Abstract
This protocol describes a workflow for utilizing large scale cross-linking with mass spectrometry (XL-MS) to make systems level structural biology measurements in complex biological samples including cells, isolated organelles and tissue samples. XL-MS is a structural biology technique that provides information on the molecular structure of proteins and protein complexes using chemical probes that report on the proximity of the probed-reactive amino acids within proteins, typically lysine residues. Information gained through XL-MS studies is often complimentary to more traditional structural biology methods such as X-ray crystallography, nuclear magnetic resonance, and cryo-electron microscopy. The use of mass spectrometry cleavable cross-linkers, including Protein Interaction Reporter (PIR) technologies, has enabled XL-MS studies on protein structures and interactions in extremely complex biological samples including intact living cells. PIR cross-linkers contain selectively cleavable bonds that when broken release the intact peptides that were cross-linked as well as a reporter ion. Conservation of mass dictates a resulting mass relationship being the sum of the two released peptide masses and the reporter mass equaling the measured precursor mass is useful for identifying cross-linked peptide pairs. Importantly release of the individual peptides allows for accurate measurement of their masses as well as independent amino acid sequence determination by tandem mass spectrometry. This allows for standard proteomics search engines such as Comet to be used for peptide sequence assignment, greatly simplifying data analysis of cross-linked peptide pairs. Search results are processed with XLinkProphet for validation and can be uploaded into XlinkDB for interaction network and structural analysis.
INTRODUCTION:
XL-MS is a powerful technique for studying the structures and interactions of proteins[1, 2]. Chemical cross-linking reagents with a variety of chemical features have been described[3]. Mass spectrometry cleavable cross-linkers contain one or more labile bonds that are selectively cleavable within the mass spectrometer upon input of energy sufficient to cause dissociation of labile bonds engineered within the cross-linker[4]. Selective release of the cross-linked peptides allows for independent MS3 analysis and overcomes the inherent combinatorial problem when assigning cross-linked peptide sequences to mass spectra obtained with non-cleavable cross-linkers limiting their usefulness to relatively low complexity systems [5, 6]. The use of mass spectrometry cleavable cross-linkers overcomes this issue and has allowed for large scale interactome measurements from extremely complex systems including virus particles, cell lysates[7, 8], isolated organelles [9–11], intact cells[12–18] and tissue samples[19]. Indeed, the rapid growth of the XL-MS field is reflected by the recent availability of multiple protocols which detail specific approaches towards XL-MS experiments taken by different laboratories [20–22]. Since 2005 our laboratory has focused on the development of Protein Interaction Reporter (PIR) technologies[23, 24] as a means of proteome wide cross-linking carried out in biological systems of extreme complexity as described above. The PIR approach utilizes chemical cross-linkers with specific engineered properties including; selectively cleavable bonds (CID or photocleavable) to release the intact cross-linked peptides and a reporter ion for mass spectrometric analysis and affinity tags (biotin) for enrichment of low abundance cross-linked products from complex protein and enzymatic digest samples. PIR cross-linkers are peptide based molecules which affords the benefits of modular synthesis based on the widespread availability of solid phase peptide synthesis chemistry and biological compatibility as they readily penetrate cellular membranes. An expanding array of chemical cross-linker options are available to researchers, including MS cleavable molecules (see Sinz for recent review)[4], photocleavable[25, 26] and electrochemical cleavable options[27, 28]. Several MS cleavable cross-linkers are now commercially available including DSSO[29] and DBSU[30] available from Thermo Scientific and CBDPS[31] available from Creative Molecules. Researches planning XL-MS experiments should carefully consider the different options available to them including the molecular features and properties of cross-linker molecules to ensure compatibility with their biological systems and experimental design. Regardless of the cross-linker used, information from these experiments generates distance restraints between surface exposed proximal amino acids that are reactive with the chemical cross-linker used (typically Lys). One differing feature of chemical cross-linker molecules is their spacer arm length, ultimately used to generate an upper bound on distance restraints for structural analysis. For example, DSSO has a reported spacer arm length of 10.3 Å, while the PIR cross-linker BDP has a spacer arm length of 29.3 Å. Despite these differences, results from large scale cross-linking experiments indicate that the distributions of observed distances from identified cross-links from either cross-linker are similar when mapped to structural models in the PDB [9, 10]. Importantly this observation highlights that molecular features of proteins themselves, such as backbone and sidechain flexibility, residue reactivity and accessibility can be more important determinants of what cross-linked sites are ultimately observed. The generated distance restraints can be used to guide molecular modeling and docking experiments for protein and protein complexes. The information generated from XL-MS experiments is often complementary to other structural techniques such as X-ray crystallography, electron microscopy (EM) and nuclear magnetic resonance spectroscopy (NMR)[2, 32]. Intermolecular cross-links provide information on protein-protein interactions, including the identity of interacting partners and regions near the interaction interface. Intramolecular cross-links provide information on the structure and conformation of proteins. For in vivo cross-linking studies where protein structures and interactions are highly dynamic and in their native molecularly crowded environment, XL-MS is able to provide a snapshot from an ensemble of protein conformations and interactions that exist in cells during cross-linking. Here we provide a protocol for applying large scale cross-linking utilizing the PIR approach.
Development of the protocol
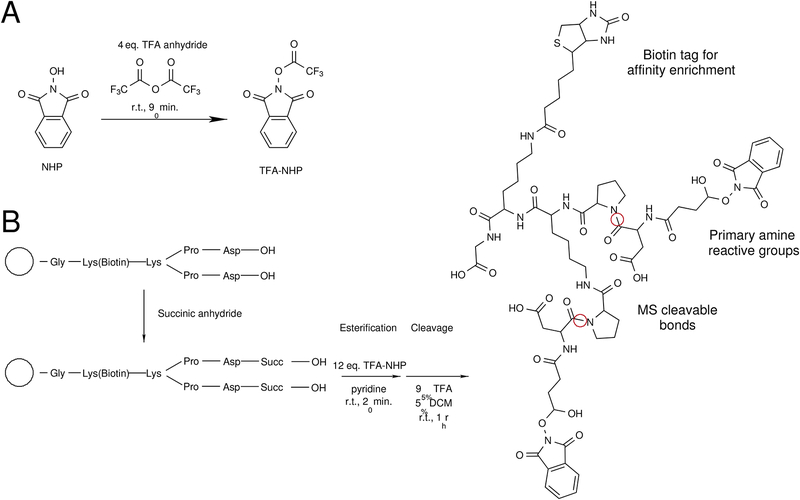
The development of the protocol described herein has been a continual process since the initial application of PIR cross-linking in 2005[23]. While numerous improvements and variations have been made in the years since the original inception of the PIR concept, the core foundation of the method remains the same. Fundamentally the approach depends on the implemented design features of the PIR cross-linker, including amine reactive functional groups, selectively labile bonds (either collisionally-activated or photo activated cleavage), and an affinity tag for enrichment (biotin) (Figure 1). Selective cleavage of the labile bonds allows mass spectrometric measurements to define a mass relationship shown in Equation 1.
| Equation 1 |
Figure 1.
Synthesis and molecular features of PIR cross-linker. a) Synthesis of N-hydroxyphthalimide trifluoroacetate ester (TFA-NHP) from N-hydroxyphthalimide and trifluoroacetic anhydride. b) Synthesis and molecular features of the PIR cross-linker BDP-NHP. The final produce is a bifunctional amine reactive cross-linker that contains a biotin affinity tag and two mass spectrometry labile Asp-Pro peptide bonds.
The affinity tag allows for enrichment of cross-linked peptide pairs, which are formed in relatively low abundance, from a complex sample such as an enzymatic digest of a complete proteome. Since the original implementation, recent improvements including addition of SCX fractionation [33], the detection of mass relationships in real-time[15], and detection and identification of the mass relationship and peptide sequences from chimeric spectra[34] have enabled routine detection and identification of thousands of non-redundant cross-linked peptide pairs from in vivo cross-linking experiments. Currently, XLinkDB [35, 36]represents the largest database of identified cross-linked peptides and contains 19,798 non-redundant cross-linked peptide pairs, a majority of which were acquired with in vivo cross-linking methods described in this protocol. Beyond identification the incorporation of isotope labels either metabolically or within the cross-linker molecule allow for extension of the method for quantitative measurements [37–40].
Application of the method
The described method has been successfully applied for large scale protein structural and interaction measurements in a number of biological systems including viruses [41, 42] [43], cultured bacterial[14, 15, 18, 44] and mammalian cells[16] [39] [38] and tissue samples[19] (Fig. 2). The method requires a range of skills in sample preparation, mass spectrometry analysis, and data analysis as with most cross-linking experiments. Our experience with collaborator laboratories indicate that successful PIR experiments can be carried out once learning the details embodied in this protocol[42, 43, 45]. Here we describe the protocol for application of large scale in vivo cross-linking to cultured bacterial and mammalian cell lines as well as murine tissue samples.
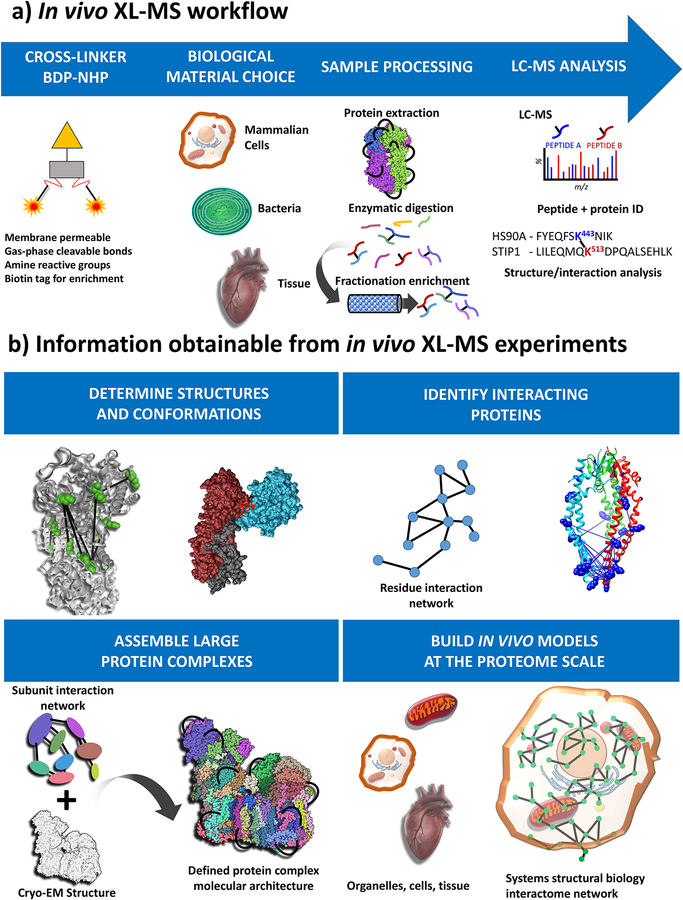
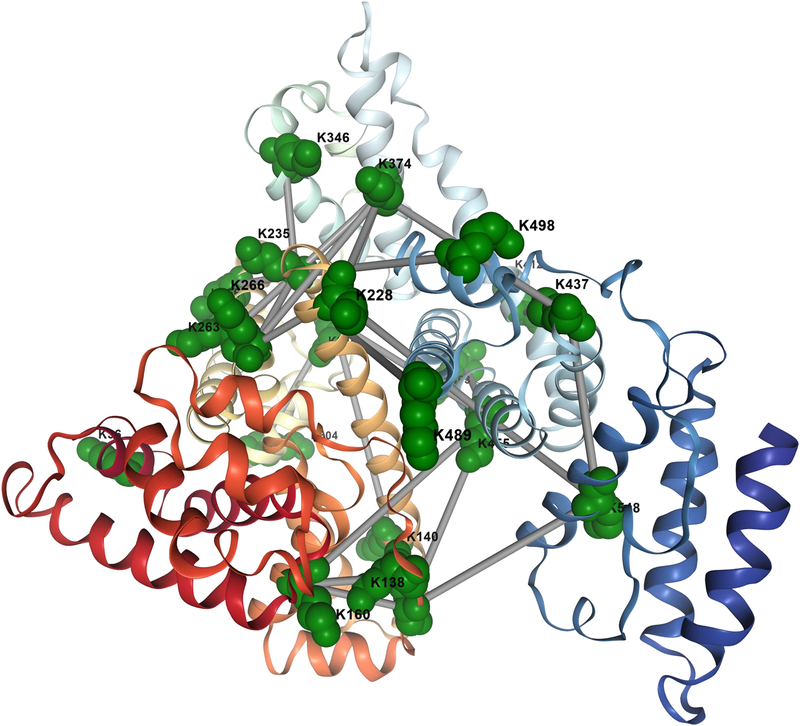
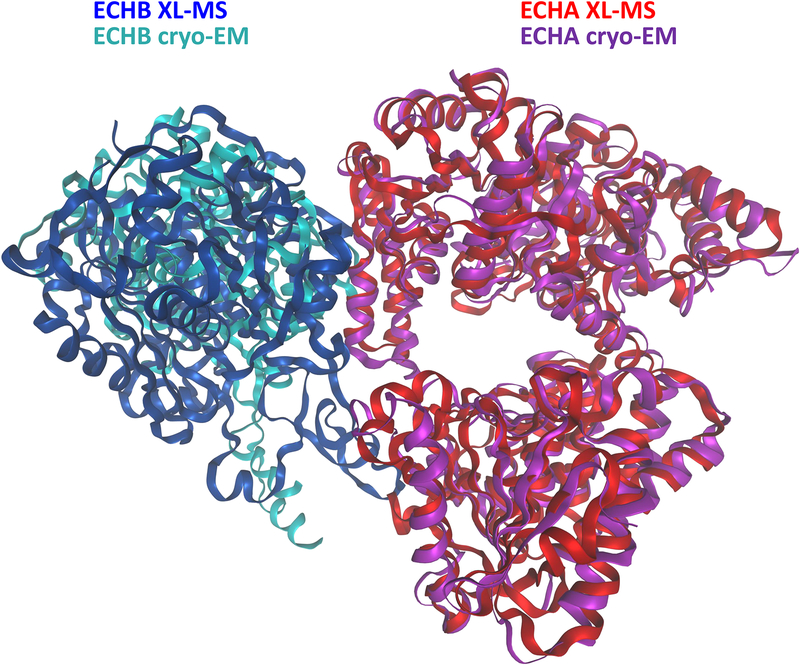
Figure 2.
Experimental overview of in vivo XL-MS. a) General workflow includes applying cross-linker (BDP-NHP) to a biological material of choice, which can include mammalian cells, bacterial cells, or animal tissue samples. This is followed by protein extraction, enzymatic digestion, fractionation and enrichment of cross-linked peptide pairs. Cross-linked peptide pairs are identified by LC-MS analysis. b) Types of information resulting from in vivo XL-MS experiments include protein structural and conformational information. XL-MS data can be integrated with data from Cryo-EM or other structural biology techniques to assemble structural models for large protein complexes. Identification of interacting proteins can be used to assemble proteome scale interactome maps for cross-linked systems including organelles, cells and tissues.
Experimental design
It is important to consider the multiple levels of information that are generated by large scale XL-MS when planning experiments. Fundamentally identification of cross-linked peptide pairs will provide information on proximal amino acid residues within protein molecules. The identities of interacting proteins are obtained by identifying cross-linked peptide sequences specific to particular protein molecules.
-. Cross-linker production
PIR cross-linkers are peptide based cross-linkers that contain Lys reactive esters, mass spectrometry cleavable bonds and a biotin affinity tag[23, 24]. Being that they are peptide based, they are readily synthesized using standard solid phase peptide synthesis protocols as described previously [15, 23], and outlined in steps 1–19. They are also available through suppliers offering custom peptide synthesis (i.e. Anaspec or others). Variations of PIR molecules have been described including MS cleavable versions employing the Rink functional group [46], or the Asp-Pro bond [16] as labile bonds that cleave under relatively low input of collisional induced dissociation (CID) energy. Photocleavable versions of PIR cross-linkers have also been developed and successfully used for large scale in vivo cross-linking experiments [26]. The final synthesis step for PIR cross-linkers is the formation of reactive ester groups on the cross-linker by reaction with a trifluoroacetate ester, typically N-hydroxysuccinimide (NHS) or N-hydroxyphthalamide (NHP) are described below in steps 20–24 and illustrated in Fig 1a. After esterification the cross-linker is cleaved from the solid phase resin using a solution of 95% TFA and 5% DCM, which also deprotects the Boc protected Asp side chains.
-. Generation of biological sample
Regardless of the type of biological sample to be studied, cultured mammalian cells, bacterial cells, or animal tissue, sample sizes that can yield approximately 5 – 10 mg of total protein are required. Cross-linked peptide pairs are formed at low abundance in the sample and this starting amount ensures enough material to form cross-linked peptides at a detectible level (femtomole to picomole sensitivity levels) with modern LC-MS instrumentation.
