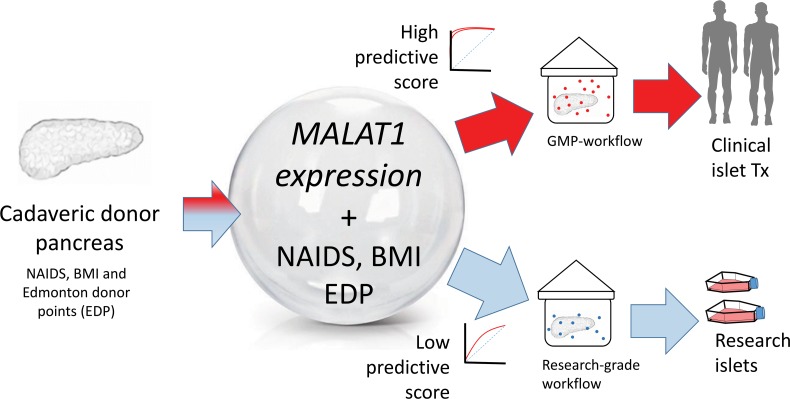
Abstract
Human islet isolation is a cost- and resource-intensive program for generating islets for cell therapy in type 1 diabetes. However, only one-third of cadaveric pancreases get to clinical transplantation because of low quality/number of islets. There is a need to identify biomarkers that predict the quality of islets, before initiating their isolation. Here, we sequenced transcriptomes from 18 human islet preparations stratified into 3 groups (group 1: best quality/transplantable islets; group 2: intermediary quality; and group 3: inferior quality/nontransplantable islets) based on routine measurements, including islet purity/viability. Machine-learning algorithms involving penalized regression analyses identified 10 long noncoding RNAs (lncRNAs) that were significantly different across all group-wise comparisons (group 1 vs. group 2, group 2 vs. group 3, and group 1 vs. group 3). Two variants of metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) lncRNA were common across all comparisons. We then confirmed RNA-Seq findings in a validation set of 75 human islet preparations. Finally, in 19 pancreas samples, we demonstrated that assessing the levels of MALAT1 variants alone (receiver operator characteristic curve AUC: 0.83) offers higher specificity in predicting postisolation islet quality, further improving the predictive potential for clinical islet transplantation when combined with Edmonton Donor Points/BMI/North American Islet Donor Score. We present this resource of islet quality–stratified lncRNA transcriptome data and identify MALAT1 as a biomarker that significantly enhances current selection methods for clinical-grade (good manufacturing practice–grade) islet isolation.
Keywords: Transplantation
Keywords: Islet cells
A signature of human islet lncRNAs that accurately predict the quality of human islets isolated from cadaveric human pancreas, prior to isolation, is presented.
Introduction
Type 1 diabetes (T1D) is currently managed via multiple daily insulin injections or continuous glucose monitors. Transplantation of human cadaveric islets is the only approved/health care–supported cell therapy for T1D in countries, such as Australia, Canada, France, Italy, Switzerland, and the United Kingdom. In the United States, although islet autotransplants (in chronic pancreatitis) are reimbursable, islet allotransplantation in individuals with T1D is performed only through clinical trials. Usually, costs of islet isolation from a human cadaveric pancreas are very high (~US$40,000/cadaveric pancreas) because of good manufacturing practice–grade (GMP-grade) reagents and workflows (1) necessary for clinical islet transplantation. Donor characteristics are important in selecting cadaveric pancreas for GMP-grade islet isolation procedures (2). Although some studies (3, 4) demonstrated donor BMI as a positive predictor of islet yield, islet viability, and insulin secretion, the Collaborative Islet Transplant Registry (CITR) data failed to validate these observations (5). Although older donors tend to yield higher numbers of islets than younger donors (6, 7), islet function appears to deteriorate with age (8). Therefore, islet isolation centers often face the difficult question of whether cost- and resource-intensive GMP-grade ($40,000) or the less pricey (~$700) research-grade reagents should be used for an available donor cadaveric pancreas. There is a need to identify biomarkers that can predict the quality and not just higher numbers of islets within donor pancreases, before undertaking this resource-, time-, and cost-intensive islet isolation procedure. Such a biomarker should not only independently predict the quality of islets before their isolation from the donor pancreas, but also add value when combined with existing parameters for donor pancreas selection.
Long noncoding RNAs (lncRNAs) constitute a group of RNA molecules more than 200 nucleotides long that are not translated into proteins. LncRNAs are associated with different diseases (9, 10), including diabetes (11–14), and are known to orchestrate biologically relevant (15) signaling networks that regulate cell- and tissue-specific gene expression (16–18). Recent studies have examined lncRNAs in human pancreatic islets (19–23); one of these landmark studies (22) uncovered human islet cell lncRNAs that are associated with key pancreatic transcription factors. A recent follow-up study from the same group (19) identified 2373 β cell lncRNAs in a pool of 41 human islet preparations. The lncRNA PDX1-AS1/PLUTO (19), although expressed at low abundance in human islets, demonstrated a positive correlation with PDX1 repression in islets from donors with type 2 diabetes (T2D) or impaired glucose tolerance (IGT). These studies describe the potential role of lncRNAs in the development of IGT or T2D.
Here, we sought to examine and compare the lncRNA profiles of isolated human islet sample preparations that were stratified into 3 groups based on their postisolation islet quality. Whole-transcriptome RNA-Seq followed by implementation of machine-learning statistical analytical algorithms identified lncRNA candidate biomarkers of islet quality. Next, we validated the expression of these lncRNAs using TaqMan-based real-time quantitative PCR (qPCR) in 75 additional islet sample preparations. Finally, the potential of these lncRNAs to predict postisolation islet quality was assessed in 19 donor pancreatic tissue samples. The lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) predicted the quality of islets within the cadaveric pancreas, before their isolation. Pancreatic MALAT1 lncRNA expression alone could predict the suitability of donor pancreases for clinical islet transplantation (receiver operator characteristic [ROC] curve AUC: 0.83), better than the current BMI-alone criterion (ROC curve AUC: 0.79). A combined mathematical model generated to predict higher islet number (BMI) and better islet quality (MALAT1 lncRNA expression) delivered higher predictive power (ROC curve AUC: 0.94) that substantially improves the predictive power over either of the currently used donor criteria (BMI, Edmonton donor points, or North American Islet Donor Score/NAIDS) alone. When combined with existing pancreas donor scores, such as the North American Islet Donor Score (NAIDS) (24) and the Edmonton Donor Points (2), MALAT1 offered the highest predictive power.
We present here our discovery (n = 18), validation (n = 75), and prediction (n = 19) data sets, which to the best of our knowledge, represent the first resource of lncRNA expression in human islets stratified on the basis of their postisolation islet quality. Because tissue quality prediction is of utmost importance in clinical islet transplantation, which currently is the only approved cell therapy for T1D, our study presents molecular tools to advance selection of cadaveric tissues for stratification into GMP-grade workflows.
Results
LncRNA expression profiles are different across groups with varying islet quality.
