Abstract
Background
The molecular genetic classification of gliomas, particularly the identification of isocitrate dehydrogenase (IDH) mutations, is critical for clinical and surgical decision-making. Raman spectroscopy probes the unique molecular vibrations of a sample to accurately characterize its molecular composition. No sample processing is required allowing for rapid analysis of tissue. The aim of this study was to evaluate the ability of Raman spectroscopy to rapidly identify the common molecular genetic subtypes of diffuse glioma in the neurosurgical setting using fresh biopsy tissue. In addition, classification models were built using cryosections, formalin-fixed paraffin-embedded (FFPE) sections and LN-18 (IDH-mutated and wild-type parental cell) glioma cell lines.
Methods
Fresh tissue, straight from neurosurgical theatres, underwent Raman analysis and classification into astrocytoma, IDH-wild-type; astrocytoma, IDH-mutant; or oligodendroglioma. The genetic subtype was confirmed on a parallel section using immunohistochemistry and targeted genetic sequencing.
Results
Fresh tissue samples from 62 patients were collected (36 astrocytoma, IDH-wild-type; 21 astrocytoma, IDH-mutated; 5 oligodendroglioma). A principal component analysis fed linear discriminant analysis classification model demonstrated 79%–94% sensitivity and 90%–100% specificity for predicting the 3 glioma genetic subtypes. For the prediction of IDH mutation alone, the model gave 91% sensitivity and 95% specificity. Seventy-nine cryosections, 120 FFPE samples, and LN18 cells were also successfully classified. Meantime for Raman data collection was 9.5 min in the fresh tissue samples, with the process from intraoperative biopsy to genetic classification taking under 15 min.
Conclusion
These data demonstrate that Raman spectroscopy can be used for the rapid, intraoperative, classification of gliomas into common genetic subtypes.
Keywords: brain tumor, intraoperative, isocitrate hydrogenase, Raman spectroscopy, personalized medicine
Key points.
Raman spectroscopy can be used for rapid, intraoperative, classification of gliomas.
Simple intraoperative protocol with results in under 15 min.
Accurate genetic classification possible with cryosections and FFPE sections.
Importance of the Study.
In this era of personalized medicine and the development of treatments targeting specific glioma genetic subtypes, it will become increasingly important for surgeons to be aware of the genetics of a tumor at the time of operation to inform their surgical strategy and to balance the survival benefit of increasing surgical resection with the risk of causing neurological deficit. In this study, we demonstrate that Raman spectroscopy can accurately and rapidly classify gliomas according to isocitrate dehydrogenase mutations and 1p/19q-codeletion. The protocol can be carried out within a neurosurgical theatre without the need for laboratory input and results are available in under 15 min.
Diffuse gliomas are the most common and aggressive primary tumors of the central nervous system in adults. The molecular genetic stratification of gliomas is important as more evidence emerges of the predictive and prognostic implications of different genetic subtypes. Surgical resection plays a key role in the management of patients with gliomas, and there is increasing evidence that the greater the extent of surgical resection, the greater the patient’s progression-free and overall survival.1–3 The diffuse nature of these tumors means that the surgeon can never achieve complete tumor resection. At present, it has been demonstrated that the clinical benefit of the extent of tumor resection does depend on the specific genetic subtypes.2 Information on the tumor genetic subtype at the time of surgery, providing real-time predictive and prognostic information, would allow the surgeon to tailor their surgical strategy to each patient’s genetic profile.
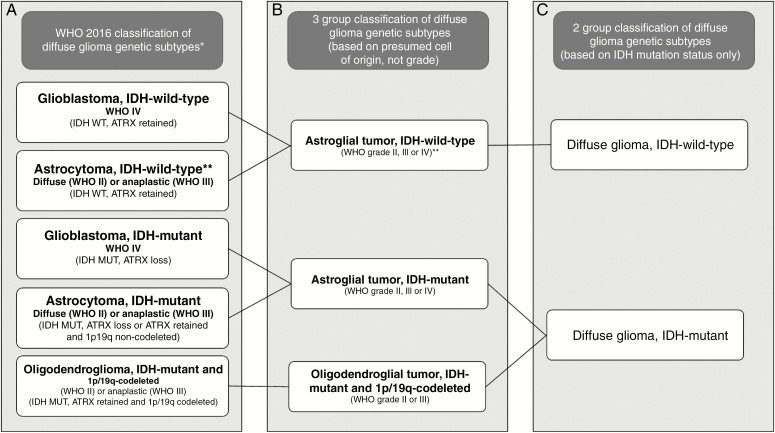
The 2016 World Health Organization (WHO) classification of gliomas introduced molecular and genetic subtyping integrated with the histological glioma type and grade4 (Figure 1). Common and clinically relevant genetic subtypes include gliomas with mutations of the isocitrate dehydrogenase (IDH) genes 1 or 2 with or without chromosome 1p and 19q (1p/19q) codeletion.5 Within most neuropathology laboratories, 1p/19q-codeletion is established using the cytogenetic technique of fluorescence in-situ hybridization (FISH). The presence of the most common IDH mutation (IDH1 R132H) can be established using immunohistochemistry (IHC) with an antibody highly sensitive and specific for this IDH mutation,6 which accounts for around 85%–90% of the mutations in gliomas.7 If there is suspicion of a rare IDH mutation, the sample would undergo targeted IDH-1/IDH-2 genetic sequencing.
Figure 1.
(A) The World Health Organization (WHO) 2016 classification of diffuse gliomas; (B) genetic subtypes for 3-group model; (C) genetic subtypes for 2-group model. (*not all subtypes are shown, only common groups; ** these are provisional categories in the classification: it is now recognized that most of these neoplasms are incipient or undersampled glioblastomas, isocitrate dehydrogenase-wild-type11).
Raman spectroscopy is a vibrational spectroscopy technique that probes the unique molecular vibrations of a sample to characterize its molecular composition.8 The significant advantage of Raman analysis is that no or minimal sample processing is required, allowing for rapid and cost-effective analysis of tissue without the need for specialist staff, time-consuming laboratory protocols, or expensive consumables.
The main aim of this study was to evaluate the ability of Raman spectroscopy to identify the most common molecular genetic subtypes of diffuse glioma in the neurosurgical theatre setting. An important objective was to develop a simple protocol, which would allow the surgeon or scrub staff to undertake Raman analysis of tissue in the operating theatre without the need for specialized technicians or laboratories. We have also evaluated the application of Raman analysis in the neuropathological diagnostic workflow of glioma tissue, including analysis of cryosections, formalin-fixed paraffin-embedded (FFPE) sections, and glioma cell lines.
