Abstract
BACKGROUND
Acute graft versus host disease (aGvHD) is a major factor that limits the successful outcomes of allogeneic hematopoietic cell transplantation (alloHSCT). Currently, there are few validated biomarkers that can help predict the risk of aGvHD in clinical settings.
METHODS
We performed an integrated metabolomics and transcriptomics study and identified biomarkers that distinguish alloHSCT recipients with aGvHD from alloHSCT recipients without aGvHD in 2 separate cohorts.
RESULTS
Pathway analysis of 38 significantly altered metabolites and 1,148 differentially expressed genes uncovered a distinctly altered glycerophospholipid (GPL) metabolism network. Subsequently, we developed an aGvHD risk score (GRS) based on 5 metabolite markers from GPL metabolism to predict the risk of aGvHD. GRS showed a positive predictive value of 92.2% and 89.6% in the training and validation cohorts, respectively. In addition, high GRS was correlated with poor overall survival. Gene expressions of GPL-related lipases were significantly altered in aGvHD samples, leading to dysregulated GPLs.
CONCLUSION
Using integrative “Omic” analysis, we unraveled a comprehensive view of the molecular perturbations underlying the pathogenesis of aGvHD. Our work represents an initial investigation of a unique metabolic and transcriptomic network that may help identify aGvHD at an early stage and facilitate preemptive therapy.
FUNDING
National Natural Science Foundation of China (NSFC; 81530047, 81870143, 81470321, 81770160, 81270567, 81270638, 81573396, 81703674) provided funding for material, processing, metabolomics, and transcriptomics studies. This work was also supported by Shanghai Sailing Program from Science and Technology Commission Shanghai Municipality (17YF1424700), the Scholarship from Shanghai Municipal Health and Family Planning Commission (2017BR012), and the Special Clinical Research in Health Industry in Shanghai (20184Y0054). The projects supporting young scholars are 17YF1424700 (to YL), 2017BR012 (to XH), and 20184Y0054 (to QC).
Keywords: Metabolism, Transplantation
Keywords: Bone marrow transplantation
Acute graft-versus-host disease has a unique metabolic signature in patients characterized by alterations in the glycerophospholipid metabolism network.
Introduction
Graft versus host disease (GvHD), caused by alloreactive donor T cells, is a major factor that limits the success of allogeneic hematopoietic cell transplantation (alloHSCT) (1). An estimated 40%–60% of alloHSCT recipients are affected by GvHD (2, 3). Currently, the diagnosis of acute GvHD (aGvHD) is mainly based on clinical symptoms pertaining to 1 or more target organ systems (4, 5). Glucocorticoids and immunosuppressive agents are used as the first-line treatment for GvHD. However, generalized immunosuppression is liable to lead to opportunistic infections. Therefore, there has been an increased interest in identifying biomarkers of aGvHD and understanding the pathogenesis of aGvHD so as to guide treatment decision-making and direct more intensive clinical surveillance of high-risk patients (6–14).
Extensive research has been dedicated to identifying blood biomarkers for aGvHD to improve diagnosis and facilitate personalized treatment. Biomarkers for aGvHD have been studied thoroughly at the facets of cytokines (15–17), immune cells (18–21), single nucleotide polymorphisms (22), miRNAs (23–27), and chemokines (12, 28–31). However, no single biomarker predicts outcomes across different transplant practices or patient characteristics. Up to now, the best combination was ST2 and REG3α. Studies have shown that ST2 and REG3α levels at the initiation of GvHD therapy and during the first month after transplantation may help improve risk stratification for treatment-resistant GvHD (11, 12, 30). An increasing body of evidence suggests a strong connection between aGvHD and metabolism (32, 33). Cell metabolism determines the fate and function of T cells, which play a key role in the pathogenesis of aGvHD. Metabolomics is an important component of system biology that allows us to measure metabolic alterations that reflect the genetic, epigenetic, and environmental determinants of cellular physiology (34, 35). The stearic acid/palmitic acid (SA/PA) ratio detected on day 7 after HSCT was shown to be a useful biomarker of aGvHD and relapse (36). However, the influence of gene changes on metabolism and the gene-metabolite networks involved in aGvHD are not well characterized. Integration of metabolomics and transcriptomics may yield further insights into the pathogenesis of aGvHD. Indeed, it may help uncover the complex regulatory networks involving genes and metabolic pathways in aGvHD than either approach alone.
In the present study, we performed global metabolic analysis in alloHSCT recipients with or without aGvHD to identify critical metabolites that may help predict aGvHD. We performed transcriptomic profiling in a cohort of patients (training set) to identify significantly altered genes between patients with and without aGvHD. Integrative analysis of metabolomics and transcriptomic data revealed that glycerophospholipid (GPL) metabolism may play an important role in the pathogenesis of aGvHD. These relevant metabolic and transcriptomic alterations were further verified in a separate cohort of patients (validation set). Subsequently, we combined the metabolic and transcriptomic data to identify significantly disturbed pathways at both the metabolic and transcriptional levels. Lastly, we developed a model including 5 highly connected GPL metabolites to predict the probability of aGvHD.
Results
Metabolic alterations in aGvHD patients.
