Abstract
Hartsfield syndrome (HS) is an ultrarare developmental disorder mainly featuring holoprosencephaly and ectrodactyly. It is caused by heterozygous or biallelic variants in FGFR1. Recently, a dominant-negative effect was suggested for FGFR1 variants associated with HS. Here, exome sequencing analysis in a 12-year-old boy with HS disclosed a novel de novo heterozygous variant c.1934C>T in FGFR1 predicted to cause the p.(Ala645Val) amino-acid substitution. In order to evaluate whether the variant, changing a highly conserved residue of the kinase domain, affects FGFR1 function, biochemical studies were employed. We measured the FGFR1 receptor activity in FGF2-treated cell lines exogenously expressing wild-type or Ala645Val FGFR1 by monitoring the activation status of FGF2/FGFR1 downstream pathways. Our analysis highlighted that RAS/ERK1/2 signaling was significantly perturbed in cells expressing mutated FGFR1, in comparison with control cells. We also provided preliminary evidence showing a modulation of the autophagic process in cells expressing mutated FGFR1. This study expands the FGFR1 mutational spectrum associated with HS, provides functional evidence further supporting a dominant-negative effect of this category of FGFR1 variants and offers initial insights on dysregulation of autophagy in HS.
Subject terms: DNA sequencing, Disease genetics
Introduction
Hartsfield syndrome (HS; MIM#615465) is an ultrarare developmental disorder mainly characterized by (various degrees of) holoprosencephaly and ectrodactyly. Ancillary findings include intellectual disability, cerebellar vermis hypoplasia, spasticity, seizures, hypothalamic dysfunction, central diabetes insipidus, and hypogonadotropic hypogonadism [1]. HS is caused by heterozygous or, more rarely, biallelic variants in FGFR1 [2, 3]. The FGFR family comprises four receptor tyrosine kinases that cooperate with extracellular fibroblast growth factors (FGFs) in the transduction of signals through the plasma membrane [4]. FGFRs consist of an extracellular region of three immunoglobulin-like (Ig-like) domains (namely, D1, D2, and D3), a single hydrophobic transmembrane domain, and a cytoplasmic tyrosine kinase domain. The extracellular portion interacts with FGFs and triggers a cascade of downstream signals influencing organogenesis, angiogenesis, metabolism, and tissue repair [5].
FGFR1 abnormalities recur in multiple developmental and acquired diseases. Germline variants have been identified in five pleiotropic disorders, including HS, Kallmann syndrome [6], nonsyndromic hypogonadotropic hypogonadism [7–9], Pfeiffer syndrome [10], and osteoglophonic dysplasia [11]. Somatic mosaicism for developmental post-zygotic variants cause encephalocraniocutaneous lipomatosis [12]. Somatic variants in FGFR1 can also occur postnatally and associate with cancer [13]. Limited genotype–phenotype correlations predict the clinical outcome among FGFR1-related developmental disorders. Gain-of-function effects are hypothesized in conditions with craniosynostosis, while loss-of-function variants typically occur in disorders with hypogonadotropic hypogonadism [2]. More recently, a single study attributed a dominant-negative effect to variants causing HS by zebrafish overexpression assays [3]. However, the molecular pathogenesis of HS remains incompletely defined.
We describe a HS patient carrying a novel c.1934C>T, p.(Ala645Val) FGFR1 heterozygous variant, identified by exome sequencing. We also provide functional evidence supporting a dominant-negative effect of this novel FGFR1 variant and offer preliminary insights on deregulation of autophagy in HS.
Materials and methods
Molecular study
A clinical diagnosis of HS was established on the proband (see Clinical report). Proband’s parents gave their informed consent for genetic testing and processing of personal data according to the Italian bioethics laws. The molecular testing (clinical exome, see below) carried out in this patient for diagnostic purposes is based on routine clinical care. Therefore, Institutional Review Boards (IRB) approval was not requested. Genomic DNA was extracted from patients’ and parents’ peripheral blood by using Bio Robot EZ1 (Qiagen). The DNA was quantified with Nanodrop 2000 C spectrophotometer (Thermo Fisher Scientific). Proband’s DNA was analyzed by whole-exome sequencing (WES) by using SureSelect Human Clinical Research Exome (Agilent Technologies) and following manufacturer's instructions. This is a combined shearing free transposase-based library prep and target enrichment solution, which enables comprehensive coverage of the entire exome. This system enables a specific mapping of reads to targets for deep coverage of target protein coding regions from RefSeq, GENCODE, CCDS, and UCSC known genes, with excellent overall exonic coverage and increased coverage of HGMD, OMIM, ClinVar, and ACMG targets. Sequencing was performed on a NextSeq 500 system (Illumina Inc.) by using the high output flow cells (300 cycles), with a minimum expected coverage depth of 70×. The average coverage obtained was 147×.