Materials and Methods
Study Participants
Ethical approval for the use of fresh, cryosection, and FFPE tissue was obtained through the Oxford Brain Bank generic Research Ethics Committee approval (15/SC/0639). All tissue used was deemed surplus to diagnostic requirements.
Pathological Diagnosis
All cases were reviewed by a consultant neuropathologist, and diagnoses were established using the European Association for Neuro-Oncology guidelines,9 which incorporates the updated (2016) WHO classification of tumors of the central nervous system.4 See Supplementary material for details of histopathological diagnostic protocols. The WHO integrated diagnosis was based on the IHC/genetic results and histological grade (Figure 1), giving following 5 possible diagnostic groups:
diffuse or anaplastic astrocytoma, IDH-mutant (grade II or III);
glioblastoma, IDH-mutant (grade IV);
oligodendroglioma or anaplastic oligodendroglioma, IDH-mutant, and 1p/19q-codeleted (grade II or III);
diffuse or anaplastic astrocytoma, IDH-wild-type (grade II or III);
glioblastoma, IDH-wild-type (grade IV).
Tissue Processing
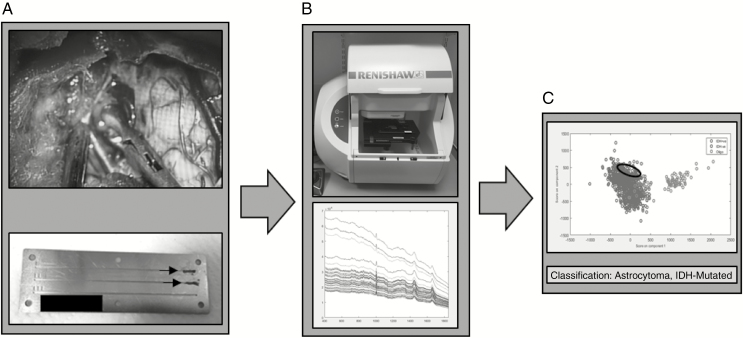
For fresh tissue samples, 1 biopsy per patient was taken by the neurosurgeon from an area of tumor bulk defined both visually and using the intraoperative neuronavigation system (Brainlab, Germany). The biopsy was immediately transferred to the neuropathologist, who cut the tissue biopsy into 2 pieces: 1 for intraoperative smear evaluation, and the adjacent piece for Raman analysis. The portion for Raman analysis measured approximately 3 mm3. The sample was immediately transferred to a grooved stainless steel slide10 (Figure 2) and compressed into the groove using a surgical scalpel creating a relatively flat surface onto which the Raman laser could be focused and compacting the tissue to limit the effect of tissue drying. The adjacent biopsy underwent intraoperative smear preparation with hematoxylin and eosin (H&E) and was reviewed by the consultant neuropathologist to confirm the presence of tumor.
Figure 2.
Fresh tissue workflow. (A) Intraoperative fresh tissue biopsy squashed into grooved stainless steel slide (arrows). (B) Slide placed in Raman spectrometer and multiple spectra acquired. (C) Spectra classified into genetic subtype using principal component analysis fed linear discriminant analysis model.
Details of cryosection, FFPE, and LN18 cell line preparation can be found in the Supplementary material. Two LN18 IDH-mutant and 2 IDH-wild-type cell lines were cultured and processed separately at 2 different time points. Cell line batches, once harvested, were processed as either cryosections or FFPE sections, with protocols to match the respective tissue sample preparations.
Raman Spectroscopy
Raman spectra were collected using a Renishaw bench-top RA800 series spectrometer (Renishaw plc., UK). For fresh tissue samples, single point spectra were obtained, widely distributed across the smeared sample, aiming for at least 100 spectra per fresh tissue sample. Two accumulations of either 0.5 s or 1 s depending on the amount of background fluorescence and resulting detector saturation for that sample. The total time to acquire data from each area was taken from the spectrometer log. The time from surgical biopsy to completion of spectral collection was also recorded. See Supplementary material for further details.
Glioma Genetic Subtype Models
For each tissue preparation type, 2 separate models were built. The first was a 2-group model, stratified based on the IDH mutation alone (Figure 1C):
IDH-mutant (including all IDH-mutant astrocytomas, glioblastomas, and oligodendrogliomas);
IDH-wild-type (including all IDH-wild-type diffuse astrocytomas and glioblastomas).
The second model was a 3-group model, stratifying gliomas into the 3 main genetic subtypes (Figure 1B) regardless of histological grade:
astrocytomas, IDH-mutant (astrocytoma [grade II or III] and glioblastoma [grade IV]);
astrocytomas, IDH-wild-type (astrocytoma [grade II or III] and glioblastoma [grade IV]);
oligodendrogliomas, IDH-mutant, and 1p/19q-codeleted (grade II or III).
All astrocytomas, IDH-wild-type were glioblastomas (grade IV)—there were no grade II or III astrocytomas, IDH-wild-type tumors; these are often undersampled glioblastomas.11
Statistical Analysis and Model Building
All data analysis was carried out using MATLAB R2015b (MathsWorks). For each genetic subtype, normalized (using Extended Multiplicative Scatter Correction12) mean spectra and the negative of the second derivative13 were plotted against the wave number shift. Statistical differences between peak intensities were evaluated using the Wilcoxon rank-sum test, and the magnitude of the resulting P value was used as an indicator of the magnitude of the difference between the intensities. Tentative molecular assignment of the identified Raman peaks was carried out based on the published literature.14–16 See Supplementary material for further details of spectral preprocessing and analysis.
Principal component analysis (PCA) fed linear discriminant analysis (LDA) was used to build the pathology classification models, which used all the spectra, after preprocessing, from each patient. In this classification method, PCA is used to reduce the data dimensionality, with the analysis of variance used to select only those PCs that describe a statistically significant difference in the data. An LDA model is then built from those significant PCs. Model performance was assessed using leave-one-patient-out cross-validation to generate model sensitivity and specificity (Supplementary material). Multi-class receiver operating characteristic (ROC) curves were generated with the area under ROC curve (AUROC) calculated as a measure of accuracy.
Results
A total of 62 fresh tissue samples were included in the study. No cases were excluded on the grounds of the absence of tumor on the adjacent H&E analysis of the smeared section. Ninety-seven cryosections and 132 FFPE sections were cut for analysis. Of these, 18 cryosections and 12 FFPE sections were excluded after review of the parallel H&E section showed no evidence of tumor. A total of 79 cryosections and 120 FFPE sections were used for analysis. Table 1 shows the number of samples analyzed in each genetic subgroup for each sample preparation method.
Table 1.
