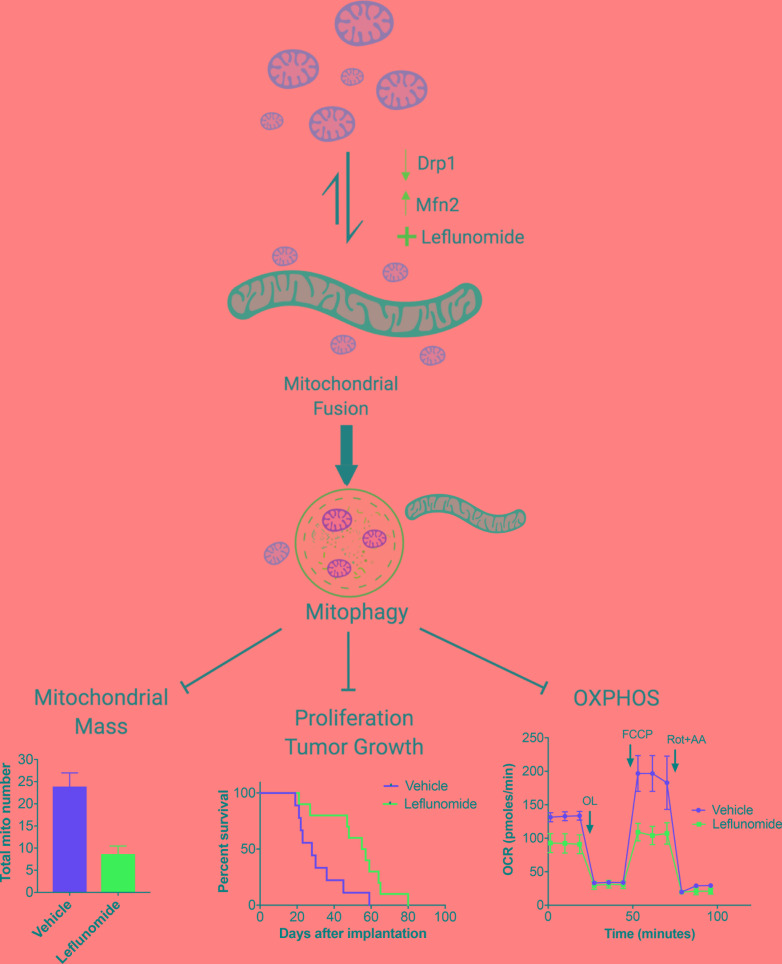
Abstract
Pancreatic ductal adenocarcinoma (PDAC) requires mitochondrial oxidative phosphorylation (OXPHOS) to fuel its growth; however, broadly inhibiting this pathway might also disrupt essential mitochondrial functions in normal tissues. PDAC cells exhibit abnormally fragmented mitochondria that are essential to the oncogenicity of PDAC, but it was unclear if this mitochondrial feature was a valid therapeutic target. Here, we present evidence that normalizing the fragmented mitochondria of pancreatic cancer via the process of mitochondrial fusion reduces OXPHOS, which correlates with suppressed tumor growth and improved survival in preclinical models. Mitochondrial fusion was achieved by genetic or pharmacologic inhibition of dynamin-related protein-1 (Drp1) or through overexpression of mitofusin-2 (Mfn2). Notably, we found that oral leflunomide, an FDA-approved arthritis drug, promoted a 2-fold increase in Mfn2 expression in tumors and was repurposed as a chemotherapeutic agent, improving the median survival of mice with spontaneous tumors by 50% compared with vehicle. We found that the chief tumor-suppressive mechanism of mitochondrial fusion was enhanced mitophagy, which proportionally reduced mitochondrial mass and ATP production. These data suggest that mitochondrial fusion is a specific and druggable regulator of pancreatic cancer growth that could be rapidly translated to the clinic.
Keywords: Gastroenterology, Oncology
Keywords: Cancer, Mitochondria, Mouse models
Mitochondrial fusion in pancreatic cancer cells induces mitophagy, which alters tumor energetics and correlates with improved survival in mouse models.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) relies on energy produced by oxidative phosphorylation (OXPHOS) from the mitochondria to grow and metastasize (1). Mitochondria within a cell can collectively alter their structures to optimize their metabolic functions in response to cellular insults (2). For instance, mitochondria join together in a process called mitochondrial fusion, which is critical for organelle quality control (3). In other cases, mitochondria fragment into smaller organelles through a controlled process called mitochondrial fission, which is often a response to oxidative stress (4). The homeostatic processes of fission and fusion in response to cellular demands is often referred to as mitochondrial dynamics.
The morphology of these networked mitochondria is regulated by a few key proteins. The mitofusin family of proteins (MFN1 and MFN2) positively regulate the fusion of mitochondria by bringing together the outer mitochondrial membranes. Dysfunction or deficits in MFN expression can lead to unopposed mitochondrial fission and possibly disease, such as Charcot-Marie-Tooth type 2A syndrome, which is caused by an autosomal dominant mutation in MFN2 (5). On the other hand, the GTPase dynamin-related protein-1 (DNM1L/DRP1) regulates mitochondrial fission, and its dysfunction may promote unregulated mitochondrial fusion (6).