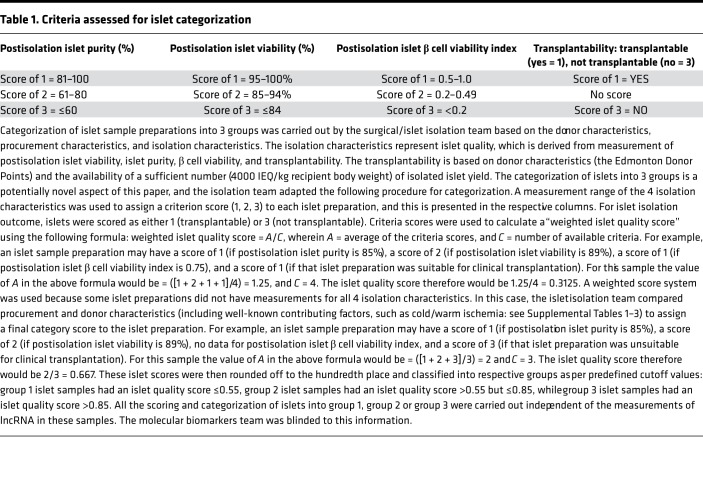
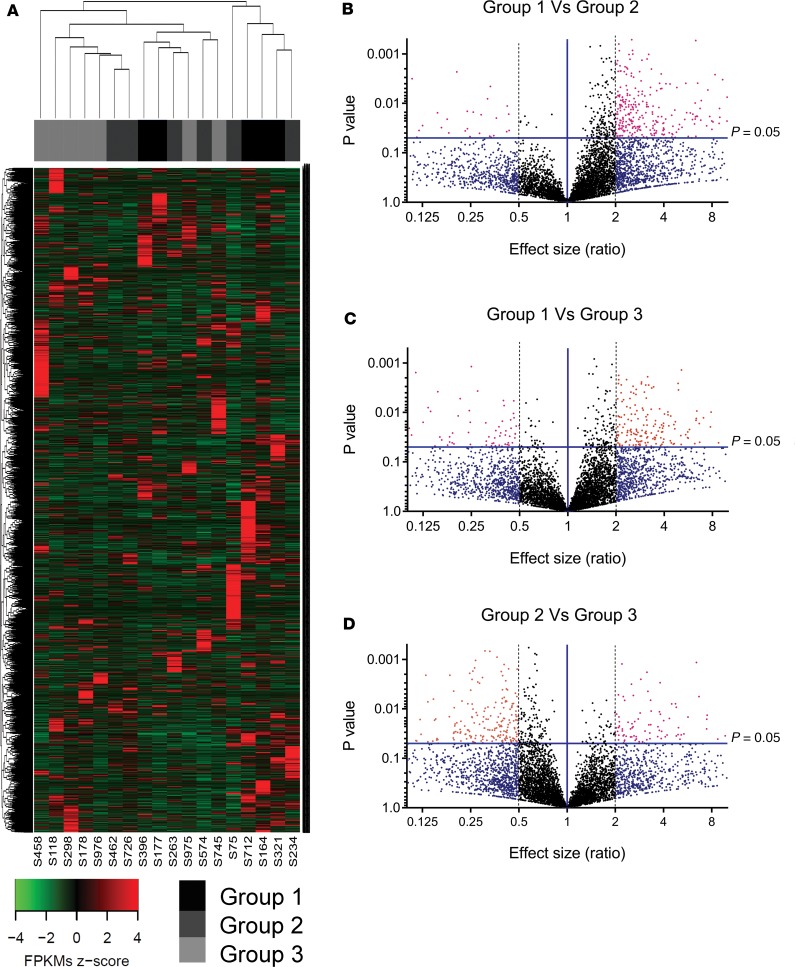
We had access to 93 pancreatic islets samples and 19 human pancreatic tissue samples from the Westmead Islet Isolation Centre (ref. 25 and Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.129299DS1). Based on the results of standard quality control protocols (26), isolated islet samples from each of the pancreases were stratified into 3 groups (group 1: high quality/transplantable; group 2: intermediary quality, meaning good quality but low yield; and group 3: inferior-quality islet preparations). This stratification was performed using the pancreas islet quality score (Supplemental Tables 1–3) derived after assessment of 4 key postisolation measurements: (a) islet purity, (b) islet viability, (c) β cell viability, and (d) transplantability (Table 1). Human islet samples within group 2 are also of high quality. These islet samples could not be transplanted in individuals with T1D because the number of islet equivalents (IEQ) from these isolations was fewer than the desired number (4000 IEQ/kg of recipient body weight) for clinical transplantation. We randomly selected 18 islet samples from 3 groups for high-depth RNA-Seq (rRNA depletion library construction) using the HiSeq4000 platform. All the samples passed postsequencing quality control (QC), and on average 77,127,891 clean reads were obtained from these samples of the discovery set. A total of 6983 annotated lncRNAs and 450 potentially novel lncRNAs were identified (Supplemental Figure 2). An unsupervised hierarchical cluster analysis performed on the annotated lncRNAs revealed that all (n = 5) of the group 1 (transplantable) islet samples clustered closely with the intermediary-quality group 2 islet samples (n = 6) but not with the inferior-quality (n = 7) group 3 islet sample preparations (Figure 1A). We then compared the levels and abundance (fragments per kilobase of transcript sequence per million base pairs [FPKM]) of all lncRNAs expressed between the 3 groups of these discovery set islet samples. A significantly large number (246–322 lncRNA candidates) was identified to be differentially expressed (>2-fold, P < 0.05) in group-wise comparisons of lncRNAs (Figure 1, B–D).
Table 1. Criteria assessed for islet categorization.
Figure 1. Discovery analyses using next-generation sequencing of human islets categorized on the basis of postisolation quality.
(A) Unsupervised hierarchical euclidean (complete) cluster heatmap of the annotated (6721) lncRNAs in n = 18 human islets, categorized into 3 groups. (B–D) Volcano plots of annotated lncRNAs in the human islets categorized group 1 versus group 2 (B), group 1 versus group 3 (C), and group 2 versus group 3 (D). The effect size differences are depicted on the x axis while the −log10 P value is depicted on the y axis. The horizontal blue line represents the significant P value of 0.05, while the vertical blue line represents no difference.
Penalized regression analyses identify key lncRNAs associated with islet quality.