Number of Each Genetic Subtype Included for Fresh Tissue, Cryosections and Formalin-Fixed Paraffin-Embedded (FFPE) Sections, Subdivided into 3-Group Model and 2-Group Model
| Genetic subtypes (WHO grade) | Tissue preparation | |||||
|---|---|---|---|---|---|---|
| Fresh tissue | Cryosections | FFPE sections | ||||
| 3-grp model | 2-grp model | 3-grp model | 2-grp model | 3-grp model | 2-grp model | |
| Astrocytoma, IDH-wild-type (All WHO IV (GB)) | 36 | 36 | 19 | 19 | 41 | 41 |
| Astrocytoma, IDH-mutant (WHO II/WHO III/WHO IV (GB)) | 21 (4/7/10) | 26 | 41 (12/10/19) | 60 | 51 (8/12/31) | 79 |
| Oligodendroglioma (WHO II/WHO III) | 5 (3/2) | 19 (8/11) | 28 (12/16) | |||
| Total | 62 | 79 | 120 |
GB = glioblastoma; grp = group; IDH = isocitrate dehydrogenase; WHO = World Health Organization.
A total of 9799 spectra were collected in the fresh tissue group, with a mean of 161 spectra per sample. The mean time for data collection was 6 min and 6 s per sample, and the whole process from intraoperative biopsy to completion of Raman spectroscopy was under 15 min in every case. See Supplementary material for cryosection and FFPE details).
Classification Model Performance
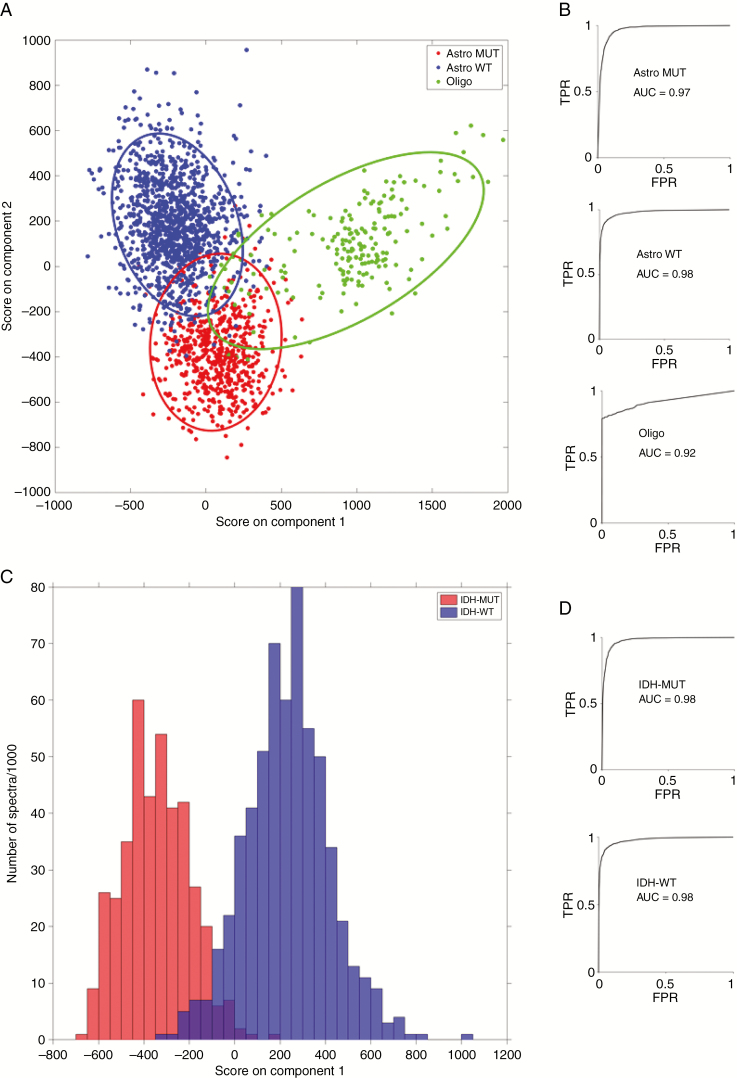
Table 2 summarizes the sensitivities and specificities for 3-group and 2-group prediction of glioma genetic subtypes using PCA-LDA classification modeling of the Raman spectra with leave-one-patient-out cross-validation. With fresh tissue samples, the models were able to classify tumors into either IDH-mutant or IDH-wild-type (2-group model) with 0.91–0.95 sensitivity and specificity and 0.98 accuracy. Fresh tissue classification into the 3 genetic subtypes (3-group model) was performed with 0.79–0.94 sensitivity, 0.89–1.00 specificity, and an accuracy of between 0.92 and 0.98 (Figure 3). Cryosection and FFPE section models classified with slightly less sensitivity and specificity than the fresh tissue models as shown in Table 2. The overall accuracy of these models remained high, with cryosection 3-group classification accuracy of 0.89–0.93, cryosection 2-group classification accuracy of 0.93 (Supplementary Figure 4), FFPE section 3-group classification accuracy of 0.87–0.93, and FFPE section 2-group classification accuracy of 0.82 (Supplementary Figure 5). LDA plots for the 3-group and 2-group models show good separation of the genetic subtypes using fresh tissue (Figure 3), cryosections (Supplementary Figure 4), and FFPE sections (Supplementary Figure 5).
Table 2.
Sensitivity and Specificity for Predicting Genetic Subtypes Using PC-Linear Discriminant Analysis (LDA) 3-Group and 2-Group Models for Fresh Tissue Samples, Cryosections, and Formalin-Fixed Paraffin-Embedded (FFPE) Sections
| Model | Genetic subtypes | Fresh tissue | Cryosections | FFPE sections | |||
|---|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Sensitivity | Specificity | Sensitivity | Specificity | ||
| 3-group | Astrocytoma, IDH-wild-type | 0.94 | 0.90 | 0.78 | 0.85 | 0.81 | 0.84 |
| Astrocytoma, IDH-mutant | 0.91 | 0.95 | 0.79 | 0.89 | 0.72 | 0.87 | |
| Oligodendroglioma | 0.79 | 1.00 | 0.74 | 0.90 | 0.79 | 0.93 | |
| 2-group | IDH-wild-type | 0.95 | 0.91 | 0.88 | 0.83 | 0.77 | 0.71 |
| IDH-mutant | 0.91 | 0.95 | 0.83 | 0.88 | 0.71 | 0.77 |
Figure 3.
Fresh tissue linear discriminant analysis score plots and receiver operating characteristic curves for 3-group model (A and B) and 2-group model (C and D). (TPR = true positive rate; FPR = false positive rate; Astro MUT = astroglial tumor isocitrate dehydrogenase [IDH]-mutant; Astro WT = astroglial tumor, IDH-wild-type; Oligo = Oligodendroglioma).
