Summary
Biobanks support medical research by facilitating access to biospecimens. Biospecimens that are linked to clinical and molecular information are particularly useful for translational biomedical research. Tracking and managing the biospecimens and their associated data are therefore crucial tasks in the functioning of a biobank. Adequate computing hardware, efficient and comprehensive biobanking software, and cost-effective data storage are needed for proper management of biospecimens. As biobanks build up extensive stores of specimens and patient data, ethical considerations also inevitably arise. Herein, we describe some basic considerations for establishing a biobanking information technology infrastructure that a beginning biobanker needs. Finally, we also discuss trends and future needs in biobanking informatics.
Keywords: Biobank, Biorepository, Information Technology, Hardware, Software, Database, Informatics
1. Introduction
Biobanks are an essential aspect of biomedical research in this new era of targeted therapy or personalized medicine. These biorepositories serve as a ready source of high quality tissue and blood specimens, sourced and stored in coordination with surgeons and pathology staff. The biospecimens may contain genomic, epigenomic, transcriptomic, proteomic or metabolomic changes that characterize the patient’s disorder or cancer. One of the most valuable uses of biobanks arises from the linking of patient clinical information with the aforementioned changes in the biospecimens. These linkages can be analyzed to determine whether specific genetic or other changes might predict response to a specific therapy. A sufficiently large number of biospecimens can provide statistical power for answering research questions. The appropriate computing or informatics infrastructure is critical for managing the data and performing analyses for these biospecimen-based studies that are a fundamental component of many modern clinical trials.
2. Hardware and basic software requirements
To manage data, a computer with the correct operating system to run the software, sufficient memory, and reasonable speed for efficient operations is necessary. An operating system should be primarily chosen based on broad compatibility with software to be used. Currently, most computers can operate satisfactorily with 8–16 gigabytes of RAM at a processor speed of 1 GHz or more. In addition, hard drives with at least 1 terabyte (Tb) should be adequate for daily work. However, these hardware requirements are likely to change every few years as software evolves- typically requiring greater processing power and more storage capacity. External drives and other forms of data storage may be necessary for additional storage and backup. Cloud storage is emerging as a dynamic and cost-effective alternative to physical forms of data storage and will be discussed later in this chapter. These computers require office and security software. An adequate office program should have word processing, spreadsheet, and presentation functionalities. Other useful software include e-mail and note taking applications. To protect sensitive patient information, security software is needed to protect against malware. Malware are software programs that can damage or cause unwanted actions in the computer. In the general public, the term malware may be used interchangeably with the term virus. However, for those in the information technology (IT) world, malware typically encompasses a number of often different but sometimes overlapping sub-types that include viruses, spyware, adware, and ransomware. Viruses are programs that can replicate in your computer and spread to other linked computers while damaging the software on them and sometimes completely incapacitating the computer. Spyware can be used to steal passwords and private information. Adware are unwanted software that project advertisements that sometimes may also have virus capabilities. Ransomware are malicious software that can lock out the end-user unless a ransom is paid. Anti-malware or antivirus software have the ability to search for the relevant malware or viruses. A complementary protective element is a firewall. A firewall monitors network traffic, i.e. data coming into the computer and leaving the computer over the ethernet. If the firewall detects anomalies known to be malicious, it can stop the transmission of data. These can be hardware based or software based. Larger companies with sophisticated IT staff and infrastructure can have hardware firewalls that protect their entire network. For smaller entities with modest budgets, a software firewall can be purchased together with the anti-malware or antivirus. The security programs should also have the capability of regularly scheduled system scans and update procedures. In addition, computer hardware and software must be updated periodically to ensure compatibility and maintain efficiency. Consequently, computing choices must remain scalable and financially feasible. A checklist of these requirements can be found in Table 1.
Table 1.