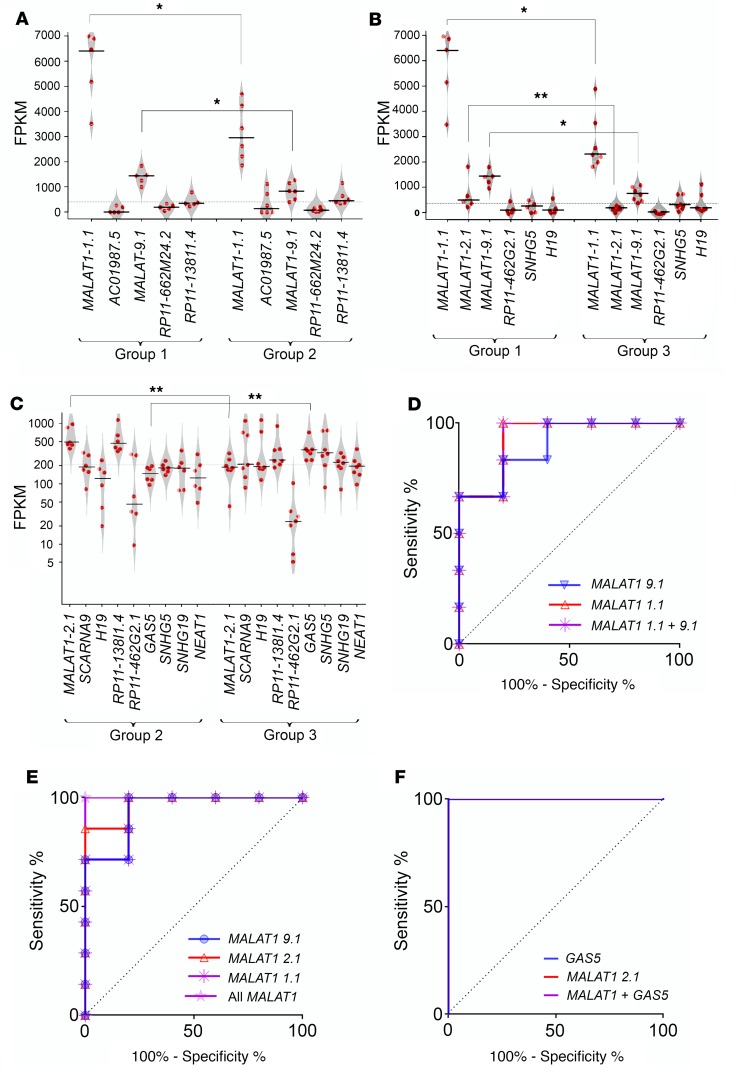
To identify key lncRNAs that possess discriminatory capacity to identify the high-quality islet sample preparations, we performed penalized logistic/linear regression (PLR) analyses (27). To confirm that the outcome of this regression analysis was not a result of the sampling bias, we implemented resampling validation using bootstrapping workflows. These analyses identified 5–10 lncRNAs in each group-wise comparison (Supplemental Table 4). Of these, MALAT1 was found to be common and significantly different across all comparisons (Figure 2, A–C). Intriguingly, 2 (of 8) splice variants of MALAT1 (ENST00000616691.1, denoted henceforth as “MALAT1-1.1,” and ENST00000619449.1, denoted henceforth as “MALAT1-9.1”) were identified to be common in each bootstrap comparing group 1 islet samples, while another MALAT1 variant (ENST00000620902.1, “MALAT1-2.1”) was identified in comparisons involving group 3 islet samples (Figure 2, A–C, and Supplemental Figure 3). ROC curves for the significantly dysregulated lncRNAs demonstrated the capacity of MALAT1 variants and variant ENST00000450589.5 (GAS5) lncRNAs to stratify islet samples into the relevant groups (Figure 2, D–F).
Figure 2. Key lncRNAs identified in islet quality stratified discovery sample set.
Penalized regression analysis and bootstrapping were performed on the (6983) annotated lncRNAs and the identified lncRNAs across all group-wise comparisons (n = 18 human islet samples, categorized into 3 groups) as presented in A–C: (A) group 1 versus group 2, (B) group 1 versus group 3, and (C) group 2 versus group 3. (See Supplemental Table 4 for details; unequal-variance Student’s t test was used. Note: The lncRNA AC010987.5 was present at very low FPKM and hence is not shown in C.) *P < 0.05; **P < 0.01. Each red dot represents individual islet preparation. The horizontal line represents the mean while the polygons represent the estimated density of data (scatter plot). Two-tailed distribution, with 2-sample unequal-variance Student’s t test, was used to identify the difference for each lncRNA between each group-wise comparison. ROC curves: (D) group 1 versus group 2: MALAT1-1.1, MALAT1-9.1, and both MALAT1 variants (9.1 + 1.1); (E) group 1 versus group 3: MALAT1-1.1, MALAT1-2.1, MALAT1-9.1, and all the above 3 MALAT1 variants (1.1 + 2.1 + 9.1); (F) group 2 versus group 3: ENST00000450589.5 (GAS5), MALAT1-2.1, and both; GAS5 + MALAT1-2.1.
We also performed penalized linear (islet purity, islet viability, β cell viability) or logistic (transplant outcome: yes/no) regression analyses and bootstrapping on lncRNAs for each of these 4 parameters. Four MALAT1 splice variants (MALAT1-9.1; MALAT1-2.1; ENST00000508832.2, “MALAT1-2.2”; and MALAT1-1.1) were identified to be common across individual penalized regression comparisons (Supplemental Table 5) between lncRNA levels and each of the 4 islet quality score criteria (outlined in Table 1). In univariate correlation analysis, the same 4 MALAT1 variants showed significant positive correlation with postisolation islet viability, islet purity, or β cell viability index (Supplemental Table 6). We also identified other potentially novel and annotated lncRNAs that showed significance (P < 0.05) in univariate analyses for each of the 4 islet quality score criteria (data not shown). However, these were significantly lower in abundance (at least 10- to 48-fold less) than the highly abundant MALAT1 splice variants (MALAT1-1.1 and MALAT1-9.1). The biological relevance of these low-abundance lncRNAs can be the subject of future investigations.
The lncRNA MALAT1 is a potential biomarker for islet isolation outcome.
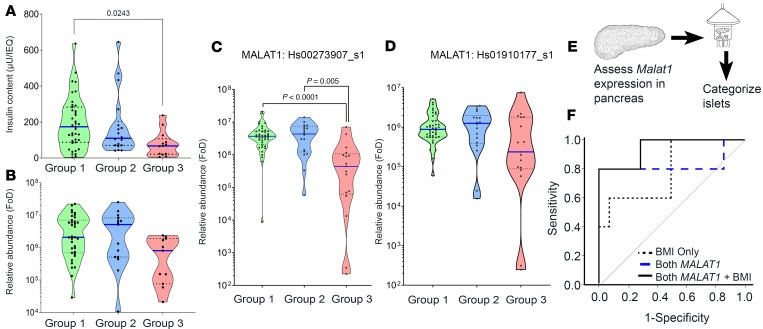
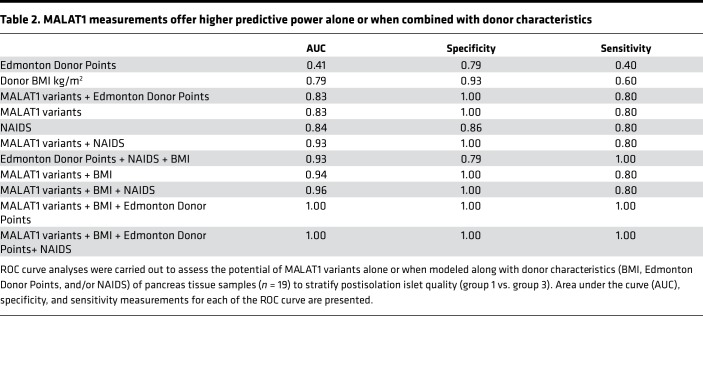
Because high expression of MALAT1-1.1 and MALAT1-9.1 was identified in group 1 islet samples, we decided to confirm our findings in a validation set of 75 human islet samples for which all the relevant information to categorize them into 3 groups (group 1, n = 40; group 2, n = 18; group 3, n = 17), as well as RNA for qPCR-based MALAT1 measurement, were available. Although group 1 islet samples had significantly higher insulin content than group 3 islet samples (Figure 3A), the levels of insulin gene transcript (Figure 3B) were similar. The inferior-quality (group 3) islet samples had significantly lower levels of MALAT1 gene transcript (assay-907) (Figure 3C), but not the other (assay-177) MALAT1 gene transcript assay (Figure 3D). To test whether these MALAT1 variants could be biomarkers of postisolation islet quality, it was essential to measure MALAT1 before islet isolation. We assessed MALAT1 levels in a set of 19 (preisolation) pancreas samples, where a small piece of tissue was stored from the middle of the pancreas at the time of transection for cannulation, usually close to the duct itself, before islet isolation. The surgical team isolated islets from each of these 19 pancreatic samples (Supplemental Figure 1 and Figure 3E) and categorized them into group 1 or group 3 islet samples (group 1: transplantable, n = 14; group 3: nontransplantable, n = 5) while the lab team assessed the levels of MALAT1 transcripts (assay-907 and assay-177) in each of the 19 deidentified pancreas samples. ROC curves, based on the pancreatic levels of the 2 MALAT1 variants and donor BMI, returned an AUC of 0.79 for BMI alone, 0.83 for both MALAT1 assays (907 and 177), and 0.94 for both MALAT1 variants plus BMI (Figure 3F and Table 2). Because islet isolation centers across the world follow several criteria, we tested whether MALAT1 measurements had any added benefit to the current donor pancreas selection score. A combination of MALAT1 measurements with any of the standard scores, such as Edmonton Donor Points or the NAIDS (24), offers the highest predictive power (Table 2).