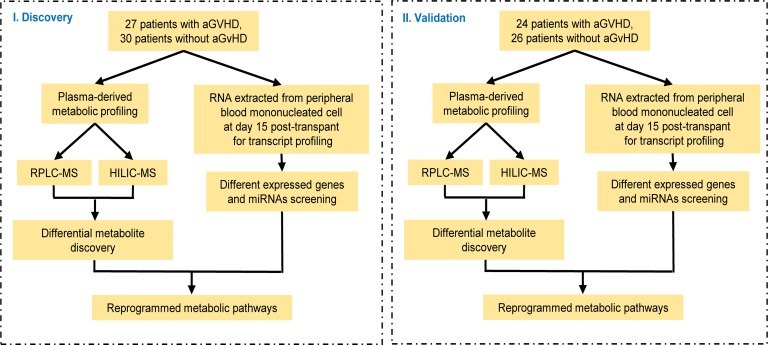
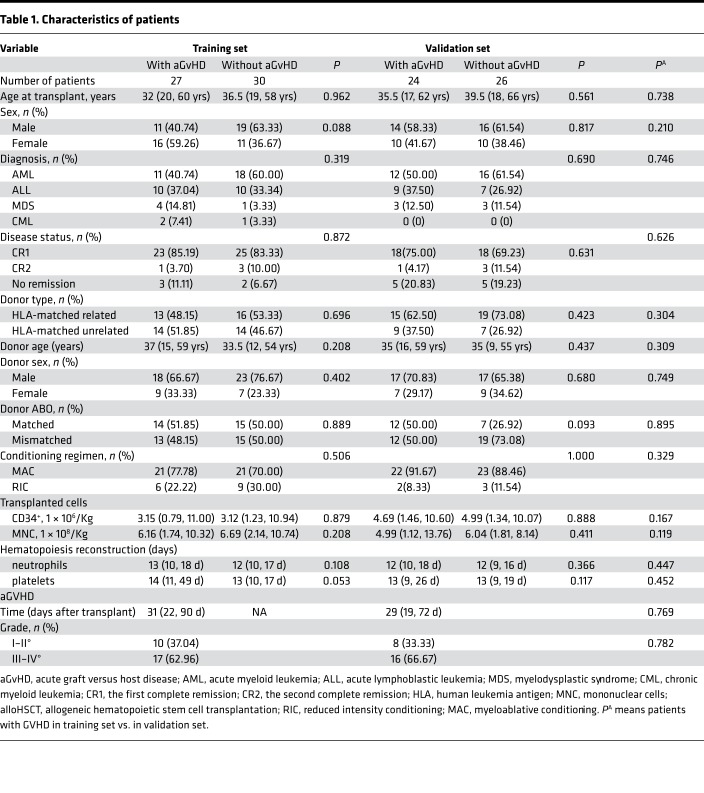
The overall study design is shown in Figure 1, and the clinical characteristics of patients are summarized in Table 1. There were no significant differences with respect to age, sex, diagnosis, disease status, conditioning regimen, or type of transplantation between the 2 sample sets or between patients with and without aGvHD in each set. To explore the metabolic profile of aGvHD, we used a metabolomic approach based on liquid chromatography−mass spectrometry (LC-MS) to identify differentially expressed metabolites in the aGvHD and control groups (without aGvHD) in the training set. The metabolomic profile of the aGvHD group at +15 days after transplantation was distinctly different from that of the control group, as evidenced by robust orthogonal partial least squares discriminant analysis (OPLS-DA) models established in the training (R2Xcum = 0.523; R2Ycum = 0.745; and Q2cum = 0.634) and validation sets (R2Xcum = 0.519; R2Ycum = 0.684; and Q2cum = 0.576). The scores plot demonstrated a clear separation between cases with and without aGvHD, and no overfitting of the OPLS-DA models were found (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.129494DS1).
Figure 1. Experimental flow chart.
Table 1. Characteristics of patients.
Based on the criteria of variables important to projection (VIP) value > 1 and P < 0.05 (2-tailed Student’s t test), 38 metabolites were identified as potential biomarkers (Supplemental Table 1). These metabolites pertained to GPL metabolism, fatty acid metabolism, amino acid metabolism, citrate cycle, sphingolipid metabolism, purine metabolism, and bile acid metabolism.
Integration of metabolomics and transcriptomics revealed altered GPL metabolism pathway during pathogenesis of aGvHD.
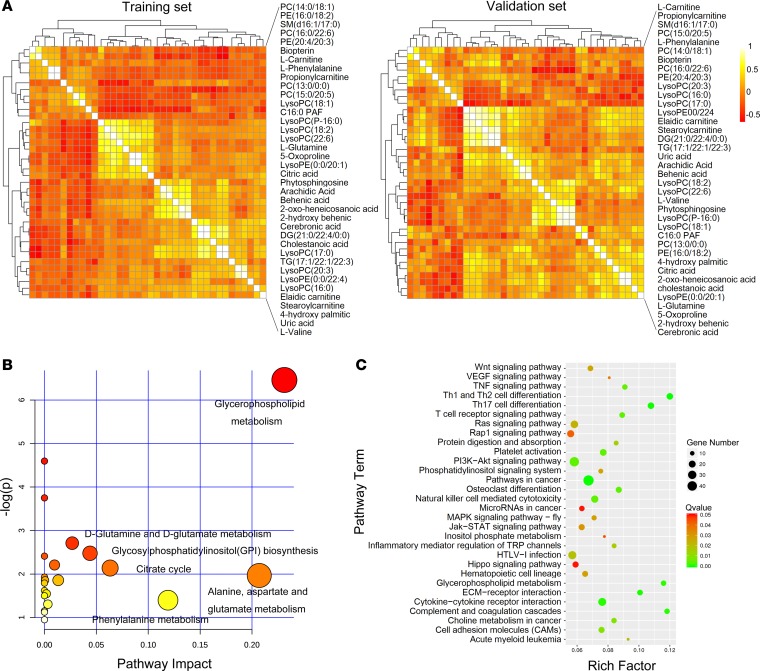
To discover the essential metabolites, data pertaining to 38 potential biomarkers in the training and validation sets were used to build correlation networks (Figure 2A). Five highly connected metabolites (lysophosphatidylcholine [LysoPC] [18:1], C16:0 PAF, LysoPC [P-16:0], LysoPC [18:2], and LysoPC [22:6]) with the brightest colors appeared in the center of the networks in both sets. Interestingly, these metabolites were all GPLs, which indicated that GPL metabolism may be notably changed in aGvHD. Figure 2B illustrates the altered pathways based on metabolic profiling data. We identified GPL metabolism as the top hit because of its lower P value and greater pathway impact; this suggested that GPL metabolism is one of the most significant pathways.
Figure 2. Integration of metabolomics and transcriptomics identified a unique set of GPL metabolism pathways in aGvHD.
(A) Correlation analysis using profiling data from 38 metabolites in the training (n = 57) and validation sets (n = 50). (B) Overview of the pathway analysis based on metabolites alteration in +15 day samples after transplantation. (C) Pathway enrichment analysis of transcriptomic profiles associated with aGvHD.
To trace the upstream variations of metabolome, we conducted transcriptional profiling of the same specimens. Of the 57,773 genes profiled, 1,148 were significantly altered in patients with aGvHD (t test, FDR corrected P < 0.05, |fold change| > 1.5). Figure 2C shows the top 30 significant pathways based on transcriptomic pathway analysis. After metabolic pathway analysis and transcriptomic pathway analysis, we found 2 differentially expressed pathways at both the metabolomic and mRNA expression levels in patients with aGvHD: GPL metabolism and inositol phosphate metabolism (Figure 2). Furthermore, transcriptomic pathway analysis (Figure 2C) showed that the GPL metabolism pathway was one of the most significant pathways with Bonferroni correction P value (Q value) < 0.001; this indicated that it is a critical pathway related to the development of aGvHD. We identified 13 significantly altered genes associated with GPL metabolism. Moreover, gene-related pathways known to play a key role in aGvHD physiopathogenesis were also differentially expressed between patients with and without aGvHD; these included Th1 and Th2 differentiation, Th17 differentiation, and MAPK signaling pathways.