Mean Spectra and Peak Assignment
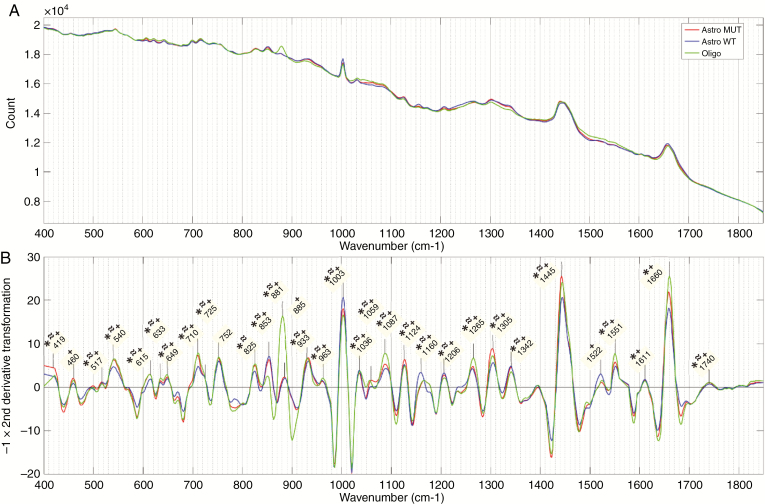
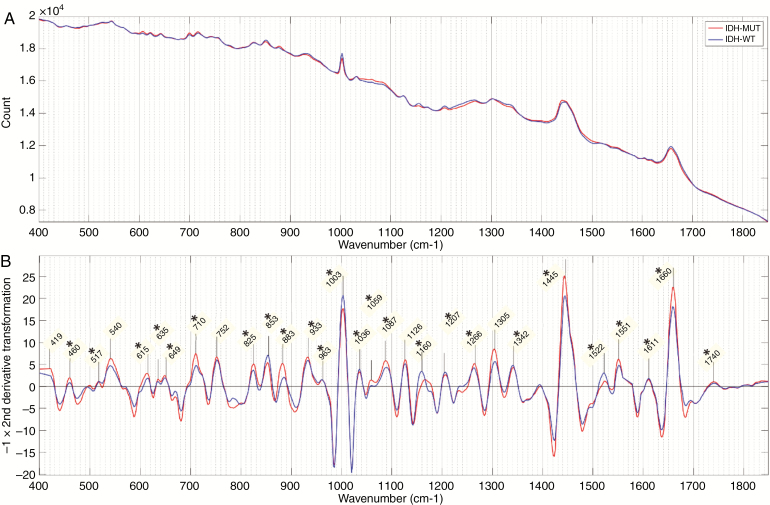
For the fresh tissue samples, mean spectra and the negative of the second derivative of the mean spectra for each genetic subgroup in the 3-group model are shown in Figure 4. Similar figures for cryosection and FFPE sections are shown in Supplementary Figure 1. For the 2-group model, mean spectra and the negative of the second derivative of the mean spectra are shown in Figure 5 for the fresh tissue samples and Supplementary Figure 2 for cryosections and FFPE sections. On all second-derivative plots, the wave numbers of prominent Raman peaks are labeled and the statistical significance of the difference between the peak intensities of each genetic subtype in that model is indicated (P < .001, Wilcoxon rank-sum test).
Figure 4.
Fresh tissue 3-group model. (A) Mean spectra and (B) negative of second derivative transformation for each genetic subtype. Prominent Raman peaks labeled in panel B. Statistical significance between peak intensities of each genetic subtype in the model is indicated (P < .01) for *isocitrate dehydrogenase (IDH)-mutant versus IDH-wild-type; ≈Astro MUT versus Oligo; and ⁺Astro wild-type (WT) versus Oligo. (Astro MUT = astroglial tumor, IDH-mutant; Astro WT = astroglial tumor, IDH-wild-type; Oligo = Oligodendroglioma).
Figure 5.
Fresh tissue 2-group model. (A) Mean spectra and (B) negative of the second derivative transformation of mean spectra for each genetic subtype. Prominent Raman peaks labeled in panel B. Statistical significance between peak intensities of each genetic subtype is indicated (P < .01) for *isocitrate dehydrogenase (IDH)-mutant versus IDH-wild-type. (IDH-MUT = IDH-mutant; IDH-WT = IDH-wild-type).
The most dominant Raman peaks in the fresh tissue sample are seen at 1445 cm–1 and 1660 cm–1. These are both a summation of multiple overlapping peaks and represent the ratio of protein and lipid content within the sample. There is also a characteristically narrow peak at 1003 cm–1, which, along with peaks at 615 cm–1 and 1036 cm–1, represent phenylalanine. Other amino acid peaks include 460 cm–1 (tryptophan/tyrosine), 635 cm–1 and 649 cm–1 (tyrosine), and 540 cm–1 (cysteine). Various peaks were assigned to classes of lipids including triglycerides/fatty acids (853 cm–1, 1059 cm–1, 1087 cm–1, 1126 cm–1, 1160 cm–1, 1266 cm–1, 1305 cm–1, 1445 cm–1, 1740 cm–1) and cholesterol (419 cm–1, 825 cm–1). Peaks assigned to proteins include 1522 cm–1 and 1551 cm–1 (amide III) and 1660 cm–1 (amide I). DNA peaks were assigned at 517 cm–1, 883 cm–1, 963 cm–1, 1207 cm–1, and 1342 cm–1. Peaks at 710 cm–1 and 933 cm–1 most likely represent glycogen.
The majority of identified peaks exhibited significant differences in intensity between the various genetic subtypes. When comparing IDH-mutant with IDH-wild-type intensities, in both the 2- and 3-group fresh tissue models, the most significant differences (lowest P values) were found in peaks representing lipid (853 cm–1, 1059 cm–1, 1087 cm–1, 1266 cm–1, 1445 cm–1, and 1660 cm–1), phenylalanine (1003 cm–1), and DNA (1207 cm–1 and 1342 cm–1). The differences observed between astrocytomas, IDH-mutant, and oligodendroglioma were very similar to those observed between astrocytomas, IDH-wild-type, and oligodendroglioma with oligodendroglioma intensities having the most significant differences at peaks assigned to phenylalanine (1003 cm–1 and 1036 cm–1), DNA (881 cm–1, 885 cm–1, and 1342 cm-–1), and protein (1522 cm–1 and 1551 cm–1). The DNA peaks around 800–900 cm–1 are the most prominent of these. In general, the peak differences between oligodendroglioma and IDH-wild-type astrocytomas were more pronounced than between oligodendroglioma and IDH-mutant astrocytomas.