Figure 3. Assessing MALAT1 lncRNA expression in the validation (islet) and prediction (pancreas) sample sets.
The 3 categorized human islet sample groups in the validation set (n = 75) were assessed for (A) insulin content levels, (B) (pro-) insulin transcript levels, and (C and D) MALAT1 lncRNA expression (using qPCR primer/probe assay Hs_00273907 and Hs_01910177, respectively) targeting 2 variants of MALAT1 lncRNA. To test the difference between the 3 categorized groups, nonparametric, 1-way ANOVA was used, and significant P values, adjusted for multiple comparisons, are reported. Each dot in the violin plot represents a different islet sample preparation. Horizontal solid blue line represents the median for each group, the horizontal dotted line represents quartiles, and the polygons represent the density of individual data points and extend to min/max values. FoD, fold over the detectable limit (55). (E) MALAT1 lncRNA qPCR was carried out before islet isolation. (F) ROC curve for MALAT1 lncRNA of pancreas tissue samples (n = 19) to stratify postisolation islet quality (group 1 vs. group 3).
Table 2. MALAT1 measurements offer higher predictive power alone or when combined with donor characteristics.
The stimulation index for group 1 islet samples appeared to be higher, although there was no statistically significant difference across the groups (Supplemental Figure 4A). Islet samples from the 3 groups were transplanted under the kidney capsule of diabetic animals, and the time for diabetes reversal was monitored to further classify the transplanted animals as early or late responders (Supplemental Figure 4B). A higher proportion of mice showed diabetes reversal within the first 3 weeks after transplantation (early responders) for group 1 and 2 islets as compared with group 3 islet recipients (Supplemental Figure 4C).
Discussion
The present study was designed to generate lncRNA profiles of human islet sample preparations that were stratified based on their postisolation islet quality. Currently, there are no predictive biomarkers of postisolation islet quality, and data from the CITR (5) have failed to validate the potential of clinical characteristics, such as BMI, in predicting the quality of islet sample preparations isolated from each cadaveric human pancreas. There is a need to identify molecular biomarkers that could allow real-time analysis and prediction of islet quality.
Islets were categorized into 3 groups based on their postisolation islet quality variables (purity, viability, β cell viability, and transplantability; Table 1). The functionality of all isolated human islets was determined using in vitro glucose-stimulated insulin secretion (GSIS) assay and in vivo islet transplantation in diabetic mice. The GSIS and in vivo mouse transplant outcomes were included to assess the function of islets from the 3 groups (Supplemental Figure 4, A–C). Intriguingly, the majority of the group 3 islet samples had a late response (>3 weeks) for diabetes reversal (Supplemental Figure 4, B and C). Although 71% of group 1 islet samples (prediction study set) were transplanted into individuals with T1D, the β scores (28) for these are not shown because most of our islet recipients had a second transplant within 3 months of receiving their first islet transplant. Thus, changes in HbA1c over 3 months after transplantation would be confounded because of the successive transplant. The current study was not designed or powered to predict islet transplantation outcome. This study presents the potential of MALAT1 variants as a biomarker to help accurately predict the quality of islets before initiating human islet isolation from the pancreas. The prediction of the outcome of islet transplant procedure is a separate question, which will be the focus of future investigation.
A recent study demonstrated the potential of assessing insulin immunopositive area in predicting islet isolation outcomes with 89% sensitivity and 76% specificity (29). Our data demonstrate that MALAT1 expression alone (AUC: 0.83, maximum specificity of 100% and a sensitivity of 80%) or along with Edmonton Donor Points, BMI, and NAIDS can accurately stratify a donor pancreas to group 1 (transplantable) or group 3 (not transplantable) before the initiation of islet isolation. In addition to MALAT1, lncRNAs such as GAS5 may add better discriminatory value to identify the effect of glucocorticoids (30) and stratify group 2 versus group 3 human islet sample preparations. MALAT1 was originally identified to be the most abundant lncRNA in the pancreas (31), and purified human islets (19), as well as being highly abundant in purified human β cells (19). Intriguingly, mouse knockout of MALAT1 showed no differences in the histology of pancreases and islets (32, 33) and did not affect proliferation or viability in hepatocellular carcinoma cells (32). It is currently unclear why MALAT1 is associated with high-quality (group 1) islet sample preparations. MALAT1 is encoded within an active enhancer cluster offering multiple binding sites for pancreatic islet transcription factors (34). Thus, a possible role of MALAT1 could be in facilitating (pro-) endocrine gene expression within human islets. Apart from the role of MALAT1 in islet cells, it is intriguing to note that MALAT1 is regulated by hypoxia (35–37). Indeed, hypoxia can be a major determinant of postisolation islet quality (38), and therefore the regulation of MALAT1 under hypoxic conditions can influence islet survival. Knockdown of MALAT1 leads to apoptosis (39), whereas increase in MALAT1 expression has been shown to inhibit apoptosis (40). It can therefore be speculated that islets that do not demonstrate the capacity to upregulate MALAT1 expression following exposure to hypoxic conditions would be more vulnerable to apoptotic cell death. Therefore, levels of MALAT1 detected in human pancreas before islet isolation may provide a measure of the postisolation islet quality. Indeed, the abundance of 2 MALAT1 variants (MALAT1-1.1 and -9.1) negatively correlated with postisolation islet cell death (Supplemental Figure 5).
Our study strengths are (a) the use of an unprejudiced high-depth sequencing platform for human islets from 3 groups (n = 18); (b) the use of sophisticated, unbiased data analysis methods with resampling validation (bootstrapping); (c) the independent replication of these findings in a separate set (n = 75) of human islet samples; and (d) the contribution of MALAT1 variants to improving the predictive power of Edmonton Donor Points, BMI, and NAIDS in classifying islet samples from a set (n = 19) of preisolation pancreatic tissue samples. The study limitations are the lack of larger number of matching high-quality discovery samples (which we have attempted to address via resampling/bootstrapping strategies) and the need to replicate our findings in independent islet isolation centers. It is essential to note that MALAT1 variants alone demonstrate better specificity to predict postisolation quality of islets than Edmonton Donor Points, BMI, and NAIDS. We do not know whether MALAT1 can predict clinical outcomes or graft functions after transplant, which would be an interesting question to pursue.