Glycerophospholipolytic metabolites and enzymes were significantly altered during aGvHD.
Next, we investigated the significant metabolic alterations in GPL metabolism. Compared with patients without aGvHD, the following 17 metabolites involved in the GPL metabolism were identified in this study: LysoPC (16:0), LysoPC (20:3), LysoPC (17:0), LysoPC (18:2), LysoPC (22:6), LysoPC (18:1), LysoPC (P-16:0), LysoPE (0:0/20:1), LysoPE (0:0/22:4), PC (16:0/22:6), PC (13:0/0:0), PC (14:0/18:1), PC (15:0/20:5), Phosphatidylethanolamine (PE) (20:4/20:3), PE (16:0/18:2), Diacylglycerol (21:0/22:4/0:0) (DG [21:0/22:4/0:0]), and platelet activating factor C16:0 (C16:0 PAF) (Supplemental Table 1). All of these were differently expressed in aGvHD plasma in both the training and validation sets (P < 0.05). Compared with patients without aGvHD, those with aGvHD exhibited significant accumulation of PCs and PEs, along with reduction of LysoPCs, LysoPEs, DG, and C16:0 PAF. Of note, the levels of metabolites were completely or partially restored to normal levels with the remission of aGvHD in both the training and validation sets (Supplemental Table 2).
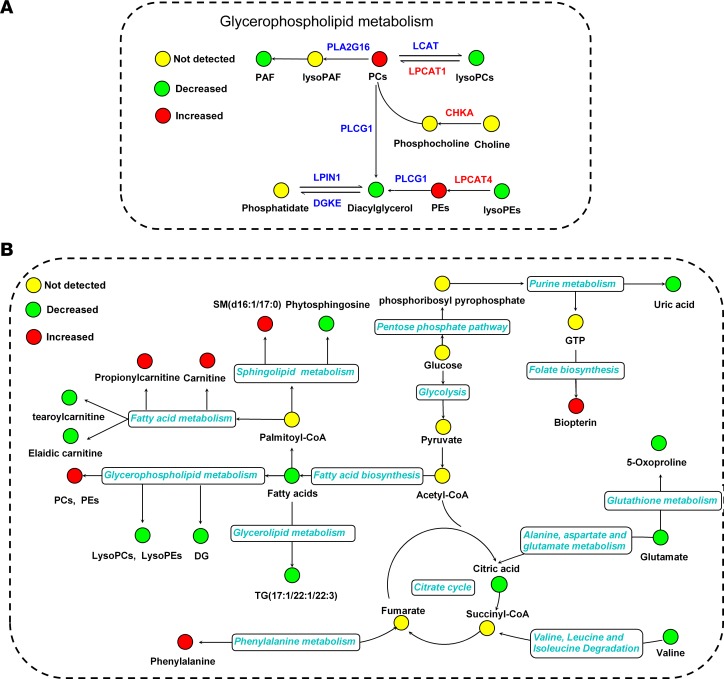
We further explored the gene expressions of these lipases involved in GPL metabolism, which included GPL lipase surrogate genes lecithin-cholesterol acyltransferase (LCAT, cholesterol esterifying enzyme), lysophosphatidylcholine acyltransferase 1 (LPCAT1, LysoPC acyltransferase 1), LPCAT4 (LysoPC acyltransferase 4), HRAS-like suppressor 3 (PLA2G16, phospholipase), PLCG1 (phosphor lipase C), phosphatidate phosphatase LPIN 1 (LPIN1, phosphatidic acid phosphohydrolase enzyme), DGKE (DG kinase), and CHKA (choline kinase) (Figure 3). The fluctuations in the relative signal intensities were in accordance with the aGvHD severity in both the training and validation sets (Figure 3, B and C). The relative gene expressions of these lipases in patients with and without aGvHD were also visualized using a heatmap (Figure 3A). As shown in Figure 4A, we observed a consistent trend in gene expressions of these lipases and the related GPLs. Downregulation of lysoPCs, PAF, and DG were associated with downregulation of LCAT, PLA2G16, PLCG1, and LPIN1; in addition, upregulation of PCs and PEs were associated with upregulation of LPAT1 and LPCAT4.
Figure 3. Gene expression of glycerophospholipolytic enzymes in 2 independent sets.
(A–C) Heatmap showing the changes in the expression of genes involved in GPL metabolism of aGvHD and control (without aGvHD) groups. Shades of red and green represent high or low expression (see color scale) (A). Each column represents a patient with aGvHD or without aGvHD. LCAT, LPCAT1, LPCAT4, PLA2G16, PLCG1, LPIN1, DGKE, and CHKA are differentially expressed in aGvHD as compared with control in training (B) and validation sets (C). In the training set, n = 13 for aGvHD and aGvHDΔ samples, n = 12 for without aGvHD samples. In validation set, n = 12 for aGvHD, aGvHDΔ, and without aGvHD samples. The comparisons were made between control vs. aGvHD and between aGvHDΔ vs. aGvHD. Box plots represent gene expression level with relative intensity (log2) of microarray data in the training set or relative Ct value normalized with endogenous control gene U6 using qPCR data in validation set. Bars indicate median value. P value refers to 1-way ANOVA. *P < 0.05, **P < 0.01. aGvHDΔ, aGvHD in remission after medication.
Figure 4. Schematic overview of the metabolites and major metabolic pathways, as well as pathway-related enzyme surrogates iterations, in aGvHD patients at day +15 after alloHSCT.
(A) Description of the glycerophospholipid metabolism pathway related metabolites and enzyme surrogate genes changes in aGvHD. (B) Dysregulation of metabolic pathways in aGvHD. The enzyme surrogates are shown in color: red text represents increased expression of enzyme surrogates, and blue text represents decreased expression of enzyme surrogates. The yellow circles represent no detected metabolites; green circles represent decreased metabolites; and red circles represent increased metabolites.
Development of a predictive model of GPL metabolite biomarkers and its association with aGvHD.