-. Cross-linking reaction and sample preparation
The cross-linking reaction needs to be carried out in an amine free, aqueous buffer. Sodium phosphate is typically used but other buffer systems including HEPES and MOPS have also been used successfully. Since the reactive ester groups on the cross-linker also react with water to form hydrolyzed products, it is important to perform the reaction on a relatively concentrated suspension of cells or tissue to help mitigate hydrolysis. In contrast with cross-linking in purified systems where over cross-linking can be a concern, in vivo cross-linking requires relatively large concentrations of cross-linker to ensure formation of cross-linked peptide pairs. This is due to the fact that in a cellular sample containing 5 mg of total protein suspended in 0.5 mL the free amine concentration from Lys residues alone is estimated to be on the order of 10 mM. This is near the solubility limit of BDP-NHP in aqueous solution and only an estimated 1% of the cross-linker will actually react with both of its arms to link Lys residues in proteins as the remainder is hydrolyzed. Thus, it is important to keep in mind that the cross-linker most often the limiting reagent with in vivo experiments and in many such cases, quenching of residual cross-linker is not necessary.
Cell lysis with 8M urea followed by sonication is generally sufficient for complete solubilization of the sample and extraction of protein. Digestion with trypsin cleaves at Arg and Lys residues but will generate internal Lys missed cleavage sites at any residue which reacted with the cross-linker. This fact can be exploited to improve sensitivity for cross-linked peptides during the database search by altering the Comet (or other search engine) parameters file (Step 118) to only consider peptide sequences with internal modified Lys residues.
-. Liquid chromatography and mass spectrometry analysis
As cross-linked peptide pairs are larger than normal tryptic peptides, the use of LC columns packed with C4 or C8 as opposed to the standard C18, have been shown to be beneficial for reversed-phase chromatographic separation of cross-linked peptides [47]. In addition the charge states of cross-linked peptide pairs tend to be higher than non-cross-linked peptides so it is beneficial to only perform MS2 on precursor ions with a charge state of 4+ or greater [15, 34]. The MS labile bonds cleave at lower CID energy (20 V normalized collision energy) than the peptide backbone which can be utilized to identify the PIR mass relationship (Equation 1). This can be exploited in real-time to only have the mass spectrometer sequence ions that fulfill this criteria, improving sensitivity for cross-linked peptide pairs in a method termed ReACT [15]. However, exploitation of mass relationship information to make downstream decisions such as triggering MS3, during LC-MS acquisition is not a standard feature available on most commercial instrumentation. To overcome this limitation we have developed a method to identify both the mass relationship and peptide fragmentation information from chimeric spectra in a post processing analysis step termed Mango [34]. As the Mango based method is able to be executed on any commercial MS instrument with a high resolving power instrument and MS2 capability and will therefore be applicable to the widest range of potential users, this protocol will focus on this method.
-. Data processing
A major benefit of using cleavable cross-linkers over non-cleavable versions is the ability to measure the individual masses of the two peptides that were linked. This reduces the problem resulting from having to consider all possible combinations of peptides being linked together. The combinatorial expansion of search space hinders use of non-cleavable cross-linkers in extremely complex samples, such as cells, where many thousands of protein sequences need to be considered. Use of cleavable cross-linkers also allows for standard open source proteomics search engines, such as Comet [48], to be used for searching the resulting MS data. An important issue to consider when processing XL-MS data is that of false discovery. Due to the nature of XL-MS data and the multiple levels of information that are gathered; cross-linked peptide sequences, cross-linked residues, cross-linked proteins, the false discovery rate or FDR estimation is more complicated that for traditional proteomics approaches [49]. Traditionally the FDR can be estimated by performing a target/decoy search where one of three possibilities arise for cross-linked peptide pairs; 1) both peptides are assigned target sequences 2) one peptide is assigned a target sequence and the other a decoy sequence and 3) both peptides are assigned decoy sequences. Only the first possibility results in a positive identification as long as both peptides receive a score, such as an expectation value (e-value), beyond some acceptable level. Because the situation can arise that one peptide in the cross-linked peptide pair receives a very good score and the other a not so good score, the conservative approach has been to assign a score to the peptide pair which is only as good as the worst scoring of the two peptides [50]. Recently a machine learning approach with increased detection sensitivity, termed XLinkProphet, was developed to model multiple types of information about cross-linked peptide pairs to compute discriminating probabilities and accurately estimate FDR [51]. This protocol describes the use of Comet (Steps 113–120) and XLinkProphet (Steps 121–123) for identification of cross-linked peptide pairs from large scale XL-MS data sets. The general data analysis workflow is illustrated in Fig 3.
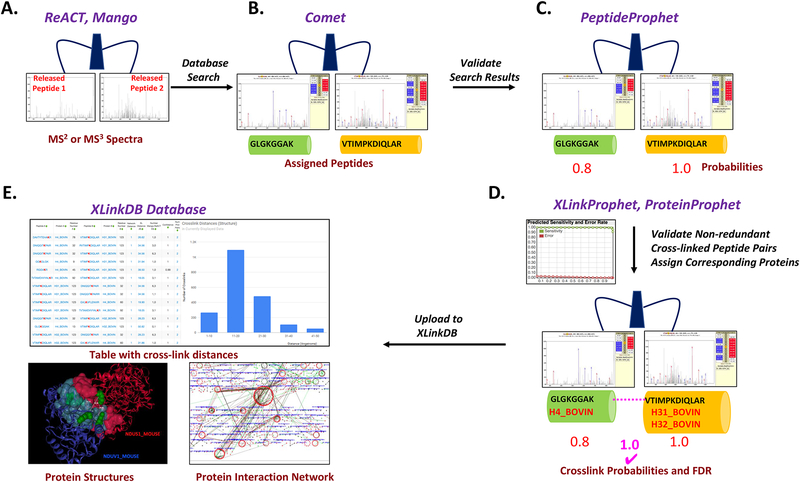
Figure 3.
Data analysis workflow. a) Mass spectra containing the mass of the intact cross-linked peptide pair, masses of the individual released peptides and fragment ions of the released peptides are acquired by ReACT or Mango methods. b) Database search is carried out by Comet to assign peptide sequences to the mass spectra. c) PeptideProphet is run on the Comet search results to assign probabilities to individual peptide sequences. d) XLinkProphet is run to validate the cross-linked peptide pair and corresponding proteins by assigning a cross-link probability and estimation of FDR. e) Validated results are uploaded into the XLinkDB database where a table of the identified cross-linked peptide pairs can be viewed in a table or interaction network format. Cross-links are mapped to existing PDB structures and homology based structural models and distance information for each cross-link is generated. Resulting protein structures with cross-links mapped onto them can be viewed.
Limitations
The application of XL-MS to complex biological samples provides low resolution structural information on proteins and protein complexes. This information can be used to guide molecular modeling and docking algorithms and combined with structural information from complementary structural biology techniques such as X-ray crystallography, NMR and cryo-EM to produce models of protein structures. The use of cross-linker molecules with two reactive groups produces binary interaction information between two reactive amino acid residues. Therefore, additional information from other sources, such as canonical complex assembly, is needed to extrapolate the XL-MS information to higher order structural information. Furthermore, cross-linker molecules can react with an ensemble of proteins and complexes present in vivo. In most cases, one cannot be certain that all identified cross-linked peptide pairs on a particular protein were formed at the same time or are compatible with a single structural model or conformation for that protein. Despite great progress made in recent years, currently XL-MS studies are still only scratching the surface of the complexity of the interactome, generating on the order of thousands of non-redundant cross-linked peptide pair ID’s involving hundreds of proteins. Continued developments will undoubtedly increase the depth and coverage providing researchers with valuable new knowledge to address increasingly challenging questions and improve understanding of biological systems.
Materials
Reagents
Cells or tissue sample of interest. In this protocol we use HeLa for the mammalian cell line, E. coli for the bacterial cell line and mouse hearts for the tissue sample.
BDP-NHP – Can be obtained from a custom peptide synthesis company (e.g. Anaspec)
Na2HPO4 (Sigma Aldrich; Cat. No. AC20651)
NaH2PO4 (Sigma Aldrich; Cat. No. S9638)
Urea (Sigma Aldrich; Cat. No. U5378)
KH2PO4 (Sigma Aldrich; Cat. No. P9791)
KCl (Sigma Aldrich; Cat. No. P9541)
Formic Acid, 99.5+%, Optima™ LC/MS Grade, Fisher Chemical (Fisher Scientific; Cat. No. A117)
Water, Optima™ LC/MS Grade, Fisher Chemical (Fisher Scientific; Cat. No. W6)
Acetonitrile, Optima™ LC/MS Grade, Fisher Chemical (Fisher Scientific; Cat. No. A955)
Acetonitrile with 0.1% Trifluoroacetic Acid (v/v), Optima™ LC/MS Grade (Fisher Scientific; Cat. No. LS121)
Water with 0.1% Trifluoroacetic Acid (v/v), Optima™ LC/MS Grade (Fisher Scientific; Cat. No. LS119)
NaCl (Sigma Aldrich; Cat. No. S 9888)
Ammonium hydroxide solution ACS reagent, 28.0–30.0% NH3 basis (Sigma Aldrich; Cat. No. 221228)
Sequencing Grade Modified Trypsin, Frozen (Promega Corp; Cat. No. V5113)
Ethylenediaminetetraacetic acid, 99%, pure, ACROS Organics™ (Fisher Scientific; Cat. No. AC118432500)
Ammonium bicarbonate (Sigma Aldrich; Cat. No. A6141)
Cryomill (Retsch Inc.; Cat. No. 20.745.0001)
TCEP-HCl (Fisher Scientific; Cat. No. 20491)
Iodoacetamide (Sigma Aldrich; Cat. No. I1149)
Trifluoroacetic Acid (Optima™ LC/MS), Fisher Chemical (Fisher Scientific; Cat. No. A116)
Pierce Monomeric Avidin UltraLink Resin (Pierce Biotechnology; Cat. No. 53146)
ReproSil-Pur 5 micron 120 A (ESI Source Solutions; Cat. No. r15.8e.0001)
MgCl2 (Sigma Aldrich; Cat. No. M8266)
CaCl2 (Sigma Aldrich; Cat. No. AC42352)
Methanol, LC-MS Grade (Fisher Scientific; Cat. No. A456)
Phosphoric Acid 49–51%, HPLC Grade (Sigma-Aldrich; Cat. No. 79607)
Bovine serum albumin (Sigma Aldrich, Cat. No. A7638)
Equipment
Extraction Manifold, 20pos, 16–75mm tubes (Waters; Cat. No. WAT200608)
nanoACQUITY UPLC Systems (Waters; Cat. No. 176816000) or equivalent nano-LC system capable of stable flow in the submicroliter/min range (eg. Ultimate 3000, Thermo Scientific; Cat No. ULTIM3000RSLCNANO or EASY nLC Thermo Scientific; Cat No. LC140)
High resolution mass spectrometer (Orbitrap or FTICR instrument) (Thermo fisher scientific; Cat. No. -)
Sep-Pak Vac C18 cartridge 3cc/500mg (Waters; Cat. No. 186004619)
Vacuum Centrifuge (e.g. EZ-2) (SP Scientific; Cat. No. -)
Ultrasonic Processor (Cole-Palmer; Cat. No. EW-04714–50)
Eppendorf® Thermomixer Compact (Sigma Aldrich; Cat. No. T1317)
Microcentrifuge (Eppendorf; Cat. No. 22620444)
Analytical/Preparative HPLC System (Agilent Technologies; Cat. No. G1311C)
SCX Column (Phenomenex; Cat. No. 00G-4398-N0)
Microfuge tubes (1.5 mL) (Fisher Scientific; Cat. No. 02-681-320)
Falcon tubes (15 mL) (Fisher Scientific; Cat. No. 14-959-53A)
Thermomixer R (Eppendorf; Cat. No. 5355)
Software
ReAdW (https://sourceforge.net/projects/sashimi/files/ReAdW%20%28Xcalibur%20converter%29/)
Mango (https://github.com/jpm369/mango)
Comet 2018.01 or forward (http://comet-ms.sourceforge.net/)
XLinkProphet (https://github.com/brucelab/xlinkprophet)
Perl v5.24.0+ (https://www.perl.org/get.html)
Reagent Set-up
Na2HPO4 170 mM pH 8.0 - Dissolve 2.61 g of NaH2PO4 and 0.16 g of Na2HPO4 in 100 mL of deionized water.
NH4HCO3 100 mM pH 8.0 - Dissolve 7.91 g of ammonium bicarbonate in 1 L of deionized water. Adjust pH to 8 by adding NH4OH.
Lysis Buffer – Dissolve 480 mg of Urea in 1 mL of 100 mM NH4HCO3
Strong Cation Exchange buffer A - 7 mM KH2PO4, pH 2.6, 30% acetonitrile. Dissolve 0.95 g KH2PO4 in 1L of 70% deionized water, 30% acetonitrile, and adjust the pH to 2.6 by adding phosphoric acid.
Strong Cation Exchange buffer B - 7 mM KH2PO4, pH 2.6, 350 mM KCl, 30% acetonitrile. Dissolve 0.95 g KH2PO4 and 24.85 g of KCl in 1L of 70% deionized water, 30% acetonitrile, and adjust the pH to 2.6 by adding phosphoric acid.
LC-MS Buffer A - 0.1 % LC-MS grade formic acid in LC-MS grade water. Add 4 mL of LC-MS grade formic acid to 4 L of LC-MS grade water.
LC-MS Buffer B - 0.1 % LC-MS grade formic acid in LC-MS grade acetonitrile. Add 4 mL of LC-MS grade formic acid to 4 L of LC-MS grade acetonitrile.
PBS – Add 8.01 g NaCl, 0.20 g KCl, 1.42 g Na2HPO4, and 0.24 g KH2PO4 to 1 L of deionized water.
Procedure
Cross-linker production – TIMING 1 day
Synthesis of BDP on resin
-
1
BDP cross-linker is a peptide based molecule synthesized by solid phase peptide synthesis (SPPS). The following steps describe how to synthesize the cross-linker using a CEM Liberty Lite peptide synthesizer. The complete succinylated form (Fig1 b) can be purchased through any custom peptide synthesis company. We have successfully obtained BDP from Anaspec. If purchasing the cross-linker on resin skip to step 20 for synthesis of the activated N-hydroxyphtalamide esters.
-
2
Weigh out amount of Fmoc-Gly-Wang resin (100–200 mesh) to obtain 0.5 mmol and transfer to reaction vessel. CRITICAL STEP: Amount to weigh out depends on resin loading capacity. Typically, the resin has a loading capacity ~ 0.8 mmol/g but will vary from lot to lot.
-
3
Weigh out 1.73 g of aspartate (Fmoc-Asp(OtBu)-OH) and dissolve in 21 mL of DMF
-
4
Weigh out 23 g of succinic anhydride and dissolve in 23 mL of DMF.
-
5
Weigh out 32 g of biotinylated lysine (Fmoc-Lys(Biotin)-OH) and dissolve in 32 mL of DMF. CRITICAL STEP: Fmoc-Lys(Biotin)-OH has limited solubility in DMF. Heat solution to 60°C with constant mixing at 700 rpm on a thermomixer for 5–10 min or until dissolved.
-
6
Weigh out 1.42 g of proline (Fmoc-Pro-OH) and dissolve in 21 mL of DMF
-
7
Weigh out 1.3 g of lysine (Fmoc-Lys(Fmoc)-OH) and dissolve in 11 mL of DMF
-
8
Fill main wash bottle with at least 400 mL of DMF
-
9
Fill deprotection bottle with at least 60 mL of 20% (w/v) of piperidine in DMF
-
10
Fill the activator bottle with at least 40 mL of 1 M dicyclohexylcarbodiimide (DIC) in DMF
-
11
Fill the activator base bottle with at least 20 mL of 1M Oxyma in DMF
-
12
Run the CEM synthesizer with the following coupling steps
-
13
Glycine - 0.5 mmol resin swelling
-
14
Biotin lysine - 0.5 mmol triple coupling CRITICAL STEP: Biotin lysine is coupled using a triple coupling step due to larger volume (32 ml) needed to dissolve the Fmoc-Lys(Biotin)-OH.
-
15
Branching lysine – 0.5 mmol single coupling
-
16
Proline – 0.5 mmol double coupling
-
17
Aspartate – 0.5 mmol double coupling
-
18
Succinic anhydride 0.5 mmol succinyl coupling. CRITICAL STEP: Succinyl coupling step does not require the addition of activators DIC or Oxyma.
-
19
Transfer the resin containing BDP from the reaction vessel to a 50 mL Eppendorf tube. Use 1 – 5 mL DMF to assist with quantitative transfer of resin.