All variants obtained from WES were annotated based on frequency, impact on the encoded protein, conservation, and expression using distinct tools, as appropriate (ANNOVAR, dbSNP, 1000 Genomes, EVS, ExAC, ESP, and KAVIAR). The deleteriousness of variants was checked by querying PolyPhen-2, SIFT, MutationAssessor, FATHMM, LRT, and CADD. Given the clinical diagnosis of HS, filtered variants were also prioritized for genes associated with holoprosencephaly, in particular: CDON (OMIM 608707), DISP1 (OMIM 607502), DLL1 (OMIM 606582), FGF8 (OMIM 600483), FGFR1 (OMIM 136350), FOXH1 (OMIM 603621), GAS1 (OMIM 139185), GLI2 (OMIM 610829), NODAL (OMIM 601265), PTCH1 (OMIM 601309), SHH (OMIM 600725), SIX3 (OMIM 603714), TDGF1 (OMIM 187395), TGFI1 (OMIM 602603), and ZIC2 (OMIM 603073). The candidate variant was confirmed by Sanger sequencing in the proband’s and parents’ DNA. PCR products (oligos indicated in Table S1) were sequenced by using BigDye Terminator v1.1 sequencing kit (Applied Biosystems) and ABI Prism 3100 Genetic Analyzer (Thermo Fisher Scientific). The novel variant has been submitted to LOVD (https://databases.lovd.nl/shared/genes/FGFR1; patient ID #00174403).
Plasmids
The plasmid encoding FGFR1 wild-type was kindly provided by Soo-Hyun Kim (University of London, UK). The Ala645Val variant was generated using the QuickChange II site-directed mutagenesis kit (Stratagene) according to the manufacturer’s instructions. The construct was confirmed by Sanger sequencing. Primer pairs used are listed in the Table S1.
ERK1/2 activation analysis
HEK293 cells were transfected with wild-type or Ala645Val FGFR1-expressing plasmids or both by using lipofectamine (Thermo Fisher Scientific) according to the manufacturer’s instruction. At 24 h after transfection, cells were grown in serum-free medium for 24 h, and then incubated in the absence or presence of 1 nM FGF2 (Peprotech) for 15 min, as previously reported in ref. no. [14]. Cells were then lysed in a buffer containing phospho STOP and proteinase inhibitor cocktail (Roche). Proteins were separated on 10% Sodium Dodecyl Sulphate-PolyAcrylamide Gel Electrophoresis (SDS-PAGE), transferred onto nitrocellulose membrane and subjected to immunoblotting with Extracellular signal–Regulated Kinases 1 and 2 (ERK1/2) and phosphorylated ERK1/2 antibodies (Cell Signalling Technology), anti-myc (Sigma), anti-actin (Santa Cruz). To analyze the reactive ERK1/2 phosphorylation level, bands intensity of phoshorylated ERK1/2 and total ERK1/2 was quantified using Image J software, and the ratio of pERK1/2 to total ERK1/2 was calculated.
Effect of FGF2 on c-Fos induction and RNA extraction
HEK293 cells were transfected with wild-type or Ala645Val FGFR1-expressing plasmids or both by using lipofectamine (Thermo Fisher Scientific) according to the manufacturer’s instruction. At 24 h after transfection, cells were cultured for 18 h in low serum medium and then incubated in the absence or presence of 0.5 nM FGF2 for 1 h, as previously reported in ref. no. [15]. Total RNA was extracted using mini RNase kit reagent (Qiagen), treated with DNase-RNase free (Qiagen) and reverse-transcribed using Quantitect Transcription kit (Qiagen) according to the manufacturer’s instructions.
Quantitative real-time PCR
Oligos for qPCR were designed using the Primer express program [16] with default parameters. GAPDH and ACTIN were used as reference genes. Primer pairs used are listed in the Table S1. Reactions were run in triplicate in 10 μl of final volume with 10 ng of sample complementary DNA, 0.3 mM of each primer, and 1XPower SYBR Green PCR Master Mix (Thermo Fisher Scientific-Applied Biosystems). Reactions were set up in a 384-wells plate format with a Biomeck 2000 (Beckmann Coulter) and run in an ABI Prism7900HT (Thermo Fisher Scientific-Applied Biosystems) with default amplification conditions. Raw Ct values were obtained using SDS 2.3 (Applied Biosystems). Calculations were carried out by the comparative Ct method as reported in [17]. Significance was determined by a two-tailed unpaired t-test for means.