There is consistency between the dominant Raman peaks across the different tissue preparations. Depending on the model, between 12 and 17 peaks of the 43 peaks identified are present in all 3 tissue preparations. Of these, 11 peaks are also present in both LN18 preparations (cryosections and FFPE sections). In general, cryosection and FFPE section spectra have more prominent DNA and amino acid peaks and less contribution from lipid and protein peaks. The cryosection spectra have a significant background in the 1200–1500 cm–1 region, which is thought to be due to water condensation on the objective lens as the cryosection dries out. There is also a strong contribution from a prominent peak at 1336 cm–1 (assigned to protein:lipid ratios) in this wave number region, which is not seen in the fresh tissue spectra. In terms of differentiating between genetic subtypes, peaks that consistently give the most significant differences between subtypes across the 3 tissue preparations are those assigned to tyrosine (635 and 649 cm–1), phenylalanine (1003 cm–1 and 1036 cm–1), lipid (825 cm–1 and 853 cm–1), protein (1551 cm–1), and lipid:protein ratios (1445 cm–1:1551 cm–1). In the 3-group models including oligodendroglioma, peaks for DNA (725 cm–1, 963 cm–1, and 1206 cm–1) are also consistent across all sample preparations.
Rare IDH
A total of 13 rare IDH mutations were present overall, in 3 fresh tissue samples, 6 cryosections, and 4 FFPE sections (Table 3). All cases were correctly classified into the IDH-mutated group (using the 2-group model), with the highest probability of correct classification occurring in the fresh tissue samples, with a mean probability of 91% for predicting IDH mutant.
Table 3.
Rare Isocitrate Dehydrogenase (IDH) Mutations and Probability of Predicting IDH Mutation with Raman Spectroscopy (GB = glioblastoma)
| Tissue preparation | Histology/grade | Rare IDH mutation confirmed with targeted genetic sequencing | Probability of predicting IDH mutation with Raman model |
|---|---|---|---|
| Fresh tissue | Astrocytoma (GB) IV | IDH1 394T | 0.95 |
| Astrocytoma (GB) IV | IDH2 G515T | 0.92 | |
| Astrocytoma II | IDH2 G515A | 0.87 | |
| Cryosections | Astrocytoma II | IDH1 R132C | 0.92 |
| Astrocytoma II | IDH1 R132C | 0.96 | |
| Astrocytoma (GB) IV | IDH1 R394T | 0.61 | |
| Oligodengroglioma II | IDH2 R172K | 0.93 | |
| Oligodengroglioma II | IDH2 R172K | 0.78 | |
| Astrocytoma III | IDH1 R132C | 0.77 | |
| FFPE sections | Astrocytoma (GB) IV | IDH1 R132C | 0.57 |
| Astrocytoma (GB) IV | IDH1 R132G | 0.82 | |
| Oligodengroglioma II | IDH2 R172K | 0.82 | |
| Oligodengroglioma III | IDH2 R172W | 0.89 |
LN18 Cells
All LN18 IDH-mutant cell lines stained strongly for R132H IDH mutation on IHC. Mean spectra and the negative of the second derivative are shown in Supplementary Figure 3. The mean spectra for the LN18 cells and key Raman peaks, which differentiate genetic subtypes correlate with the tissue preparation spectra. Table 4 gives the sensitivity, specificity, and accuracy (AUROC) using PCA-LDA for each mutant and wild-type cell lines processed as cryosections or FFPE sections.
Table 4.
LN18 Cell Lines. Sensitivity and Specificity for Predicting Isocitrate Dehydrogenase (IDH) Mutation Using PC-Linear Discriminant Analysis for Different Sample Preparation Methods Using Spectra From Individual Cell Line Batches and Combined Batches
| Preparation method, cell line | Sensitivity | Specificity | Accuracy |
|---|---|---|---|
| Cryosection, 1 | 0.85 | 0.70 | 0.82 |
| Cryosection, 2 | 0.91 | 0.99 | 0.98 |
| Cryosection, combined | 0.78 | 0.55 | 0.66 |
| FFPE, 1 | 0.99 | 0.99 | 0.99 |
| FFPE, 2 | 0.83 | 0.85 | 0.92 |
| FFPE combined | 0.95 | 0.89 | 0.97 |
FFPE = formalin-fixed paraffin-embedded.
Discussion
We have demonstrated that Raman spectroscopy can determine the most common genetic subtypes of adult diffuse gliomas using fresh tissue straight from the operating theatre, cryosections, and FFPE sections. Most significantly, our fresh tissue workflow requires no sample processing, and results are available in under 15 min, making this technique ideal for rapid intraoperative diagnosis.
Surgery plays an important role in the treatment of gliomas. There is mounting evidence that the greater the extent of surgical resection the longer the progression-free and overall survival.1–3 The aim of surgery is to remove all the contrast-enhancing tumor in high-grade tumors or all the T2-FLAIR signal in low-grade gliomas, but, due to their diffuse nature, it is known that tumor cells will have infiltrated beyond these boundaries. Removal of the brain surrounding the tumor may result in neurological disability for the patient. The crucial balance for the surgeon is to maximize resection of the tumor whereas minimizing the risk of disability. Surgeons vary widely as to how aggressively they pursue tumor resection and the techniques they use (such as awake surgery and white matter tractography) to define this tumor:functional boundary. This is reflected in a wide range of 15%–70% complete resection rates reported in the literature.2,17 In this era of personalized medicine, the ability to perform accurate genetic subtyping of gliomas at the time of surgery will allow the surgeon to adapt their surgical strategy with some knowledge of the likely prognostic and treatment potential for the lesions. In this study, we have shown that Raman spectroscopy can accurately identify the 3 most common and clinically relevant glioma groups. The presence of an IDH mutation is a strong predictor of longer progression-free survival18–20 and overall survival18–25 when compared with IDH-wild-type tumors. 1p/19q-codeletion in the context of an IDH mutation is the genetic hallmark of oligodendrogliomas26 and is a strong favorable prognostic factor for overall survival20,23,27 and a strong predictor of response to radiochemotherapy.28–31 More relevant to intraoperative decision-making, Wijnenga et al.2 examined the impact on overall survival of the extent of surgical resection, measured as volume of residual tumor seen on postoperative contrast magnetic resonance imaging, in molecularly stratified low grade gliomas (WHO grade II astrocytomas and oligodendrogliomas). They reported that the effect of less tumor residual volume on overall survival was much greater in IDH-mutant astrocytomas compared with oligodendrogliomas. In the IDH-mutant astrocytoma group, even a small amount of residual tumor of 0.1 cm3 affected negatively on survival compared with 0 cm3 residual. This was not seen in the oligodendroglioma group, with a small residual having no significant effect on overall survival. Similar finding was reported by Kawaguchi et al.32 who found a significant survival advantage of gross total resection over subtotal resection in IDH-mutant astrocytoma but no significant survival advantage for oligodendroglioma or IDH-wild-type astrocytoma. Consequently, pushing the boundaries of surgical resection when there is a possible increased risk of disability might be in the patient’s best interest if they have an IDH-mutant astrocytoma but would not be in their interest if they have an oligodendroglioma: intraoperative information of the glioma genetic subtyping is therefore essential. As we learn more about glioma subtypes and develop novel therapies tailored for their treatment, including the insertion of therapy agents at the time of surgery, the need for accurate genetic subtype information at the time of surgery will increase. We, and others, have shown that preoperative non-invasive magnetic resonance spectroscopy at 3- or 7-Tesla may provide the neurosurgeon with information on IDH status of diffuse gliomas33; however, to the best of our knowledge, this currently does not allow distinction between IDH-mutant astrocytomas and oligodendrogliomas.