Synthesis of–N-hydroxyphthalimide trifluoroacetate ester (TFA-NHP) see Fig. 1a
-
20
Weigh out 5.86 g N-hydroxyphthalimide (NHP, 163.139 g/mol) and place in 50 mL round bottom flask. CRITICAL STEP: N-hydroxysuccinimide can be substituted for N-hydoxyphthalamide here if desired.
-
21
Add a 4-fold molar excess of Trifluoroacetic acid anhydride (210.03 g/mol, 1.511 g/mL) = 20 mL. CRITICAL STEP: Both acid anhydrides and the esterified cross-linker product are moisture sensitive and readily hydrolyze upon exposure to water. Store in a dry environment and prepare using fresh, dry reagents. CAUTION: Trifluoroacetic acid anhydride is highly corrosive. Use only in a fume hood while wearing proper PPE, including gloves, goggles, and a lab coat.
-
22
Allow reaction to proceed under a dry N2 atmosphere with constant mixing for 1.5 hrs.
-
23
Evaporate excess TFA anhydride under vacuum until dry white crystalline product obtained in quantitative yield.
-
24
Transfer product to 2 mL Eppendorf tubes and store at −20 °C or use for esterification of BDP in the following steps.
Esterification and purification of BDP
-
25
Swell BDP resin with a minimum saturating volume of DMF.
-
26
Weigh out a 12-fold molar excess (1.55 g for 0.5 mmol of BDP) of TFA-NHP.
-
27
Dissolve TFA-NHP in 10 mL of dry pyridine.
-
28
Immediately transfer TFA-NHP pyridine solution to BDP resin and mix for 20 min at 1400 rpm at room temperature using a thermomixer.
-
29
Transfer reaction mixture to a Bio-Rad poly-prep chromatography column.
-
30
Filter away pyridine solution.
-
31
Wash resin 3 times with 20 mL DMF.
-
32
Incubate resin in 20 mL DMF for 20 min.
-
33
Wash resin 3 times with 20 mL DCM.
-
34
Incubate resin in 20 mL DCM for 20 min.
-
35
Cleave the PIR cross-linker from the resin by incubating for 3 h in 5 mL cleavage solution 95% TFA, 5% DCM.
-
36
Collect and save cleavage solution.
-
37
Wash beads with 5 mL cleavage solution incubating for 5 min.
-
38
Collect cleavage solution and pool with collected solution in step 13.
-
39
Precipitate cross-linker by slowly adding cleavage solution to 150 mL ice cold diethyl ether.
-
40
Pellet cross-linker by centrifugation at 3400 g for 30 min at 4°C.
-
41
Decant diethyl ether and dry cross-linker under vacuum using a vacuum centrifuge.
-
42
Weigh product (>80% yield is expected based on resin loading capacity) and dissolve using dry DMSO to obtain desired stock concentration, typically, 200–300 mM CRITICAL STEP: The purity of cross-linker can be checked by direct infusion ESI-MS analysis of a 1:1000 dilution of the concentrated stock solution into a solution of 49% methanol, 49% water, 2% acetic acid. A purity of 80–90% should be observed by the relative intensity of the pseudo-molecular ion peak for the PIR cross-linker ([M+H]+= 1472.53 m/z) Fig. 4.
-
43
Measure concentration of BDP-NHP by UV-Vis absorbance CRITICAL STEP: The concentration of BDP-NHP can be measured by UV-Vis absorbance. Dilute a small aliquot (~1 μL) of concentrated stock solution of cross-linker into 0.1 M NH4HCO3 pH = 8.0 to achieve an estimated concentration of ~ 0.5 mM. This will quench the cross-linker and release free NHP which has a yellow color. Measure the absorbance of NHP at 300 nm and/or 410 nm and compare to a calibration curve for free NHP in 0.1 M NH4HCO3 pH = 8.0 spanning the range from 0.1 mM to 2 mM to calculate the concentration of released NHP. The concentration of BDP cross-linker in the stock solution is half the calculated concentration of NHP multiplied by the dilution factor.
-
44
Use cross-linker stock immediately for cross-linking biological samples or aliquot and store at −80°C for up to 1 year. CRITCAL STEP: The esterified cross-linker appears to be stable for long periods of time when stored at −80°C, however repeated freeze, thaw cycles should be avoided due to the absorption of water into the solution which can hydrolyze the activated esters in the cross-linker.
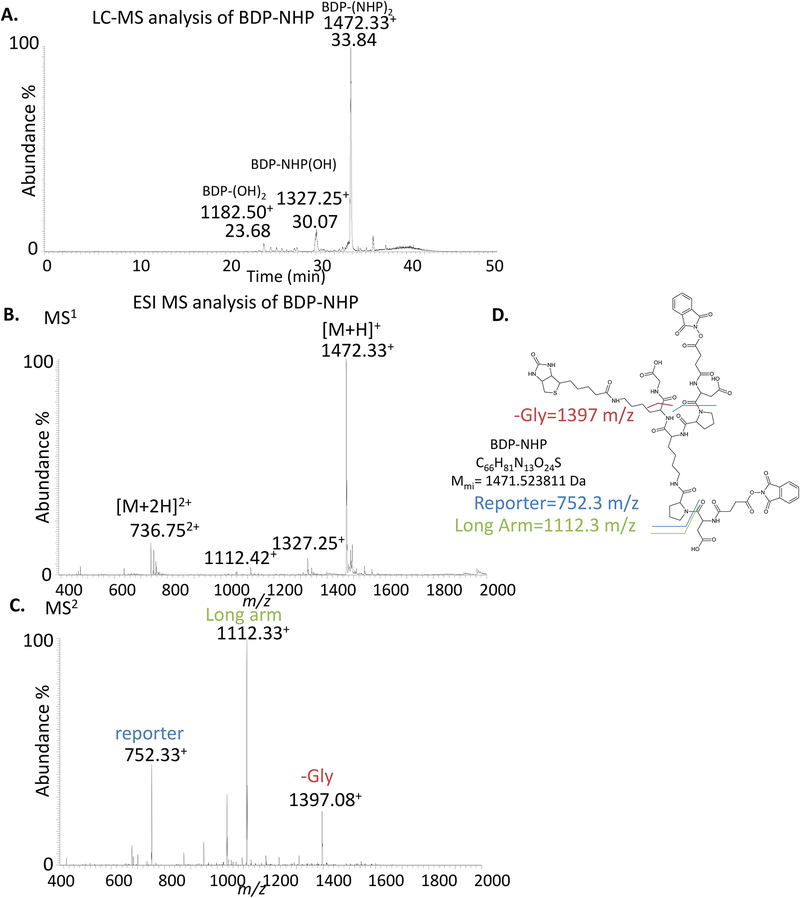
Figure 4.
LC-ESI-MS analysis of the BDP-NHP cross-linker. a) LC-MS chromatogram of BDP-NHP. The desired product containing two NHP-ester reactive groups elutes at a retention time of 33.84 min with a m/z value of 1472.33+. The peak at 30.07 min results from hydrolysis of a single NHP group with a m/z value of 1327.25+. The peak at 23.68 min results from hydrolysis of both NHP groups with a m/z value of 1182.50+. Some hydrolysis products during this analysis are expected due to the aqueous solvents. b) Direct infusion generated ESI MS1 spectrum of BDP-NHP. The single protonated pseudo-molecular ion [M+H]+ is observed at 1472.33 m/z and the doubly protonated form [M+2H]2+ is observed at 736.75 m/z. These peaks should be the most abundant and constitute >80% of the total signal indicating acceptable purity of the cross-linker. The peak at 1327.25 m/z represents hydrolysis of one of the NHP reactive groups. The peak at 1112.42 m/z results from in-source fragmentation of one of the Asp-Pro amide bonds in the cross-linker. c) MS2 spectrum of the precursor ion 1472.33 m/z. Major fragment ions are observed at 1112.33 m/z (long arm) and 752.33 m/z (reporter) resulting from cleavage of one or two of the Asp-Pro amide bonds respectively. The ion observed at 1397.08 results from loss of the C-terminal Gly. d) Chemical structure of BDP-NHP with the bonds resulting in the major fragment ions observed in panel b highlighted. The chemical formula and theoretical monoisotopic mass (Mmi) are indicated.
Mammalian Cell Growth – TIMING 5 days
-
45
Establish culture of cells on appropriate growth medium (DMEM with 10% FBS and 1% (v/v) penicillin/streptomycin. CRITICAL STEP: We have successfully used this approach for HeLa and H292 cells. Growth media and conditions may need to be altered for different cell lines.
-
46
Subculture and expansion should be performed as needed.
-
47
Grow cells in 15 cm plates using 20 mL growth medium until cells are 80–90% confluent. CRITICAL STEP: This yields approximately 2E7 cells and approximately 5 mg of protein for HeLa cells. Adjust the initial amount of starting material depending on the growth density of the selected cell line.
-
48
Wash cells with PBS.
-
49
Harvest cells by incubating with 5 mL PBS containing 20 mM EDTA and returning plates to incubator for 5–10 min. Collect cells and pellet by centrifugation at 300 g for 3 min. CRITICAL STEP: Detaching adherent cells with an EDTA containing buffer is preferred to trypsin to avoid cleavage of cell surface proteins. If growing cells in a suspension, this step can be skipped.
-
50
Suspend cell pellet in PBS containing 1 mM Ca2+ and 1 mM Mg2+.
-
51
Pellet at 300 g for 3 minutes at room temperature and discard the supernatant.
-
52
Repeat steps 50 and 51 two more times.
-
53
GO TO 64
Bacterial Growth – TIMING 1 Days
-
54
Grow bacterial cells to desired density in 250–500 mL of growth media. Transfer to several 50 mL conical tubes. CRITICAL STEP: We have successfully used this approach for E. coli, A. baumanii and P. aeruginosa. Growth conditions may need to be altered for other bacterial cell types.
-
55
Harvest bacteria by centrifuging at 1500g for 10 minutes at room temperature. Discard the supernatant.
-
56
Gently resuspend bacterial pellet in 30 mL PBS by pipetting. Centrifuge again at 1500g for 10 minutes at room temperature and discard the supernatant. Repeat until the supernatant is clear. CRITICAL STEP: Incomplete washing will leave residual growth media behind which contains primary amines that will interfere with cross-linking efficiency.
-
57
Pellet at 1500g for 10 minutes at room temperature and discard the supernatant.
-
58
GO TO 64
Mouse Tissue – TIMING 1–3 hours
-
59
Anesthetize mouse with pentobarbital at dose > 270mg/kg intraperitoneally CRITICAL STEP: Confirm deep anesthesia by lack of response to toe pinch and decreased respiratory rate
-
60
Excise tissue of interest.
-
61
Wash all traces of blood from tissue with 170 mM Na2HPO4 pH 8.0 or other cross-linker compatible (amine-free) buffer of choice. CRITICAL STEP: We have successfully used this approach for heart tissue. Other tissues may need additional washing or preparation prior to cross-linking.
-
62
Mince tissue into 1 mm3 pieces with a razor blade.
-
63
GO TO 64
Cross-linking reaction TIMING 1–2 hours
-
64
Resuspend cell pellet or minced tissue sample in one pellet volume of 170 mM Na2HPO4 to get a 50 % suspension. CRITICAL STEP: Gently pipette up and down several times to resuspend. Pipetting quickly can lead to premature cell lysis.
-
65
Add appropriate volume of PIR cross-linker stock to reach desired final concentration, typically 10 mM final concentration for live cell cross-linking. CRITICAL STEP: Cross-linker may aggregate after initial addition but will homogenize after shaking for a few minutes. A negative control sample can be generated by adding an equivalent volume of DMSO instead of PIR cross-linker to a second sample. This sample can be taken through all remaining steps of the protocol and the resulting MS signal compared to that of the cross-linked sample. A positive control sample can be generated by reacting 1mL of bovine serum albumin (BSA) at 1 mg/mL in 170 mM Na2HPO4 with 1 mM PIR cross-linker and following through the protocol from step 76.
-
66
Allow reaction to proceed for 30 minutes at room temperature in a thermomixer while shaking at 600 rpm. The solution will turn yellow as the cross-linker reacts. CRITICAL STEP: The yellow color results from the NHP released from the cross-linker and can be used as a visual indicator of reaction progress. The half-life of the reactive ester groups of the cross-linker is ~ 8 min in aqueous buffer at pH 8 and ~ 2 min in the presence of 1 mg/mL bovine serum albumin (BSA). After 30 min all of the reactive cross-linker has been depleted and there is no need to quench the reaction.
-
67
Add one pellet volume of 0.1 M NH4HCO3 to quench any remaining active cross-linker.
-
68
Pellet cells by centrifugation at 300 g for 3 min (1500g for 10 minutes for bacterial cells) and remove the supernatant.
-
69
Resuspend cells in one pellet volume of fresh 170 mM Na2HPO4.
-
70
Pellet cells by centrifugation at 300 g for 3 min (1500g for 10 minutes for bacterial cells) and remove the supernatant.
-
71
Resuspend cells in one pellet volume of PBS.
-
72
Pellet cells by centrifugation at 300 g for 3 min (1500g for 10 minutes for bacterial cells) and remove the supernatant.
-
73
Repeat steps 71 and 72 until supernatant is no longer visibly yellow, typically about 3 washes.
Lysis, protein extraction, and digestion TIMING 16 hours
-
74
Suspend cross-linked cell pellet in cell lysis buffer (8 M urea, 0.1 M NH4CO3, pH 8.0). CRITICAL STEP: A minimal volume (0.1–1 mL) should be used to resuspend the cell, as the volume will increase 10-fold prior to digestion.
-
75
Lyse cells by sonication using an ultrasonic processor until sample is no longer viscous. CRITICAL STEP: The conditions of lysis will depend upon the biological sample used. Typically, using a GE – 130 ultrasonic processor, five pulses at amplitude 40 for 5 s each is enough to reduce sample viscosity of cultured human cells. Sample container should be kept in ice to prevent excessive heat buildup. For samples which are difficult to lyse (ex: tissue samples and gram negative bacteria) cryogrinding using a mixer mill (Retch MM 400) can be used using 5 × 3 min grinding cycles at 30 Hz, to generate a frozen powdered lysate prior to ultrasonic processing.
-
76
Reduce protein disulfide bonds by adding TCEP to a final concentration of 5 mM and incubate at room temperature for 30 min.
-
77
Alkylate reduced thiols with 10 mM final concentration of iodoacetamide (IAA) for 30 min at room temp. CRITICAL STEP: IAA is light sensitive, so the reaction should be shielded from light during the alkylation reaction.
-
78
Dilute sample 10-fold with 0.1 M NH4HCO3 to reduce urea concentration below 1 M. It may be necessary to transfer sample to a 15mL tube. CRITICAL STEP: If urea concentration is not sufficiently reduced, trypsin digestion will be highly inefficient.
-
79
Perform a Bradford or BCA assay to determine the protein content of the sample.
-
80
Add 1:200 ratio of sequencing grade modified trypsin to sample and digest overnight at 37°c in a thermomixer shaking at 650 rpm.
Desalting TIMING 4 hours
-
81
Acidify digest to pH approximately 3 by adding trifluoracetic acid to a 1% (v/v) final concentration. CRITICAL STEP: If sample pH is not below 3, then the sample will not bind to the Sep-Pak cartridge, resulting in sample loss.
-
82
Centrifuge the sample at 16,000 g for 15 minutes at room temperature.
-
83
Select a Sep-Pak of sufficient capacity for the total protein determined in step X (protein assay). 50mg size cartridges (Column volume = 1 mL) can be used for up to 5mg of protein, while the 500mg cartridges (Column volume = 3 mL) can be used for samples exceeding 5mg of input protein.
-
84
Place column into a vacuum manifold.
-
85
Condition the Sep-Pak by adding 1 column volume of methanol, and drawing it through the column by vacuum. CAUTION: A pressure difference of 5–10 PSI is sufficient to draw liquid through the cartridge. Higher pressures cause faster flow rates that may inhibit adsorption to the cartridge. The maximum flowrate should be approximately 1 drop per second. Always verify that the vacuum manifold being used is rated to the pressure that will be applied and monitor the pressure difference using a pressure gauge. CRITICAL STEP: Do not allow the cartridge to ever become completely dried. Leave a minimal volume of solvent above the top of the packing material. If the cartridge becomes completely dried it may crack, causing sample loss.
-
86
Condition the column by adding 1 column volume of 0.1% TFA in acetonitrile and pulling it through the cartridge. Repeat this step 2 additional times.
-
87
Equilibrate the column by adding 1 column volume of 0.1% TFA in water and pulling it through the cartridge. Repeat this step 2 additional times.
-
88
Add supernatant from step 82 to the column up to maximum volume allowed by the cartridge size and draw it through. Repeat this step until all protein has been added, up to the maximum allowed by the cartridge. CRITICAL STEP: Collect and save the flow-through from this step. If the pH of the sample is too high, then this flow-through will contain unbound peptides.
-
89
Repeat step 87 to wash away salts.