Autophagy assay
HEK293 cells were transfected with wild-type or Ala645Val FGFR1-expressing plasmids by using lipofectamine (Thermo Fisher Scientific) according to the manufacturer’s instruction. At 24 h after transfection, cells were cultured in the absence or presence of 25 ng/ml FGF2 for 2 h, as previously reported in [18]. Cells were then lysed in a buffer containing proteinase inhibitor cocktail (Roche). Autophagy was analyzed by detecting the LC3-II and Beclin protein markers. GAPDH was used as reference protein. Twelve percent of SDS-PAGE gels were used to clearly separate the LC3-I and LC3-II bands.
Results
Clinical report
The patient was a 12-year-old boy from healthy and unrelated parents. He was born at term from an uneventful pregnancy. Birth parameters were unknown. Neonatal period was unremarkable, but the mother reported that some minor foot deformities were noted at birth. He said first words at 12 months, but language development was delayed. For this, he requested speech therapy and psychomotricity since the 2 years of age, and started to say simple sentences at 8 years. At the time of examination, he still showed poor language and dysarthria. Autonomous walking appeared at 15 months. The patient suffered of moderate intellectual disability. Recurrent episodes of unexplained laughing were registered by the parents and were also appreciated at examination. Sleep problems were never reported. Primary dentition was normal, but only five permanent teeth were erupted at 12 years. Absence of multiple (>6) tooth gems was evident at orthopanoramic X-rays (oligodontia of the permanent dentition). Diabetes insipidus was first suspected at 4 months, but a final diagnosis of hypodipsia–hypernatremia syndrome was established at 10 years. Full endocrinological assessment excluded any other disorder of the hypothalamic–hypophyseal axis at 12 years. Febrile convulsions and infections of the upper airways recurred several times during infancy. At 10 years, the patient underwent bilateral orchidopexis.
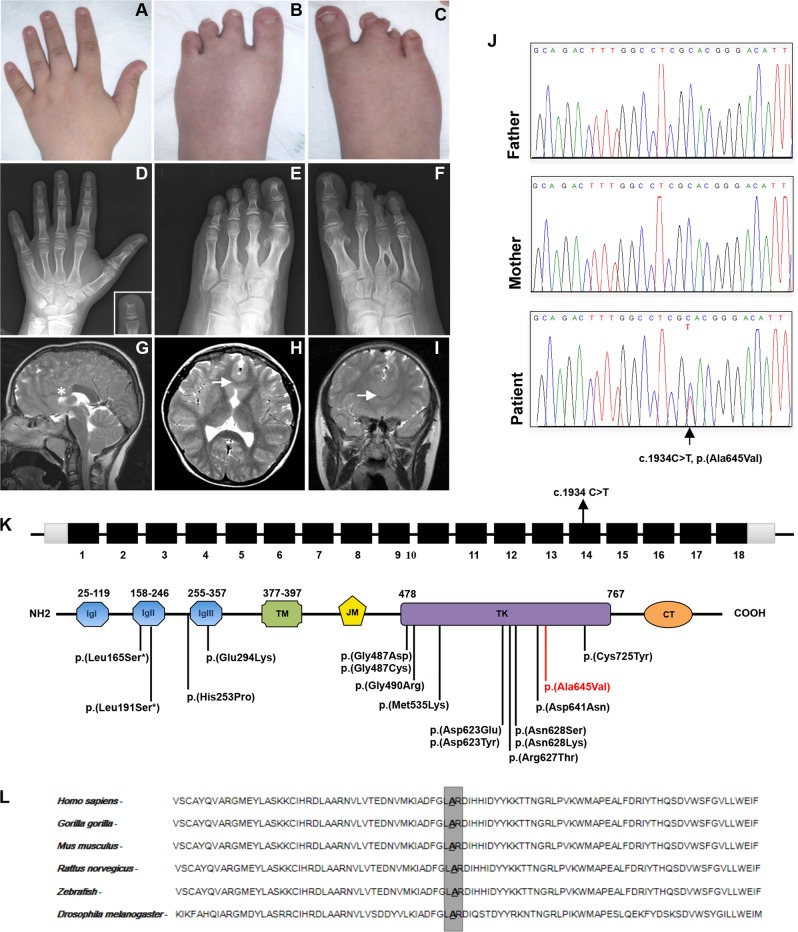
At examination, height was 138.6 cm (3rd–10th centile), weight 41 kg (25–50° centile), and head circumference 52 cm (10th centile). Facial gestalt was unremarkable. Oral cavity exploration showed retention of multiple primary teeth and amelogenesis imperfecta. Hard and soft palate was intact. Both hands had five rays. Fingers were short with broad tips and small nails (Fig. 1a). The left foot had four apparently well-formed, but slightly shortened toes, and partial cutaneous syndactyly of the second and third toes (Fig. 1b). Central toes of the right foot appeared rudimentary and partially fused (Fig. 1c). Testes and penis were small. Gait was ataxic with a broad base. Plain radiographs of hands and feet showed brachydactyly of hands with short and broad metacarpals, shortened middle phalanx of the fifth fingers, bifid appearance of the distal phalanx of the left third finger, four rays on the left foot, inverted “Y” synostosis of the third and fourth metatarsals and rudimentary bones of the third toe of the right foot (Fig. 1d–f). Brain magnetic resonance imaging (MRI) demonstrated agenesis of the anterior part of the corpus callosum and partial fusion of the frontal lobes, compatible with lobar holoprosencephaly (Fig. 1g–i). The suspicion of HS was put forward.