Pancreatic cancer cells exhibit highly fragmented mitochondria (7), which suggests basal mitochondrial dynamics that favor mitochondrial fission. We hypothesized that shifting the balance of mitochondrial dynamics toward mitochondrial fusion in PDAC cells would normalize their function and reduce oncogenicity, as has been suggested in previous cell culture studies (8). We also reasoned that such an approach may have a favorable therapeutic ratio, since mitochondrial fusion is more frequently observed in normal cells (9). Indeed, we found that the genetic or pharmacologic activation of mitochondrial fusion reduces PDAC growth and improves survival in mouse models of pancreatic cancer. Moreover, we found that the induction of mitochondrial fusion correlates with less mitochondrial mass and reduced OXPHOS compared with controls. We provide evidence that mitochondrial fusion induces mitophagy in pancreatic cancer cells, which may selectively reduce the functional mitochondrial mass in tumors. These proof-of-principle experiments demonstrate the critical nature of mitochondrial dynamics and how they could potentially be exploited therapeutically against PDAC.
Results
Genetic or pharmacological inhibition of mitochondrial fission promotes mitochondrial fusion and suppresses OXPHOS.
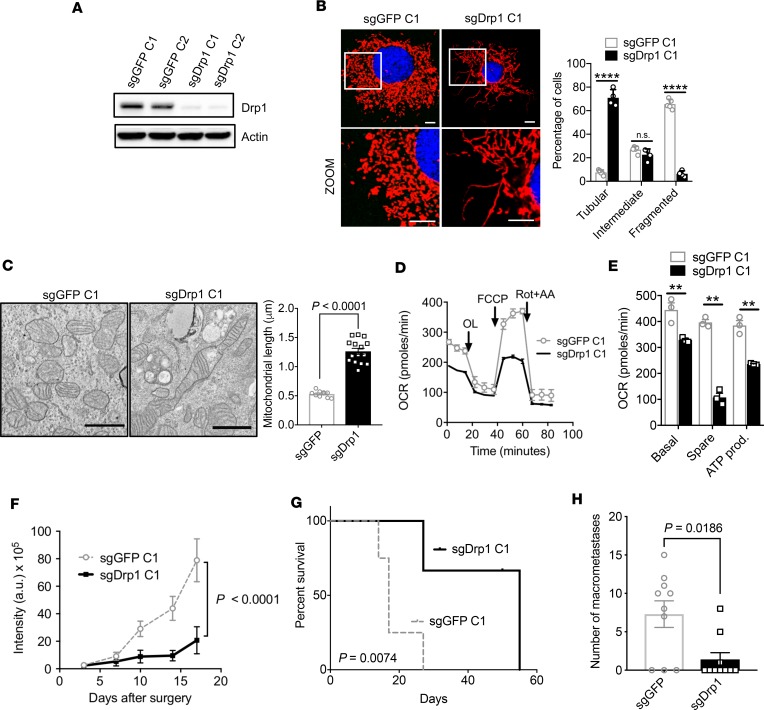
Pancreatic cancer is frequently driven by oncogenic KRAS, the downstream signaling of which activates DRP1 and promotes mitochondrial fission (10). Whether disrupting mitochondrial fission would have a therapeutic effect in pancreatic cancer was unknown. Toward this end, we used CRISPR/Cas9 methodology to edit the endogenous locus of Dnm1l/Drp1 to ablate expression of DRP1 in murine KPC cells syngeneic to C57BL/6 that also expressed a luciferase transgene. We confirmed a nearly quantitative abrogation of DRP1 by Western blot (Figure 1A). This loss of DRP1 expression led to unopposed mitochondrial fusion, as observed with confocal microscopy with MitoTracker staining, which selectively stains live mitochondria in cells (Figure 1B; for other clones see Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.126915DS1). Control KPC cells edited with guide RNAs to GFP (sgGFP) retained highly fragmented morphology (Figure 1B), but sgDrp1 cells normalized the mitochondria in more than 70% of the cells (Figure 1B and Supplemental Figure 1A). These data were confirmed by transmission electron microscopy (TEM), where we found that the average mitochondrial length after Drp1 knockout doubled compared with sgGFP controls (Figure 1C; sgGFP, 0.53 μm vs. sgDrp1, 1.26 μm, P < 0.0001).
Figure 1. Genetic inhibition of mitochondrial fission suppresses mitochondrial OXPHOS and improves survival.
(A) CRISPR/Cas9 knockout of Drp1 (sgDrp1) or GFP control (sgGFP) in KPC cells. (B) Representative (original magnification, ×60) confocal image with MitoTracker Red CMXRos staining, with morphology quantified, n = 100–200 cells. Scale bar: 10 μm. Red fluorescence, mitochondria; blue fluorescence, DAPI-labeled nucleus. ****P < 0.0001 by unpaired t test. (C) Mitochondrial morphology by TEM imaging, average mitochondrial length in μm quantified, and significance by unpaired t test. Scale bar: 1 μm. (D) Mito Stress assay showing (E) decreased basal respiration, spare respiration, and ATP production in sgDrp1 cells compared with controls. **P < 0.01 by unpaired t test. (F) Orthotopic tumor growth in C57BL/6J mice (n = 5 per cohort). Statistical analysis by unpaired t test. (G) Kaplan-Meier survival curves of sgDrp1 or sgGFP implanted orthotopically (n = 5 per cohort, analysis by log rank). (H) Quantification of macrometastases after knockout of Drp1 (n = 10). Statistical analysis by Wilcoxon rank-sum test. Data are presented as mean ± SEM.