The present study identifies lncRNA biomarkers such as MALAT1, which could facilitate the prediction of islet isolation quality, within minutes of receiving the cadaveric pancreas. Currently, commercially available kits have been demonstrated (41) to offer the capacity to isolate and directly measure RNA in minutes. Advances in nanotechnology further enable reliable as well as cost- and time-efficient assessment of RNA quantity (42) and can be used to measure biomarker levels (43). Cross-disciplinary technological advances can be easily used to develop a rapid detection kit, similar to a pregnancy test, for clinical biomarkers (such as MALAT1 variants). Platforms to measure microRNAs and lncRNAs using nanosensor-based (44) and photonics-based technologies (45) can detect ultralow (subpicomolar to attomolar) levels of ncRNAs from cell lysates. Because MALAT1 is a highly abundant lncRNA in the human pancreas, the development of such rapid detection tests for MALAT1 variants is achievable and is indeed one of our research foci in the coming years. Nanotechnology- and photonics-based platforms offer a rapid (2–5 minutes) and low-cost (<$1) method that could improve donor selection and enhance the development of tests that ultimately help us stratify donors with high accuracy, so as to meet the release criteria for clinical islet transplantation. Because the time from cross-clamp at retrieval of a donor pancreas to initiating islet isolation is around 9.8 ± 0.179 (mean ± SEM) hours (46), the identification of MALAT1 lncRNA variants allows us a sufficient window to use this test that could change current practice. Measurement of MALAT1 variants will improve donor selection while offering the other (nonselected) pancreas for research-grade isolation workflows. Our study underscores the importance of existing donor pancreas selection criteria (Edmonton Donor Points, BMI, and NAIDS) and presents the added predictive power offered by the new lncRNA biomarkers. As discussed above, although existing criteria (such as donor BMI) may be good estimators of islet number, the measurement of MALAT1 lncRNA variants in the pancreas provides a “readout” of their quality, substantially increasing the predictive power for selection of donor pancreases in clinical islet transplantation.
Study impact.
The present study provides the capacity to make an informed choice for research or clinical islet isolation workflow. Clinical islet isolation costs using GMP-grade facilities and reagents and involving screening, organ recovery, transportation, and islet preparation are estimated around US$40,000 per preparation (1). Based on CITR data, around US$97,000,000 was spent on 2421 isolations (47), of which only 750 preparations (48) were deemed suitable for human islet transplantation. The current study provides the means to not only direct the best donor tissues for clinical transplants but also make appropriate decisions for the less cost-intensive research-grade islet isolation workflows. Our data corroborate previous findings indicating the high level of MALAT1 lncRNA in islets. The stratification of islets based on the quality of islet preparations helps in understanding the role of MALAT1 lncRNA variants as biomarkers for predicting islet isolation outcome. We also provide the first report to our knowledge of human islet lncRNA expression profiles in a quality-stratified set of islet samples to the research community through this publication.
Methods
Human pancreatic islets
Pancreatic tissue samples were taken from the middle of the pancreas at the time of transection of the pancreas for cannulation, usually close to the duct itself. Islets were isolated following the standard protocol carried out by the National Islet Transplantation Unit at the Westmead Hospital, University of Sydney (25). A standard set of donor characteristics were recorded by the team and were used in calculations of the islet quality score (Table 1), Edmonton Donor Points (2, 6), and the NAIDS (24). Islet samples were categorized into 3 groups based on their islet quality score and presented in this study as group 1 (high quality), group 2 (intermediary quality), and group 3 (inferior quality) islet sample preparations. To compare the predictive power of lncRNA biomarkers along with existing clinical and donor characteristics, we also compared the classification of islet quality based on Edmonton Donor Points as well as NAIDS.
Islet quality score
Islet quality score (presented in Supplemental Tables 1–3) was mathematically computed as the average score divided by the number of available criteria (Table 1) from the scoring of each criterion listed in Table 1. The methodology for each of these assessments is provided below.
Postisolation islet purity.
Islet purity was assessed as described elsewhere (49). Briefly, postisolation islet purity was assessed by using Dithizone stain (MilliporeSigma) (3 mg/mL). Triplicate aliquots of known volumes were sampled from the final islet cell preparation. The total number of islets was counted using standard criteria and percentage of acinar tissue quantified and scored in each of the aliquots (IEQ > 4000 IEQ/kg recipient body weight) as detailed elsewhere (49).
Postisolation islet viability.
Postisolation islet viability was determined using fluorescent labeling of cell preparations using DNA-binding dyes, so as to differentiate between live and dead cells. Fluorescein diacetate (FDA; 24 μM) and propidium iodide (PI; 750 μM) solutions (MilliporeSigma) were used to stain 10 replicates of cells with compromised membranes. Westmead Hospital utilizes a cutoff of 70% viability as a minimum for release of the product for clinical transplantation. FDA diffuses passively across the cell membrane and is converted to fluorescein by nonspecific esterases in the cytoplasm, causing live cells to fluoresce green under a 490-nm excitation wavelength. Dead or dying cells have compromised cell membranes and do not show cytoplasmic esterase activity and therefore do not fluoresce green. Counterstaining with PI allows better identification of these dead and dying cells because these cells (nuclei) will take up PI, fluorescing red at 545 nm. Triplicate aliquots of a known volume were sampled from the final islet cell preparation and stained with FDA and PI. A total of at least 100 islets were stained and cells counted and quantified as to the percentages of viable (green) versus dead (red) cells. A total accumulated score was then calculated and the mean value taken as the viability score.
Postisolation islet β cell viability index.
Flow cytometric assessment was carried out on the human islet cells following an established method developed by Ichii et al. (50). This method simultaneously determines the β cell composition, viability, and apoptotic cell percentage in enzymatically dispersed single cells from islets. Zinc-binding dye Newport Green (NPG) (Thermo Fisher Scientific) was used to determine β cell composition (NPG-positive staining), while apoptotic cells were probed using tetramethylrhodamine ethyl ester and the membrane-impermeant 7-aminoactinomycin D staining (50). Our center at Westmead Hospital conducts flow cytometric analysis on islet cells after culture to determine this β cell viability index, defined as (% β cells × % viable β cells)/10,000, with indices of 0.5 or higher considered satisfactory for release for transplantation.
Transplantability.
The analysis of the suitability of the islet preparation for human transplantation was carried out using the following criteria: the release criteria formally accepted for our program are based on islet count per recipient weight (5000–20,000 IEQ/kg for the first transplant and 3000–20,000 for following transplants), with purity at least 30%, viability at least 70%, endotoxin concentration less than 5 endotoxin units/kg recipient weight, and no detectable organisms in a Gram stain before transplant, in addition to a glucose stimulation index (ratio of stimulated insulin secretion/basal insulin secretion) greater than 1 and a β cell viability index of 0.5 or higher.
Islet insulin content
Insulin content was measured in 2 replicates for each postisolation islet preparation. One mL Azol was added to each replicate before sonication. Sonicated aliquots were then mixed with FSA solution and appropriately processed and diluted for total insulin content analysis using the manufacturer’s method for immunoassay on the Architect (Abbott Diagnostics). Data were normalized to DNA content (Quant-IT Pico Green dsDNA Assay Kit, Life Technologies). Values are presented in total insulin content for the entire islet preparation, after normalizing for the amount of DNA.