One aim of our study was to build up the training dataset for model development and therefore true intraoperative classification was not possible until enough data had been acquired. Once a classification model was developed, samples could be analyzed and classified immediately intraoperatively. This was piloted in the last 5 cases collected and yielding similar classification performances to the results presented. The major advantage of Raman spectroscopy compared with conventional techniques is the lack of sample preparation required. We developed a protocol that could easily be replicated by anyone in the operating theatre. The fresh tissue samples simply require the biopsy to be smeared into a grooved slide and then placed on the Raman microscope stage (Figure 2). In our study protocol, the sample was split into 2 sections by the neuropathologist before Raman analysis, as adjacent intraoperative or permanent tissue staining was essential to establish the quality of the biopsy and tumor content, and spectral collection was supervised by a researcher. These steps would not be required as part of a Raman diagnostic pathway, particularly with the straightforward automation of the process, which, at the press of a button, would perform spectrometer quality control, laser autofocusing, spectral acquisition, preprocessing, and classification. This sets Raman spectroscopy apart from other techniques that have been used to acquire intraoperative genetic subtyping, such as polymerase chain reaction-based rapid genotyping assays.34,35 More traditional techniques can give genetic stratification within the timeframe of an operation and are more accurate than Raman spectroscopy, but they all require complex laboratory-based sample processing and analysis by qualified laboratory technicians. They have significant consumables costs per test, compared with the minimal cost of the grooved stainless steel slide used for Raman analysis, which after the procedure could be sterilized, reused, and become part of the standard neurosurgical instrument set. Others have reported the use of Raman and attenuated total reflectance-Fourier transform infrared spectroscopy spectroscopy to distinguish IDH-mutant versus IDH-wild-type but have used snap frozen sections before spectroscopy, which still requires laboratory-based sample preparation.36,37 These vibrational spectroscopy studies have not attempted to distinguish oligodendrogliomas as a separate group, which is essential for clinical decision-making. Raman spectroscopy has also been used intraoperatively in the form of a probe device to attempt to differentiate tumor boundaries.38 A probe device is essential for boundary detection but adds no benefit to the aim of this study: as the tumor is being removed by the surgeon anyway, in- or near-operating theatre analysis is sufficient and allows for more sensitive Raman spectrometry than would be achievable via a fiber optic probe setup.
A barrier to the translation of Raman spectroscopy into the clinical setting is that clinicians are skeptical about the reliance on a change in spectral patterns to define different pathological entities. The spectral characteristics of glioma found in this study add to a body of published evidence, which consistently defines similar key spectral features of glioma tissue.15,16,36,39,40 Moreover, the most significant spectral differences seen between genetic subgroups are broadly consistent with our understanding of the biology of these tumors. IDH-mutant and IDH-wild-type gliomas are known to have significant differences in lipid metabolism,41–44 collagen maturation,45 and DNA methylation,7,46 which correlates with the most differentiating Raman peaks assigned to lipids, phenylalanine (involved in collagen synthesis), and DNA. The 881 cm–1 peak is markedly different in the fresh tissue oligodendroglioma spectrum. We currently do not have an experimentally validated explanation for this; however, although superficially similar (regarding high levels of 2HG as a result of an IDH mutation), IDH-mutant astrocytomas and oligodendrogliomas are otherwise clinically and molecularly distinct. It is also known that certain metabolic features are associated with the 1p/19q codeletion rather than IDH mutation (eg, cystathionine47). As altered phospholipid metabolism is known to occur in subsets of glioma,48 we cannot rule out that the increased peak at 881 cm–1 reflects these metabolic changes rather than an increased amount of DNA. It is important to note that this study did not aim to investigate proteomic or metabolomic profiles of different genetic subtypes—there are more suitable techniques to do this than Raman spectroscopy.
There is considerable consistency of Raman spectral peaks across all of the different sample preparation methods and cell lines used in this study. As might be expected, the performance of the classification model decreases as the tissue becomes more manipulated by the sample preparation, with fresh tissue models performing better than cryosection, which in turn classifies better than FFPE sections. Spectra of cryosections and FFPE sections have fewer contributions from proteins and lipid, differentiating genetic subtypes more on DNA and amino acid peaks. This is consistent with the known effect of protein denaturing due to freezing, cross-linking of proteins as part of formalin fixation, and the loss of lipid due to paraffin embedding.49
The LN18 cell line classification was very accurate within individual cell line preparations. In the cryosection preparations, the performance dropped off when the model includes spectra from both cell lines. This may be the result of large intragroup variability, as there are only 2 groups of each mutation and wild-type cells in the model. The performance would be likely to increase when the number of cell line batches is much greater, causing a decrease in intragroup variability and an increase the intergroup variability.
This study has also shown that Raman spectroscopy has application in the neuropathology and research laboratory settings, offering rapid genetic subtyping of gliomas using cryosections, FFPE sections, and glioma cell lines. The minimal sample preparation and rapid analysis advantages of Raman spectroscopy are less critical within a laboratory setting. However, routine IHC testing for IDH mutation is only sensitive to the common IDH1 mutation,7 whereas Raman spectroscopy measures the downstream biological consequences of the mutation, most likely associated with the intratumor accumulation of 2-hydroxyglutarate, which is common to all glioma-associated IDH1 and IDH2 mutations.50 In our study, the IHC test for the common IDH1-R132H mutation was, as expected, negative in those cases harboring rare IDH1 or IDH2 mutations (Table 3). This amounted to a significant proportion of our cohort (5%) and all cases were correctly classified as IDH-mutant in the Raman analysis. High-throughput assays based on next-generation sequencing or microarray methylation chip techniques are likely to replace single marker assessment by IHC, Sanger sequencing, or FISH, but these are not currently widely available and have a 5- to 14-day turnaround. Raman analysis may provide a faster and lower-cost alternative, certainly for initial tissue screening in environments where access to molecular diagnostics may not be available.