The induction of mitochondrial fusion by DRP1 knockout decreased oxygen consumption rates (OCR), as determined by extracellular flux assay (Figure 1D). In addition, we observed lower basal respiration, spare respiration capacity, and ATP production in sgDrp1 cells compared with controls (Figure 1E; repeated in other clones in Supplemental Figure 1, B and C). This reduced OCR was correlated with reduced growth in vitro (Supplemental Figure 1D) and increased G1 arrest (Supplemental Figure 1E). To determine the significance of these metabolic changes, we orthotopically implanted Drp1-knockout cells into the pancreata of recipient C57BL/6 mice and observed impaired in vivo growth (Figure 1F), as estimated by luciferase signal. The loss of DRP1 more than tripled the median survival from 17 days to 55 days (Figure 1G; sgGFP vs. sgDrp1, P = 0.0074). These data were confirmed in one other sgDrp1 clone and sgGFP control (Supplemental Figure 1F). Knockout of DRP1 was retained in orthotopic tumors, as we verified via immunoblots (Supplemental Figure 1G). We also assessed the metastatic burden in these animals and found that sgDrp1 tumors were approximately 5 times less likely to develop macrometastases compared with sgGFP controls (Figure 1H; P = 0.0186).
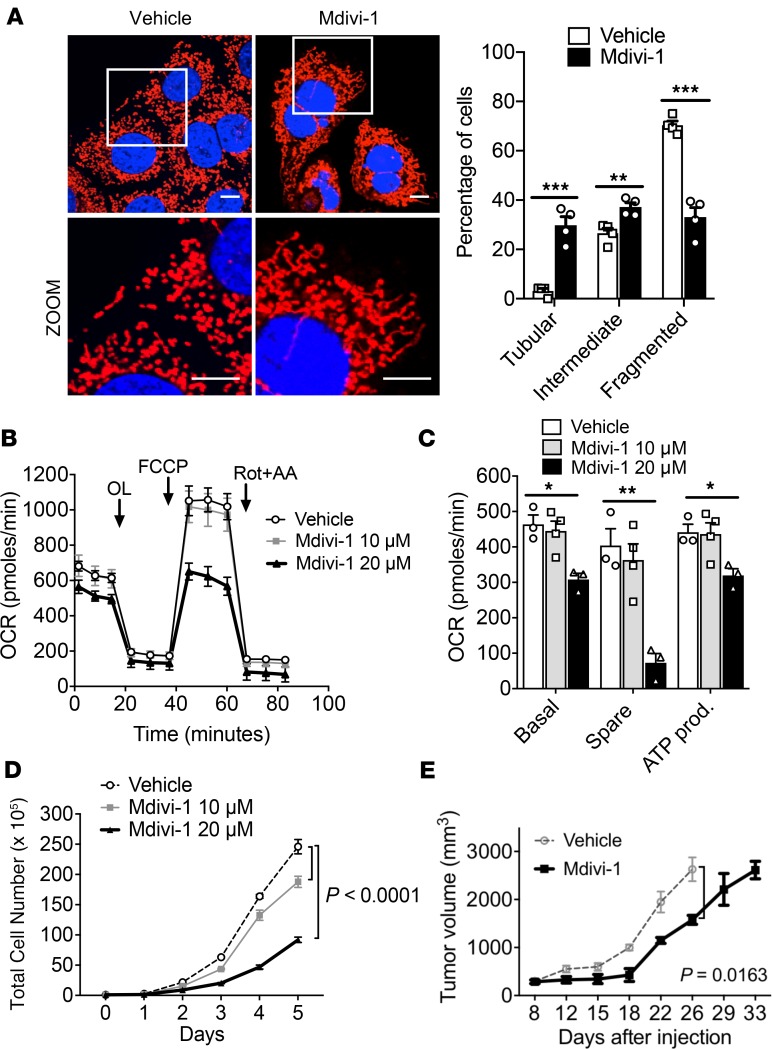
We complemented these genetic studies with pharmacologic inhibition of DRP1 by the small-molecule DRP1 inhibitor, Mdivi-1 (11). Similar to DRP1 knockouts, Mdivi-1 promoted mitochondrial fusion, as confirmed by confocal microscopy with MitoTracker staining (Figure 2A). These treated KPC cells also exhibited decreased OCR (Figure 2B) as well as basal respiration, spare respiration capacity, and ATP production in a dose-dependent manner (Figure 2C). Furthermore, these metabolic changes corresponded to a dose-dependent decrease of in vitro cell growth (Figure 2D) and increased apoptosis, as determined by altered sub G0/G1 population on cell cycle analysis (Supplemental Figure 2A) and TUNEL staining (Supplemental Figure 2, B and C). We also observed reduced in vivo pancreatic cancer growth in a syngeneic flank model (Figure 2E).
Figure 2. Suppression of mitochondrial OXPHOS by pharmacologic inhibition of mitochondrial fission improves survival.
(A) Confocal microscopy image (original magnification, ×60) of mitochondrial morphology of KPC cells treated with Mdivi-1 quantified; n = 100–200 cells. Scale bar: 10 μm. Red fluorescence, mitochondria; blue fluorescence; DAPI-labeled nucleus. ***P = 0.0006 for tubular, **P = 0.007 for intermediate, ***P = 0.0003 for fragmented by unpaired t test. (B) Mdivi-1 decreases OCR, quantified in C, and reduces cell proliferation in a dose-dependent manner (D). *P < 0.05; **P < 0.01 by unpaired t test (C); statistical analysis by 1-way ANOVA (D). (E) In vivo tumor suppression of pancreatic tumors treated with vehicle or 10 mg/kg Mdivi-1 significantly slowed tumor growth; n = 10 per cohort. Statistical analysis by 2-way ANOVA. Data are presented as mean ± SEM.
Direct expression of MFN2 promotes mitochondrial fusion and decreases OXPHOS.