Human islet transplantation into mice
Athymic mice (Animal Resources Centre) were rendered diabetic with streptozotocin, before transplantation of between 2000 and 3000 IEQ of (after culture) islets beneath the kidney capsule. Blood sugar levels were monitored after transplant to determine the success of the islet transplant procedure, defined as either a reduction in nonfasting blood glucose levels to under 11 mmol/L (200 mg/dL) or halving in nonfasting blood glucose levels from the diabetic state, in 2 successive measurements 48–72 hours apart. An intraperitoneal glucose tolerance test was conducted 3–4 weeks after transplant to assess glucose clearance and hence determine whether success in islet function was achieved after transplant.
Human islet stimulation index
The functional capacity of human islets was assessed by measuring insulin secretion in response to glucose. Briefly, (after culture) islets were incubated at 37°C and 5% CO2 for 1 hour in RPMI 1640 (Thermo Fisher Scientific) with either 2.8 mM glucose or 25 mM glucose. For each islet preparation, 6 replicates were exposed to 2.8 mM glucose and 25 mM glucose. Supernatants were collected and their insulin content was measured using an Architect (Abbott Diagnostics). The stimulation index was calculated by dividing the insulin content in the 25-mM glucose incubation sample by the insulin content in the 2.8-mM glucose incubation sample. Higher values indicate a better insulin secretion response to glucose stimulation and therefore a higher functional capacity (26, 51).
RNA isolation and QC
Total RNA was isolated from 2000 to 5000 IEQ using the manufacturer’s TRIzol RNA isolation (Thermo Fisher Scientific) protocol with minor modifications (52). Details and QC of the selected samples (discovery set, n = 18) for RNA-Seq are provided in supplemental material (Supplemental Table 7). RNA integrity number (RIN) was assessed for every sample, and only those with an RIN value of at least 7.4 were taken for discovery analysis.
High-depth RNA-Seq
High-depth RNA-Seq (rRNA depletion library construction) was carried out using the HiSeq4000, 150 paired-end (PE) reads platform (Novogene), on n = 18 human pancreatic islet preparations (Supplemental Table 7). An input of 3 μg RNA per sample was used for sequencing. Epicentre Ribo-Zero rRNA Removal Kit and ethanol precipitation were used to remove the rRNA and rRNA-free residues for each sample, respectively. Subsequently, sequencing libraries were generated using the rRNA-depleted RNA with the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB) following the manufacturer’s protocol. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using HiSeq PE Cluster Kit cBot-HS (Illumina) following the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina HiSeq platform and PE reads were generated. FPKM sequenced were calculated for both lncRNAs and coding genes in each sample using Cuffdiff (v2.1.1). FPKM are the means mapped and calculated based on the length of the fragments and read count mapped to these fragments (53). Results are presented in normalized FPKM.
RNA-Seq analysis
In-house perl scripts were used to remove reads containing adapter, reads containing poly-N, and low-quality reads from the raw data (reads), leaving only the clean data (reads). The index of the reference human GRCh38 genome (http://ftp.ensembl.org/pub/release-82/fasta/homo_sapiens/dna/) and the GTF files (ftp://ftp.ensembl.org/pub/release-82/gtf/homo_sapiens/) were used for annotation (obtained from the genome website) and built using Bowtie v2.0.6. TopHat v2.0.9 was used to align PE clean reads to the reference genome. On average 77,127,891 clean reads were obtained from each of the 18 samples that passed QC in this discovery set. Phylogenetic codon substitution frequency (phyloCSF; v20121028) was used to identify and distinguish the characteristics to align conserved coding regions (54). Multispecies genome sequence alignments were built and run on phyloCSF with default parameters. Transcripts predicted with coding potential (by all or either of the 4 tools CMCI, CPS, Pfam-scan, and phyloCSF) were removed, while transcripts without coding potential were identified to be the candidate set of lncRNAs.
Real-time qPCR
The High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific) was used followed by TaqMan real-time qPCR for assays. Briefly, synthesis of cDNA from RNA was carried out using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). TaqMan real-time qPCR was performed in 5-μL reactions using 96-well plates with 33.3 ng input cDNA with TaqMan Fast Universal PCR Master Mix (Thermo Fisher Scientific). Selected TaqMan primer/probe gene expression assays are provided in Supplemental Table 8. We used 2 MALAT1 gene transcript assays (907 and 177), which span different regions of the human MALAT1 gene. MALAT1 gene transcript assay-907 captures variants MALAT1-9.1 and -2.1, while MALAT1 gene transcript assay-177 covers only MALAT1-1.1. Results were normalized to 18s rRNA values. Real-time qPCR was carried using the ViiA7 platform (Thermo Fisher Scientific).
Data availability
We intend to replicate these findings at other islet isolation centers and welcome collaborative or independent assessment of our predictive algorithms. All data from our lncRNA-Seq studies were uploaded to the Gene Expression Omnibus database and are available through the study accession number GSE134068. We welcome future collaborations to analyze and validate these in other centers. Such studies through multiple islet isolation centers would help in confirming the potential of these MALAT1 lncRNA variants for improving the prediction of pancreatic islet isolation procedure outcomes.
Statistics
All statistical analyses were performed using GraphPad Prism v7 (GraphPad Software), SPSS, R glmnet, penalized, or corrplot packages. A 2-tailed distribution, with 2-sample unequal-variance Student’s t test, was used to compare specific lncRNAs between 2 groups for the RNA-Seq data. To identify a subset of lncRNAs presenting the strongest associations with the 3 categorized groups of islet preparations, L1-PLR (Lasso) techniques were used, as described elsewhere (27). The lncRNAs identified by Lasso analyses were confirmed by resampling validation/bootstrapping analyses (for 1000 iterations). During bootstrapping, about 37% of samples were randomly removed and the same number of different (randomly selected) samples duplicated so that the total number of samples remained the same in each of the 1000 iterations. Resampling validation was an important part of the process, so as to eliminate any sampling bias. A Wilcoxon P value cutoff of P > 0.5 was also applied in each iteration. Univariate logistic and linear regression was carried for each comparison to examine the association of each independent variable with the outcome. A 1-way ANOVA test with multiple-comparisons correction was used for all group comparisons for the qPCR data. R scripts for the above machine-learning algorithms will be made available following publication.
Study approval
Human tissue samples.
Use of research-consented human tissues was approved through the human research ethics committee (HREC) approval X16-0289 (previously X12-0176) and the HREC/12/RPAH/282 at the University of Sydney, Sydney Local Health District, and tissues from donors who gave informed research consent were obtained through the National Islet Transplantation Program at the Westmead Hospital, University of Sydney, Australia (AU RED/HREC/15/WMEAD/284).
Animal studies.
Animal studies were approved by the animal ethics committee at the Western Sydney Local Health District (protocols 4198.10.12 and 5146.10.17).
Author contributions
WKMW performed data analysis, experimentation, write-up, and revision. GJ, AES, RCM, and LTD were responsible for biostatistics, data analysis/validation, and data interpretation. CLM, YVC, DL, LW, PJO, and WJH performed data analysis, data interpretation, and islet isolation/banking. AAH and MVJ were responsible for study design, acted as guarantors of the work, and performed data analysis, data interpretation, and manuscript writing and revision. All authors agreed on the final revised draft.