In summary, this study has demonstrated the potential for Raman spectroscopy to be used in a straightforward way within the neurosurgical and neuropathological glioma diagnostic pathway to provide genetic subtyping of gliomas within 15 min of the biopsy. The resulting data give the surgeon the opportunity to tailor the surgical strategy to the patient’s specific tumor genetics. Despite this being the largest study of its kind published to date, the small sample size is a limitation. Further multicenter and larger studies need to be undertaken to expand the training data set and to evaluate the effect of inter-user and inter-machine variability on the predictive model. Finally, the predictive model needs to be tested with a prospective independent cohort of patients to robustly validate the technique.
Supplementary Material
Funding
The Clinical Research Training Fellowship of L.J.L. was funded by Cancer Research UK (CRUK) Oxford Centre. The Oxford Brain Bank is supported by funds from the Medical Research Council (MRC), Brains for Dementia Research (BDR) (Alzheimer Society and Alzheimer Research UK) and the National Institute of Health Research (NIHR) Oxford Biomedical Research Centre (to O.A.). The views expressed are those of the authors and not necessarily those of the National Health Service (NHS), the NIHR or the Department of Health. This work uses data provided by patients and collected by the NHS as part of their care and support and would not have been possible without access to this data. The NIHR recognizes and values the role of patient data, securely accessed and stored, both in underpinning and leading to improvements in research and care. Spectroscopic instrumentation was loaned by Renishaw plc. (UK).
Conflict of interest statement.
M.I. and I.M.B. are full-time employees of Renishaw plc. Spectroscopy Products Division. Spectroscopic instrumentation was loaned by Renishaw plc. Renishaw did not provide any additional funding for the project or individually to the other authors.
Authorship statement:
Study conception and design: Laurent James Livermore, Martin Isabelle, Ian Mac Bell, Claire Vallance, Olaf Ansorge. Sample collection and processing: Laurent James Livermore, Connor Scott, John Walsby-Tickle, Joan Gannon. Analysis and interpretation of data: Laurent James Livermore, Martin Isabelle, Ian Mac Bell, Claire Vallance, Olaf Ansorge. Writing, review and/or editing manuscript: Laurent James Livermore, Connor Scott, Martin Isabelle, Ian Mac Bell, John Walsby-Tickle, Joan Gannon, Puneet Plaha, Claire Vallance, Olaf Ansorge. Study supervision: Claire Vallance, Olaf Ansorge.
References
- 1. Brown TJ, Brennan MC, Li M, et al. . Association of the extent of resection with survival in glioblastoma: a systematic review and meta-analysis. JAMA Oncol. 2016;2(11):1460–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wijnenga MMJ, French PJ, Dubbink HJ, et al. . The impact of surgery in molecularly defined low-grade glioma: an integrated clinical, radiological, and molecular analysis. Neuro Oncol. 2018;20(1):103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. D’Amico RS, Englander ZK, Canoll P, Bruce JN. Extent of resection in glioma-A review of the cutting edge. World Neurosurg. 2017;103:538–549. [DOI] [PubMed] [Google Scholar]
- 4. Louis DN, Perry A, Reifenberger G, et al. . The 2016 world health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 5. Aquilanti E, Miller J, Santagata S, Cahill DP, Brastianos PK. Updates in prognostic markers for gliomas. Neuro Oncol. 2018;20 (suppl_7):vii17–vii26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Capper D, Weissert S, Balss J, et al. . Characterization of R132H mutation-specific IDH1 antibody binding in brain tumors. Brain Pathol. 2010;20(1):245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Waitkus MS, Diplas BH, Yan H. Isocitrate dehydrogenase mutations in gliomas. Neuro Oncol. 2016;18(1):16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Butler HJ, Ashton L, Bird B, et al. . Using raman spectroscopy to characterize biological materials. Nat Protoc. 2016;11(4):664–687. [DOI] [PubMed] [Google Scholar]
- 9. Weller M, van den Bent M, Tonn JC, et al. ; European Association for Neuro-Oncology (EANO) Task Force on Gliomas. European association for neuro-oncology (EANO) guideline on the diagnosis and treatment of adult astrocytic and oligodendroglial gliomas. Lancet Oncol. 2017;18(6):e315–e329. [DOI] [PubMed] [Google Scholar]
- 10. Lewis AT, Gaifulina R, Isabelle M, et al. . Mirrored stainless steel substrate provides improved signal for raman spectroscopy of tissue and cells. J Raman Spectrosc. 2017;48(1):119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brat DJ, Aldape K, Colman H, et al. . Cimpact-NOW update 3: recommended diagnostic criteria for “diffuse astrocytic glioma, IDH-wildtype, with molecular features of glioblastoma, WHO grade IV”. Acta Neuropathol. 2018;136(5):805–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Afseth NK, Kohler A. Extended multiplicative signal correction in vibrational spectroscopy, a tutorial. Chemometr Intell Lab Syst. 2012;117:92–99. [Google Scholar]
- 13. Téllez Soto CA, Medeiros-Neto LP, dos Santos L, et al. . Infrared and confocal Raman spectroscopy to differentiate changes in the protein secondary structure in normal and abnormal thyroid tissues. J Raman Spectrosc. 2018;49(7):1165–1173. [Google Scholar]
- 14. Movasaghi Z, Rehman S, Rehman IU. Raman spectroscopy of biological tissues. Appl Spectrosc Rev. 2007;42(5):493–541. [Google Scholar]
- 15. Krafft C, Sobottka SB, Schackert G, Salzer R. Near infrared raman spectroscopic mapping of native brain tissue and intracranial tumors. Analyst. 2005;130(7):1070–1077. [DOI] [PubMed] [Google Scholar]
- 16. Kalkanis SN, Kast RE, Rosenblum ML, et al. . Raman spectroscopy to distinguish grey matter, necrosis, and glioblastoma multiforme in frozen tissue sections. J Neurooncol. 2014;116(3):477–485. [DOI] [PubMed] [Google Scholar]
- 17. Li YM, Suki D, Hess K, Sawaya R. The influence of maximum safe resection of glioblastoma on survival in 1229 patients: can we do better than gross-total resection? J Neurosurg. 2016;124(4):977–988. [DOI] [PubMed] [Google Scholar]
- 18. Sanson M, Marie Y, Paris S, et al. . Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol. 2009;27(25):4150–4154. [DOI] [PubMed] [Google Scholar]
- 19. Weller M, Felsberg J, Hartmann C, et al. . Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: a prospective translational study of the german glioma network. J Clin Oncol. 2009;27(34):5743–5750. [DOI] [PubMed] [Google Scholar]