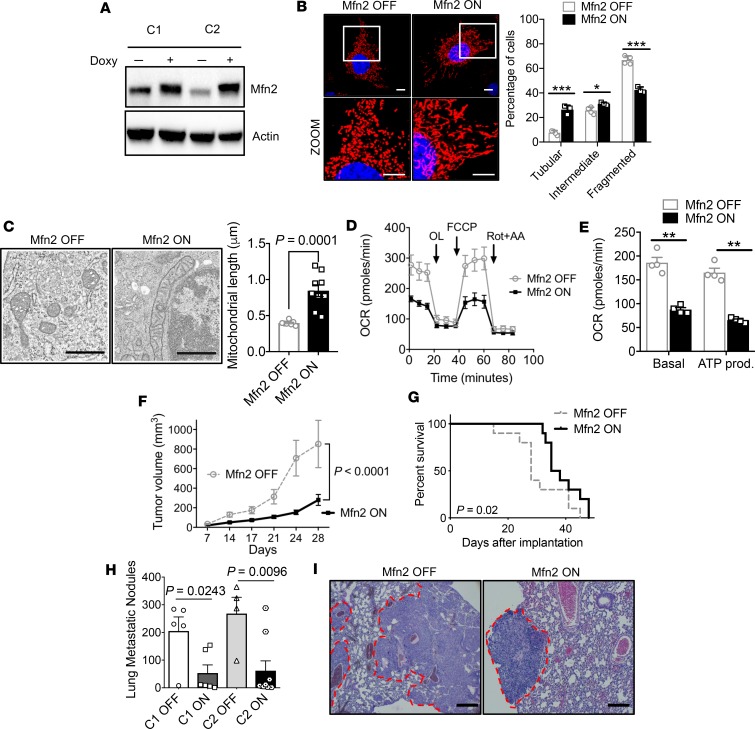
To ensure that this phenotype was due specifically to the induction of mitochondrial fusion and not a pleiotropic alteration caused by DRP1 knockout or Mdivi-1 (12), we overexpressed MFN2 in KPC cells in a doxycycline-dependent manner (Tet-On-Mfn2 KPC, Figure 3A). After 48 hours, mitochondria were more elongated by MitoTracker staining compared with PBS controls (Figure 3B and Supplemental Figure 3A). To verify that these changes in mitochondrial morphology could also be observed in vivo, we implanted these syngeneic Tet-On-Mfn2 KPC cells into the flanks of C57BL/6 mice and analyzed the resultant tumors by electron microscopy (Figure 3C). Mitochondrial fusion was readily apparent in MFN2-overexpressing tumors, with a doubling of the average length of mitochondria (Figure 3C; Mfn2 OFF, 0.39 μm vs. Mfn2 ON, 0.84 μm, P = 0.0001). These data suggest that MFN2 expression directly regulates mitochondrial dynamics in vivo and are not limited to in vitro studies.
Figure 3. Direct induction of mitochondrial fusion by Mfn2 overexpression suppresses pancreatic cancer growth.
(A) Immunoblot of 2 independent clones of Tet-On-Mfn2 KPC cells showing doxycycline-inducible expression of Mfn2. (B) Mfn2 overexpression induces fusion (original magnification, ×60; scale bar: 10 μm); mitochondrial morphology was quantified; n = 100–200 cells. Red fluorescence, mitochondria; blue fluorescence, DAPI-labeled nucleus. ***P = 0.0004 for tubular, *P = 0.025 for intermediate, ***P = 0.0003 for fragmented by unpaired t test. (C) TEM image of tumors grown in vivo with Mfn2 overexpression. Note that Mfn2 overexpression shows elongated mitochondria; average mitochondrial length in μm was quantified, compared by unpaired t test. Scale bar: 800 nm. (D) Reduced OCR with Mfn2 overexpression and quantified parameters in E. **P < 0.01 by unpaired t test. (F) Orthotopic tumor volume in C57BL/6J mice, with n = 10 per cohort. Statistical analysis by unpaired t test. (G) Kaplan-Meier survival curves of C57BL/6J mice bearing orthotopic pancreatic cancer tumor; n = 10 per cohort. (H) Lung metastatic nodules are significantly reduced with Mfn2 overexpression in 2 different clones; n = 5–10 per cohort. P value by unpaired t test. (I) Representative H&E staining (original magnification, × 20) of the lungs with metastatic nodules. Scale bar: 100 μm. Data are presented as mean ± SEM.
Direct mitochondrial fusion by MFN2 expression reduced OCR (Figure 3D), basal respiration, and ATP production (Figure 3E) compared with controls. This reduction in OXPHOS by MFN2 overexpression correlated with decreased cell proliferation in vitro (Supplemental Figure 3B) and enhanced G1 arrest (Supplemental Figure 3C). Furthermore, MFN2 overexpression reduced tumor volume (Figure 3F) and improved survival in a syngeneic orthotopic model (Figure 3G). Doxycycline alone exhibited no significant effects on OCR and did not decrease KPC tumor growth over a range of concentrations (Supplemental Figure 3, D–F). MFN2 expression in implanted tumors was confirmed by Western blot (Supplemental Figure 3G). MFN2 expression also decreased metastatic lung colonization after tail vein injection by more than 4-fold using 2 different Tet-On-Mfn2 clones (Figure 3H; representative H&E in Figure 3I).
Leflunomide activates MFN2 expression and improves survival in multiple mouse models of pancreatic cancer.