Fig. 1.
Clinical features and molecular findings. a Brachydactyly with broad fingertips and short nails. b Left foot with four toes, brachydactyly II–IV and partial cutaneous syndactyly II–III. c Right foot presenting cutaneous syndactyly and severe brachydactyly II–IV. d Radiograph of the left hand showing short metacarpals, shortened middle phalanx of the fifth finger, and bifid distal phalanx of the third finger. e Radiograph of the left foot with four rays, malformed metatarsal of the second toe, absence of the middle phalanx of the fifth toe, and partial synostosis of the middle and distal phalanges of the second toe. f Radiograph of the right foot with inverted “Y” synostosis of the third and fourth metatarsal, rudimentary bones of the third toe, and absence of the middle phalanx of the fifth toe. Brain MRI with absence of the anterior part of the corpus callosum on the sagittal scan (g; asterisk indicates the absence of the genu and rostrum of the corpus callosum), and partial fusion of the frontal lobes on axial (h) and coronal scans (i; arrows indicate midline fusion of the frontal lobes in both scans). j Sanger sequencing showing the c.1934C>T nucleotide change in FGFR1. k Diagram showing the structure of FGFR1 and the secondary structure of the protein. Coding regions are in black, UTR sequences are in gray, introns are not to scale. Previously identified and the novel variants associated with Hartsfield syndrome are located on the protein structure. The amino-acid change identified in this work is in red. CT C-terminal-tail, IgI–III, immunoglobulin-like domains, JM juxtamembrane domain, TD transmembrane domain, TK tyrosine kinase domain. l Conservation of the involved amino-acid (Ala645) among species (color figure online)
Identification of a novel missense FGFR1 variant
WES detected a novel heterozygous missense variant in exon 14 of FGFR1 (FGFR1: NM_023110.2: c.1934C>T), predicted to cause the p.(Ala645Val) amino-acid substitution (Fig. 1j, k). This result was confirmed by Sanger sequencing of proband’s and parents’ DNA. The variant originated de novo as it was not detected in both parents. The c.1934C>T variant is not reported in major databases, including EcAC and gnomAD and this suggests that the identified variant is a rare event. Variants identified in other genes associated with holoprosencephaly were filtered and excluded according to American College of Medical Genetics and Genomics guidelines (Table S2).