Supplementary Material
Acknowledgments
This study was supported by the Marian and E.H. Flack Trust (to WJH; FLACK-2016-17), Juvenile Diabetes Research Foundation (JDRF) Australia T1D CRN (to AAH; 4-CDA-2016-228-M-B), JDRF International (to MVJ; 3-APF-2016-178-A-N), Australian Research Council (to AAH; FT110100254), Sydney Medical Foundation (to AAH and MVJ), Danish Diabetes Academy funded by the Novo Nordisk Foundation (to AAH, LTD, and AES). In addition, we acknowledge the Postdoctoral Fellowship Scheme, The Chinese University of Hong Kong (to GJ) and the University of Sydney postgraduate award and top-up scholarship from the JDRF Australia (to WKMW).
Version 1. 07/30/2019
In-Press Preview
Version 2. 08/22/2019
Electronic publication
Funding Statement
Career Development Award from the JDRF Australia T1D Clinical Research Network
JDRF International Advanced Post-doctoral Fellowship to MVJ
Danish Diabetes Academy Visiting Professorship to AAH
Footnotes
Conflict of interest: AAH, MVJ, WKMW, and WJH have intellectual property (2019202308: Tissue assessment method).
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(16):e129299. https://doi.org/10.1172/jci.insight.129299.
Contributor Information
Wilson K.M. Wong, Email: wilson.wong@ctc.usyd.edu.au.
Guozhi Jiang, Email: gzjiang@cuhk.edu.hk.
Anja E. Sørensen, Email: elaine@ruc.dk.
Yi Vee Chew, Email: yi.chew@sydney.edu.au.
Cody Lee-Maynard, Email: cody-lee.maynard@ctc.usyd.edu.au.
David Liuwantara, Email: david@aust-biosearch.com.au.
Lindy Williams, Email: lindy.williams@sydney.edu.au.
Louise T. Dalgaard, Email: ltd@ruc.dk.
Ronald C. Ma, Email: rcwma@cuhk.edu.hk.
Wayne J. Hawthorne, Email: wayne.hawthorne@sydney.edu.au.
Mugdha V. Joglekar, Email: mugdha.joglekar@ctc.usyd.edu.au.
Anandwardhan A. Hardikar, Email: anand.hardikar@ctc.usyd.edu.au.
References
- 1.Guignard AP, et al. Cost analysis of human islet transplantation for the treatment of type 1 diabetes in the Swiss-French Consortium GRAGIL. Diabetes Care. 2004;27(4):895–900. doi: 10.2337/diacare.27.4.895. [DOI] [PubMed] [Google Scholar]
- 2.O’Gorman D, et al. The standardization of pancreatic donors for islet isolations. Transplantation. 2005;80(6):801–806. doi: 10.1097/01.tp.0000172216.47547.d5. [DOI] [PubMed] [Google Scholar]
- 3.Brandhorst H, Brandhorst D, Hering BJ, Federlin K, Bretzel RG. Body mass index of pancreatic donors: a decisive factor for human islet isolation. Exp Clin Endocrinol Diabetes. 1995;103(Suppl 2):23–26. doi: 10.1055/s-0029-1211388. [DOI] [PubMed] [Google Scholar]
- 4.Nano R, et al. Islet isolation for allotransplantation: variables associated with successful islet yield and graft function. Diabetologia. 2005;48(5):906–912. doi: 10.1007/s00125-005-1725-3. [DOI] [PubMed] [Google Scholar]
- 5.Balamurugan AN, et al. Islet product characteristics and factors related to successful human islet transplantation from the Collaborative Islet Transplant Registry (CITR) 1999-2010. Am J Transplant. 2014;14(11):2595–2606. doi: 10.1111/ajt.12872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lakey JR, et al. Variables in organ donors that affect the recovery of human islets of Langerhans. Transplantation. 1996;61(7):1047–1053. doi: 10.1097/00007890-199604150-00010. [DOI] [PubMed] [Google Scholar]
- 7.Toso C, et al. Factors affecting human islet of Langerhans isolation yields. Transplant Proc. 2002;34(3):826–827. doi: 10.1016/S0041-1345(01)02925-6. [DOI] [PubMed] [Google Scholar]
- 8.Niclauss N, et al. Influence of donor age on islet isolation and transplantation outcome. Transplantation. 2011;91(3):360–366. doi: 10.1097/TP.0b013e31820385e6. [DOI] [PubMed] [Google Scholar]
- 9.de Almeida RA, Fraczek MG, Parker S, Delneri D, O’Keefe RT. Non-coding RNAs and disease: the classical ncRNAs make a comeback. Biochem Soc Trans. 2016;44(4):1073–1078. doi: 10.1042/BST20160089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18(1):5–18. doi: 10.1038/nrc.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guay C, Jacovetti C, Nesca V, Motterle A, Tugay K, Regazzi R. Emerging roles of noncoding RNAs in pancreatic β-cell function and dysfunction. Diabetes Obes Metab. 2012;14(Suppl 3):12–21. doi: 10.1111/j.1463-1326.2012.01654.x. [DOI] [PubMed] [Google Scholar]
- 12.Bunch H. Gene regulation of mammalian long noncoding RNA. Mol Genet Genomics. 2018;293(1):1–15. doi: 10.1007/s00438-017-1370-9. [DOI] [PubMed] [Google Scholar]
- 13.Nie L, et al. Long noncoding RNAs: versatile master regulators of gene expression and crucial players in cancer. Am J Transl Res. 2012;4(2):127–150. [PMC free article] [PubMed] [Google Scholar]
- 14.Mirza AH, Kaur S, Pociot F. Long noncoding RNAs as novel players in β cell function and type 1 diabetes. Hum Genomics. 2017;11(1):17. doi: 10.1186/s40246-017-0113-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pelechano V, Steinmetz LM. Gene regulation by antisense transcription. Nat Rev Genet. 2013;14(12):880–893. doi: 10.1038/nrg3594. [DOI] [PubMed] [Google Scholar]
- 16.Harrow J, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22(9):1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cabili MN, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25(18):1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mattick JS, Makunin Non-coding RNA. Hum Mol Genet. 2006;15(Suppl 1):R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 19.Akerman I, et al. Human pancreatic β cell lncRNAs control cell-specific regulatory networks. Cell Metab. 2017;25(2):400–411. doi: 10.1016/j.cmet.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benner C, van der Meulen T, Cacéres E, Tigyi K, Donaldson CJ, Huising MO. The transcriptional landscape of mouse beta cells compared to human beta cells reveals notable species differences in long noncoding RNA and protein-coding gene expression. BMC Genomics. 2014;15:620. doi: 10.1186/1471-2164-15-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li B, et al. RNA-seq methods for identifying differentially expressed gene in human pancreatic islet cells treated with pro-inflammatory cytokines. Mol Biol Rep. 2014;41(4):1917–1925. doi: 10.1007/s11033-013-3016-2. [DOI] [PubMed] [Google Scholar]
- 22.Morán I, et al. Human β cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab. 2012;16(4):435–448. doi: 10.1016/j.cmet.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singer RA, Sussel L. Islet long noncoding RNAs: a playbook for discovery and characterization. Diabetes. 2018;67(8):1461–1470. doi: 10.2337/dbi18-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang LJ, et al. A multicenter study: North American islet donor score in donor pancreas selection for human islet isolation for transplantation. Cell Transplant. 2016;25(8):1515–1523. doi: 10.3727/096368916X691141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Connell PJ, et al. Multicenter Australian trial of islet transplantation: improving accessibility and outcomes. Am J Transplant. 2013;13(7):1850–1858. doi: 10.1111/ajt.12250. [DOI] [PubMed] [Google Scholar]
