: a new lysosomal storage disorder. Arch Neurol. 1979;36:88–94. doi: 10.1001/archneur.1979.00500380058006. [DOI] [PubMed] [Google Scholar]
- 33.Renlund M, Aula P, Raivio KO, Autio S, Sainio K, Rapola J, et al. Salla disease: a new lysosomal storage disorder with disturbed sialic acid metabolism. Neurology. 1983;33:57–66. doi: 10.1212/WNL.33.1.57. [DOI] [PubMed] [Google Scholar]
- 34.Kure S, Takayanagi M, Narisawa K, Tada K, Leisti J. Identification of a common mutation in Finnish patients with nonketotic hyperglycinemia. J Clin Invest. 1992;90:160–4. doi: 10.1172/JCI115831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peltonen L, Palotie A, Lange K. Use of population isolates for mapping complex traits. Nat Rev Genet. 2000;1:182–90. doi: 10.1038/35042049. [DOI] [PubMed] [Google Scholar]
- 36.Norio R. Finnish disease heritage I: characteristics, causes, background. Hum Genet. 2003;112:441–56. doi: 10.1007/s00439-002-0875-3. [DOI] [PubMed] [Google Scholar]
- 37.Polvi A, Linturi H, Varilo T, Anttonen AK, Byrne M, Fokkema IF, et al. The Finnish disease heritage database (FinDis) update-a database for the genes mutated in the Finnish disease heritage brought to the next-generation sequencing era. Hum Mutat. 2013;34:1458–66. doi: 10.1002/humu.22389. [DOI] [PubMed] [Google Scholar]
- 38.Romero Delfina M., Bahi-Buisson Nadia, Francis Fiona. Genetics and mechanisms leading to human cortical malformations. Seminars in Cell & Developmental Biology. 2018;76:33–75. doi: 10.1016/j.semcdb.2017.09.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.