Affected residue is evolutionarily conserved (GERP++_RS score = 5.9; phyloP100way_vertebrate score = 10.003; phyloP100way_mammalian score = 0.902; phastCons100way_vertebrate score = 1,000; SiPhy_29way_logOdds = 20.270) and is located in the FGFR1 intracellular kinase domain (Fig. 1k, l) [2].
Ala645Val FGFR1 has dominant-negative effect
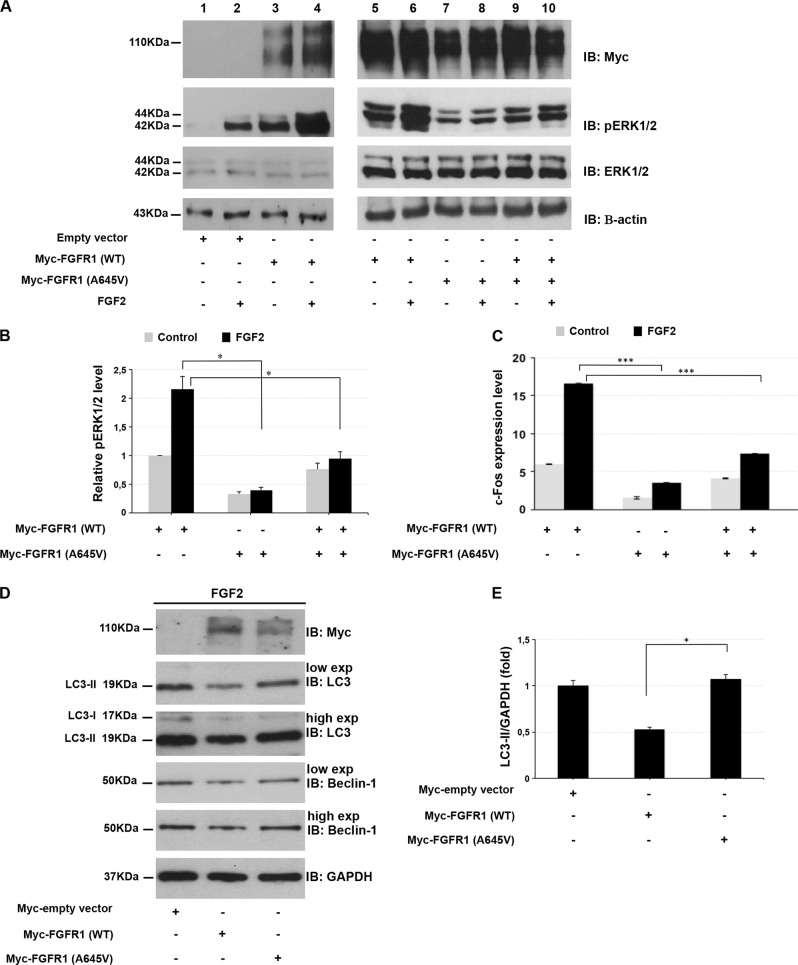
Under normal conditions, FGFR1 signaling is triggered by growth factors, such as FGF2, leading to receptor dimerization and transphosphorylation of FGFR1. Activated FGFR1 induces several downstream pathways including the RAS/ERK1/2 signaling, a serine/threonine-selective protein kinase involved in the regulation of fundamental biological processes such as proliferation, survival, and differentiation. In order to evaluate whether the Ala645Val variant affects the function of FGFR1, we measured the receptor activity in HEK293 cell lines exogenously expressing wild-type or Ala645Val FGFR1 by monitoring the phosphorylation level of endogenous ERK1/2. As expected, the phosphorylation of ERK1/2 induced by FGF2 in cells transfected with wild-type FGFR1-expressing vector (Fig. 2a, lane 6; Fig. 2b) was higher than in cells transfected with an empty vector and in non-treated cells (Fig. 2a, lanes 2 and 5, respectively; Fig. 2b). Interestingly, the FGF2-induced ERK1/2 phosphorylation in cells transfected with Ala645Val FGFR1-expressing plasmid (Fig. 2a, lane 8; Fig. 2b) was lower than in cells transfected with wild-type FGFR1 (Fig. 2a, lane 6; Fig. 2b). Indeed, the phosphorylation of ERK1/2 elicited by FGF2 was significantly reduced when Ala645Val FGFR1 was co-expressed with the wild-type FGFR1 (Fig. 2a, lane 10; Fig. 2b), compared with cells transfected with wild-type FGFR1 (Fig. 2a, lane 6; Fig. 2b). These data suggested that Ala645Val FGFR1 expression resulted in a receptor activity deregulation, probably acting in a dominant-negative manner. To better address whether Ala645Val FGFR1 variant affects the activity of the receptor, we profiled the endogenous expression level of c-Fos, a direct known transcriptional target of FGF2/FGFR1 axis [19], in HEK293 cell lines expressing wild-type or Ala645Val FGFR1 or wild-type plus Ala645Val FGFR1. We found that in response to FGF2 treatment, c-Fos expression in mutated FGFR1-expressing cells was significantly lower than in cells transfected with wild-type FGFR1 (Fig. 2c). A significant reduction of c-Fos transcriptional level was also detected in cells co-transfected with both wild-type and mutated FGFR1. Altogether, our data suggested that Ala645Val FGFR1 might interfere with the function of wild-type receptor in inducing ERK1/2 phosphorylation.
Fig. 2.