Computing requirements in establishing a biobank. Each category includes minimum and optimal considerations as well as common functionalities. See Table 2 for BIMS requirements
| Biobank Computing Requirement Checklist | Considerations | |
|---|---|---|
| Workstation | Desktop computer | Laptop computer |
| Operating System | Windows 10, Apple OS X | Must be compatible with biobanking software |
| RAM | Minimum: 8 GB | Better: 16 GB |
| Hard Drive Space | Minimum: 500 GB | Better: 1 – 2 TB |
| Hard Drive Speed | Minimum: 7200 RPM |
Better: 10,000 – 15,000 RPM |
| Internet Speed | Minimum: 10 Mbps | Better: 100–200 Mbps |
| External Drive Space | Minimum: 500 GB | Better: 1–2 TB |
| Cloud Database Considerations | Security, Encryption | Audited by third-party |
| Redundancy | Stable customer base/finances | |
| Office Software Functions | Spreadsheets | E-mail & Note log |
| Word processors | Presentations | |
| Security Software Functions | Firewall | Anti-malware/anti-virus |
| Scheduled scans | Protection against phishing | |
2.1. Backing up data
Direct Attached Storage (DAS), Network Attached Storage (NAS), and file servers are the 3 major ways to backup data on your computer. Direct attached storage is typically an external drive attached to the computer via a Universal Serial Bus (USB) connection. Currently, external drives with USB 3.0 connections that allow significantly faster data transfers than the original USB connections are common. One should ensure that the drive connection is compatible with the computer’s ports for such connections. The external drives should be encrypted, and password protected. Network attached storage are, as the name implies, storage accessible over a network. It is essentially, a collection of hard drives connected to a network that the biobank staffs’ computers can access. NAS is ideal for simple file storage. File servers or servers are similar to network attached storage except that they are essentially computers with hard drives giving them more capabilities to partition storage, to control different tiers of access, and to run shared programs. In short, NAS is less complex to manage than servers but has less functionality. The IT staff at your institution will likely have a preferred mode of providing backup storage. We use an encrypted external drive to back-up data in our own laboratory space and also store data in folders on servers at a remote location provided by our departmental IT staff. Having a local drive is helpful in that, sometimes when the network is down, one can still work from the local files. In addition, if the computer it is attached to is not functioning, the external drive can be easily moved to another computer,
2.2. Redundant Servers
In times of power outages or main server failures, redundant servers are necessary to maintain biobank server functionality. With the same specifications and applications as the original servers, redundant servers come online when the dedicated servers are down and continue to provide support until normal server function can be restored (1). Typically, data must be encrypted en route to the server and on the server, and governmental privacy and security requirements must be met. The secondary server should ideally have a location different from the primary server. The secondary server can be set up to mirror the primary server. A server status page is used to check the primary server on a regular schedule for an expected response. There should be a failover service to automatically switch to the secondary server when the primary server fails to return the expected response. The failover service should switch back to the primary server once it is functional. Multiple backup servers can also be stringed together to provide multiple levels of redundancy, in case even the secondary server is out of service. This redundancy should be provided by your IT department.
3. Biobank Information Management System (BIMS)
3.1. Biobank Information Management System (BIMS), a form of Laboratory Information Management System (LIMS)
With the immense amounts of data with which biobanks are associated, biobank information management systems (BIMS) are powerful if not necessary tools. LIMS are data management software programs that manage the various types of information in laboratory environments. A BIMS is essentially a LIMS that is adapted for biobanks. As each biobank, from the informatics point of view, is essentially a large workflow, a BIMS support the multiple processes involved to assist personnel in tracking and managing samples. However, not all BIMS are the same, as each configuration is designed to best support the processes of a particular laboratory with its own unique workflows and data set types. In general, a BIMS serve a set of core functions: storage and registration of a sample and its corresponding data, tracking of the sample throughout the laboratory workflow, storage locations, organization and analysis of data, and auditing of sample data. It is important that the BIMS keep a running custody log for each sample. The custody log would be a chronological record of staff handling each specimen at each step of the workflow. In case of a missing biospecimen, the custody trail can help in tracking down a biospecimen and provide a window into how to improve the standard operating procedure.
3.2. Considerations for biospecimen labeling and registration in the BIMS
In order to effectively navigate through this extensive database, the ideal BIMS requires a user-friendly front end offering flexible search criteria (2). An index of labels (categories) and their respective abbreviations should be available for anyone using the BIMS to be able to properly categorize biospecimens and to conduct efficient searches. With any new categories, a standard abbreviation should be chosen and included in the index for future use. A well-established BIMS often has a large and practical ontology or hierarchical nomenclature for categorizing biospecimens typically available from pick lists. For example, the BIMS would have options for type of biospecimen such as tissue, blood, and cerebrospinal fluid as well as source such as lung, brain, heart etc. There might be further options to characterize source such as left upper lobe, right kidney or left temporal lobe. In addition, the BIMS should have the capability to specify materials derived from the biospecimens such as cell lines, DNA, RNA, and protein etc., and analytical data such as quality assurance metrics like RNA integrity number (RIN).