- 26.Ricordi C. Quantitative and qualitative standards for islet isolation assessment in humans and large mammals. Pancreas. 1991;6(2):242–244. doi: 10.1097/00006676-199103000-00018. [DOI] [PubMed] [Google Scholar]
- 27.Goeman JJ. L1 penalized estimation in the Cox proportional hazards model. Biom J. 2010;52(1):70–84. doi: 10.1002/bimj.200900028. [DOI] [PubMed] [Google Scholar]
- 28.Ryan EA, Paty BW, Senior PA, Lakey JR, Bigam D, Shapiro AM. Beta-score: an assessment of beta-cell function after islet transplantation. Diabetes Care. 2005;28(2):343–347. doi: 10.2337/diacare.28.2.343. [DOI] [PubMed] [Google Scholar]
- 29.Berkova Z, et al. Combining donor characteristics with immunohistological data improves the prediction of islet isolation success. J Diabetes Res. 2016;2016:4214328. doi: 10.1155/2016/4214328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lucafò M, et al. Role of the long non-coding RNA growth arrest-specific 5 in glucocorticoid response in children with inflammatory bowel disease. Basic Clin Pharmacol Toxicol. 2018;122(1):87–93. doi: 10.1111/bcpt.12851. [DOI] [PubMed] [Google Scholar]
- 31.Ji P, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage nonsmall cell lung cancer. Oncogene. 2003;22(39):8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 32.Eißmann M, et al. Loss of the abundant nuclear noncoding RNA MALAT1 is compatible with life and development. RNA Biol. 2012;9(8):1076–1087. doi: 10.4161/rna.21089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakagawa S, et al. Malat1 is not an essential component of nuclear speckles in mice. RNA. 2012;18(8):1487–1499. doi: 10.1261/rna.033217.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pasquali L, et al. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat Genet. 2014;46(2):136–143. doi: 10.1038/ng.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brock M, et al. Analysis of hypoxia-induced noncoding RNAs reveals metastasis-associated lung adenocarcinoma transcript 1 as an important regulator of vascular smooth muscle cell proliferation. Exp Biol Med (Maywood) 2017;242(5):487–496. doi: 10.1177/1535370216685434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lelli A, et al. Induction of long noncoding RNA MALAT1 in hypoxic mice. Hypoxia (Auckl) 2015;3:45–52. doi: 10.2147/HP.S90555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sallé-Lefort S, et al. Hypoxia upregulates Malat1 expression through a CaMKK/AMPK/HIF-1α axis. Int J Oncol. 2016;49(4):1731–1736. doi: 10.3892/ijo.2016.3630. [DOI] [PubMed] [Google Scholar]
- 38.Lau J, Henriksnäs J, Svensson J, Carlsson PO. Oxygenation of islets and its role in transplantation. Curr Opin Organ Transplant. 2009;14(6):688–693. doi: 10.1097/MOT.0b013e32833239ff. [DOI] [PubMed] [Google Scholar]
- 39.Zhao M, Wang S, Li Q, Ji Q, Guo P, Liu X. MALAT1: a long noncoding RNA highly associated with human cancers. Oncol Lett. 2018;16(1):19–26. doi: 10.3892/ol.2018.8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xin JW, Jiang YG. Long noncoding RNA MALAT1 inhibits apoptosis induced by oxygen-glucose deprivation and reoxygenation in human brain microvascular endothelial cells. Exp Ther Med. 2017;13(4):1225–1234. doi: 10.3892/etm.2017.4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Peer G, Mestdagh P, Vandesompele J. Accurate RT-qPCR gene expression analysis on cell culture lysates. Sci Rep. 2012;2:222. doi: 10.1038/srep00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dimov IK, et al. Discriminating cellular heterogeneity using microwell-based RNA cytometry. Nat Commun. 2014;5:3451. doi: 10.1038/ncomms4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharma TK, et al. Aptamer-mediated ‘turn-off/turn-on’ nanozyme activity of gold nanoparticles for kanamycin detection. Chem Commun (Camb) 2014;50(100):15856–15859. doi: 10.1039/C4CC07275H. [DOI] [PubMed] [Google Scholar]
- 44.Joshi GK, et al. Label-free nanoplasmonic-based short noncoding RNA sensing at attomolar concentrations allows for quantitative and highly specific assay of microRNA-10b in biological fluids and circulating exosomes. ACS Nano. 2015;9(11):11075–11089. doi: 10.1021/acsnano.5b04527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vaisocherová H, et al. Rapid and sensitive detection of multiple microRNAs in cell lysate by low-fouling surface plasmon resonance biosensor. Biosens Bioelectron. 2015;70:226–231. doi: 10.1016/j.bios.2015.03.038. [DOI] [PubMed] [Google Scholar]
- 46.Andres A, et al. Clinical islet isolation and transplantation outcomes with deceased cardiac death donors are similar to neurological determination of death donors. Transpl Int. 2016;29(1):34–40. doi: 10.1111/tri.12650. [DOI] [PubMed] [Google Scholar]
- 47. Collaborative Islet Transplant Registry Coordinating Center and Investigators. CITR 9th Annual Report. Collaborative Islet Transplant Registry Coordinating Center; 2016. [Google Scholar]
- 48. Bruni A, McCall M, Shapiro AMJ. Islet cell transplantation. In: Ledbetter D, Johnson P, eds. Endocrine Surgery in Children. Berlin, Germany: Springer; 2018:181–196. [Google Scholar]
- 49.Hawthorne WJ. Necessities for a clinical islet program. Adv Exp Med Biol. 2016;938:67–88. doi: 10.1007/978-3-319-39824-2_6. [DOI] [PubMed] [Google Scholar]
- 50.Ichii H, et al. A novel method for the assessment of cellular composition and beta-cell viability in human islet preparations. Am J Transplant. 2005;5(7):1635–1645. doi: 10.1111/j.1600-6143.2005.00913.x. [DOI] [PubMed] [Google Scholar]
- 51.Ricordi C, Lacy PE, Finke EH, Olack BJ, Scharp DW. Automated method for isolation of human pancreatic islets. Diabetes. 1988;37(4):413–420. doi: 10.2337/diab.37.4.413. [DOI] [PubMed] [Google Scholar]
- 52.Joglekar MV, Hardikar AA. Isolation, expansion, and characterization of human islet-derived progenitor cells. Methods Mol Biol. 2012;879:351–366. doi: 10.1007/978-1-61779-815-3_21. [DOI] [PubMed] [Google Scholar]
- 53.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin MF, Jungreis I, Kellis M. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011;27(13):i275–i282. doi: 10.1093/bioinformatics/btr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hardikar AA, Farr RJ, Joglekar MV. Circulating microRNAs: understanding the limits for quantitative measurement by real-time PCR. J Am Heart Assoc. 2014;3(1):e000792. doi: 10.1161/JAHA.113.000792. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We intend to replicate these findings at other islet isolation centers and welcome collaborative or independent assessment of our predictive algorithms. All data from our lncRNA-Seq studies were uploaded to the Gene Expression Omnibus database and are available through the study accession number GSE134068. We welcome future collaborations to analyze and validate these in other centers. Such studies through multiple islet isolation centers would help in confirming the potential of these MALAT1 lncRNA variants for improving the prediction of pancreatic islet isolation procedure outcomes.