Ala645Val FGFR1 impairs ERK1/2 signaling pathway and autophagy flux. a HEK293 cells were transfected with empty vector (lanes 1, 2), wild-type FGFR1 (lanes 3–6) and Ala645Val FGFR1-expressing plasmids (lanes 7, 8) or wild-type plus Ala645Val FGFR1 plasmids (lanes 9, 10). At 24 h after transfection, cells were grown in serum-free medium for 24 h, and then incubated in the absence (lanes 1, 3, 5, 7, 9) or presence of 1 nM FGF2 (lanes 2, 4, 6, 8, 10) for 15 min. After incubation with FGF2, the phosphorylated ERK1/2 was analyzed by SDS-polyacrylamide gel electrophoresis using indicated antibodies. Actin was used as loading control. b Quantitative analysis of ERK1/2 phosphorylation in cells transfected with indicated vectors. Protein levels were quantified by densitometry. The relative ERK1/2 phosphorylation level in cells transfected with the vector expressing wild-type FGFR1 was set as 1. Bar represents the average of three independent experiments and scale bars represent standard errors. *P < 0.05. c qPCR was performed to measure the c-Fos endogenous expression in HEK293 cells transfected with indicated plasmids and cultured in the absence or presence of 0.5 nM FGF2 for 1 h. The relative c-Fos expression in cells transfected with the empty vector was set as 1. Scale bars represent standard errors. ***P < 0.01. d Whole protein lysates of HEK293 cells transfected with empty vector, wild-type or Ala645Val FGFR1-expressing plasmids and cultured in the absence or presence of 25 ng/ml FGF2 for 2 h, were separated on 12% SDS-gel and subjected to immunoblotting with LC3 and Beclin-1 antibodies (low and high exposure). The autophagy flux was monitored by the conversion of LC3-I to its lipidated form, LC3-II. GAPDH was used as loading control. The same lysates were separated on 7.5% SDS-gel for immunoblotting with anti-Myc. e Quantification of LC3-II levels. Graph shows averages calculated on two different experiments and scale bars represent standard errors. *P < 0.05
Ala645Val FGFR1 role in autophagy process
Recent findings provide a novel direct role of FGF2/FGFR1 signaling in the regulation of autophagy flux in cancer. In particular, FGF2/FGFR1 inhibition contributes to the induction of autophagy by Beclin-1 upregulation, which is mediated, at least in part, through inhibiting ERK1/2 pathway [18]. In this regard, to better explore the biological impact of the novel variant, we investigated whether Ala645Val FGFR1 affects autophagy flux by exploring its effect on the levels of the autophagosome-associated lipidated form of LC3 and Beclin-1, which are the major autophagy-related proteins, in FGF2-treated HEK293 cells. In our model, FGF2 stimulation resulted in an increase of LC3-II lipidated form (50%) and a slight enhancement of Beclin-1 (20%) in cells expressing Ala645Val FGFR1 in comparison with cells containing exogenous wild-type FGFR1 (Fig. 2d, e, S1). According to Yuan et al. [18], our preliminary finding suggests a functional implication of FGFR1 in the autophagic process.
Discussion
We report a further case of HS caused by a novel FGFR1 variant falling within the cytoplasmic tyrosine kinase domain. Compared with previously published patients, our case shows the typical features pathognomonic of HS [1].
To date, 20 HS individuals (including ours) from 17 families with variants in FGFR1 have been described [2, 3, 20–25] (Table S3). Biallelic variants compatible with autosomal recessive inheritance were demonstrated in two cases [2], whereas heterozygosity for a single variant is observed in the remaining. De novo/sporadic origin was registered in 13 of the 15 dominant families, whereas germinal mosaicism explained recurrence in one pedigree [20, 22]. The heterozygous c.1460G>A (p.(Gly487Asp)) variant inherited from an unaffected parent is described in two further sibs, in whom synergy (digenic inheritance?) with a concurrent FGF8 15-bp deletion transmitted by the other unaffected parent is proposed [3]. Fifteen FGFR1 variants were novel, while a single variant recurred in two families [3, 20]. The residues Gly487, Asp623, and Asn628 might be mutational hot-spots, as they were involved twice by different amino-acid changes. Twelve (75%) variants, including ours, affect the tyrosine kinase domain and four (25%) the Ig-like domains; among the latter, two fell in the Ig-like domain D2, one in the domain D3 and the remaining close to domain D3 (Fig. 1k). Available data do not allow any genotype–phenotype correlations. In particular, no significant difference is appreciated among individuals carrying variants within the cytoplasmic tyrosine kinase domain compared with those with variants in the extracellular Ig domains (Fig. 1k; Table S3). However, the two individuals with recessive variants were more severely affected and died at the age of 4–5 years [1]. It will be interesting to see if this clustering within the FGFR1 structure and the lack of genotype–phenotype correlation will persist in publications of other HS cases.
Our functional studies support a dominant-negative effect for Ala645Val FGFR1 variant. In fact, they highlighted that the phosphorylation status of ERK1/2 and the expression profile of c-Fos were significantly perturbed in FGF2-treated cells expressing Ala645Val FGFR1 or co-expressing Ala645Val FGFR1 with the wild-type one, in comparison with control cells. This suggests that an altered Ala645Val FGFR1 receptor activity is the possible molecular mechanism underlying HS in our case. Our findings are perfectly in line with an elegant study showing that the HS-specific FGFR1 variants, which cluster in the kinase domain, exhibit a dominant-negative function when modeled in zebrafish; a fact which well explains the craniofacial, brain, and limb anomalies typical of HS [3].
In our study, we also suggested, for the first time, that deregulation of autophagy might underlie a developmental disorder linked to FGFR1. Autophagy is a catabolic process that plays a fundamental part in tissue homeostasis [26]. Therefore, its involvement in the pathogenesis of different birth defects is probable. Accordingly, several studies have shown that FGF/FGFR signaling axis regulates autophagy and, in this way, plays a crucial role in cell differentiation, heart development, bone growth, and musculoskeletal system [27–30].