A BIMS should be able to integrate multiple types of inputs into a single searchable framework. For example, whole slide digital images, photos, molecular data, and scanned documents may be attached to a biospecimen. Integration into the singular framework also eliminates duplicates to streamline data access (3). Free texting for categorization should be avoided as that may be lead to inconsistencies in the data entry through typographical or formatting errors. When any new data is entered into the BIMS, ideally there should be a second party present to audit all the newly entered data. Practically, total and contemporaneous audit is difficult and only a subset of data entered is typically audited. Some software requires data entry of an important data element in duplicate; i.e. the data must be entered twice, and the data must match. If a biospecimen already has associated data imbedded in bar codes or radio frequency identification (RFID) tags, bar code or RFID scanners linked to the BIMS can be used to capture the associated data. These steps limit simple data mis entries that can have profound consequences due to biospecimen misidentification.
If the appropriate consent for research has been obtained, the patient’s name, date of birth, medical record number, diagnosis, and other clinical information can be collected and associated with the biospecimen. For the best protection of the patient’s privacy, all specimens should be assigned a research identifier that can add a layer of separation from the patient’s name and clinical identifiers (date of birth, medical record number etc.). An identifier unique to the patient and a second identifier unique to the biospecimen are necessary. Having only a patient identifier is inadequate as the patient may have more than one biospecimen over time. Each specimen must have a date of collection in order to begin creating a chronological record for the specimen. Under some protocols, deidentified tissue is collected such that only basic information such as tissue or cancer type is provided to the biobank.
3.3. Aliquots, chain of custody, and location tracking
Once in the biobank, each biospecimen is tracked by the biobanking software with constant updating of biospecimen quantity, storage location, storage method, and storage conditions. It is imperative that a custody log be maintained meticulously (4). Every specimen should also have a genealogy that gives a record of aliquots and derivatives and their quantities. Aliquots are smaller volumes of the original specimen. Often the original specimen is divided into several aliquots to store in suitably sized containers or to create several different derivatives or to provide to researchers. Derivatives may be thought of as materials extracted or derived from the original biospecimen. Examples of derivatives include: Nucleic acids extracted from tissue, cell lines grown out from cancer biospecimens, formalin fixed paraffin embedded blocks made from tissue, white blood cells or serum collected from blood etc. Maintaining a comprehensive genealogy also allows researchers to track availability for each specimen so as to prevent depleting irreplaceable and unique biospecimens.
With each specimen also follows a research history, as the inherent value of any biobank comes from the variety of research efforts it is able to support. A typical experimental history for a specimen would entail the specific proposed research study in need of the sample, grant funding that the proposal has received, IRB approval, experimental procedures performed on the sample, relevant data and results from the experiment, any consequent publication history, and possible clinical trials supported by the research. Finally, as biobanks often work with other institutions, samples must be sent out for collaborative research efforts. This requires industry standard hazard classifications, destination, the courier service employed, and tracking numbers. Table 2 organizes these multiple layers of data for each biospecimen.
Table 2.
General information for each biospecimen to be stored in a biobank
| Preliminary Information | ||
|---|---|---|
| Patient Information | Name | Date of Birth |
| Medical record number | Diagnosis | |
| Research consent | Date of specimen release | |
| Research Identifier | ||
| Biologic Type | Solid Biologics | Fluid Biologics |
| Tissue | Blood | |
| Organ | Cerebrospinal fluid | |
| Other solid | Other bodily fluid | |
| Quality Control | ||
| Biobank Specimen Information | ||
| Storage | History of custody | Current location |
| Current method of storage | Storage conditions | |
| Genealogy | Derivatives (Materials derived from the biospecimen) | Aliquots (Smaller samples of the original biospecimen, e.g. A 5 ml tube of blood may be aliquoted into five 1 ml cryovials) |
| FFPE blocks and slides | ||