-
90
Place an open microfuge tube under the Sep-Pak cartridge outlet to collect the flow-through for the next step.
-
91
Elute peptides from Sep-Pak cartridge into the open microfuge tube by adding 1 column volume of 80% acetonitrile/0.1% TFA and drawing it through.
-
92
Concentrate sample to less than 100 μL by vacuum centrifugation.
PAUSE POINT
Strong cation exchange fractionation TIMING 2 hours initial set-up + 2 hours per sample
-
93
Dilute sample to 500 μL volume with SCX solvent A (5 mM KH2PO4, pH 2.6, 30% ACN).
-
94
Centrifuge sample at 16,000 g for 15 min at room temperature.
-
95
Transfer supernatant to a LC vial.
-
96
Inject sample onto LC system equipped with SCX column (Phenomenex Luna SCX column). CRITICAL STEP: Verify that injection volume is compatible with the sample loop in use on the HPLC system. Additionally verify that protein content of the injection does not exceed the loading capacity of the column, which is 10mg for the column specified here.
-
97
Sample is fractionated using a flow rate of 1.5 mL/min and a 97.5 min gradient of an increasing percentage of SCX solvent B (7 mM KH2PO4, pH 2.6, 30% ACN, 350 mM KCl) as follows: 0% B at 0 min, 5% B at 7.5 min, 60% B at 47.5 min, 100% B at 67.5 min, 100% B at 77.5 min, 0% B at 77.51 min, 0% B at 97.5 min. Fractions are collected every 5 min starting at 17.5 min.
-
98
Fractions are pooled (6 total) as follows: 1–5, 6–7, 8, 9, 10, 11–14. Fraction pools were then dried to a final volume of ~2 mL in a vacuum centrifuge and pH adjusted to a pH of 8.0 with 1.5 M NaOH. CRITICAL STEP: Avidin binding efficiency is greatly reduced if the pH is too low.
Biotin capture TIMING 4 hours
-
99
Enrich PIR-labeled peptides by adding 200 μL monomeric avidin bead slurry (Ultralink, Pierce) to each pooled fraction, fractions 1–5 are unlikely to contain a significant number of cross-linked peptides.
-
100
Incubate for 30 min at room temp on a Thermomixer (1000 rpm).
-
101
Wash avidin bound, cross-linked peptides 5 times with 2 mL 0.1 M NH4HCO3, pH 8.0.
-
102
Incubate the avidin beads with 1 mL of 70% ACN, 1% formic acid for 5 minutes and collect the eluates.
-
103
Concentrate sample to approximately 10 μL in a vacuum centrifuge.
-
104
PAUSE POINT: Samples can be stored at −80C for several months.
LC-MS Analysis TIMING 3 or 5 hours per fraction injection depending on gradient;
-
105
Add 20–50 μl of 0.1% formic acid to reconstitute cross-linked peptide samples. CRITICAL STEP: If peptides were dried to completion, first add a minimal volume of 50% Acetonitrile/50% Water/0.1 formic acid to redissolve the peptdes, then dilute with 0.1% FA to the final desired volume. Ensure that the final acetonitrile concentration is less than 5% to ensure efficient trapping and separation by reverse-phase LC.
-
106
Centrifuge at 16,000 g for 10 min to pellet any particulate.
-
107
Transfer supernatant to a LC-MS vial.
-
108
Inject 1–5 μl of sample into nano LC system (Waters NanoAcquity UPLC).
-
109
Load peptides onto a trap column (3 cm × 100 μm inner diameter fused silica trap column packed with a stationary phase consisting of ReproSil-Pur C8, 5 μm diameter, 120 Å pore size particles) with a flow rate of 2 μL/min of mobile phase consisting of 98% solvent A (H2O containing 0.1% formic acid) and 2% solvent B (ACN containing 0.1% formic acid) for 10 minutes.
-
110
Fractionate peptides over a 60 cm × 75 μm inner diameter fused silica analytical column packed with ReproSil-Pur C8, 5 μm diameter, 120 Å pore size particles by applying a linear gradient from 95% solvent A, 5% solvent B to 60% solvent A, 40% solvent B over either 120 or 240 minutes at a flow rate of 300 nL/min. The analytical column can be heated to 40 °C to enhance the separation if a column heater is available.
-
111
Ionize eluting peptides by ESI with a voltage of 2–2.5 kV to the laser pulled fused silica spray tip at the end of the column.
-
112
Operate the mass spectrometer using a data dependent analysis method where one high-resolution MS1 (70,000 resolving power at 200 m/z, AGC = 1e6) followed by 20 high-resolution MS2 scans selecting ions of 4+ to 8+ (70,000 resolving power at 200 m/z, AGC = 5e4, Maximum ion time = 100 ms, isolation window = 3 Da, collision energy = 30 NCE, dynamic exclusion 30 seconds).
Cross-link search – TIMING 5–60 Minutes per fraction
-
113
Convert instrument data files (e.g. .raw riles) to the open .mzXML format. This can be done using either vendor specific software, or a tool such as msconvert or ReAdW.
-
114
Download the current release of mango from https://github.com/jpm369/mango. The repository includes 64-bit binaries for both Windows and Linux operating systems, as well as detailed usage instructions and parameter descriptions.
-
115
Generate a mango.params.new file. Command: mango.exe -p
-
116
Adjust the settings in the mango.params.new file, save it, then rename it as mango.params. The default parameters in the file work for BDP-NHP cross-linked samples run using the mass spectrometer method described in step X.
-
117
Run mango.exe on an .mzXML file, this will return an .ms2 file containing individual precursor masses of released peptides for each spectrum, as well as a .peaks file containing all relationships within tolerance. Command: mango.exe file.mzXML
-
118
Using the included comet binary, generate a comet.params.new file. Command: comet.exe -p
-
119
Adjust the settings in the comet.params.new file to match your experiment. A sample high resolution comet.params file is included in the TestCase folder of the mango repository. Details on the various parameters used by comet can be found at http://comet-ms.sourceforge.net/parameters/parameters_201801/. Set the output_txtfile parameter to 1 to return an easy to parse output. Once parameters have been set, rename the file to comet.params.
-
120
Run comet.exe on the .ms2 file generated by mango, this will return a .pep.xml file containing unpaired results. Command: comet.exe file.ms2
Post-processing – TIMING 5–60 Minutes per fraction
-
121
Run PeptideProphet [52] on the Comet search results to validate with assigned probabilities. Command: xinteract -OEAdP -p0 -PPM -l5 search_result1.pep.xml search_result2.pep.xml,….. where search_result*.pep.xml are the search result files to be included in the analysis. Note that when the search database has decoys with a unique protein prefix (e.g. ‘rev_’), that information should be passed as an additional parameter -dprefix (e.g. – drev_). Command: xinteract -OEAdP -p0 -PPM -l5–dprefix search_result1.pep.xml search_result2.pep.xml,…..
-
122
Run iProphet [53] to further refine PeptideProphet probabilities based on additional information. Command: InterProphetParser interact.pep.xml iprophet.pep.xml
-
123
Run XLinkProphet [51] (available at https://github.com/brucelab/xlinkprophet) to validate Mango results based on iProphet search result probabilities and crosslink properties. Note that the Mango peaks files should be in the same directory locations as the Comet search result pepXML files. Command: XLinkProphet.pl iprophet.pep.xml MIN_PEPLEN=5 CROSSLINK_MODS=K:329.152492 REPORTERMASS=xxxx where K:329.152492 indicates the amino acid and modification mass of the crosslinker stump used to define the site of crosslink attachment to the peptide (this should be substituted with value or comma delimited values appropriate for the analyzed data), and xxx is to be substituted by the actual mass of the crosslink reporter relevant to the data.
Automated interaction network and structural modeling/docking – TIMING 1 hour
-
124
Resulting data from XLinkProphet can be uploaded into XLinkDB at http://xlinkdb.gs.washington.edu/
-
125
Arrange the cross-link identifications in a tab delimited text file with the following six columns: Peptide A Protein A | Labeled position A || Peptide B | Protein B | Labeled position B. A template file is available at http://xlinkdb.gs.washington.edu/xlinkdb/Help.php
-
126
Upon successful upload an interactive interaction network will be generated and cross-linked proteins will automatically submitted for structural modeling and molecular docking algorithms producing structural models in PDB format which are viewable directly from the website.
TROUBLESHOOTING
Troubleshooting advice can be found in Table 1.
Table 1.
CEM synthesizer coupling steps
| Step | Problem | Possible Reason | Solution |
|---|---|---|---|
| 23 | Low yield of TFA-NHP | Degraded or contaminated reagents used during synthesis. | Use fresh, high purity reagents. Trifluoroacetic acid anhydride is prone to hydrolysis and should be used within a week of opening. |
| 42 | Low yield of cross-linker during resin cleavage, resin has uncleaved cross-linker remaining. | Residual DMF on cross-linker resin inhibits cleavage. | Wash resin extensively with DCM, and then incubate the resin in DCM for 20 minutes, pipetting occasionally to improve solvent accessibility. Make a fresh stock of cleavage solution and repeat the incubation in cleavage solution. |
| 42 | Impure cross-linker observed during direct infusion ESI-MS analysis. | Degraded or contaminated reagents used during synthesis. | Make sure all solvents and reagents used for synthesis are freshly made and of high purity. Solvents should be ACS grade or higher. DMF should not be more than 6 months old. Amino acid and DIC solutions are stable for up to two weeks. The piperidine solution is stable for one month. |
| 44 | Cross-linker has hydrolyzed. | Cross-linker stock solution has become contaminated with water. | Verify hydrolysis by ESI-MS anlaysis. See Fig. 4. |
| 65 | Cross-linker precipitated in pipet tip. | Pipet tip was immersed into aqueous buffer prior to expelling the concentrated cross-linker solution causing it to precipitate | Do not immerse pipet tip in the solution when adding cross-linker. Instead hold the tip above the solution and proceed with pushing the cross-linker solution out. |
| 66 | No yellow color observed during cross-linking reaction. | pH of solution is too low. | Check the pH of the reaction solution. It should be between 7–8. If it is lower adjust it within range by titrating with 1M NaOH or use a higher concentration of buffer. |
| 73 | Cell pellet is small or solution is sticky and viscous. | Cells have lysed during pelleting and resuspension. | Check centrifugation speed to make sure it is appropriate for the sample type used. Be extremely gental while using a pipet to resuspend cells only using a minimal number of cycles to suspend the cell pellet. |
| 75 | Tissue samples not lysing during cryogrinding. | Cryoginding ball was added to liquid sample prior to freezing in liquid nitrogen and is now encased in ice and not able to grind sample. | Add cryogriding ball after sample solution has been frozen in liquid N2 to ensure it is free to move during shaking. |
| 80 | Large amount of precipitation in tryptic digest sample. | Typsin failed to digest proteins into peptides. | Sample solution conditions were not compatible with trypsin. Check pH to ensure it is between 7–8. Ensure urea concentration is less than 1 M. Correct solution conditions and add fresh trypsin to repeat step 80. |
| 81 | Large amount of precipitation observed after acidifying sample. | Large amount of residual undigested protein due to inefficient trypsin digestion | Check solution conditions for trypsin digestion. Make sure the pH is between 7–8, urea concentration is less than 1 M and fresh trypsin is used. Repeat steps 80–81. |
| 97 | No signal or low signal observed in SCX absorbance traces. | Loss of protein during desalting due to poor binding to the packing material. | Check pH of the input sample for the Sep-Pak. Reacidify if above 3, and repeat the desalting procedure. |
| 111 | Spray is sputtering or droplets forming at the tip of the column. | Incorrect spray voltage or tip position, blunt, broken or dirty spray tip. | Adjust the applied spray voltage and tip position until stable spray is obtained. If tip appears blunt or dirty replace with a new column. |
| 112 | No signal or low signal observed in chromatogram on mass spectrometer. | Poor binding of sample to monomeric avidin resin. | Verify that the pH of the SCX fractions after base addition are approximately 8. These samples can be recaptured again after the pH is corrected. |
| Sample not injected by autosampler on LC | Verify that autosampler is delivering the requeseted volume of sample onto the LC column. | ||
| Mass spectrometer is out of calibration or has low sensitivity due to dirty ion optics. | Clean and calibrate mass spectrometer verifying sensitivity with a standard sample. | ||
| Multiple contributing factors. | Use a cross-linked purified protein such as BSA as a positive control sample and ensure that results similar to those presented Fig. 5 and Table 2 are obtained. | ||
| 120 | No significant identifications in .pep.xml file. | Incorrect database or search parameters. | Check that the correct protein database was used and the the comet search parameters match the experimental setup. |
ANTICIPATED RESULTS
The workflow described in this protocol can be used to synthesize PIR cross-linker and make large scale structural biology measurements on protein structures and interactions within cells. The cross-linker should be obtained in high purity (>80%) as evaluated by LC-ESI-MS and direct infusion ESI-MS analysis (Fig. 4). Successful use of this protocol will result in the identification of thousands of non-redundant cross-linked peptide pairs resulting from hundreds of proteins when filtered to an estimated FDR of 1% or lower. Since the relative frequency of proximal intra-protein lysine residues is unity while that of inter-protein lysine residues can only approach unity, even for the most highly stable interactions, it stands to reason that more than 50% of identified in vivo cross-linked peptides should be anticipated to result from intra-protein links. Consistent with this, between 70 to 80% of the identified in vivo cross-linked peptide pairs are typically intra-protein links, while 20–30% are inter-protein links. Despite the observation that many non-redundant cross-linked peptide pairs are identified from highly abundant proteins, cross-linked peptides from proteins localized in all cellular compartments are normally identified. Moreover, cross-linked peptides from lower abundance proteins are often identified, albeit with fewer numbers of pairs as illustrated elsewhere[38]. As for experimental controls, a non-cross-linked control sample should not yield any significant cross-linked peptide pair identifications. Results from cross-linked BSA should yield results similar to the 37 non-redundant cross-linked peptide pairs listed in Table 2 and displayed on the structure of BSA in Fig. 5. If anticipated results are not obtained, please consult the troubleshooting guide (Table 1).
Table 2.–
XLinkProphet output table for cross-linked peptide pairs from a BSA positive control sample. The table includes the raw unfiltered results. Positive identifications were filtered to less than 1% FDR resulting in 37 non-redundant cross-linked peptide sequences.