In a recent study on cancer, Yuan and co-authors showed that autophagy is induced in FGFR1-amplified non-small cell lung cancer cells after pharmacological or genetic inhibition of FGFR1 [18]. This effect is dependent on Beclin-1 through suppressing ERK/Mitogen-Activated Protein Kinase (MAPK) pathway. In line with these results, our preliminary functional studies in mammalian cell lines demonstrated that mutated Ala645Val FGFR1 overexpression resulted in an increase of Beclin-1 levels and autophagosome-associated lipidated form of LC3, when compared with wild-type FGFR1. These data allow us to hypothesize that key amino-acid changes in the tyrosine kinase domain of FGFR1 might deregulate autophagy and, thus, contribute to generate the developmental anomalies characterizing HS.
In conclusion, we identified a further HS case caused by a novel de novo variant, which expands the FGFR1 mutational spectrum and associates with a dominant-negative effect by in vitro studies. This is also the first report providing initial insights on autophagy as a molecular mechanism possibly underlying HS. If confirmed by further studies with different experimental approaches and patient cell lines, these data could open the path to novel research projects exploring tailored therapeutic approaches for HS.
Supplementary information
Acknowledgements
We thank the patient and his family for participation in this study. We acknowledge Professor Soo-Hyun Kim (University of London, UK) for providing myc-FGFR1 wild-type vector, and G. Zampi (IBBR, Naples, Italy) and M.P. Leone (Fondazione IRCCS-Casa Sollievo della Sofferenza, San Giovanni Rotondo, Italy) for technical assistance. This work was supported by Ricerca Corrente 2018 granted by the Italian Ministry of Health to LM, Ricerca Finalizzata 2011 granted by the Italian Ministry of Health to LM (GR2011–02349694) and M Carella (RF2011–02350693). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
PP, LM, and M Castori designed the study and wrote the manuscript. PP, M Carella, and TB performed exome sequencing and bioinformatic analysis. LM and GN carried out the functional assays. LM, RM, M Castori, and EDS interpreted functional data. M Castori, AP, and MCS provided clinical evaluation of the patient. All authors contributed to the writing and reviewing and approved the main manuscript text.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41431-019-0350-4) contains supplementary material, which is available to authorized users.
References
- 1.Dhamija R, Babovic-Vuksanovic D. Hartsfield syndrome. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, Amemiya A, editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2018 (last updated: 3 Mar 2016).
- 2.Simonis N, Migeotte I, Lambert N. FGFR1 mutations cause Hartsfield syndrome, the unique association of holoprosencephaly and ectrodactyly. J Med Genet. 2013;50:585–92. doi: 10.1136/jmedgenet-2013-101603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hong S, Hu P, Marino J, et al. Dominant-negative kinase domain mutations in FGFR1 can explain the clinical severity of Hartsfield syndrome. Hum Mol Genet. 2016;25:1912–22. doi: 10.1093/hmg/ddw064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ornitz DM, Itoh N. The fibroblast growth factor signaling pathway. Wiley Interdisciplinary Rev Dev Biol. 2015;4:215–66. doi: 10.1002/wdev.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teven CM, Farina EM, Rivas J, et al. Fibroblast growth factor (FGF) signaling in development and skeletal diseases. Genes Dis. 2014;1:199–213. doi: 10.1016/j.gendis.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dode C, Levilliers J, Dupont JM, et al. Loss- of- function mutations in FGFR1 cause autosomal dominant Kallmann syndrome. Nat Genet. 2013;33:463–5. doi: 10.1038/ng1122. [DOI] [PubMed] [Google Scholar]
- 7.Villanueva C, Jacobson-Dickman E, Xu C, et al. Congenital hypogonadism with split hand/foot malformation: a clinical entity with high frequency of FGFR1 mutations. Med Genet. 2015;17:651–9. doi: 10.1038/gim.2014.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tommiska J, Känsäkaski J, Christiansen P, et al. Genetics of congenital hypogonadotropic hypogonadism in Denmark. Eur J Med Genet. 2014;57:345–8. doi: 10.1016/j.ejmg.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 9.Hero M, Laitinen EM, Varimo T, et al. Childhood growth of females with Kallmann syndrome and FGFR1 mutations. Clin Endocrinol (Oxf) 2015;82:122–6. doi: 10.1111/cen.12504. [DOI] [PubMed] [Google Scholar]