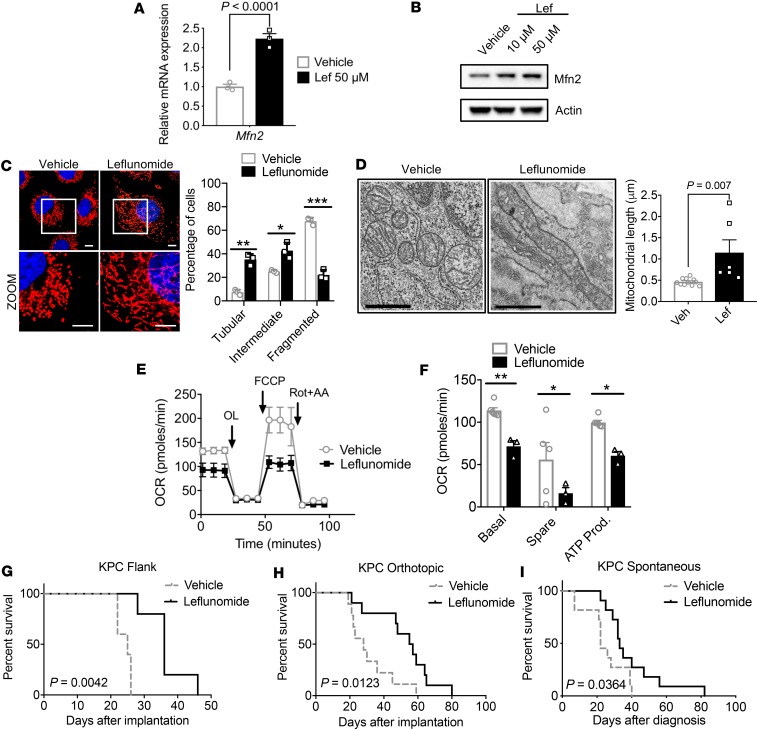
Recently, the FDA-approved anti-arthritis drug, leflunomide, was found to also enhance the expression of MFN2 and promote mitochondrial fusion in HeLa cells (13). To determine whether leflunomide had similar activity in pancreatic cancer, we treated KPC cells with the drug and observed a 2-fold enhancement of Mfn2 mRNA (Figure 4A) and MFN2 protein levels (Figure 4B). Interestingly, we did not observe any changes in dihydroorotate dehydrogenase (DHODH) expression by leflunomide treatment (Supplemental Figure 4A). DHODH expression was also unchanged in sgDrp1 (Supplemental Figure 4B) and Tet-On-Mfn2 cells compared with controls (Supplemental Figure 4C). Fused mitochondria were more frequently observed in KPC cells (Figure 4C) and in implanted syngeneic KPC tumors (Figure 4D) after leflunomide treatment, which led to a doubling of the average mitochondrial length observed compared to vehicle (Figure 4D; vehicle, 0.46 μm vs. leflunomide, 1.14 μm, P = 0.007). This increased mitochondrial fusion was associated with reduced OCR (Figure 4E) and decreased basal respiration, spare respiratory capacity, and ATP production (Figure 4F). Leflunomide also suppressed in vitro growth of KPC cells and correlated with G2 cell cycle arrest (Supplemental Figure 5, A and B). Interestingly, we did not detect any changes in apoptosis by TUNEL staining (Supplemental Figure 5, C and D).
Figure 4. Pharmacological induction of Mfn2 expression suppresses pancreatic growth and improves survival.
Leflunomide (Lef) increases (A) mRNA and (B) protein levels of Mfn2. Statistical analysis by unpaired t test. (C) Leflunomide elongated mitochondria, with morphology quantified; n = 100–200 counted cells. Red fluorescence, mitochondria; blue fluorescence, DAPI-labeled nucleus. Original magnification, ×60. **P = 0.0015 for tubular, *P = 0.011 for intermediate, ***P = 0.0003 for fragmented by unpaired t test. (D) TEM image of KPC tumors treated with leflunomide displaying elongated mitochondria. Scale bars: 800 nm. Average mitochondrial length quantified in μm, compared by unpaired t test. Leflunomide decreases (E and F) mitochondrial basal respiration, spare respiration, and ATP production. *P < 0.05, **P < 0.01 by unpaired t test. Leflunomide (20 mg/kg) treatment improves survival in a (G) flank tumor model (n = 5 per cohort), (H) orthotopic model (n = 10 per cohort), and (I) spontaneous KPC model (n = 11 per cohort). P values by log-rank test. Data are presented as mean ± SEM.
Oral leflunomide can be used to treat rheumatoid arthritis, but its effects on pancreatic tumors have not been studied extensively. Thus, we used leflunomide to treat pancreatic cancer in 3 different cancer models. First, we heterotopically implanted KPC cells into the flanks of recipient C57BL/6 mice and gavaged mice with leflunomide at a dose of 20 mg/kg per day and found that it decreased tumor growth and weight (Supplemental Figure 5, E–G) and improved median survival by almost 50% (Figure 4G; 25 vs. 36 days, vehicle vs. leflunomide, P = 0.004). None of the leflunomide animals exhibited overt toxicity to treatment, which is in contradistinction to previous studies that observed significant toxicity with intravenous leflunomide (14). We then tested leflunomide efficacy in a syngeneic orthotopic model, where the drug doubled the median survival of the treatment cohort compared to vehicle (Figure 4H; 28 vs. 56 days, vehicle vs. leflunomide, P = 0.012). Finally, orally administered leflunomide also demonstrated single-agent efficacy in an aggressive autochthonous KPC model of pancreatic cancer (Figure 4I), increasing the median survival by 50% (22 vs. 33 days, vehicle vs. leflunomide, P = 0.036).