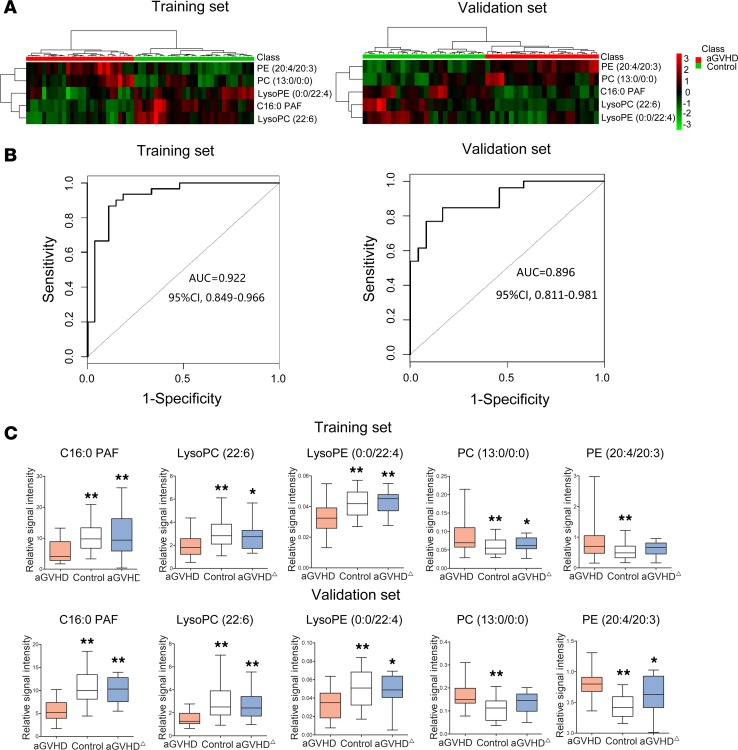
An ideal biomarker should be detectable before the onset of aGvHD. We analyzed the metabolites in plasma samples collected from aGvHD patients before the onset of aGvHD (median 15 days [range: 4–75] before aGvHD diagnosis). A distinct GPL metabolism signature was uncovered in aGvHD plasma, as evidenced by significant alteration of 17 of 38 identified plasma metabolites and their corresponding lipases in this pathway. We hypothesized that dysregulated GPL metabolism is an early event in the activation of alloreactive T cells during aGvHD pathogenesis. We further used least absolute shrinkage and selection operator (LASSO) logistic regression model to build aGvHD risk score (GRS) in the training set and identified 5 metabolites from the 17 GPL metabolites (Figure 5 and Supplemental Figure 2). We evaluated the predictive value of these 5 metabolites for the development of aGvHD (Supplemental Figure 3, A and B). Hierarchical clustering analysis revealed significantly altered metabolite concentrations (Figure 5A). Subsequently, we calculated GRS for each patient based on their individual expression levels of the 5 metabolites, where GRS = 2.2974 × v1 + 20.8171 × v2 – 102.8055 × v3 – 0.0939 × v4 – 1.1977 × v5 (Supplemental Table 3). The receiver operating characteristic (ROC) curves based on the GRS yielded satisfactory results in the training and validation sets, respectively (Figure 5B). The metabolism signature had an AUC value of 0.922 (95% CI, 0.849–0.996) in the training set and 0.896 (95% CI, 0.811–0.981) in the validation set. The optimal cut-off point of GRS in training set (–4.74) was associated with sensitivity of 88.9% and specificity of 86.7%. Using this cut-off point, patients were divided into high and low GRS groups. Patients with high GRS had OR of 32.8 for aGvHD compared with low GRS. The 5-biomarker model successfully identified HLA-matched related or unrelated alloHSCT recipients with a high risk for aGvHD (83.02% in the high-risk group and 12.96% in the low-risk group) (P < 0.0001). The algorithm performed equally well in the training set (85.71% vs. 10.34%, P < 0.0001) and the validation set (80.00% vs. 16.00%, P = 0.00002). There were no significant differences between these 2 groups with respect to clinical characteristics (Supplemental Table 4).
Figure 5. Five metabolites exhibited a strong correlation with the development and severity of aGvHD.
(A) The heatmap of 5 metabolites (C16:0 PAF, LysoPC [22:6], PE [20:4/20:3], PC [13:0/0:0], LysoPE [0:0/22:4]) selected from 17 GPL metabolites. (B) ROC curve analysis of the ability of plasma metabolites, including C16:0 PAF, LysoPC (22:6), PE (20:4/20:3), PC (13:0/0:0), and LysoPE (0:0/22:4), to predict the development of aGvHD based on both the training (n = 57) and validation sets (n = 50). The area under the ROC curve was 0.922 (95% CI, 0.849–0.966) in the training set, and 0.896 (95% CI, 0.811–0.981) in the validation set. (C) Box plots showing fluctuations in relative signal intensities of 5 metabolites. Five metabolites within the panel were restored to normal levels at varying degrees after aGvHD in remission after medication. The comparisons were made between control vs. aGvHD, and aGvHDΔ vs. aGvHD, respectively. Bars indicate median value. P value refers to 1-way ANOVA. *P < 0.05, **P < 0.01. aGvHDΔ, aGvHD in remission after medication.
Further, we observed that the levels of 38 metabolites were largely restored to normal with amelioration of aGvHD clinical scores in both the training and validation sets (Supplemental Table 2 and Figure 5C). In particular, the levels of C16:0 PAF, LysoPC (22:6), PC (13:0/0:0), and LysoPE (0:0/22:4) in the panel were completely restored to normal after aGvHD remission. The other metabolites were also restored to varying degrees in both sets.
Subsequently, we investigated whether the metabolites exhibit aGvHD target organ specificity. LysoPE (0:0/22:4) and PC (13:0/0:0) showed a positive correlation with liver and skin, respectively. PC (13:0/0:0) showed a negative correlation with gastrointestinal aGvHD (Supplemental Figure 3, C–E).
To develop a clinically applicable method that could help predict an individual’s risk of aGvHD, we developed a nomogram based on the panel of these 5 metabolites of all patients in the 2 sets (Figure 6A). Calibration plots of the nomograms for predicting the probability of aGvHD showed that the nomograms did well as compared with an ideal model. The results indicated the reliability of the model based on GRS (Figure 6B).
Figure 6. Nomogram based on training set for predicting risk/probability of aGvHD.
(A) Nomogram based on training set (n = 57) for predicting the risk/probability of aGvHD. (B) The calibration plot comparing predicted outcomes with actual outcome. Dashed line is the 45° reference line, indicating an ideal nomogram. The dotted line is the apparent accuracy of the nomogram without correcting for overfit. The solid line is the bootstrap-corrected performance of the nomogram with a scatter estimate for future accuracy. Calibration plots showed that the nomograms did well compared with an ideal model.
Lastly, we assessed whether the GRS can be used as a prognostic tool in alloHSCT. One hundred seven alloHSCT recipients were divided into high GRS and low GRS groups. Kaplan-Meier survival analysis suggested that patients in the high GRS group had significantly poorer OS as compared with those in the low GRS group (P = 0.0106, Supplemental Figure 4). On multivariate Cox regression analysis with GRS and clinical characteristics, GRS was an independent unfavorable prognostic factor for OS (P = 0.014, hazard ratio = 2.696; Supplemental Table 5).