- 10.Muenke M, Schell U, Hehr A, et al. A common mutation in the fibroblast growth factor receptor 1 gene in Pfeiffer syndrome. Nat Genet. 1994;8:269–74. doi: 10.1038/ng1194-269. [DOI] [PubMed] [Google Scholar]
- 11.White KE, Cabral JM, Davis SI, et al. Mutations that cause osteoglophonic dysplasia define novel roles for fgfr1 in bone elongation. Am J Human Genet. 2005;76:361–7. doi: 10.1086/427956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bennett JT, Tan TY, Alcantara D, et al. Mosaic activating mutations in fgfr1 cause encephalocraniocutaneous lipomatosis. Am J Human Genet. 2016;98:579–87. doi: 10.1016/j.ajhg.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brady N, Chuntova P, Bade LK, et al. The FGF/FGFR axis as a therapeutic target in breast cancer. Expert Rev Endocrinol Metab. 2013;110:e29–39. doi: 10.1161/CIRCRESAHA.111.255950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo H, Zheng R, Zhao Y, et al. A dominant negative FGFR1 mutation identified in a Kallmann syndrome patient. Gene. 2017;621:1–4. doi: 10.1016/j.gene.2017.04.017. [DOI] [PubMed] [Google Scholar]
- 15.Lopez ME, Korc M. A novel type I fibroblast growth factor receptor activates mitogenic signaling in the absence of detectable tyrosine phosphorylation of FRS2. J Biol Chem. 2000;275:15933–9. doi: 10.1074/jbc.M909299199. [DOI] [PubMed] [Google Scholar]
- 16.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–86. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 17.Micale L, Loviglio MN, Manzoni M, et al. A fish-specific transposable element shapes the repertoire of p53 target genes in zebrafish. PLoS ONE. 2012;7:e46642. doi: 10.1371/journal.pone.0046642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan H, Li ZM, Shao J, et al. FGF2/FGFR1 regulates autophagy in FGFR1-amplified non-small cell lung cancer cells. J Exp Clin Cancer Res. 2017;36:72. doi: 10.1186/s13046-017-0534-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weekes D, Kashima TG, Zandueta C, et al. Regulation of osteosarcoma cell lung metastasis by the c-Fos/AP-1 target FGFR1. Oncogene. 2016;35:2852–61. doi: 10.1038/onc.2015.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dhamija R, Kirmani S, Wang X, et al. Novel de novo heterozygous FGFR1 mutation in two siblings with Hartsfield syndrome: a case of gonadal mosaicism. Am J Human Genet. 2014;164A:2356–9. doi: 10.1002/ajmg.a.36621. [DOI] [PubMed] [Google Scholar]
- 21.Prasad R, Brewer C, Burren CP. Hartsfield syndrome associated with a novel heterozygous missense mutation in FGFR1 and incorporating tumoral calcinosis. Am J Human Genet. 2016;170:2222–5. doi: 10.1002/ajmg.a.37731. [DOI] [PubMed] [Google Scholar]
- 22.Shi Y, Dhamija R, Wren C, et al. Detection of gonadal mosaicism in Hartsfield syndrome by next generation sequencing. Am J Human Genet. 2016;170:3359. doi: 10.1002/ajmg.a.37869. [DOI] [PubMed] [Google Scholar]
- 23.Takagi M, Miyoshi T, Nagashima Y, et al. Novel heterozygous mutation in the extracellular domain of FGFR1 associated with Hartsfield syndrome. Hum Genome Var. 2016;3:16034. doi: 10.1038/hgv.2016.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lansdon LA, Bernabe HV, Nidey N, et al. The use of variant maps to explore domain-specific mutations of FGFR1. J Dent Res. 2017;96:1339–45. doi: 10.1177/0022034517726496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oliver JD, Menapace DC, Cofer SA. Otorhinolaryngologic manifestations of Hartsfield syndrome: case series and review of literature. Int J Pediatr Otorhinolaryngol. 2017;98:4–8. doi: 10.1016/j.ijporl.2017.04.035. [DOI] [PubMed] [Google Scholar]
- 26.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–32. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 27.Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368:651–62. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- 28.Cinque L, Forrester A, Bartolomeo R, et al. FGF signalling regulates bone growth through autophagy. Nature. 2015;528:272–5. doi: 10.1038/nature16063. [DOI] [PubMed] [Google Scholar]
- 29.Wang Z, Huang J, Zhou S, et al. Loss of Fgfr1 in chondrocytes inhibits osteoarthritis by promoting autophagic activity in temporomandibular joint. J Biol Chem. 2018;293:8761–74. doi: 10.1074/jbc.RA118.002293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J, Liu J, Huang Y, et al. FRS2alpha-mediated FGF signals suppress premature differentiation of cardiac stem cells through regulating autophagy activity. Circ Res. 2012;110:e29–e39. doi: 10.1161/CIRCRESAHA.111.255950. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.