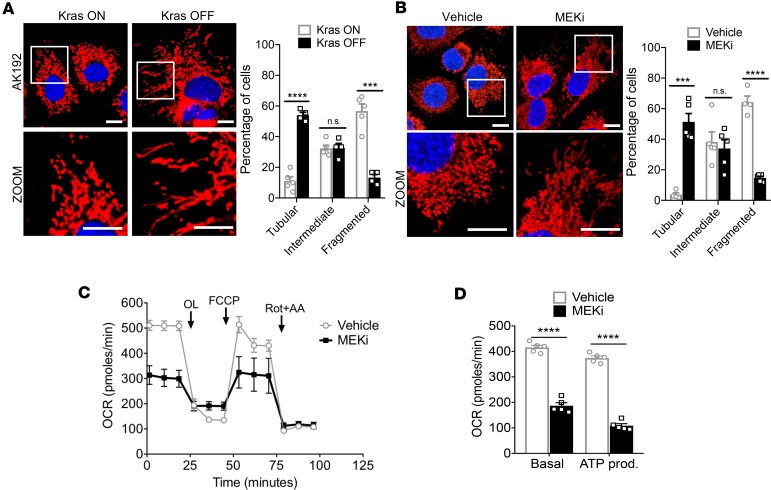
Mitochondrial dynamics are dependent on Kras activity. Oncogenic KRAS plays a critical role in the maintenance of pancreatic tumor progression (15). To gain more insights into whether mitochondrial dynamics are regulated by the RAS/MAPK pathway, we used a doxycycline-inducible Kras (iKras) cell line (AK192), derived from iKras tumors (15). Induced Kras expression (Kras ON) caused a 4-fold increase in fragmented mitochondria (Figure 5A, 13.3% in Kras OFF vs. 56.7% in Kras ON, P = 0.0001). Similarly, treatment of KPC cells with MEK inhibitor (MEKi), PD0325901, yielded a 13-fold increase in elongated mitochondria compared with controls (3.7% vs. 51.3%; vehicle vs. MEKi, Figure 5B, P = 0.0002). MEKi treatment also decreased OCR (Figure 5C) as well as basal respiration and ATP production compared with vehicle controls (Figure 5D).
Figure 5. Mitochondrial dynamics are regulated by the RAS/MAPK pathway.
(A) Inducible-Kras cell line AK192 activated with doxycycline exhibits punctate mitochondria, with quantification; n = 100–200 cells. ****P < 0.0001 for tubular, ***P = 0.0002 for fragmented by unpaired t test. Original magnification, ×60. Scale bar: 10 μm. (B) Treatment with MEK inhibitor, PD0325901, induces mitochondrial fusion in KPC cells, with quantification; n = 100–200 cells. ***P = 0.0002 for tubular, ****P < 0.0001 for fragmented by unpaired t test. Original magnification, ×60. Scale bar: 10 μm. (C) Mito Stress assay of KPC cells treated with MEK inhibitor and quantified (D). ****P < 0.0001 by unpaired t test. Data are presented as mean ± SEM.
Kras-dependent response of leflunomide in patient-derived pancreatic cancer cells.
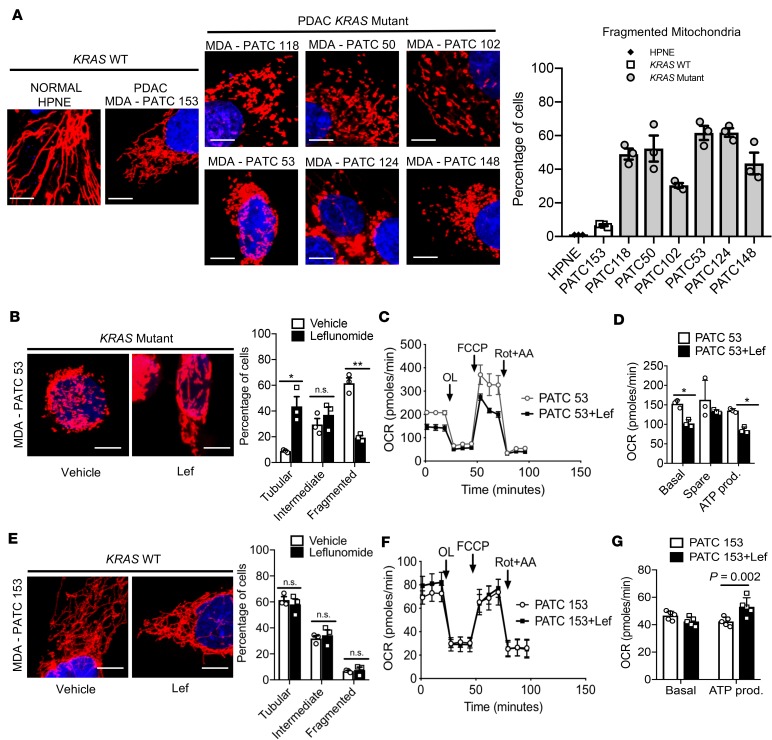
To determine whether human pancreatic cancer patients also exhibited the KRAS-dependent mitochondrial morphology change we observed in murine KPC cells, we characterized 7 different patient-derived pancreatic cell lines (PATC), which were generated from resected pancreatic tumors at the MD Anderson Cancer Center, as has been described (16–18). We performed sequencing on these PATC lines and described the mutations in Supplemental Figure 6A. We noted that nearly all the PATC lines had an expected KRAS mutation. However, one cell line, MDA-PATC153, was KRAS WT with alternative driver mutations in TP53 and PIK3CA.