Discussion
In this study, we employed metabolic analysis, along with transcriptome analysis, to identify critical biomarkers that distinguish alloHSCT recipients with aGvHD from alloHSCT recipients without aGvHD in 2 separate cohorts. Pathway analysis of 38 significantly altered metabolites and 1,148 differentially expressed gene surrogates revealed distinctly altered GPL metabolism. Subsequently, we developed a GRS based on 5 highly connected GPLs to diagnose aGvHD (AUC 0.922 in training set and 0.896 in validation set). In addition, high GRS was correlated with poor overall survival (OS). Moreover, GPL enzyme surrogate genes also exhibited an ability to predict the development of aGvHD (Supplemental Table 6). Collectively, our data suggest that the plasma GPL signature is an independent predictive factor for aGvHD. GRS on day 15 after alloHSCT may facilitate risk stratification of HLA-matched alloHSCT recipients for aGvHD.
Recent studies in the aGvHD model revealed that alloreactive T cells may employ fatty acid or glucose to fuel their activation (32, 33), which might be related to the altered GPL metabolism observed in our results. Lipids, such as GLPs, cholesterol, and glycolipids, are the most abundant molecular components of cell membranes. With the increased need of membrane building blocks, pathogenic T cells were shown to depend on de novo fatty acid synthesis and the underlying glycolytic-lipogenic metabolic pathway for their development (37, 38). Consistent with these findings, we observed increased GPL and fatty acid metabolism at +15 days after alloHSCT in aGvHD patients prior to the onset of clinical symptoms (Figure 4B), which indicated the proliferation of alloreactive T cells. Once activation, T cells need to transit from quiescence to fast expansion. In the meantime, lipid metabolism of T cells switched from energy generation through fatty acid oxidation to fatty acid biosynthesis for membranes and signaling molecules (38–40). A previous study also reported that GPL metabolism and fatty acid metabolism were accelerated in response to the need for rapid proliferation of T cells (39), which is in accordance with our results. During activation of T cells, beta-oxidation of fatty acids is decreased while other metabolic pathways increase. As shown in Supplemental Table 1, free fatty acids (the ingredient for synthesis of GPLs) were reduced in aGvHD plasma, which indicated increased consumption of fatty acids for GPL synthesis by activated cells. Interestingly, it was reported that lipid metabolism reversed to resting state conditions and proliferation of T cells was inhibited immediately after removing the stimulus in proliferating T cells (41), which demonstrated that lipid metabolism was closely related to T cell proliferation. Our results show that the acute graft versus host reaction is already in progress by day 15 and that the increased biomarker levels may precede clinical symptoms by days or weeks. The median day for aGvHD onset was 31 days in the training set and 29 days in the validation set (Table 1). The fluctuations in relative signal intensities were in accordance with the alterations of metabolites. Because of the scarcity of T cells in peripheral blood or BM on +15 days after transplantation, we were unable to conduct a metabolomic analysis for T cells in this study.
Active proliferation of T cells entails synthesis of several cellular constituents such as membranes. As a major structural lipid component of cellular membranes, enhanced GPL biosynthesis in activated T cells has been reported in the setting of inflammatory and autoimmune diseases (42, 43); these findings are consistent with our data (Figure 4 and Supplemental Table 1). Increased plasma levels of PEs and PCs in aGvHD plasma were observed in patients with aGvHD (Figure 4B), which may suggest increased proliferative activity in alloreactive T cells. LysoPCs and LysoPEs are intermediates in the biosynthesis pathway of PCs and PEs. Reduced LysoPCs and LysoPEs and increased activity of lipases LPCAT1 and LPCAT4 in aGvHD plasma suggested increased synthesis of PC and PE (Figure 4). Downregulated expressions of lipases LCAT, PLA2G16, PLCG1, and LPIN1 eventually led to reduced levels of LysoPCs, LysoPEs, PAF, and DG. Overall, GPL levels showed a positive association with gene expressions of these lipases in plasma samples of patients with aGvHD, which suggested that dysregulation of the GPL network appeared in the very early stage of aGvHD (Figure 4). These alterations of GPL metabolism directly reflect the host histocompatibility antigens to which donor T cells respond within days of graft infusion and were significant predictors of aGvHD in univariate and multivariate analysis.
In summary, based on metabolic and transcriptomic investigation combined with pathway analysis of differentially expressed metabolites and genes, we discovered distinctive altered GPL metabolism in the early stage of aGvHD pathogenesis. Gene expressions of GPL-related lipases were significantly altered in aGvHD samples, leading to dysregulated GPLs. The GRS generated from 5 GPL metabolites had the ability to predict the development of aGvHD with an AUC of 0.922 in the training set and 0.896 in the validation set. In addition, our data suggest that high GRS on day 15 after alloHSCT was a unique feature of patients who had a high risk for aGvHD before the overt onset of aGvHD (median = 15.0 days before diagnosis). There were several limitations in our study. First, we enrolled HLA-matched alloHSCT patients to maximize the homogeneity of the study. Thus, the GRS could only be applied in HLA-matched alloHSCT settings. For other kinds of alloHSCT, further identification and verification are needed. Second, small sample size was a major limitation in our study, which made it difficult to comprehensively analyze the prediction ability of GRS in populations from different backgrounds. However, the GRS showed powerful prediction ability for aGvHD with high AUC and discrimination ability with high OR. A prospective, multicenter clinical trial is needed in the future.
To the best of our knowledge, this is the first study that identifies GRS as a potential warning signal that may help improve risk stratification for aGvHD. By integrative “Omic” analysis, we have offered both a comprehensive view of the molecular perturbations underlying the pathogenesis of aGvHD and an initial investigation of a unique metabolic and transcriptomic network that may facilitate identification of aGvHD patients at a very early stage and help institute preemptive therapy. However, owing to the small sample size in our study, the results await further validation using a larger and independent cohort.
Methods
Patients and sample collection.
This study was designed to assess whether plasma-derived metabolites, measured at the 15th day after transplant, could predict aGvHD. All patients (n = 128) who underwent HLA-matched alloHSCT at the Changhai Hospital between September 2014 and June 2018 were enrolled in this study. The inclusion criteria are presented in Supplemental Methods. Twenty-one patients relapsed within 6 months after alloHSCT. The study comprised a training set and a validation set. The median duration of follow-up was 33.43 months (range, 13.60–53.77), and the last date of follow-up was December 31, 2018. The median age at alloHSCT was 36 years (range, 17–66); 43.93% patients were female. In brief, 27 allogeneic transplanted patients with aGvHD and 30 patients without aGvHD (controls) were enrolled as the training cohort, while 24 allogeneic transplanted patients with aGvHD and 26 patients without aGvHD (controls) were enrolled as the validation cohort for the metabolomics study.