| probability | spectrum | mango_query | peptide1 | peptide2 | protein1 | protein2 | charge1 | charge2 | probability1 | probability2 | peptide1_mass | peptide2_mass | parent_charge | parent_mass | composite_probability | composite_id | decoy | decoy-decoy | decoy_expect_fdr | decoy_prob_fdr | massdiff_ppm | max_expect | peptide1_len | peptide2_len | peptide_pair | product_probability | homopeptide | intra | joint_score | massdiff_bin | nrx | nsx | total_charge | fdr | orig_nsx |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.13468.13468.1 |
1 | ALK[325.13]AWSVAR | LVTDLTK[325.13]VHK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1197.614 | 1349.719 | 4 | 3298.746 | 1 | ALK[325.13] AWSVAR_LVTDLTK[325.13]VHK |
0 | 0 | 0 | 0 | 2.4303 | 7.85E-03 | 9 | 10 | ALK[325.13]AWSVAR_LVTDLTK[325.13]VHK | 0.99 | 0 | 1 | 3 | 0 | 1 | 1 | 2 | 0 | 36 |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.09740.09740.1 |
1 | LSQK[325.13]FPK | EK[325.13]VLTSSAR | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1043.529 | 1186.583 | 4 | 2981.511 | 1 | LSQK[325.13]FPK_EK[325.13]VLTSSAR | 0 | 0 | 0 | 0 | −1.9862 | 1.38E-02 | 7 | 9 | LSQK[325.13]FPK_EK[325.13]VLTSSAR | 0.99 | 0 | 1 | 3 | 0 | 1 | 1 | 2 | 0 | 36 |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.14022.14022.1 |
1 | ALK[325.13]AWSVAR | LAK[325.13]EYEATLEECCAK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1197.614 | 2010.855 | 4 | 3959.88 | 1 | ALK[325.13]AWSVAR_LAK[325.13]EYEATLEECCAK | 0 | 0 | 0 | 0 | 1.2978 | 3.28E-03 | 9 | 15 | ALK[325.13]AWSVAR_LAK[325.13]EYEATLEECCAK | 0.99 | 0 | 1 | 3 | 0 | 1 | 1 | 2 | 0 | 36 |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.13155.13155.1 |
1 | CASIQK[325.13]FGER | LAK[325.13]EYEATLEECCAK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1391.614 | 2010.855 | 4 | 4154.864 | 1 | CASIQK[325.13]FGER_LAK[325.13]EYEATLEECCAK | 0 | 0 | 0 | 0 | 2.3353 | 2.38E-04 | 10 | 15 | CASIQK[325.13]FGER_LAK[325.13]EYEATLEECCAK | 0.99 | 0 | 1 | 3 | 0 | 1 | 1 | 2 | 0 | 36 |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.09613.09613.1 |
1 | DTHK[325.13]SEIAHR | FK[325.13]DLGEEHFK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1389.627 | 1445.646 | 4 | 3586.687 | 1 | DTHK[325.13]SEIAHR_FK[325.13]DLGEEHFK | 0 | 0 | 0 | 0 | 2.3989 | 1.93E-04 | 10 | 10 | DTHK[325.13]SEIAHR_FK[325.13]DLGEEHFK | 0.99 | 0 | 1 | 3 | 0 | 1 | 1 | 2 | 0 | 36 |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.13562.13562.1 |
2 | ALK[325.13]AWSVAR | LVTDLTK[325.13]VHK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1197.614 | 1349.719 | 5 | 3298.746 | ALK[325.13]AWSVAR_LVTDLTK[325.13]VHK | 0 | 0 | 0 | 0 | 2.2484 | 1.72E-04 | 9 | 10 | ALK[325.13]AWSVAR_LVTDLTK[325.13]VHK | 0.99 | 0 | 1 | 3 | 0 | 1 | 1 | 2 | 0 | 36 | |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.10627.10627.1 |
1 | SLGK[325.13]VGTR | LSQK[325.13]FPK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1013.514 | 1043.529 | 4 | 2808.449 | 1 | SLGK[325.13]VGTR_LSQK[325.13]FPK | 0 | 0 | 0 | 0 | 0.3867 | 2.20E-03 | 8 | 7 | SLGK[325.13]VGTR_LSQK[325.13]FPK | 0.99 | 0 | 1 | 3 | 0 | 1 | 1 | 2 | 0 | 36 |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.13952.13952.1 |
2 | EK[325.13]VLTSSAR | GACLLPK[325.13]IETM[147.04]R | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1186.583 | 1600.759 | 4 | 3538.745 | 1 | EK[325.13]VLTSSAR_GACLLPK[325.13]IETM[147.04]R | 0 | 0 | 0 | 0 | −0.4691 | 2.82E-03 | 9 | 12 | EK[325.13]VLTSSAR_GACLLPK[325.13]IETM[147.04]R | 0.99 | 0 | 1 | 3 | 0 | 1 | 1 | 2 | 0 | 36 |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.10737.10737.1 |
1 | SLGK[325.13]VGTR | LSQK[325.13]FPK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1013.514 | 1043.529 | 4 | 2808.451 | SLGK[325.13]VGTR_LSQK[325.13]FPK | 0 | 0 | 0 | 0 | 1.0255 | 2.70E-03 | 8 | 7 | SLGK[325.13]VGTR_LSQK[325.13]FPK | 0.99 | 0 | 1 | 3 | 0 | 1 | 1 | 2 | 0 | 36 | |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.13802.13802.1 |
1 | ALK[325.13]AWSVAR | LAK[325.13]EYEATLEECCAK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1197.614 | 2010.855 | 4 | 3959.871 | ALK[325.13]AWSVAR_LAK[325.13]EYEATLEECCAK | 0 | 0 | 0 | 0 | −0.7478 | 2.51E-03 | 9 | 15 | ALK[325.13]AWSVAR_LAK[325.13]EYEATLEECCAK | 0.99 | 0 | 1 | 3 | 0 | 1 | 1 | 2 | 0 | 36 | |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.10571.10571.1 |
1 | LSQK[325.13]FPK | EK[325.13]VLTSSAR | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1043.529 | 1186.583 | 4 | 2981.518 | LSQK[325.13]FPK_EK[325.13]VLTSSAR | 0 | 0 | 0 | 0 | 0.3616 | 3.68E-03 | 7 | 9 | LSQK[325.13]FPK_EK[325.13]VLTSSAR | 0.99 | 0 | 1 | 3 | 0 | 1 | 1 | 2 | 0 | 36 | |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.13911.13911.1 |
1 | ALK[325.13]AWSVAR | LAK[325.13]EYEATLEECCAK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1197.614 | 2010.855 | 4 | 3959.882 | ALK[325.13]AWSVAR_LAK[325.13]EYEATLEECCAK | 0 | 0 | 0 | 0 | 1.9038 | 4.14E-05 | 9 | 15 | ALK[325.13]AWSVAR_LAK[325.13]EYEATLEECCAK | 0.99 | 0 | 1 | 3 | 0 | 1 | 1 | 2 | 0 | 36 | |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.14063.14063.1 |
2 | EK[325.13]VLTSSAR | GACLLPK[325.13]IETM[147.04]R | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1186.583 | 1600.759 | 4 | 3540.771 | EK[325.13]VLTSSAR_GACLLPK[325.13]IETM[147.04]R | 0 | 0 | 0 | 0 | 6.7595 | 1.50E-02 | 9 | 12 | EK[325.13]VLTSSAR_GACLLPK[325.13]IETM[147.04]R | 0.99 | 0 | 1 | 3 | 1 | 1 | 1 | 2 | 0 | 36 | |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.15826.15826.1 |
1 | SLGK[325.13]VGTR | K[325.13]QTALVELLK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1013.514 | 1338.739 | 4 | 3103.667 | 1 | SLGK[325.13]VGTR_K[325.13]QTALVELLK | 0 | 0 | 0 | 0 | 2.6952 | 9.04E-04 | 8 | 10 | SLGK[325.13]VGTR_K[325.13]QTALVELLK | 0.99 | 0 | 1 | 3 | 1 | 1 | 1 | 2 | 0 | 36 |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.09549.09549.1 |
3 | LSQK[325.13]FPK | EK[325.13]VLTSSAR | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1043.529 | 1186.583 | 4 | 2981.517 | LSQK[325.13]FPK_EK[325.13]VLTSSAR | 0 | 0 | 0.0161 | 0 | −0.0409 | 4.31E-01 | 7 | 9 | LSQK[325.13]FPK_EK[325.13]VLTSSAR | 0.99 | 0 | 1 | 3 | 0 | 2 | 1 | 2 | 0 | 36 | |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.09721.09721.1 |
1 | DTHK[325.13]SEIAHR | FK[325.13]DLGEEHFK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1389.627 | 1445.646 | 4 | 3586.678 | DTHK[325.13]SEIAHR_FK[325.13]DLGEEHFK | 0 | 0 | 0 | 0 | −0.1383 | 3.31E-03 | 10 | 10 | DTHK[325.13]SEIAHR_FK[325.13]DLGEEHFK | 0.99 | 0 | 1 | 3 | 0 | 2 | 1 | 2 | 0 | 36 | |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.10410.10410.1 |
2 | LSQK[325.13]FPK | EK[325.13]VLTSSAR | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1043.529 | 1186.583 | 4 | 2981.51 | LSQK[325.13]FPK_EK[325.13]VLTSSAR | 0 | 0 | 0 | 0 | −2.3552 | 9.80E-03 | 7 | 9 | LSQK[325.13]FPK_EK[325.13]VLTSSAR | 0.99 | 0 | 1 | 3 | 0 | 2 | 1 | 2 | 0 | 36 | |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.10462.10462.1 |
1 | LSQK[325.13]FPK | EK[325.13]VLTSSAR | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1043.529 | 1186.583 | 4 | 2981.505 | LSQK[325.13]FPK_EK[325.13]VLTSSAR | 0 | 0 | 0.0161 | 0 | −3.9987 | 1.66E-01 | 7 | 9 | LSQK[325.13]FPK_EK[325.13]VLTSSAR | 0.99 | 0 | 1 | 3 | −1 | 1 | 1 | 2 | 0 | 36 | |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.09682.09682.1 |
3 | LSQK[325.13]FPK | EK[325.13]VLTSSAR | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1043.529 | 1186.583 | 4 | 2983.531 | LSQK[325.13]FPK_EK[325.13]VLTSSAR | 0 | 0 | 0.1266 | 0 | 4.7166 | 3.13E+00 | 7 | 9 | LSQK[325.13]FPK_EK[325.13]VLTSSAR | 0.99 | 0 | 1 | 3 | 1 | 2 | 1 | 2 | 0 | 36 | |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.12296.12296.1 |
2 | ALK[325.13]AWSVAR | VHK[325.13]ECCHGDLLECADDRADLAK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 3 | 1 | 1 | 1197.614 | 2808.19 | 7 | 4757.212 | 1 | ALK[325.13]AWSVAR_VHK[325.13]ECCHGDLLECADDRADLAK | 0 | 0 | 0 | 0 | 0.5749 | 1.38E-01 | 9 | 22 | ALK[325.13]AWSVAR_VHK[325.13]ECCHGDLLECADDRADLAK | 0.99 | 0 | 1 | 3 | 0 | 1 | 1 | 4 | 0 | 36 |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.12245.12245.1 |
1 | ALK[325.13]AWSVAR | VHK[325.13]ECCHGDLLECADDRADLAK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 3 | 1 | 0.9998 | 1197.614 | 2808.19 | 5 | 4758.214 | ALK[325.13]AWSVAR_VHK[325.13]ECCHGDLLECADDRADLAK | 0 | 0 | 0.4788 | 0 | 0.8728 | 3.11E+01 | 9 | 22 | ALK[325.13]AWSVAR_VHK[325.13]ECCHGDLLECADDRADLAK | 0.9898 | 0 | 1 | 3 | 0 | 1 | 1 | 4 | 0 | 36 | |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.08552.08552.1 |
1 | SLGK[325.13]VGTR | CCTK[325.13]PESER | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 0.9996 | 1013.514 | 1362.518 | 4 | 3128.411 | 1 | SLGK[325.13]VGTR_CCTK[325.13]PESER | 0 | 0 | 0 | 0 | 8.4305 | 3.11E-02 | 8 | 9 | SLGK[325.13]VGTR_CCTK[325.13]PESER | 0.9896 | 0 | 1 | 3 | 2 | 1 | 1 | 2 | 0 | 36 |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.11361.11361.1 |
3 | LAK[325.13]EYEATLEECCAK | VHK[325.13]ECCHGDLLECADDR | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 2 | 1 | 1 | 2010.855 | 2309.91 | 5 | 5073.164 | 1 | LAK[325.13]EYEATLEECCAK_VHK[325.13]ECCHGDLLECADDR | 0 | 0 | 0 | 0 | 1.2785 | 1.21E-13 | 15 | 17 | LAK[325.13]EYEATLEECCAK_VHK[325.13]ECCHGDLLECADDR | 0.99 | 0 | 1 | 3 | 0 | 1 | 1 | 3 | 0 | 36 |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.19591.19591.1 |
1 | FWGK[325.13]YLYEIAR | DDSPDLPK[325.13]LKPDPNTLCDEFK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 2 | 1 | 1 | 1641.783 | 2640.19 | 5 | 5034.365 | 1 | FWGK[325.13]YLYEIAR_DDSPDLPK[325.13]LKPDPNTLCDEFK | 0 | 0 | 0 | 0 | 2.5332 | 4.23E-05 | 11 | 21 | FWGK[325.13]YLYEIAR_DDSPDLPK[325.13]LKPDPNTLCDEFK | 0.99 | 0 | 1 | 3 | 1 | 1 | 1 | 3 | 0 | 36 |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.12072.12072.1 |
1 | ALK[325.13]AWSVAR | VHK[325.13]ECCHGDLLECADDR | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 2 | 1 | 1 | 1197.614 | 2309.91 | 6 | 4259.956 | 1 | ALK[325.13]AWSVAR_VHK[325.13]ECCHGDLLECADDR | 0 | 0 | 0 | 0 | 6.3555 | 1.04E-03 | 9 | 17 | ALK[325.13]AWSVAR_VHK[325.13]ECCHGDLLECADDR | 0.99 | 0 | 1 | 3 | 1 | 1 | 1 | 3 | 0 | 36 |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.11410.11410.1 |
1 | CASIQK[325.13]FGER | VHK[325.13]ECCHGDLLECADDR | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 2 | 1 | 1 | 1391.614 | 2309.91 | 5 | 4454.914 | 1 | CASIQK[325.13]FGER_VHK[325.13]ECCHGDLLECADDR | 0 | 0 | 0 | 0 | 3.4548 | 6.61E-05 | 10 | 17 | CASIQK[325.13]FGER_VHK[325.13]ECCHGDLLECADDR | 0.99 | 0 | 1 | 3 | 1 | 1 | 1 | 3 | 0 | 36 |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.09699.09699.2 |
1 | DTHK[325.13]SEIAHR | FK[325.13]DLGEEHFK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 2 | 2 | 1 | 1 | 1389.627 | 1445.646 | 7 | 3586.679 | DTHK[325.13]SEIAHR_FK[325.13]DLGEEHFK | 0 | 0 | 0 | 0 | −0.0268 | 2.10E-06 | 10 | 10 | DTHK[325.13]SEIAHR_FK[325.13]DLGEEHFK | 0.99 | 0 | 1 | 3 | 0 | 2 | 1 | 4 | 0 | 36 | |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.09688.09688.2 |
1 | DTHK[325.13]SEIAHR | FK[325.13]DLGEEHFK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 2 | 2 | 1 | 1 | 1389.627 | 1445.646 | 6 | 3586.679 | DTHK[325.13]SEIAHR_FK[325.13]DLGEEHFK | 0 | 0 | 0 | 0 | −0.0268 | 3.06E-08 | 10 | 10 | DTHK[325.13]SEIAHR_FK[325.13]DLGEEHFK | 0.99 | 0 | 1 | 3 | 0 | 2 | 1 | 4 | 0 | 36 | |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.11445.11445.1 |
1 | LSQK[325.13]FPK | LCVLHEK[325.13]TPVSEK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 2 | 1 | 1 | 1043.529 | 1735.845 | 5 | 3530.764 | 1 | LSQK[325.13]FPK_LCVLHEK[325.13]TPVSEK | 0 | 0 | 0.0704 | 0 | −4.3359 | 1.45E+00 | 7 | 13 | LSQK[325.13]FPK_LCVLHEK[325.13]TPVSEK | 0.99 | 0 | 1 | 3 | −1 | 1 | 1 | 3 | 0 | 36 |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.09589.09589.2 |
1 | DTHK[325.13]SEIAHR | FK[325.13]DLGEEHFK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 2 | 2 | 1 | 1 | 1389.627 | 1445.646 | 7 | 3588.709 | DTHK[325.13]SEIAHR_FK[325.13]DLGEEHFK | 0 | 0 | 0 | 0 | 8.4624 | 1.75E-03 | 10 | 10 | DTHK[325.13]SEIAHR_FK[325.13]DLGEEHFK | 0.99 | 0 | 1 | 3 | 2 | 1 | 1 | 4 | 0 | 36 | |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.11736.11736.1 |
1 | SLGK[325.13]VGTR | CASIQK[325.13]FGER | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1013.514 | 1391.614 | 4 | 3158.531 | 1 | SLGK[325.13]VGTR_CASIQK[325.13]FGER | 0 | 0 | 0 | 0 | 0.7355 | 3.58E-03 | 8 | 10 | SLGK[325.13]VGTR_CASIQK[325.13]FGER | 0.99 | 0 | 1 | 3 | 0 | 0 | 1 | 2 | 0 | 36 |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.21809.21809.1 |
1 | ALK[325.13]AWSVAR | AEFVEVTK[325.13]LVTDLTK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1197.614 | 1888.967 | 4 | 3837.978 | 1 | ALK[325.13]AWSVAR_AEFVEVTK[325.13]LVTDLTK | 0 | 0 | 0 | 0 | −2.2869 | 2.03E-03 | 9 | 15 | ALK[325.13]AWSVAR_AEFVEVTK[325.13]LVTDLTK | 0.99 | 0 | 1 | 3 | 0 | 0 | 1 | 2 | 0 | 36 |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.21196.21196.1 |
1 | ALK[325.13]AWSVAR | NYQEAK[325.13]DAFLGSFLYEYSR | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1197.614 | 2497.107 | 4 | 4446.134 | 1 | ALK[325.13]AWSVAR_NYQEAK[325.13]DAFLGSFLYEYSR | 0 | 0 | 0 | 0 | 1.6673 | 6.72E-04 | 9 | 19 | ALK[325.13]AWSVAR_NYQEAK[325.13]DAFLGSFLYEYSR | 0.99 | 0 | 1 | 3 | 0 | 0 | 1 | 2 | 0 | 36 |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.09556.09556.1 |