We performed MitoTracker staining on these PATC lines and used human pancreatic epithelial cells (HPNE) as a control for the morphology of normal tissues. We found that, as expected, normal HPNE cells exhibited fused, elongated mitochondria (Figure 6A). We stratified the rest of our analysis by KRAS mutation status. Interestingly, the KRAS WT line, MDA-PATC153, exhibited mitochondrial morphology similar to the normal HPNE cells and was not statistically different after quantification (Figure 6A and Supplemental Figure 6, B and C). The other PATC lines that harbored mutations in KRAS, however, exhibited marked mitochondrial fragmentation (Figure 6A, MDA-PATC 118, 50, 102, 53, 124, and 148).
Figure 6. Patient-derived pancreatic cancer cells respond to leflunomide in a Kras-dependent fashion.
(A) Kras WT patient samples of PDAC exhibited elongated mitochondria similar to HPNE cells, while Kras mutant patient samples appeared punctate and fragmented. The mitochondria morphology from each genotype was quantified; n = 100–200. Quantification for intermediate and tubular mitochondrial morphology can be found in Supplemental Figure 6, B and C. (B) Leflunomide treatment on Kras mutant MDA-PATC53 cell line induces mitochondrial fusion, with quantification; n = 100–200 cells. *P = 0.02 for tubular, **P = 0.002 for fragmented by unpaired t test. (C) Mito Stress assay of MDA-PATC53 cells treated with 50 μM leflunomide, with quantification in D. *P = 0.04 by unpaired t test for basal and ATP production. Original magnification, ×60; scale bars: 10 μm. Data are presented as mean ± SEM. (E) Leflunomide treatment does not affect morphology in Kras WT MDA-PATC153 cell line, with quantification; n = 100–200 cells. (F) Mito Stress assay of MDA-PATC153 cells treated with 50 μM leflunomide, with quantification in G. Statistical analysis by unpaired t test. Data are presented as mean ± SEM.
To determine whether PATC cell lines respond to leflunomide treatment in a KRAS-dependent fashion, we first treated KRAS mutant MDA-PATC53 with leflunomide and found that the drug induced mitochondrial fusion, with 5 times more tubular mitochondria by MitoTracker staining in leflunomide-treated cells compared to vehicle controls (Figure 6B, 8.86% vs. 43.61%, vehicle vs. leflunomide, P = 0.02). Consistent with our previous findings, this stimulation of mitochondrial fusion decreased OCR (Figure 6C) and basal and spare respiration in addition to ATP production (Figure 6D) When we interrogated the KRAS WT cell line, MDA-PATC153, with leflunomide, we observed no significant changes in mitochondrial morphology (Figure 6E). By the same token, we did not observe any significant changes in OCR, although ATP production increased by about 25% (Figure 6, F and G).
Mitochondrial fusion promotes mitophagy, which proportionally reduces functional mitochondrial mass.
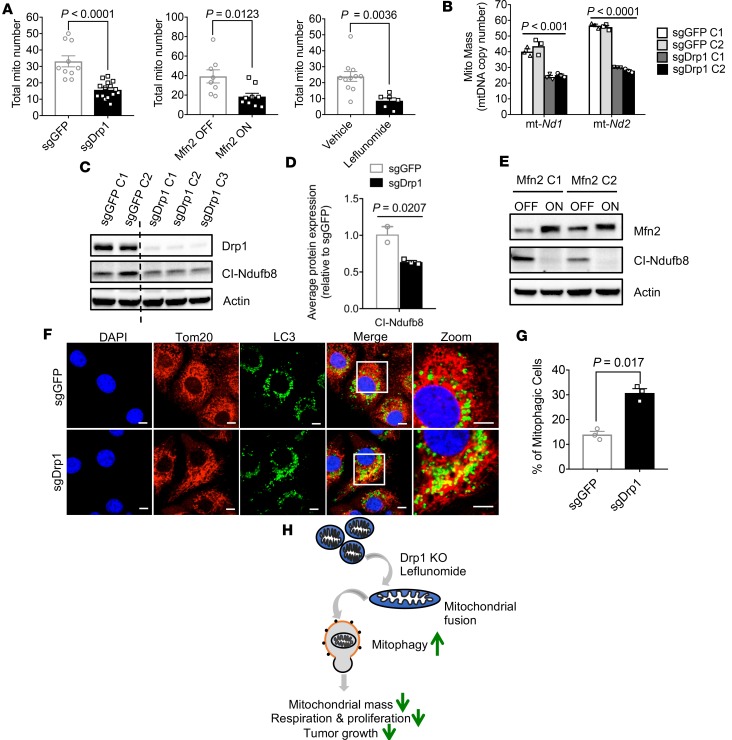
Mitochondrial fusion is a known mechanism of organelle quality control (3); thus, we hypothesized that our treatments might be inhibiting OXPHOS simply by reducing mitochondrial mass. We calculated the total mitochondria present by electron microscopy and found that the average number of mitochondria decreased by at least 50% when we induced mitochondrial fusion. For instance, the mitochondrial number decreased after DRP1 ablation (33.1 vs. 15.7 in sgGFP vs. sgDrp1, P < 0.0001), overexpression of MFN2 (39.2 vs. 18.4 in Mfn2 OFF vs. Mfn2 ON, P = 0.01), and pharmacological promotion of fusion by leflunomide (23.9 vs. 8.6 in vehicle vs. leflunomide treatment, P = 0.003, Figure 7A).