| DNA, RNA, protein, cell lines | ||
| Experimental History | Proposed research study | Grant funding |
| IRB approval | Experimental procedures | |
| Data obtained | Results | |
| Publication history | Trials supported | |
| Shipping | Hazard classifications | Destination |
| Courier service | Tracking number | |
3.4. Freezer Maps
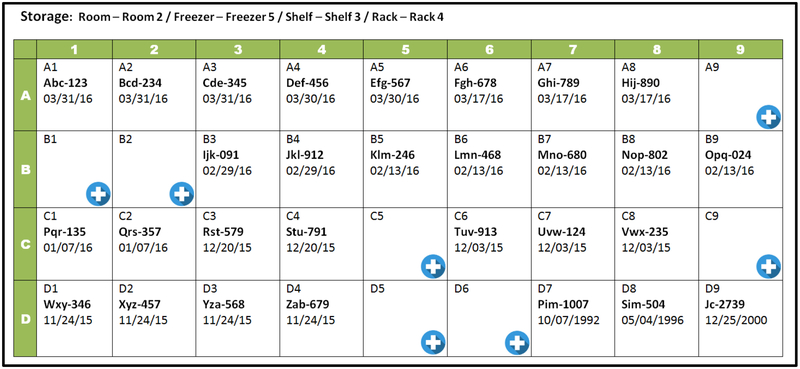
Freezer mapping creates comprehensive and updated location inventories of biospecimens and their corresponding aliquots. Freeze maps can greatly expedite research efforts by reducing time spent in finding specific samples. The freezer software should also allow users to create their own defined fields as searchable categories to navigate the variety of specimens available in storage; at the minimum, there should be localization as the level of the shelf of the freezer or rack of a liquid nitrogen vat. Fig. 1 shows a typical grid that would be displayed by the freezer software program when searching for a specific specimen or aliquot according to particular criteria among multiple freezers and multiple divisions within each freezer. In addition, tracking storage conditions such as temperature and humidity are desirable to ensure specimen integrity. A sensor (or multiple sensors) within each freezer tracks and logs internal conditions that are recorded into the freezer software often via a wireless network connection. In case of freezer failure, specimen degradation can occur very quickly as temperatures rise and samples are exposed to moisture. For timely response in transferring affected samples to functioning freezers, alarms should be present to notify appropriate staff of any malfunction. Each freezer can be equipped with a physical audible alarm, and the freezer software can be configured to provide notifications if freezer conditions deviate from the norm. Certain freezer software programs also feature labeling functions along with label printers for marking vials and slides. Labels should remain adherent and be waterproof to prevent loss of identification under freezing and thawing conditions, and they can be printed according to set templates.
Figure 1.
Example of freezer software interface displaying sample locations, relevant specimen information, and options for adding new specimens.
3.5. Radio frequency identification (RFID) tags
Radio Frequency Identification (RFID) technology offers numerous advantages in reducing errors while identifying, tracking, and archiving biospecimens (5). RFID tags can be scanned without direct alignment. Multiples tags can be scanned simultaneously, and each tag possesses a relatively high data storage capacity compared to most bar codes. Furthermore, RFID systems are capable of data transmission, essential in tracking storage conditions such as temperature, and multiple cycle of read-write processes can be performed on each tag to keep a running log of any changes. However, implementing RFID systems can be difficult in terms of high equipment and software setup costs, as well as inevitable technological obsolescence necessitating periodic software updates and new hardware. There also exist security concerns in employing RFID systems, in which radio communication channels remain open and vulnerable to unwarranted access. This privacy concern can be overcome with encryption, use of research identifiers, shielding, and limiting access to biospecimen storage areas. A cost benefit analysis is advised prior to implementation.
3.6. Biobanking Example
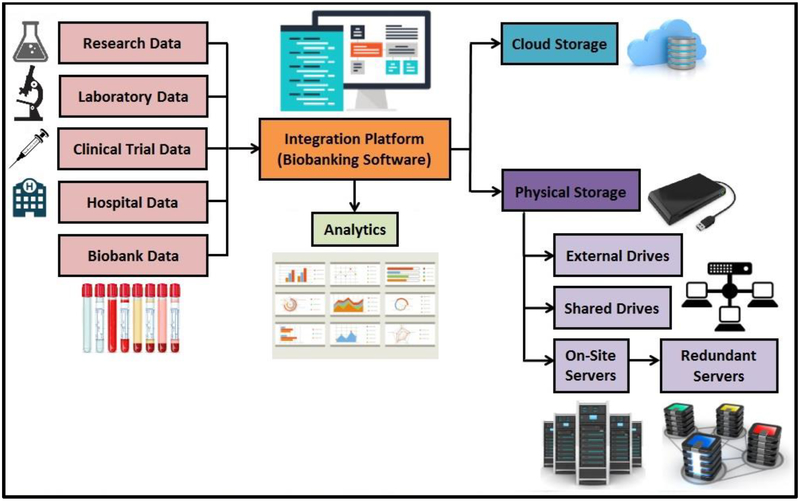
Mr. John Doe is a patient who has been diagnosed with glioblastoma multiforme (GBM). His records show his birthdate to be 1/1/1970, and he has been assigned a medical record number: MRN X-01234. Mr. John Doe has given research consent ahead of time for the tumor to be used in research studies. Mr. John Doe then undergoes surgery on 1/1/2016, and the GBM is removed. Up to this point, patient and surgical information is logged by the electronic health record program authorized by the institution. When the GBM is obtained by clinical pathology personnel, the BIMS assigns the specimen its research identifier: R-5678. Since it is a tumor, specimen R-5678 is categorized as a solid biologic obtained from brain, right parietel lobe, and it then undergoes quality control histologic assessments, such as tumor percentage, necrosis percentage, or cancer biomarkers. The quality control results are then logged in before the specimen can be stored away. Once released by pathology staff to researchers, the history of custody for specimen R-5678 begins within the biobanking software database. After proper labeling, the specimen is assigned a slot within a specific storage freezer. The method of storage (frozen), and specific storage conditions (temperature and humidity) are tracked by the freezer software. As samples