1 | LSQK[325.13]FPK | EK[325.13]VLTSSAR | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1043.529 | 1186.583 | 5 | 2982.514 | LSQK[325.13]FPK_EK[325.13]VLTSSAR | 0 | 0 | 0.0704 | 0 | 1.0743 | 1.34E+00 | 7 | 9 | LSQK[325.13]FPK_EK[325.13]VLTSSAR | 0.99 | 0 | 1 | 3 | 0 | 0 | 1 | 2 | 0 | 36 | |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.19314.19314.1 |
1 | FWGK[325.13]YLYEIAR | LK[325.13]PDPNTLCDEFK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1641.783 | 1772.793 | 4 | 4165.987 | 1 | FWGK[325.13]YLYEIAR_LK[325.13]PDPNTLCDEFK | 0 | 0 | 0 | 0 | 1.5266 | 1.55E-05 | 11 | 13 | FWGK[325.13]YLYEIAR_LK[325.13]PDPNTLCDEFK | 0.99 | 0 | 1 | 3 | 0 | 0 | 1 | 2 | 0 | 36 |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.17748.17748.1 |
1 | K[325.13]QTALVELLK | LK[325.13]PDPNTLCDEFK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1338.739 | 1772.793 | 4 | 3864.931 | 1 | K[325.13]QTALVELLK_LK[325.13]PDPNTLCDEFK | 0 | 0 | 0 | 0 | 1.6518 | 3.47E-03 | 10 | 13 | K[325.13]QTALVELLK_LK[325.13]PDPNTLCDEFK | 0.99 | 0 | 1 | 3 | 0 | 0 | 1 | 2 | 0 | 36 |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.09714.09714.1 |
1 | SLGK[325.13]VGTR | LSQK[325.13]FPK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1013.514 | 1043.529 | 4 | 2809.451 | SLGK[325.13]VGTR_LSQK[325.13]FPK | 0 | 0 | 0 | 0 | 1.0251 | 6.76E-04 | 8 | 7 | SLGK[325.13]VGTR_LSQK[325.13]FPK | 0.99 | 0 | 1 | 3 | 0 | 0 | 1 | 2 | 0 | 36 | |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.09582.09582.2 |
1 | DTHK[325.13]SEIAHR | FK[325.13]DLGEEHFK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 2 | 1 | 1 | 1 | 1389.627 | 1445.646 | 5 | 3587.714 | DTHK[325.13]SEIAHR_FK[325.13]DLGEEHFK | 0 | 0 | 0 | 0 | 9.7338 | 6.36E-04 | 10 | 10 | DTHK[325.13]SEIAHR_FK[325.13]DLGEEHFK | 0.99 | 0 | 1 | 3 | 2 | 1 | 1 | 3 | 0 | 36 | |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.14989.14989.1 |
2 | K[325.13]VPQVSTPTLVEVSR | HLVDEPQNLIK[325.13]QNCDQFEK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1835.963 | 2551.165 | 5 | 5141.564 | 1 | K[325.13]VPQVSTPTLVEVSR_HLVDEPQNLIK[325.13]QNCDQFEK | 0 | 0 | 0 | 0 | 5.9764 | 4.48E-08 | 15 | 19 | K[325.13]VPQVSTPTLVEVSR_HLVDEPQNLIK[325.13]QNCDQFEK | 0.99 | 0 | 1 | 3 | 1 | 0 | 1 | 2 | 0 | 36 |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.15713.15713.1 |
1 | SLGK[325.13]VGTR | K[325.13]QTALVELLK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1013.514 | 1338.739 | 4 | 3105.651 | SLGK[325.13]VGTR_K[325.13]QTALVELLK | 0 | 0 | 0.0161 | 0 | 2.5248 | 2.69E-01 | 8 | 10 | SLGK[325.13]VGTR_K[325.13]QTALVELLK | 0.99 | 0 | 1 | 3 | 1 | 0 | 1 | 2 | 0 | 36 | |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.12947.12947.1 |
1 | ALK[325.13]AWSVAR | VTK[325.13]CCTESLVNR | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1197.614 | 1662.734 | 4 | 3612.771 | 1 | ALK[325.13]AWSVAR_VTK[325.13]CCTESLVNR | 0 | 0 | 0 | 0 | 4.8262 | 1.33E-03 | 9 | 12 | ALK[325.13]AWSVAR_VTK[325.13]CCTESLVNR | 0.99 | 0 | 1 | 3 | 1 | 0 | 1 | 2 | 0 | 36 |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.14583.14583.1 |
2 | K[325.13]VPQVSTPTLVEVSR | LAK[325.13]EYEATLEECCAK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1835.963 | 2010.855 | 4 | 4600.251 | 1 | K[325.13]VPQVSTPTLVEVSR_LAK[325.13]EYEATLEECCAK | 0 | 0 | 0 | 0 | 6.0566 | 4.82E-04 | 15 | 15 | K[325.13]VPQVSTPTLVEVSR_LAK[325.13]EYEATLEECCAK | 0.99 | 0 | 1 | 3 | 1 | 0 | 1 | 2 | 0 | 36 |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.13599.13599.1 |
1 | ALK[325.13]AWSVAR | LAK[325.13]EYEATLEECCAK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1197.614 | 2010.855 | 4 | 3961.888 | ALK[325.13]AWSVAR_LAK[325.13]EYEATLEECCAK | 0 | 0 | 0 | 0 | 3.3163 | 1.94E-03 | 9 | 15 | ALK[325.13]AWSVAR_LAK[325.13]EYEATLEECCAK | 0.99 | 0 | 1 | 3 | 1 | 0 | 1 | 2 | 0 | 36 | |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.13402.13402.1 |
2 | EK[325.13]VLTSSAR | LK[325.13]PDPNTLCDEFK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1186.583 | 1772.793 | 4 | 3710.79 | 1 | EK[325.13]VLTSSAR_LK[325.13]PDPNTLCDEFK | 0 | 0 | 0 | 0 | 2.5124 | 9.03E-03 | 9 | 13 | EK[325.13]VLTSSAR_LK[325.13]PDPNTLCDEFK | 0.99 | 0 | 1 | 3 | 1 | 0 | 1 | 2 | 0 | 36 |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.14553.14553.1 |
2 | LSQK[325.13]FPK | GACLLPK[325.13]IETM[147.04]R | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1043.529 | 1600.759 | 4 | 3395.71 | 1 | LSQK[325.13]FPK_GACLLPK[325.13]IETM[147.04]R | 0 | 0 | 0 | 0 | 5.024 | 1.09E-02 | 7 | 12 | LSQK[325.13]FPK_GACLLPK[325.13]IETM[147.04]R | 0.99 | 0 | 1 | 3 | 1 | 0 | 1 | 2 | 0 | 36 |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.08501.08501.1 |
3 | EK[325.13]VLTSSAR | CCTK[325.13]PESER | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1186.583 | 1362.518 | 4 | 3302.491 | 1 | EK[325.13]VLTSSAR_CCTK[325.13]PESER | 0 | 0 | 0 | 0 | 4.5971 | 1.21E-02 | 9 | 9 | EK[325.13]VLTSSAR_CCTK[325.13]PESER | 0.99 | 0 | 1 | 3 | 1 | 0 | 1 | 2 | 0 | 36 |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.15147.15147.1 |
4 | EK[325.13]VLTSSAR | GACLLPK[325.13]IETMR | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1186.583 | 1584.764 | 4 | 3523.771 | 1 | EK[325.13]VLTSSAR_GACLLPK[325.13]IETMR | 0 | 0 | 0 | 0 | 5.3448 | 1.74E-02 | 9 | 12 | EK[325.13]VLTSSAR_GACLLPK[325.13]IETMR | 0.99 | 0 | 1 | 3 | 1 | 0 | 1 | 2 | 0 | 36 |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.13046.13046.1 |
1 | CASIQK[325.13]FGER | LAK[325.13]EYEATLEECCAK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1391.614 | 2010.855 | 4 | 4155.891 | CASIQK[325.13]FGER_LAK[325.13]EYEATLEECCAK | 0 | 0 | 0 | 0 | 4.0403 | 3.34E-03 | 10 | 15 | CASIQK[325.13]FGER_LAK[325.13]EYEATLEECCAK | 0.99 | 0 | 1 | 3 | 1 | 0 | 1 | 2 | 0 | 36 | |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.13448.13448.1 |
1 | ALK[325.13]AWSVAR | LVTDLTK[325.13]VHK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1197.614 | 1349.719 | 5 | 3298.749 | ALK[325.13]AWSVAR_LVTDLTK[325.13]VHK | 0 | 0 | 0 | 0 | 3.2185 | 1.87E-03 | 9 | 10 | ALK[325.13]AWSVAR_LVTDLTK[325.13]VHK | 0.99 | 0 | 1 | 3 | 1 | 0 | 1 | 2 | 0 | 36 | |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.13808.13808.1 |
2 | EK[325.13]VLTSSAR | GACLLPK[325.13]IETM[147.04]R | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1186.583 | 1600.759 | 4 | 3540.771 | EK[325.13]VLTSSAR_GACLLPK[325.13]IETM[147.04]R | 0 | 0 | 0 | 0 | 6.7595 | 7.26E-02 | 9 | 12 | EK[325.13]VLTSSAR_GACLLPK[325.13]IETM[147.04]R | 0.99 | 0 | 1 | 3 | 1 | 0 | 1 | 2 | 0 | 36 | |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.12313.12313.1 |
1 | LSQK[325.13]FPK | LAK[325.13]EYEATLEECCAK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1043.529 | 2010.855 | 4 | 3806.811 | 1 | LSQK[325.13]FPK_LAK[325.13]EYEATLEECCAK | 0 | 0 | 0.0161 | 0 | 5.7775 | 4.38E-01 | 7 | 15 | LSQK[325.13]FPK_LAK[325.13]EYEATLEECCAK | 0.99 | 0 | 1 | 3 | 1 | 0 | 1 | 2 | 0 | 36 |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.13552.13552.1 |
1 | SLGK[325.13]VGTR | LK[325.13]PDPNTLCDEFK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 0.9999 | 1 | 1013.514 | 1772.793 | 4 | 3538.722 | 1 | SLGK[325.13]VGTR_LK[325.13]PDPNTLCDEFK | 0 | 0 | 0.0571 | 0 | 2.8629 | 1.02E+00 | 8 | 13 | SLGK[325.13]VGTR_LK[325.13]PDPNTLCDEFK | 0.9899 | 0 | 1 | 3 | 1 | 0 | 1 | 2 | 0 | 36 |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.17873.17873.1 |
1 | K[325.13]QTALVELLK | K[325.13]VPQVSTPTLVEVSR | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 0.9929 | 1 | 1338.739 | 1835.963 | 4 | 3926.115 | 1 | K[325.13]QTALVELLK_K[325.13]VPQVSTPTLVEVSR | 0 | 0 | 0.0704 | 0 | 1.8708 | 1.60E+00 | 10 | 15 | K[325.13]QTALVELLK_K[325.13]VPQVSTPTLVEVSR | 0.983 | 0 | 1 | 3 | 0 | 0 | 1 | 2 | 0 | 36 |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.09833.09833.1 |
3 | SLGK[325.13]VGTR | EK[325.13]VLTSSAR | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1013.514 | 1186.583 | 4 | 2951.49 | 1 | SLGK[325.13]VGTR_EK[325.13]VLTSSAR | 0 | 0 | 0 | 0 | −4.0434 | 7.04E-02 | 8 | 9 | SLGK[325.13]VGTR_EK[325.13]VLTSSAR | 0.99 | 0 | 1 | 3 | −1 | 0 | 1 | 2 | 0 | 36 |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.12229.12229.1 |
1 | ALK[325.13]AWSVAR | VHK[325.13]ECCHGDLLECADDRADLAK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 3 | 1 | 1 | 1197.614 | 2808.19 | 6 | 4759.216 | ALK[325.13]AWSVAR_VHK[325.13]ECCHGDLLECADDRADLAK | 0 | 0 | 0 | 0 | 1.4448 | 9.52E-02 | 9 | 22 | ALK[325.13]AWSVAR_VHK[325.13]ECCHGDLLECADDRADLAK | 0.99 | 0 | 1 | 3 | 0 | 0 | 1 | 4 | 0 | 36 | |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.11333.11333.1 |
1 | LAK[325.13]EYEATLEECCAK | VHK[325.13]ECCHGDLLECADDR | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 3 | 1 | 1 | 2010.855 | 2309.91 | 5 | 5075.164 | LAK[325.13]EYEATLEECCAK_VHK[325.13]ECCHGDLLECADDR | 0 | 0 | 0 | 0 | 1.278 | 2.96E-06 | 15 | 17 | LAK[325.13]EYEATLEECCAK_VHK[325.13]ECCHGDLLECADDR | 0.99 | 0 | 1 | 3 | 0 | 0 | 1 | 4 | 0 | 36 | |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.13129.13129.1 |
1 | LSQK[325.13]FPK | CASIQK[325.13]FGER | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1043.529 | 1391.614 | 4 | 3185.521 | 1 | LSQK[325.13]FPK_CASIQK[325.13]FGER | 0 | 0 | 0.0161 | 0 | −8.3832 | 5.70E-01 | 7 | 10 | LSQK[325.13]FPK_CASIQK[325.13]FGER | 0.99 | 0 | 1 | 3 | −2 | 0 | 1 | 2 | 0 | 36 |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.12049.12049.1 |
1 | LSQK[325.13]FPK | ECCDK[325.13]PLLEK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 0.9693 | 1043.529 | 1487.627 | 4 | 3284.571 | 1 | LSQK[325.13]FPK_ECCDK[325.13]PLLEK | 0 | 0 | 0.0571 | 0 | 2.9903 | 1.08E+00 | 7 | 10 | LSQK[325.13]FPK_ECCDK[325.13]PLLEK | 0.9596 | 0 | 1 | 3 | 1 | 0 | 1 | 2 | 0 | 36 |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.17844.17844.1 |
1 | SLGK[325.13]VGTR | FWGK[325.13]YLYEIAR | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1013.514 | 1641.783 | 4 | 3408.731 | 1 | SLGK[325.13]VGTR_FWGK[325.13]YLYEIAR | 0 | 0 | 0 | 0 | 8.4773 | 1.36E-02 | 8 | 11 | SLGK[325.13]VGTR_FWGK[325.13]YLYEIAR | 0.99 | 0 | 1 | 3 | 2 | 0 | 1 | 2 | 0 | 36 |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.18444.18444.2 |
1 | FWGK[325.13]YLYEIAR | LK[325.13]PDPNTLCDEFKADEK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 2 | 2 | 1 | 1 | 1641.783 | 2215.994 | 5 | 4612.214 | 1 | FWGK[325.13]YLYEIAR_LK[325.13]PDPNTLCDEFKADEK | 0 | 0 | 0 | 0 | 6.8221 | 7.57E-04 | 11 | 17 | FWGK[325.13]YLYEIAR_LK[325.13]PDPNTLCDEFKADEK | 0.99 | 0 | 1 | 3 | 1 | 0 | 1 | 4 | 0 | 36 |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.09576.09576.2 |
1 | DTHK[325.13]SEIAHR | FK[325.13]DLGEEHFK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 2 | 2 | 1 | 1 | 1389.627 | 1445.646 | 6 | 3587.656 | DTHK[325.13]SEIAHR_FK[325.13]DLGEEHFK | 0 | 0 | 0 | 0 | 6.2311 | 4.60E-03 | 10 | 10 | DTHK[325.13]SEIAHR_FK[325.13]DLGEEHFK | 0.99 | 0 | 1 | 3 | 1 | 0 | 1 | 4 | 0 | 36 | |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.12071.12071.1 |
1 | ALK[325.13]AWSVAR | VHK[325.13]ECCHGDLLECADDR | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 2 | 1 | 1 | 1197.614 | 2309.91 | 5 | 4258.937 | ALK[325.13]AWSVAR_VHK[325.13]ECCHGDLLECADDR | 0 | 0 | 0 | 0 | 1.8627 | 3.94E-05 | 9 | 17 | ALK[325.13]AWSVAR_VHK[325.13]ECCHGDLLECADDR | 0.99 | 0 | 1 | 3 | 0 | 0 | 1 | 3 | 0 | 36 | |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.19581.19581.1 |
1 | FWGK[325.13]YLYEIAR | DDSPDLPK[325.13]LKPDPNTLCDEFK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 2 | 1 | 1 | 1641.783 | 2640.19 | 5 | 5035.385 | FWGK[325.13]YLYEIAR_DDSPDLPK[325.13]LKPDPNTLCDEFK | 0 | 0 | 0 | 0 | 1.4194 | 1.10E-05 | 11 | 21 | FWGK[325.13]YLYEIAR_DDSPDLPK[325.13]LKPDPNTLCDEFK | 0.99 | 0 | 1 | 3 | 0 | 0 | 1 | 3 | 0 | 36 | |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.11224.11224.1 |
1 | LSQK[325.13]FPK | LCVLHEK[325.13]TPVSEK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 2 | 1 | 1 | 1043.529 | 1735.845 | 5 | 3531.764 | LSQK[325.13]FPK_LCVLHEK[325.13]TPVSEK | 0 | 0 | 0.5847 | 0 | 4.3347 | 3.95E+01 | 7 | 13 | LSQK[325.13]FPK_LCVLHEK[325.13]TPVSEK | 0.99 | 0 | 1 | 3 | 1 | 0 | 1 | 3 | 0 | 36 | |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.11372.11372.1 |
1 | CASIQK[325.13]FGER | VHK[325.13]ECCHGDLLECADDR | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 2 | 1 | 1 | 1391.614 | 2309.91 | 5 | 4454.912 | CASIQK[325.13]FGER_VHK[325.13]ECCHGDLLECADDR | 0 | 0 | 0 | 0 | 3.9303 | 3.24E-05 | 10 | 17 | CASIQK[325.13]FGER_VHK[325.13]ECCHGDLLECADDR | 0.99 | 0 | 1 | 3 | 1 | 0 | 1 | 3 | 0 | 36 | |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.07398.07398.1 |
1 | FK[325.13]DLGEEHFK | FK[325.13]DLGEEHFK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1445.646 | 1445.646 | 5 | 3643.698 | 1 | FK[325.13]DLGEEHFK_FK[325.13]DLGEEHFK | 0 | 0 | 0 | 0 | 0.0549 | 8.11E-07 | 10 | 10 | FK[325.13]DLGEEHFK_FK[325.13]DLGEEHFK | 0.99 | 1 | * | 3 | 0 | 1 | * | 2 | 0 | |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.11046.11046.1 |