Plasma samples were collected at predetermined time points for metabolomic analysis using citrate as an anticoagulant and centrifuged at 1,590 g for 15 minutes at room temperature. The peripheral blood mononucleated cells and plasma samples were collected for transcriptomic analysis at 15 days after transplantation. aGvHD and amelioration of aGvHD were diagnosed according to the generally accepted criteria (44). All patients received prophylactic therapy with cyclosporine A, mycophenolate mofetil, and short-term methotrexate (45). Clinical information, including aGvHD grade, is shown in Table 1. All plasma samples and mononucleated cells were stored at –80°C and in liquid nitrogen before analysis.
Metabolic profiling with LC-MS.
Metabolic profiles of all plasma samples were obtained using reversed-phase–MS (RPLC-MS) and hydrophilic interaction chromatography–MS (HILIC-MS) platforms as described elsewhere (46). Samples were randomized prior to LC-MS analysis to decrease experimental drifts. An in-house quality control (QC) mechanism was established by pooling and mixing equal amounts of plasma samples from all enrolled subjects; these were used to control intra- and interbatch variability. Details pertaining to sample preparation and LC-MS analysis methods are provided in the Supplemental Methods.
After nonlinear alignment of the data in the time domain, automatic integration, and extraction of the peak intensities, raw data containing retention time, intensity, and m/z pairs were obtained. Then, LC-MS data acquired from RP and HILIC columns were combined as a new data set and were used to perform metabolomics profiling analysis.
RNA isolation and transcriptome profiling.
Total RNA was isolated from the peripheral blood mononucleated cells and then quantified using a Bioanalyzer (Agilent Technologies). Whole transcriptome sequencing libraries (3 pairs of aGvHD and controls, 6 libraries in total) were prepared according to the manufacturer’s instructions for the Whole Transcriptome Sample Prep Kit (Illumina) by GENEWIZ Company. After cutting off the adaptor sequence and deleting low-quality reads, all sequencing reads were mapped to the human genome using TopHat. The mapped results were processed using Cufflinks, producing a fragments per kilobase of transcript per million mapped reads (RPKM) data matrix and, with htseq-count, producing the raw hit counts data matrix. The raw hit counts gene matrices were transformed using DESeq variance stabilization algorithm and normalized using the quantile normalization function built into the Bioconductor Limma package. The normalized data matrix was then analyzed using the Limma and SAMR algorithms. The gene expression data has been deposited in the National Center for Biotechnology Information’s (NCBI) Gene Expression Omnibus (GEO) with accession number GSE131897.
GO-TermFinder was used to identify gene ontology (GO) terms that annotate a list of enriched genes with a 1.5-fold change cutoff and P ≤ 0.05. Gene function was then annotated on KAAS (KEGG Automatic Annotation Server). For pathway enrichment analysis, we used scripts in house to enrich significant differential expression gene in KEGG pathways.
Quantitative PCR.
RNA from plasma samples was extracted using standard TRIzol (Invitrogen) protocol. cDNA was reverse transcribed from total RNA, and quantitative PCR (qPCR) was performed using standard TaqMan chemistry and a laser-equipped thermal cycler to detect changes in fluorescence in real time (Applied Biosystems). cDNA concentrations were calculated according to the ΔCt method, corrected for differences in PCR efficiency, and normalized to expression levels of U6. All primers for qPCR in the present study were purchased from Applied Biosystems (Supplemental Table 7).
Development of GRS.
In order to construct a signature with selected GPL metabolite biomarkers for predicting aGvHD, we adopted penalized logistic regression model with the LASSO penalty to achieve shrinkage and variable selection simultaneously (47). The LASSO has been widely used for building prognostic signatures (48, 49), and the use of the LASSO logistic regression model allowed us to integrate multiple GPL metabolites into a single tool; this significantly improved the predictive accuracy as compared with accuracy using single metabolite alone. LASSO logistic regression model analysis was conducted using the “glmnet” package of the R software (version 3.4.3). The penalty parameter λ controls the amount of shrinkage. Usually, the optimal values of λ is the value that gives minimum mean cross-validated error, and a larger λ could be chosen for greater penalty at accepted cross-validated errors (50). Using the regression coefficients from LASSO logistic regression model based on the training set, we developed a GRS.
Data processing and statistical analysis.
The metabolomic data were normalized using L-2-chlorophenylalanine as the internal standard. The multivariate statistical model of orthogonal partial least square discriminate analysis (OPLS-DA) was constructed with the software SIMCA-P+ (version 11.0, Umetrics). A detailed description of the identification of biomarker metabolites is provided in the Supplemental Methods. Metabolic pathway analysis was performed via MetaboAnalyst 3.5 to identify, analyze, and visualize the affected metabolic pathways. We investigated the predictive accuracy of GRS using ROC curve analysis; AUC was used to measure the accuracy. The optimal cut-off point of GRS (associated with maximal sensitivity plus specificity) was used to divide patients into 2 groups: low and high GRS. To provide the clinician with a quantitative method to predict the risk of aGvHD, we constructed a nomogram, and the performance of the nomogram was explored graphically by calibration plots. OS was calculated from the time of transplant until death from any cause. Estimated probabilities for OS were calculated by the Kaplan-Meier method, and the log-rank test was used to compare survival curves. Univariate and multivariate analysis were constructed using the Cox proportional hazard regression model. All statistical analyses were performed using the SPSS 20.0; “rms” package of R 3.4.3 was used to prepare nomogram. Two-tailed Student’s t tests were used for comparisons of 2 groups, and 1-way ANOVA was used for comparisons of multiple groups. Two-sided P values less than 0.05 were considered statistically significant. Fold changes were calculated as the average mass response (area) ratio between 2 groups.
Study approval.
All procedures complied with the Helsinki Declaration standards and were approved by the Changhai Hospital IRB. The requirement for written informed consent was waived because this study used retrospective data from medical records and there were no interventions in the patients.
Author contributions
XH, JW, and YC designed and supervised the study. YL, AH, QC, XC, XH, JW, and YC developed the methodology. YL, AH, QC, YF, XZ, ZH, and ZZ acquired the data. YL, AH, QC, XC, XH, JW, and YC analyzed and interpreted the data. WZ, JY, XH, and JW treated patients. YL, AH, QC, XH, JW, and YC wrote, reviewed, and/or revised the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Acknowledgments
This work was supported by NSFC (81530047, 81870143, 81470321, 81770160, 81270567, 81270638, 81573396, 81703674), which provided funding for material, processing, metabolomics, and transcriptomics studies. YL was sponsored by Shanghai Sailing Program from Science and Technology Commission Shanghai Municipality (17YF1424700). XXH was sponsored by Scholarship from Shanghai Municipal Health and Family Planning Commission (2017BR012). QC was sponsored by Special Clinical Research in Health Industry in Shanghai (20184Y0054). 17YF1424700 (to YL), 2017BR012 (to XH), and 20184Y0054 (to QC) are the projects supporting young scholars.