of specimen R-5678 are requested, its genealogy of derived samples within the BBS keeps track of all FFPE slides and block requests from various research staff. If research personnel decide to use specimen R-5678 for glioblastoma research, there must first exist records of the research project proposal, proper grant funding, and IRB approval for the project linked to the specimen within the BBS database. Any experimental procedures performed on the tumor specimen or any of its derivatives, all data, and results obtained from those procedures are logged into the BBS as well. Furthermore, any publications resulting from the research project as well as clinical trials developed in accordance with the research are continuously logged for specimen R-5678 (Fig. 2).
Figure 2.
Flowchart mapping data movement into and out of a typical Biobanking system. This flowchart shows possible types of data inputted to the Biobanking software, as well as different types of cloud and physical data storage.
4. Biobank collaborations and web-based portals
Perhaps the greatest value of biobanks lies in the potential access to large numbers of biospecimens from multiple centers where often each center alone would not have sufficient material to power a study. For researchers at different sites around the world to access specimens, a consortium of biobanks can provide a single web-based portal that permits searching of their libraries of biospecimens. If the biobanks use the same BIMS, access to the shared data is facilitated. Often however, a separate database is created requiring data entry from the diverse biobanks and the web portal provides access to the central database. Regardless, through a web-based portal, collaborators can obtain pertinent information on the variety of samples available in a given biobank consortium. With proper access privileges granted ahead of time (based on an application and a relevant documented IRB-approved protocol), researchers can search the content of the BIMS through most web browsers, providing the liberty to acquire data from anywhere with an internet connection. Different levels of search access can be provided depending on the approved research protocol. Once the researcher has identified a set of biospecimens that they are interested in, they can submit a request to a central oversight committee that coordinates with the individual biobanks for shipping. A slightly different model is one where the researcher submits a request for biospecimens, e.g. lung carcinomas from patients that have been treated with a specific drug, to a central site which then runs a search across the associated biobank’s BIMS databases either directly themselves or indirectly by requesting the individual center to run the searches.
5. Commercial Cloud Data Storage
Biobanking data can be stored in cloud-based infrastructure. That is, instead of storing the data on local servers, the BIMS data can be stored remotely “in the cloud” with servers provided by the BIMS vendor or with a commercial data storage entity. There are several criteria by which a good cloud storage provider might be selected. A reliable provider should have an expansive customer base of business clients that can attest to the provider’s trusted cloud infrastructure as well as its profitable and stable finances, ensuring the provider is successful in handling large databases. To establish databases of sensitive information, cloud providers need to have security programs and multiple levels of encryption methods to prevent data breaches and ensure privacy of patient information. Most cloud storage providers are validated by third-party auditors to ensure security protocols meet international industry standards (6). Moreover, providers should be approved by the institution and have a strong record of services operating under HIPAA or relevant national privacy compliance requirements. Cloud storage offers numerous advantages over conventional practices of maintaining physical on-site data servers. In terms of financial cost and scalability, cloud storage is attractive. Establishing physical on-site data servers requires institutions to make large financial and personnel investments in acquiring data storage hardware and dedicating IT staff to set up such extensive data systems. Also, the institution may have to periodically purchase new hardware or schedule major overhauls to roll out new software. On the other hand, cloud licenses can be obtained with relative ease as the physical infrastructure and relevant software are already established by the service provider. In addition, expansions and updates are managed and executed by the commercial service providers that typically have efficiencies of scale. Consequently, some authors argue that cloud storage is the best solution to cater to rapidly expanding biobanking needs (7).