3 | LSQK[325.13]FPK | LSQK[325.13]FPK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1043.529 | 1043.529 | 4 | 2839.451 | 1 | LSQK[325.13]FPK_LSQK[325.13]FPK | 0 | 0 | 0 | 0 | 4.1233 | 6.09E-03 | 7 | 7 | LSQK[325.13]FPK_LSQK[325.13]FPK | 0.99 | 1 | * | 3 | 1 | 1 | * | 2 | 0 | |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.10191.10191.1 |
2 | FK[325.13]DLGEEHFK | FK[325.13]DLGEEHFK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1445.646 | 1445.646 | 5 | 3643.702 | FK[325.13]DLGEEHFK_FK[325.13]DLGEEHFK | 0 | 0 | 0 | 0 | 1.2625 | 3.69E-04 | 10 | 10 | FK[325.13]DLGEEHFK_FK[325.13]DLGEEHFK | 0.99 | 1 | * | 3 | 0 | 2 | * | 2 | 0 | ||
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.11420.11420.1 |
1 | LSQK[325.13]FPK | LSQK[325.13]FPK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1043.529 | 1043.529 | 4 | 2836.445 | LSQK[325.13]FPK_LSQK[325.13]FPK | 0 | 0 | 0 | 0 | −6.2409 | 6.79E-04 | 7 | 7 | LSQK[325.13]FPK_LSQK[325.13]FPK | 0.99 | 1 | * | 3 | −1 | 1 | * | 2 | 0 | ||
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.11310.11310.1 |
5 | LSQK[325.13]FPK | LSQK[325.13]FPK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1043.529 | 1043.529 | 4 | 2837.451 | LSQK[325.13]FPK_LSQK[325.13]FPK | 0 | 0 | 0 | 0 | −4.1262 | 1.88E-03 | 7 | 7 | LSQK[325.13]FPK_LSQK[325.13]FPK | 0.99 | 1 | * | 3 | −1 | 1 | * | 2 | 0 | ||
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.07451.07451.2 |
1 | FK[325.13]DLGEEHFK | FK[325.13]DLGEEHFK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 2 | 2 | 1 | 1 | 1445.646 | 1445.646 | 7 | 3643.695 | FK[325.13]DLGEEHFK_FK[325.13]DLGEEHFK | 0 | 0 | 0 | 0 | 0.8782 | 9.56E-03 | 10 | 10 | FK[325.13]DLGEEHFK_FK[325.13]DLGEEHFK | 0.99 | 1 | * | 3 | 0 | 1 | * | 4 | 0 | ||
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.09649.09649.2 |
1 | FK[325.13]DLGEEHFK | FK[325.13]DLGEEHFK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 2 | 2 | 1 | 1 | 1445.646 | 1445.646 | 6 | 3642.699 | FK[325.13]DLGEEHFK_FK[325.13]DLGEEHFK | 0 | 0 | 0 | 0 | 0.3843 | 1.15E-02 | 10 | 10 | FK[325.13]DLGEEHFK_FK[325.13]DLGEEHFK | 0.99 | 1 | * | 3 | 0 | 1 | * | 4 | 0 | ||
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.09595.09595.2 |
2 | FK[325.13]DLGEEHFK | FK[325.13]DLGEEHFK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 2 | 2 | 1 | 1 | 1445.646 | 1445.646 | 5 | 3645.714 | FK[325.13]DLGEEHFK_FK[325.13]DLGEEHFK | 0 | 0 | 0 | 0 | 4.3662 | 1.93E-04 | 10 | 10 | FK[325.13]DLGEEHFK_FK[325.13]DLGEEHFK | 0.99 | 1 | * | 3 | 1 | 2 | * | 4 | 0 | ||
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.09541.09541.2 |
2 | FK[325.13]DLGEEHFK | FK[325.13]DLGEEHFK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 2 | 2 | 0.9998 | 0.9998 | 1445.646 | 1445.646 | 6 | 3644.716 | FK[325.13]DLGEEHFK_FK[325.13]DLGEEHFK | 0 | 0 | 0.0704 | 0 | 5.1145 | 1.97E+00 | 10 | 10 | FK[325.13]DLGEEHFK_FK[325.13]DLGEEHFK | 0.9896 | 1 | * | 3 | 1 | 2 | * | 4 | 0 | ||
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.11066.11066.1 |
1 | LSQK[325.13]FPK | LSQK[325.13]FPK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 1 | 1043.529 | 1043.529 | 4 | 2840.451 | LSQK[325.13]FPK_LSQK[325.13]FPK | 0 | 0 | 0 | 0 | 4.1219 | 5.26E-03 | 7 | 7 | LSQK[325.13]FPK_LSQK[325.13]FPK | 0.99 | 1 | * | 3 | 1 | 0 | * | 2 | 0 | ||
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.08535.08535.1 |
1 | SLGK[325.13]VGTR | CCTK[325.13]PESER | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 1 | 1 | 1 | 0.8441 | 1013.514 | 1362.518 | 4 | 3127.412 | SLGK[325.13]VGTR_CCTK[325.13]PESER | 0 | 0 | 0.0161 | 0 | −8.1755 | 1.93E-01 | 8 | 9 | SLGK[325.13]VGTR_CCTK[325.13]PESER | 0.8357 | 0 | 1 | 2 | −2 | 0 | 1 | 2 | 0 | 36 | |
| 1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.07384.07384.2 |
1 | FK[325.13]DLGEEHFK | FK[325.13]DLGEEHFK | sp|P02769|ALBU_BOVIN | sp|P02769|ALBU_BOVIN | 2 | 2 | 1 | 1 | 1445.646 | 1445.646 | 6 | 3643.707 | FK[325.13]DLGEEHFK_FK[325.13]DLGEEHFK | 0 | 0 | 0 | 0 | 2.5523 | 1.06E-04 | 10 | 10 | FK[325.13]DLGEEHFK_FK[325.13]DLGEEHFK | 0.99 | 1 | * | 3 | 1 | 0 | * | 4 | 0 | ||
| 0 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.18415.18415.1 |
1 | RYAINASK[325.13]LK | LAK[325.13]EYEATLEECCAK | sp|P39630|RMLB_BACSU | sp|P02769|ALBU_BOVIN | 1 | 1 | 0.0012 | 1 | 1359.715 | 2010.855 | 4 | 4122.971 | 0 | RYAINASK[325.13]LK_LAK[325.13]EYEATLEECCAK | 0 | 0 | 0.0779 | 0.026 | 0.9481 | 2.29E+00 | 10 | 15 | RYAINASK[325.13]LK_LAK[325.13]EYEATLEECCAK | 0.0012 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0.0128 | 0 |
| 0 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.12640.12640.1 |
1 | LK[325.13]GM[147.04]EKR | CASIQK[325.13]FGER | sp|C0SPB4|YHAQ_BACSU | sp|P02769|ALBU_BOVIN | 1 | 1 | 0.0002 | 1 | 1073.518 | 1391.614 | 4 | 3218.531 | 0 | LK[325.13]GM[147.04]EKR_CASIQK[325.13]FGER | 0 | 0 | 0.3113 | 0.0385 | 1.7645 | 1.53E+01 | 7 | 10 | LK[325.13]GM[147.04]EKR_CASIQK[325.13]FGER | 0.0002 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0.0253 | 0 |
| 0 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.12618.12618.1 |
1 | LK[325.13]GM[147.04]EKR | CASIQK[325.13]FGER | sp|C0SPB4|YHAQ_BACSU | sp|P02769|ALBU_BOVIN | 1 | 1 | 0.0001 | 1 | 1073.518 | 1391.614 | 4 | 3215.531 | LK[325.13]GM[147.04]EKR_CASIQK[325.13]FGER | 0 | 0 | 0.5398 | 0.0506 | −1.7661 | 3.62E+01 | 7 | 10 | LK[325.13]GM[147.04]EKR_CASIQK[325.13]FGER | 0.0001 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0.0375 | 0 | |
| 0 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.16165.16165.1 |
1 | K[325.13]PNSMVAK | GACLLPK[325.13]IETMR | sp|Q04810|DPAB_BACSU | sp|P02769|ALBU_BOVIN | 1 | 1 | 0.0001 | 1 | 1070.507 | 1584.764 | 4 | 3408.691 | 0 | K[325.13]PNSMVAK_GACLLPK[325.13]IETMR | 0 | 0 | 0.2688 | 0.0506 | 4.4504 | 1.03E+01 | 8 | 12 | K[325.13]PNSMVAK_GACLLPK[325.13]IETMR | 0.0001 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | 0.0494 | 0 |
| −1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.10737.10737.1 |
2 | GIK[325.13]SHK | K[325.13]VIAYSGEK | sp|P96583|TOP3_BACSU | sp|P39216|TLPA_BACSU | 1 | 2 | 0 | 0 | 865.4294 | 1190.582 | 4 | 2808.451 | 0 | GIK[325.13]SHK_K[325.13]VIAYSGEK | 0 | 0 | 0.7841 | 0.7644 | 12.2897 | 1.45E+02 | 6 | 9 | GIK[325.13]SHK_K[325.13]VIAYSGEK | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 3 | 0.061 | 0 |
| −1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.11445.11445.1 |
2 | K[325.13]ELSEK | DVLK[325.13]KIDGYTSVGTR | sp|O31489|YDCI_BACSU,sp|P94583|RAPD_BACSU,sp|O34574|YEFB_BACSU,sp|O31522|YESS_BACSU,sp|O05409|SIGZ_BACSU,rev_sp|Q02115|LYAT_BACSU | sp|C0SP94|YHFQ_BACSU | 1 | 2 | 0 | 0 | 929.4342 | 1847.927 | 5 | 3530.764 | 0 | K[325.13]ELSEK_DVLK[325.13]KIDGYTSVGTR | 0 | 0 | 0.6127 | 0.7644 | 0.612 | 4.63E+01 | 6 | 15 | K[325.13]ELSEK_DVLK[325.13]KIDGYTSVGTR | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0.0723 | 0 |
| −1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.13599.13599.1 |
2 | K[325.13]IFGGALGSR | LAK[325.13]EYEATLEECCAK | sp|O34607|SDHAA_BACSU | sp|P02769|ALBU_BOVIN | 1 | 1 | 0 | 1 | 1201.609 | 2010.855 | 4 | 3961.888 | 0 | K[325.13]IFGGALGSR_LAK[325.13]EYEATLEECCAK | 0 | 0 | 0.5847 | 0.7644 | −247.805 | 4.05E+01 | 10 | 15 | K[325.13]IFGGALGSR_LAK[325.13]EYEATLEECCAK | 0 | 0 | 0 | 0 | −5 | 0 | 0 | 2 | 0.0833 | 0 |
| −1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.10410.10410.1 |
1 | SLGK[325.13]VGTR | K[325.13]ELSFHEK | sp|P02769|ALBU_BOVIN | sp|P96738|CAPA_BACSU | 1 | 1 | 0.9992 | 0 | 1013.514 | 1213.562 | 4 | 2981.51 | 0 | SLGK[325.13]VGTR_K[325.13]ELSFHEK | 0 | 0 | 0.4218 | 0.7644 | 9.7367 | 2.64E+01 | 8 | 8 | SLGK[325.13]VGTR_K[325.13]ELSFHEK | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 2 | 0.0941 | 0 |
| −1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.13808.13808. |
1 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 3540.771 | 0.7644 | 0.1047 | |||||||||||||||||||||||
| −1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.08552.08552. |
2 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 3128.411 | 0.7644 | 0.1149 | |||||||||||||||||||||||
| −1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.11046.11046.1 |
5 | SSK[325.13]KER | LLTK[325.13]LMNK | sp|P27632|CPXM_BACPZ,rev_sp|O34990|PURU_BACSU | sp|P25144|CCPA_BACSU | 1 | 1 | 0 | 0 | 930.4407 | 1156.616 | 4 | 2839.451 | 0 | SSK[325.13]KER_LLTK[325.13]LMNK | 0 | 0 | 0.6296 | 0.7644 | 3.8934 | 5.45E+01 | 6 | 8 | SSK[325.13]KER_LLTK[325.13]LMNK | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | 0.125 | 0 |
| −1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.15147.15147. |
3 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 3523.771 | 0.7644 | 0.1348 | |||||||||||||||||||||||
| −1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.13562.13562.1 |
3 | EAPRVPK[325.13]GGK | DVK[325.13]IRITDR | sp|P09122|DPO3X_BACSU | sp|P94441|YFIM_BACSU | 1 | 2 | 0 | 0 | 1234.631 | 1311.678 | 5 | 3298.746 | 0 | EAPRVPK[325.13]GGK_DVK[325.13]IRITDR | 0 | 0 | 0.7841 | 0.7644 | 9.6528 | 1.32E+02 | 10 | 9 | EAPRVPK[325.13]GGK_DVK[325.13]IRITDR | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 3 | 0.1444 | 0 |
| −1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.08501.08501. |
2 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 3302.491 | 0.7644 | 0.1538 | |||||||||||||||||||||||
| −1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.11310.11310. |
1 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 2837.451 | 0.7644 | 0.163 | |||||||||||||||||||||||
| −1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.09714.09714. |
3 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 2809.451 | 0.7644 | 0.172 | |||||||||||||||||||||||
| −1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.11310.11310.1 |
2 | K[325.13]PIFPK | GSIMK[325.13]LGEK | sp|O32057|YRZF_BACSU,sp|O07546|YHEE_BACSU | sp|P16971|RECA_BACSU | 1 | 1 | 0 | 0 | 925.4909 | 1158.559 | 4 | 2837.451 | 0 | K[325.13]PIFPK_GSIMK[325.13]LGEK | 0 | 0 | 0.5847 | 0.7644 | 1.4732 | 4.11E+01 | 6 | 9 | K[325.13]PIFPK_GSIMK[325.13]LGEK | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0.1809 | 0 |
| −1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.11046.11046.2 |
2 | MGK[325.13]GDPK | LLTK[325.13]LMNK | sp|P10103|HMGB1_BOVIN | sp|P25144|CCPA_BACSU | 2 | 1 | 0 | 0 | 928.396 | 1156.616 | 4 | 2839.451 | 0 | MGK[325.13]GDPK_LLTK[325.13]LMNK | 0 | 0 | 0.6873 | 0.7644 | 11.8287 | 7.44E+01 | 7 | 8 | MGK[325.13]GDPK_LLTK[325.13]LMNK | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 3 | 0.1895 | 0 |
| −1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.13562.13562.1 |
1 | K[325.13]LFRLK | K[325.13]LTAAENLPEHK | sp|O34879|YKUO_BACSU,sp|P71006|ALBF_BACSU | sp|P94593|YWQA_BACSU | 1 | 1 | 0 | 0 | 1000.571 | 1546.763 | 5 | 3298.746 | 0 | K[325.13]LFRLK_K[325.13]LTAAENLPEHK | 0 | 0 | 0.7841 | 0.7644 | 2.2487 | 7.77E+02 | 6 | 12 | K[325.13]LFRLK_K[325.13]LTAAENLPEHK | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0.1979 | 0 |
| −1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.11046.11046.1 |
1 | K[325.13]PIFPK | QK[325.13]VTDLMK | sp|O32057|YRZF_BACSU,sp|O07546|YHEE_BACSU | sp|P21881|ODPA_BACSU | 1 | 1 | 0 | 0 | 925.4909 | 1158.559 | 4 | 2839.451 | 0 | K[325.13]PIFPK_QK[325.13]VTDLMK | 0 | 0 | 0.5909 | 0.7644 | 1.4721 | 4.27E+01 | 6 | 8 | K[325.13]PIFPK_QK[325.13]VTDLMK | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0.2062 | 0 |
| −1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.08501.08501. |
1 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 3302.491 | 0.7644 | 0.2143 | |||||||||||||||||||||||
| −1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.11310.11310.1 |
3 | SLQK[325.13]MK | LLTK[325.13]LMNK | sp|O34904|YEBD_BACSU,rev_sp|P37946|ILVA_BACSU | sp|P25144|CCPA_BACSU | 1 | 1 | 0 | 0 | 930.4481 | 1156.616 | 4 | 2837.451 | 0 | SLQK[325.13]MK_LLTK[325.13]LMNK | 0 | 0 | 0.6296 | 0.7644 | −6.5016 | 5.48E+01 | 6 | 8 | SLQK[325.13]MK_LLTK[325.13]LMNK | 0 | 0 | 0 | 0 | −1 | 0 | 0 | 2 | 0.2222 | 0 |
| −1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.17748.17748. |
2 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 3864.931 | 0.7644 | 0.23 | |||||||||||||||||||||||
| −1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.14553.14553. |
1 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 3395.71 | 0.7644 | 0.2376 | |||||||||||||||||||||||
| −1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.09833.09833.2 |
1 | TIGAK[325.13]VNGK | K[325.13]LGPASSEK | sp|O54408|RELA_BACSU | sp|O34538|YCDA_BACSU | 2 | 1 | 0 | 0 | 1083.556 | 1112.535 | 4 | 2951.49 | 0 | TIGAK[325.13]VNGK_K[325.13]LGPASSEK | 0 | 0 | 0.4788 | 0.7644 | 1.9604 | 3.10E+01 | 9 | 9 | TIGAK[325.13]VNGK_K[325.13]LGPASSEK | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0.2451 | 0 |
| −1 | /net/gs/vol4/shared/brucelab/search/ keller/ControlDataset/ MANGO2/112817_BSA_BDP_mango_2hr _1.11310.11310.1 |
6 | SGAK[325.13]AYR | K[325.13]AITLGHAK | sp|P71089|YGAJ_BACSU | sp|P42420|IOLJ_BACSU | 1 | 1 | 0 | 0 | 948.4301 | 1134.603 | 4 | 2837.451 | 0 | SGAK[325.13]AYR_K[325.13]AITLGHAK | 0 | 0 | 0.7841 | 0.7644 | 4.3638 | 2.56E+02 | 7 | 9 | SGAK[325.13]AYR_K[325.13]AITLGHAK | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | 0.2524 | 0 |
Figure 5.