Version 1. 07/25/2019
In-Press Preview
Version 2. 08/22/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(16):e129494.https://doi.org/10.1172/jci.insight.129494.
Contributor Information
Yue Liu, Email: lyue2015@163.com.
Aijie Huang, Email: huangaijie66@163.com.
Qi Chen, Email: cqchenqi1989@163.com.
Xiaofei Chen, Email: Xfchen2010@163.com.
Yang Fei, Email: Feiyang420@126.com.
Xiaoming Zhao, Email: 978705217@qq.com.
Weiping Zhang, Email: zhangwp@medmail.com.cn.
Zhanying Hong, Email: hongzhy001@163.com.
Zhenyu Zhu, Email: zzyzyfzhu@163.com.
Jianmin Yang, Email: chyangjianmin@163.com.
Yifeng Chai, Email: yfchai@smmu.edu.cn.
Jianmin Wang, Email: jmwangch@139.com.
Xiaoxia Hu, Email: Hu_xiaoxia@126.com.
References
- 1.Ferrara JL, Levine JE, Reddy P, Holler E. Graft-versus-host disease. Lancet. 2009;373(9674):1550–1561. doi: 10.1016/S0140-6736(09)60237-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jagasia M, et al. Risk factors for acute GVHD and survival after hematopoietic cell transplantation. Blood. 2012;119(1):296–307. doi: 10.1182/blood-2011-06-364265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Majhail NS, et al. Significant improvement in survival after unrelated donor hematopoietic cell transplantation in the recent era. Biol Blood Marrow Transplant. 2015;21(1):142–150. doi: 10.1016/j.bbmt.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Przepiorka D, et al. 1994 Consensus Conference on Acute GVHD Grading. Bone Marrow Transplant. 1995;15(6):825–828. [PubMed] [Google Scholar]
- 5.Rowlings PA, et al. IBMTR Severity Index for grading acute graft-versus-host disease: retrospective comparison with Glucksberg grade. Br J Haematol. 1997;97(4):855–864. doi: 10.1046/j.1365-2141.1997.1112925.x. [DOI] [PubMed] [Google Scholar]
- 6.Levine JE, et al. A prognostic score for acute graft-versus-host disease based on biomarkers: a multicentre study. Lancet Haematol. 2015;2(1):e21–e29. doi: 10.1016/S2352-3026(14)00035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D’Asaro M, Dieli F, Caccamo N, Musso M, Porretto F, Salerno A. Increase of CCR7- CD45RA+ CD8 T cells (T(EMRA)) in chronic graft-versus-host disease. Leukemia. 2006;20(3):545–547. doi: 10.1038/sj.leu.2404079. [DOI] [PubMed] [Google Scholar]
- 8.Abrahamsen IW, Sømme S, Heldal D, Egeland T, Kvale D, Tjønnfjord GE. Immune reconstitution after allogeneic stem cell transplantation: the impact of stem cell source and graft-versus-host disease. Haematologica. 2005;90(1):86–93. [PubMed] [Google Scholar]
- 9.Yamashita K, et al. Severe chronic graft-versus-host disease is characterized by a preponderance of CD4(+) effector memory cells relative to central memory cells. Blood. 2004;103(10):3986–3988. doi: 10.1182/blood-2003-09-3286. [DOI] [PubMed] [Google Scholar]
- 10.Rosenzwajg M, et al. Regulatory T cell content in the bone marrow graft does not predict the occurrence of acute GVHD. Biol Blood Marrow Transplant. 2011;17(2):265–269. doi: 10.1016/j.bbmt.2010.07.024. [DOI] [PubMed] [Google Scholar]
- 11.Vander Lugt MT, et al. ST2 as a marker for risk of therapy-resistant graft-versus-host disease and death. N Engl J Med. 2013;369(6):529–539. doi: 10.1056/NEJMoa1213299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrara JL, et al. Regenerating islet-derived 3-alpha is a biomarker of gastrointestinal graft-versus-host disease. Blood. 2011;118(25):6702–6708. doi: 10.1182/blood-2011-08-375006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris AC, et al. Plasma biomarkers of lower gastrointestinal and liver acute GVHD. Blood. 2012;119(12):2960–2963. doi: 10.1182/blood-2011-10-387357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lia G, et al. Extracellular vesicles as potential biomarkers of acute graft-vs-host disease. Leukemia. 2018;32(3):765–773. doi: 10.1038/leu.2017.277. [DOI] [PubMed] [Google Scholar]
- 15.Holtan SG, et al. Low EGF in myeloablative allotransplantation: association with severe acute GvHD in BMT CTN 0402. Bone Marrow Transplant. 2017;52(9):1300–1303. doi: 10.1038/bmt.2017.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sauer S, et al. A prospective study on serum Cytokeratin (CK)-18 and CK18 fragments as biomarkers of acute hepato-intestinal GVHD. Leukemia. 2018;32(12):2685–2692. doi: 10.1038/s41375-018-0183-0. [DOI] [PubMed] [Google Scholar]
- 17.Liu S, et al. Soluble interleukin-27 receptor alpha is a valuable prognostic biomarker for acute graft-versus-host disease after allogeneic haematopoietic stem cell transplantation. Sci Rep. 2018;8(1):10328. doi: 10.1038/s41598-018-28614-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drokov MY, et al. Level of Granzyme B-positive T-regulatory cells is a strong predictor biomarker of acute Graft-versus-host disease after day +30 after allo-HSCT. Leuk Res. 2017;54:25–29. doi: 10.1016/j.leukres.2017.01.014. [DOI] [PubMed] [Google Scholar]
- 19.Budde H, et al. Prediction of graft-versus-host disease: a biomarker panel based on lymphocytes and cytokines. Ann Hematol. 2017;96(7):1127–1133. doi: 10.1007/s00277-017-2999-5. [DOI] [PubMed] [Google Scholar]
- 20.Hueso T, et al. Citrulline and Monocyte-Derived Macrophage Reactivity before Conditioning Predict Acute Graft-versus-Host Disease. Biol Blood Marrow Transplant. 2017;23(6):913–921. doi: 10.1016/j.bbmt.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 21.Min SS, et al. Composite biomarker panel for prediction of severity and diagnosis of acute GVHD with T-cell-depleted allogeneic stem cell transplants-single centre pilot study. J Clin Pathol. 2017;70(10):886–890. doi: 10.1136/jclinpath-2017-204399. [DOI] [PubMed] [Google Scholar]
- 22.Karabon L, et al. A CT60G>A polymorphism in the CTLA-4 gene of the recipient may confer susceptibility to acute graft versus host disease after allogeneic hematopoietic stem cell transplantation. Immunogenetics. 2015;67(5-6):295–304. doi: 10.1007/s00251-015-0840-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao B, et al. Plasma microRNA signature as a noninvasive biomarker for acute graft-versus-host disease. Blood. 2013;122(19):3365–3375. doi: 10.1182/blood-2013-06-510586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stickel N, et al. MiR-146a regulates the TRAF6/TNF-axis in donor T cells during GVHD. Blood. 2014;124(16):2586–2595. doi: 10.1182/blood-2014-04-569046. [DOI] [PubMed] [Google Scholar]