6. Patient Privacy and Ethical Considerations
As with any collection of patient information, biobanks must follow strict legal and ethical guidelines. Foremost, any patient data can only be obtained from participants who have given consent for corresponding specimens to be used in research studies. All samples should be anonymized, using a coding system with research identifiers so as to prevent anyone from being able to track a specimen back to the original patient. Such research identifiers should only be given out to collaborators on a need-to-know basis, to further minimize patient information from being compromised. Under the direction of the institution, all personnel should receive computer security and HIPAA compliance training to be prepared against potential security breaches and phishing attacks that may compromise privacy of patient data. This may include learning to recognize spam e-mails, create strong passwords, report suspicious notifications, and adhere to privacy and ethical guidelines. Protecting patient privacy in this manner is not only a legal requirement of research compliance but also works to maintain research integrity. Restricting researchers from matching specimens to individuals also ensures that researcher cannot manipulate their results to produce expected results in support of their clinical procedures or experiments (8). However, unique complications arise with genomic data. Even with de-identification, the risk of privacy breach and information exposure still exists as genotypes are very specific to each individual. While these issues can be mitigated with complete disassociation of data from patient identities, this significantly detracts from the value of the biobank as it does not allow any way of updating clinical records. The utility of specimens increases with the amount of data to which they can be associated, and if that database becomes too disjointed and partitioned for the sake of privacy, the biobank’s value to research, society and the biospecimen donor is diminished and can be rendered useless (9).
7. Biobanking Going Forward
Though increasing numbers of biobanks are emerging, there have still yet to be any widely accepted industry-wide standards or international registries. Establishing standardized protocols should greatly increase the efficiency of mining clinical data as various institutions employ identical methodologies in characterizing and annotating specimens stored in their respective biobanks. Standardization can be expedited with automated systems that would organize specimens into predetermined categories, whether based on tissue type, molecular markers, or preservation methods (10). Consequently, researchers would be able to trace specimens with ease between multiple projects using a single standardized code system. Currently, most biobanks work in complete independence, each operating under their individual standards of specimen organization and data collection. This can make collaborations between biobanks difficult, requiring additional methods to convert different data catalogues into a single registry for comparisons or pooling data together. A number of initiatives are underway to increase interoperability. A global registry to which all biobanks can subscribe would usher in a promising future of biobanking, defined by comprehensive metadata and extensive collaboration in furthering medical research.
Acknowledgement
This work was supported in part by NIH:NCI P50-CA211015, NIH:NIMH U24 MH100929, the Art of the Brain Foundation, and the Henry E. Singleton Brain Cancer Research Program.
References
- 1.Helpton S (2013) How to create a server failover solution. http://www.howto-expert.com/how-to-create-a-server-failover-solution/. Accessed 15 Mar 2016.
- 2.Schreier P (2008) Biosample Storage. http://www.scientific-computing.com/features/feature.php?feature_id=195. Accessed 2 Mar 2016.
- 3.Ball L, Brunner BJ, Chandrasekaran S (2015) Building an Intelligent Biobank to Power Research Decision-Marking. http://docplayer.net/3853972-Building-an-intelligent-biobank-to-power-research-decision-making.html. Accessed 3 Mar 2016.
- 4.Joyce JR (2010) Will Biobanking Change the World? http://www.scientificcomputing.com/articles/2010/06/will-biobanking-change-world. Accessed 2 Mar 2016
- 5.Lou JJ, Andrechak G, Riben M, Yong WH (2011) A review of radio frequency identification technology for the anatomic pathology or biorepository laboratory: Much promise, some progress, and more work needed. J Pathol Inform 2: 34. doi: 10.4103/2153-3539.83738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooks C (2014) 8 Tips on Picking the Right Cloud Storage Provider. http://www.businessnewsdaily.com/6375-tips-on-picking-cloud-storage.html. Accessed 29 Mar 2016.
- 7.Eydeler K (2013) Why Cloud-Based LIMS is Ideal for Biobanking. http://www.shonan-village.co.jp/anrrc2013/pdf/S2-1.pdf. Accessed 3 Mar 2016.
- 8.Blackman G (2010) A Booming Banking Sector. http://www.scientific-computing.com/features/feature.php?feature_id=270. Accessed 3 Mar 2016
- 9.Baker M (2012) Biorepositories: Building Better Biobanks. Nature 486: 141–146. doi: 10.1038/486141a. [DOI] [PubMed] [Google Scholar]
- 10.May M (2013) Better Features for Biobanks. http://www.biosciencetechnology.com/articles/2013/01/better-features-biobanks Accessed 4 Mar 2016.