- 20. van den Bent MJ, Dubbink HJ, Marie Y, et al. . IDH1 and IDH2 mutations are prognostic but not predictive for outcome in anaplastic oligodendroglial tumors: a report of the european organization for research and treatment of cancer brain tumor group. Clin Cancer Res. 2010;16(5):1597–1604. [DOI] [PubMed] [Google Scholar]
- 21. Yan H, Parsons DW, Jin G, et al. . IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nobusawa S, Watanabe T, Kleihues P, Ohgaki H. IDH1 mutations as molecular signature and predictive factor of secondary glioblastomas. Clin Cancer Res. 2009;15(19):6002–6007. [DOI] [PubMed] [Google Scholar]
- 23. Gravendeel LA, Kouwenhoven MC, Gevaert O, et al. . Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 2009;69(23):9065–9072. [DOI] [PubMed] [Google Scholar]
- 24. Wick W, Hartmann C, Engel C, et al. . NOA-04 randomized phase III trial of sequential radiochemotherapy of anaplastic glioma with procarbazine, lomustine, and vincristine or temozolomide. J Clin Oncol. 2009;27(35):5874–5880. [DOI] [PubMed] [Google Scholar]
- 25. Dubbink HJ, Taal W, van Marion R, et al. . IDH1 mutations in low-grade astrocytomas predict survival but not response to temozolomide. Neurology. 2009;73(21):1792–1795. [DOI] [PubMed] [Google Scholar]
- 26. Wesseling P, van den Bent M, Perry A. Oligodendroglioma: pathology, molecular mechanisms and markers. Acta Neuropathol. 2015;129(6):809–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zetterling M, Berhane L, Alafuzoff I, Jakola AS, Smits A. Prognostic markers for survival in patients with oligodendroglial tumors; a single-institution review of 214 cases. PLoS One. 2017;12(11):e0188419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cairncross G, Wang M, Shaw E, et al. . Phase III trial of chemoradiotherapy for anaplastic oligodendroglioma: long-term results of RTOG 9402. J Clin Oncol. 2013;31(3):337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. van den Bent MJ, Brandes AA, Taphoorn MJ, et al. . Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: long-term follow-up of EORTC brain tumor group study 26951. J Clin Oncol. 2013;31(3):344–350. [DOI] [PubMed] [Google Scholar]
- 30. Buckner JC, Shaw EG, Pugh SL, et al. . Radiation plus procarbazine, CCNU, and vincristine in low-grade glioma. N Engl J Med. 2016;374(14):1344–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Baumert BG, Hegi ME, van den Bent MJ, et al. . Temozolomide chemotherapy versus radiotherapy in high-risk low-grade glioma (EORTC 22033-26033): a randomised, open-label, phase 3 intergroup study. Lancet Oncol. 2016;17(11):1521–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kawaguchi T, Sonoda Y, Shibahara I, et al. . Impact of gross total resection in patients with WHO grade III glioma harboring the IDH ½ mutation without the 1p/19q co-deletion. J Neurooncol. 2016;129(3):505–514. [DOI] [PubMed] [Google Scholar]
- 33. Suh CH, Kim HS, Jung SC, Choi CG, Kim SJ. 2-hydroxyglutarate MR spectroscopy for prediction of isocitrate dehydrogenase mutant glioma: a systemic review and meta-analysis using individual patient data. Neuro Oncol. 2018;20(12):1573–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shankar GM, Francis JM, Rinne ML, et al. . Rapid intraoperative molecular characterization of glioma. JAMA Oncol. 2015;1(5):662–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ohka F, Yamamichi A, Kurimoto M, et al. . A novel all-in-one intraoperative genotyping system for IDH1-mutant glioma. Brain Tumor Pathol. 2017;34(2):91–97. [DOI] [PubMed] [Google Scholar]
- 36. Uckermann O, Yao W, Juratli TA, et al. . IDH1 mutation in human glioma induces chemical alterations that are amenable to optical raman spectroscopy. J Neurooncol. 2018;139(2):261–268. [DOI] [PubMed] [Google Scholar]
- 37. Uckermann O, Juratli TA, Galli R, et al. . Optical analysis of glioma: fourier-transform infrared spectroscopy reveals the IDH1 mutation status. Clin Cancer Res. 2018;24(11):2530–2538. [DOI] [PubMed] [Google Scholar]
- 38. Jermyn M, Mok K, Mercier J, et al. . Intraoperative brain cancer detection with raman spectroscopy in humans. Sci Transl Med. 2015;7(274):274ra19. [DOI] [PubMed] [Google Scholar]
- 39. Kast RE, Auner GW, Rosenblum ML, et al. . Raman molecular imaging of brain frozen tissue sections. J Neurooncol. 2014;120(1):55–62. [DOI] [PubMed] [Google Scholar]
- 40. Uckermann O, Galli R, Tamosaityte S, et al. . Label-free delineation of brain tumors by coherent anti-stokes raman scattering microscopy in an orthotopic mouse model and human glioblastoma. PLoS One. 2014;9(9):e107115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Esmaeili M, Hamans BC, Navis AC, et al. . IDH1 R132H mutation generates a distinct phospholipid metabolite profile in glioma. Cancer Res. 2014;74(17):4898–4907. [DOI] [PubMed] [Google Scholar]
- 42. Maus A, Peters GJ. Glutamate and α-ketoglutarate: key players in glioma metabolism. Amino Acids. 2017;49(1):21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Strickland M, Stoll EA. Metabolic reprogramming in glioma. Front Cell Dev Biol. 2017;5:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tönjes M, Barbus S, Park YJ, et al. . BCAT1 promotes cell proliferation through amino acid catabolism in gliomas carrying wild-type IDH1. Nat Med. 2013;19(7):901–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sasaki M, Knobbe CB, Itsumi M, et al. . D-2-hydroxyglutarate produced by mutant IDH1 perturbs collagen maturation and basement membrane function. Genes Dev. 2012;26(18):2038–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Turcan S, Rohle D, Goenka A, et al. . IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Branzoli F, Pontoizeau C, Tchara L, et al. . Cystathionine as a marker for 1p/19q codeleted gliomas by in vivo magnetic resonance spectroscopy. Neuro Oncol. 2019;21(6):765–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fack F, Tardito S, Hochart G, et al. . Altered metabolic landscape in IDH-mutant gliomas affects phospholipid, energy, and oxidative stress pathways. EMBO Mol Med. 2017;9(12):1681–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ali SM, Bonnier F, Tfayli A, et al. . Raman spectroscopic analysis of human skin tissue sections ex-vivo: evaluation of the effects of tissue processing and dewaxing. J Biomed Opt. 2013;18(6):061202. [DOI] [PubMed] [Google Scholar]
- 50. Dang L, White DW, Gross S, et al. . Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.