- 25.Ranganathan P, et al. Regulation of acute graft-versus-host disease by microRNA-155. Blood. 2012;119(20):4786–4797. doi: 10.1182/blood-2011-10-387522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leonhardt F, et al. Inflammatory neovascularization during graft-versus-host disease is regulated by αv integrin and miR-100. Blood. 2013;121(17):3307–3318. doi: 10.1182/blood-2012-07-442665. [DOI] [PubMed] [Google Scholar]
- 27.Crossland RE, et al. Serum and Extracellular Vesicle MicroRNAs miR-423, miR-199, and miR-93* As Biomarkers for Acute Graft-versus-Host Disease. Front Immunol. 2017;8:1446. doi: 10.3389/fimmu.2017.01446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huffman AP, et al. Pharmacodynamic Monitoring Predicts Outcomes of CCR5 Blockade as Graft-versus-Host Disease Prophylaxis. Biol Blood Marrow Transplant. 2018;24(3):594–599. doi: 10.1016/j.bbmt.2017.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hori T, et al. CCL8 is a potential molecular candidate for the diagnosis of graft-versus-host disease. Blood. 2008;111(8):4403–4412. doi: 10.1182/blood-2007-06-097287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hartwell MJ, et al. An early-biomarker algorithm predicts lethal graft-versus-host disease and survival. JCI Insight. 2018;3(16):124015. doi: 10.1172/jci.insight.124015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanakry CG, et al. Plasma-derived proteomic biomarkers in human leukocyte antigen-haploidentical or human leukocyte antigen-matched bone marrow transplantation using post-transplantation cyclophosphamide. Haematologica. 2017;102(5):932–940. doi: 10.3324/haematol.2016.152322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Byersdorfer CA, et al. Effector T cells require fatty acid metabolism during murine graft-versus-host disease. Blood. 2013;122(18):3230–3237. doi: 10.1182/blood-2013-04-495515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen HD, et al. Metabolic reprogramming of alloantigen-activated T cells after hematopoietic cell transplantation. J Clin Invest. 2016;126(4):1337–1352. doi: 10.1172/JCI82587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicholson JK, Lindon JC, Holmes E. ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29(11):1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 35.Fiehn O. Metabolomics--the link between genotypes and phenotypes. Plant Mol Biol. 2002;48(1-2):155–171. [PubMed] [Google Scholar]
- 36.Wu X, et al. Prediction of acute GVHD and relapse by metabolic biomarkers after allogeneic hematopoietic stem cell transplantation. JCI Insight. 2018;3(9):99672. doi: 10.1172/jci.insight.99672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berod L, et al. De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. Nat Med. 2014;20(11):1327–1333. doi: 10.1038/nm.3704. [DOI] [PubMed] [Google Scholar]
- 38.Yang Z, Matteson EL, Goronzy JJ, Weyand CM. T-cell metabolism in autoimmune disease. Arthritis Res Ther. 2015;17:29. doi: 10.1186/s13075-015-0542-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robichaud PP, Boulay K, Munganyiki JÉ, Surette ME. Fatty acid remodeling in cellular glycerophospholipids following the activation of human T cells. J Lipid Res. 2013;54(10):2665–2677. doi: 10.1194/jlr.M037044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen HD, Kuril S, Bastian D, Yu XZ. T-Cell Metabolism in Hematopoietic Cell Transplantation. Front Immunol. 2018;9:176. doi: 10.3389/fimmu.2018.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bettens F, Kristensen F, Walker C, Bonnard GD, de Weck AL. Lymphokine regulation of human lymphocyte proliferation: formation of resting G0 cells by removal of interleukin 2 in cultures of proliferating T lymphocytes. Cell Immunol. 1984;86(2):337–346. doi: 10.1016/0008-8749(84)90388-5. [DOI] [PubMed] [Google Scholar]
- 42.Zeng C, et al. Lipidomics profiling reveals the role of glycerophospholipid metabolism in psoriasis. Gigascience. 2017;6(10):1–11. doi: 10.1093/gigascience/gix087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huse M, Le Floc’h A, Liu X. From lipid second messengers to molecular motors: microtubule-organizing center reorientation in T cells. Immunol Rev. 2013;256(1):95–106. doi: 10.1111/imr.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harris AC, et al. International, Multicenter Standardization of Acute Graft-versus-Host Disease Clinical Data Collection: A Report from the Mount Sinai Acute GVHD International Consortium. Biol Blood Marrow Transplant. 2016;22(1):4–10. doi: 10.1016/j.bbmt.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang WP, et al. Allogeneic peripheral blood stem cell transplantation is a promising and safe choice for the treatment of refractory/relapsed acute myelogenous leukemia, even with a higher leukemia burden. Biol Blood Marrow Transplant. 2013;19(4):653–660. doi: 10.1016/j.bbmt.2013.01.015. [DOI] [PubMed] [Google Scholar]
- 46.Liu Y, et al. NMR and LC/MS-based global metabolomics to identify serum biomarkers differentiating hepatocellular carcinoma from liver cirrhosis. Int J Cancer. 2014;135(3):658–668. doi: 10.1002/ijc.28706. [DOI] [PubMed] [Google Scholar]
- 47.Tibshirani R. Regression shrinkage and selection via the lasso: A retrospective. Journal of the Royal Statistical Society. 2011;73(3):273–282. [Google Scholar]
- 48.Zhang JX, et al. Prognostic and predictive value of a microRNA signature in stage II colon cancer: a microRNA expression analysis. Lancet Oncol. 2013;14(13):1295–1306. doi: 10.1016/S1470-2045(13)70491-1. [DOI] [PubMed] [Google Scholar]
- 49.Smith BA, et al. A basal stem cell signature identifies aggressive prostate cancer phenotypes. Proc Natl Acad Sci USA. 2015;112(47):E6544–E6552. doi: 10.1073/pnas.1518007112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw. 2010;33(1):1–22. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.