Structure of BSA (PDB = 3V03) with cross-links listed in table 2 mapped on. The backbone is displayed as a cartoon ribbon colored with a gradient from red and the N-terminus to blue at the C-terminus. Cross-linked lysine residues are displayed as green space-filled residues and cross-links between them are grey bars.
Comparison of the identified cross-linked sites with existing structural data in the PDB results in high agreement with ~ 95% of the links with Euclidean Cα- Cα distances of less than 35 Å [38]. This observation indicates that cross-linking derived distance constraints can be useful for generating structural models for proteins and protein complexes for which there is little to no existing structural data. Cross-link guided molecular docking utilizing distance restraint data from large scale XL-MS experiments can produce molecular models for protein complexes that closely resemble models produced by other structural biology techniques including cryo-EM. For example, large scale cross-linking data resulting from XL-MS analysis of intact mitochondria produced a cross-link derived model for the mitochondrial oxidative phosphorylation (OXPHOS) supercomplex consisting Complex I and Complex III. This model was found to be within 1.3 – 2.4 angstroms root mean square deviation (rmsd) of a cryo-EM derived orthologous model [9]. In another example, 28 cross-linked peptide pairs identified by XL-MS analysis of mouse heart tissue, was used to construct a molecular model for the interaction between subunits ECHA and ECHB of murine trifunctional protein important for beta oxidation of fatty acids [19]. The cross-linked-directed EHCA-EHCB model structure was observed to be very similar (rmsd = 3.6 Å for ECHA, 7.9 Å for ECHB) to a subsequent model structure resultant cryo-EM measurements of the human ortholog complex [54] (Fig. 6).
Figure 6.
Comparison of structural models for the mitochondrial trifunctional enzyme complex generated by XL-MS guided docking and Cryo-EM. The trifunctional enzyme consists of alpha (ECHA, red = XL-MS or magenta = Cryo-EM) and beta (ECHB, dark blue = XL-MS or teal blue = Cryo-EM) subunits is shown in, PDB 5zqz). The XL-MS interaction model was generated by cross-link guided molecular docking using 28 Lys-Lys distance restraints identified from large scale XL-MS analysis of mouse heart tissue [19], the Cryo-EM model is the human sequence, PDB 5zpz published by Liang el al. [54].
In vivo XL-MS can also provide structural and interaction information on previously unknown complexes. In bacterial systems, in vivo XL-MS measurements enabled identification of protein interactions important in both virulence and drug resistance in a clinical isolate of an ESKAPE pathogen. For instance, in vivo XL-MS studies with A. baumannii AB-5075 isolated from a patient at Walter Reed Army Medical Center [55], identified interactions between the carbapenemase Oxa23 and a series of outer membrane porin proteins, including carbapenem associated resistance protein (CarO). These cross-linked peptides uncovered new mechanistic details on increased carbapenem resistance in this multidrug resistant highly virulent pathogen [18]. XL-MS can also be used to characterize host-pathogen molecular interactions as demonstrated in a model of bacterial infection using A. baumannii AB-5075 and the human bronchial epithelial cell line H292. In those studies, a key virulence factor in A baumannii, outer membrane protein A (OmpA) was identified cross-linked to nuclear, mitochondrial and desmosomal proteins [13]. These are a few specific examples demonstrating the potential to utilize large-scale XL-MS data to derive useful new structural data on protein complexes as they reside within their native biological system. However, as more labs begin to implement in vivo cross-linking experiments with this or other protocols, the utility of identified cross-linked peptides will grow beyond a single research lab or even beyond a single research field. Once identified, each cross-linked peptide pair can serve as useful in vivo probe for a particular protein conformation or interaction. This information can be exploited in targeted quantitative experiments using MS methods such as parallel reaction monitoring (PRM) to quantify cross-link levels between differing biological states or perturbations such as drug treatment in cells. The generality of this approach was demonstrated in a cross-laboratory study utilizing PRM to measure drug-induced structural and interaction changes of heat shock protein 90 (Hsp90) [37]. Thus, each identified cross-linked peptide pair resultant from application of this protocol will not only provide the individual researcher with new insight on their own biological questions and further increase the collective knowledge-base of in vivo interactomes, but also contribute to a massive array of PRM assays useful for in vivo studies on thousands of proteins and complexes with any type of perturbation.
REFERENCES
- 1.Holding AN, XL-MS: Protein cross-linking coupled with mass spectrometry. Methods, 2015. 89: p. 54–63. [DOI] [PubMed] [Google Scholar]
- 2.Leitner A, et al. , Crosslinking and Mass Spectrometry: An Integrated Technology to Understand the Structure and Function of Molecular Machines. Trends Biochem Sci, 2016. 41(1): p. 20–32. [DOI] [PubMed] [Google Scholar]
- 3.Paramelle D, et al. , Chemical cross-linkers for protein structure studies by mass spectrometry. Proteomics, 2013. 13(3–4): p. 438–56. [DOI] [PubMed] [Google Scholar]
- 4.Sinz A, Divide and conquer: cleavable cross-linkers to study protein conformation and protein-protein interactions. Anal Bioanal Chem, 2017. 409(1): p. 33–44. [DOI] [PubMed] [Google Scholar]
- 5.Anderson GA, et al. , Informatics strategies for large-scale novel cross-linking analysis. J Proteome Res, 2007. 6(9): p. 3412–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leitner A, et al. , Probing native protein structures by chemical cross-linking, mass spectrometry, and bioinformatics. Mol Cell Proteomics, 2010. 9(8): p. 1634–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu F, et al. , Proteome-wide profiling of protein assemblies by cross-linking mass spectrometry. Nat Methods, 2015. 12(12): p. 1179–84. [DOI] [PubMed] [Google Scholar]
- 8.Tan D, et al. , Trifunctional cross-linker for mapping protein-protein interaction networks and comparing protein conformational states. Elife, 2016. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schweppe DK, et al. , Mitochondrial protein interactome elucidated by chemical cross-linking mass spectrometry. Proc Natl Acad Sci U S A, 2017. 114(7): p. 1732–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu F, et al. , The interactome of intact mitochondria by cross-linking mass spectrometry provides evidence for coexisting respiratory supercomplexes. Mol Cell Proteomics, 2018. 17(2): p. 216–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fasci D, et al. , Histone interaction landscapes visualized by crosslinking mass spectrometry in intact cell nuclei. Mol Cell Proteomics, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu X, et al. , Dynamic Proteome Response of Pseudomonas aeruginosa to Tobramycin Antibiotic Treatment. Mol Cell Proteomics, 2015. 14(8): p. 2126–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schweppe DK, et al. , Host-Microbe Protein Interactions during Bacterial Infection. Chem Biol, 2015. 22(11): p. 1521–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Navare AT, et al. , Probing the protein interaction network of Pseudomonas aeruginosa cells by chemical cross-linking mass spectrometry. Structure, 2015. 23(4): p. 762–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weisbrod CR, et al. , In vivo protein interaction network identified with a novel real-time cross-linked peptide identification strategy. J Proteome Res, 2013. 12(4): p. 1569–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chavez JD, et al. , Protein interactions, post-translational modifications and topologies in human cells. Mol Cell Proteomics, 2013. 12(5): p. 1451–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaake RM, et al. , A new in vivo cross-linking mass spectrometry platform to define protein-protein interactions in living cells. Mol Cell Proteomics, 2014. 13(12): p. 3533–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu X, et al. , In vivo protein interaction network analysis reveals porin-localized antibiotic inactivation in Acinetobacter baumannii strain AB5075. Nat Commun, 2016. 7: p. 13414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chavez JD, et al. , Chemical Crosslinking Mass Spectrometry Analysis of Protein Conformations and Supercomplexes in Heart Tissue. Cell Syst, 2018. 6(1): p. 136–141 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klykov O, et al. , Efficient and robust proteome-wide approaches for cross-linking mass spectrometry. Nat Protoc, 2018. 13(12): p. 2964–2990. [DOI] [PubMed] [Google Scholar]
- 21.Iacobucci C, et al. , A cross-linking/mass spectrometry workflow based on MS-cleavable cross-linkers and the MeroX software for studying protein structures and protein-protein interactions. Nat Protoc, 2018. 13(12): p. 2864–2889. [DOI] [PubMed] [Google Scholar]
- 22.Orban-Nemeth Z, et al. , Structural prediction of protein models using distance restraints derived from cross-linking mass spectrometry data. Nat Protoc, 2018. 13(3): p. 478–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang X, et al. , Mass spectrometry identifiable cross-linking strategy for studying protein-protein interactions. Anal Chem, 2005. 77(1): p. 311–8. [DOI] [PubMed] [Google Scholar]
- 24.Tang X and Bruce JE, A new cross-linking strategy: protein interaction reporter (PIR) technology for protein-protein interaction studies. Mol Biosyst, 2010. 6(6): p. 939–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petrotchenko EV, et al. , BiPS, a photocleavable, isotopically coded, fluorescent cross-linker for structural proteomics. Mol Cell Proteomics, 2009. 8(2): p. 273–86. [DOI] [PubMed] [Google Scholar]
- 26.Yang L, et al. , In vivo application of photocleavable protein interaction reporter technology. J Proteome Res, 2012. 11(2): p. 1027–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng Q, et al. , Probing Protein 3D Structures and Conformational Changes Using Electrochemistry-Assisted Isotope Labeling Cross-Linking Mass Spectrometry. J Am Soc Mass Spectrom, 2016. 27(5): p. 864–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng Q, et al. , Cross-linking electrochemical mass spectrometry for probing protein three-dimensional structures. Anal Chem, 2014. 86(18): p. 8983–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kao A, et al. , Development of a novel cross-linking strategy for fast and accurate identification of cross-linked peptides of protein complexes. Mol Cell Proteomics, 2011. 10(1): p. M110 002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muller MQ, et al. , A universal matrix-assisted laser desorption/ionization cleavable cross-linker for protein structure analysis. Rapid Commun Mass Spectrom, 2011. 25(1): p. 155–61. [DOI] [PubMed] [Google Scholar]
- 31.Petrotchenko EV, Serpa JJ, and Borchers CH, An isotopically coded CID-cleavable biotinylated cross-linker for structural proteomics. Mol Cell Proteomics, 2011. 10(2): p. M110 001420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmidt C and Urlaub H, Combining cryo-electron microscopy (cryo-EM) and cross-linking mass spectrometry (CX-MS) for structural elucidation of large protein assemblies. Curr Opin Struct Biol, 2017. 46: p. 157–168. [DOI] [PubMed] [Google Scholar]
- 33.Rinner O, et al. , Identification of cross-linked peptides from large sequence databases. Nat Methods, 2008. 5(4): p. 315–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohr JP, et al. , Mango: A General Tool for Collision Induced Dissociation-Cleavable Cross-Linked Peptide Identification. Anal Chem, 2018. 90(10): p. 6028–6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng C, et al. , XLink-DB: database and software tools for storing and visualizing protein interaction topology data. J Proteome Res, 2013. 12(4): p. 1989–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schweppe DK, et al. , XLinkDB 2.0: Integrated, large-scale structural analysis of protein crosslinking data. Bioinformatics, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chavez JD, et al. , A General Method for Targeted Quantitative Cross-Linking Mass Spectrometry. PLOS ONE, 2016. 11(12): p. e0167547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chavez JD, et al. , In Vivo Conformational Dynamics of Hsp90 and Its Interactors. Cell Chem Biol, 2016. 23(6): p. 716–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chavez JD, et al. , Quantitative interactome analysis reveals a chemoresistant edgotype. Nat Commun, 2015. 6: p. 7928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhong X, et al. , Large-Scale and Targeted Quantitative Cross-Linking MS Using Isotope-Labeled Protein Interaction Reporter (PIR) Cross-Linkers. Journal of Proteome Research, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chavez JD, et al. , Cross-linking measurements of the Potato leafroll virus reveal protein interaction topologies required for virion stability, aphid transmission, and virus-plant interactions. J Proteome Res, 2012. 11(5): p. 2968–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DeBlasio SL, et al. , Visualization of Host-Polerovirus Interaction Topologies Using Protein Interaction Reporter Technology. J Virol, 2016. 90(4): p. 1973–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alexander MM, et al. , Insights in luteovirid structural biology guided by chemical cross-linking and high resolution mass spectrometry. Virus Res, 2017. 241: p. 42–52. [DOI] [PubMed] [Google Scholar]
- 44.Zhang H, et al. , In vivo identification of the outer membrane protein OmcA-MtrC interaction network in Shewanella oneidensis MR-1 cells using novel hydrophobic chemical cross-linkers. J Proteome Res, 2008. 7(4): p. 1712–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramsey JS, et al. , Protein interaction networks at the host-microbe interface in Diaphorina citri, the insect vector of the citrus greening pathogen. R Soc Open Sci, 2017. 4(2): p. 160545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng C, et al. , Cross-linking measurements of in vivo protein complex topologies. Mol Cell Proteomics, 2011. 10(10): p. M110 006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh P, Panchaud A, and Goodlett DR, Chemical cross-linking and mass spectrometry as a low-resolution protein structure determination technique. Anal Chem, 2010. 82(7): p. 2636–42. [DOI] [PubMed] [Google Scholar]
- 48.Eng JK, Jahan TA, and Hoopmann MR, Comet: an open-source MS/MS sequence database search tool. Proteomics, 2013. 13(1): p. 22–4. [DOI] [PubMed] [Google Scholar]
- 49.Fischer L and Rappsilber J, Quirks of Error Estimation in Cross-Linking/Mass Spectrometry. Anal Chem, 2017. 89(7): p. 3829–3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trnka MJ, et al. , Matching cross-linked peptide spectra: only as good as the worse identification. Mol Cell Proteomics, 2014. 13(2): p. 420–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keller A, Chavez JD, and Bruce JE, Increased Sensitivity with Automated Validation of XL-MS Cleavable Peptide Crosslinks. Bioinformatics, 2018: p. bty720–bty720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keller A, et al. , Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem, 2002. 74(20): p. 5383–92. [DOI] [PubMed] [Google Scholar]
- 53.Shteynberg D, et al. , iProphet: multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Mol Cell Proteomics, 2011. 10(12): p. M111 007690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liang K, et al. , Cryo-EM structure of human mitochondrial trifunctional protein. Proc Natl Acad Sci U S A, 2018. 115(27): p. 7039–7044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jacobs AC, et al. , AB5075, a Highly Virulent Isolate of Acinetobacter baumannii, as a Model Strain for the Evaluation of Pathogenesis and Antimicrobial Treatments. MBio, 2014. 5(3): p. e01076–14. [DOI] [PMC free article] [PubMed] [Google Scholar]