Figure 7. Loss of Drp1 reduces mitochondrial mass by increased mitophagy.
(A) Total mitochondria per cell decreased after induction of mitochondrial fusion, as measured by TEM. P values by unpaired t test are shown in the figure. (B) Decreased mitochondrial mass after Drp1 knockout. Statistical analysis by unpaired t test. (C) Immunoblot for mitochondrial complex I proteins in sgDrp1 KPC cells, with quantification in D and P values by unpaired t test. (E) Decreased complex I expression with Mfn2 expression. (F) Increased colocalization of Tom20 and LC3 in sgDrp1 cells, indicating enhanced mitophagy. Scale bar: 10 μm. Original magnification, ×60. (G) Quantified mitophagic cells from F, compared by unpaired t test. (H) Mitochondrial fusion heralds mitophagy, decreasing OXPHOS and impairing oncogenic proliferation and growth. Data are presented as mean ± SEM.
We complemented our microscopic data by estimating mitochondrial mass with mitochondrial DNA (mtDNA) copy number (19). This common approach uses a quantitative PCR assay to estimate total mtDNA by copy numbers of genes transcribed from mtDNA, such as mt-Nd1 and mt-Nd2, the gene products of with which reside in complex I (Figure 7B and Methods). Mitochondrial fusion was associated with a 39% reduction in the complex I marker mt-Nd1 (41.4 vs. 25.1, sgGFP vs. sgDrp1; P = 0.0008) and a 51% reduction in mt-Nd2 (56.4 vs. 28.5, sgGFP vs. sgDrp1, P = 0.00001). We confirmed this reduction in mitochondrial gene expression by Western blot for a mtDNA-encoded NADH dehydrogenase subunit of complex I called mt-Ndufb8, which exhibited a 48% decrease in expression (Figure 7, C and D; P = 0.007, sgGFP vs. sgDrp1). Importantly, mitochondrial fusion caused by direct MFN2 overexpression strongly suppressed complex I expression, as detected by immunoblot of the complex I protein, mt-Ndufb8 (Figure 7E).
We hypothesized that this observed loss of mitochondrial mass could have come from reduced mitochondrial biogenesis or altered turnover. To assess the former possibility, we measured expression of Ppargc1a, which is a critical regulator of this process (20). We found that Ppargc1a expression levels were unchanged by loss of Drp1 or overexpression of Mfn2 (Supplemental Figure 7, A and B). We additionally found no significant differences in expression of nuclear encoded genes involved in the electron transport chain, such as Ndufs3 and Tfam, which indicated that mitochondrial fusion was likely not inducing global problems with mitochondrial gene transcription (Supplemental Figure 7, C and D). Together, these data suggest that global defects in mitochondrial biogenesis do not explain the reduced mitochondrial mass after mitochondrial fusion.
Since mitochondrial biogenesis appeared to be unaffected, we investigated the possibility that mitochondrial fusion might stimulate mitochondrial turnover, possibly through mitophagy (21). We estimated mitophagy using an immunofluorescence assay, as has been described previously (20). Mitochondria from sgDrp1 or control sgGFP cells were stained with Tom20 and colocalized with an antibody for the autophagosome marker, microtubule-associated proteins 1A/1B light chain 3B (LC3). We found that enhancing mitochondrial fusion by loss of DRP1 was associated with a more than 2-fold increase in colocalization of Tom20/LC3 in sgDrp1 cells compared to controls (Figure 7, F and G; 12.8% vs. 29.5%, sgGFP vs. sgDrp1; P = 0.017). These data suggest that mitochondrial fusion promotes mitophagy, which then culls the excess mitochondria in pancreatic cancer cells (Figure 7H).
Discussion
Despite great strides in the basic genetics and cell biology of pancreatic cancer, patient survival in this disease remains unacceptably low (22). One of the chief reasons for this therapeutic recalcitrance may be that many driver mutations, such as KRAS, TP53, CDKN2A, and SMAD4, are not easily druggable (23). Approximately 90% of PDAC have mutations in KRAS, which drives many of the aggressive features of this cancer (24), and we confirm that KRAS drives the fragmentation of PDAC mitochondria in both genetically modified murine models (Figure 5) and patient-derived tumor lines (Figure 6), which supports previous findings (10). Here, we demonstrate that promoting mitochondrial fusion exploits this unique mitochondrial biology of PDAC to induce mitophagy, which is a known quality control process resulting from mitochondrial dynamics (21). The subsequent loss of mitochondrial mass reduced OXPHOS, suppressed tumor growth, and extended survival in multiple models (summarized in Figure 7H).
Mitochondrial dynamics have recently been correlated with treatment response of PDAC to gemcitabine (25) or combined autophagy/MEK inhibition (26), but here we established a direct link to tumor suppression by utilizing multiple different means of achieving mitochondrial fusion. These include direct enhancement of MFN2 expression or inhibiting DRP1 both genetically and pharmacologically. We demonstrate that targeting DRP1 with the small-molecule Mdivi-1 was efficacious but also that Mdivi-1 is not a great candidate for clinical trials, since it is known to have many undesirable off-target effects (27) and has poor lipophilicity and low specificity (12, 27, 28). Thus, the use of Mdivi-1 to promote mitochondrial fusion should be considered as a proof of concept rather than a definitive approach for inducing mitochondrial fusion in pancreatic cancer.
Of the several pharmacologic interventions to promote mitochondrial fusion, oral leflunomide offers the quickest path to the clinic, as it is already FDA approved for use in rheumatoid arthritis. Here, we demonstrated that oral leflunomide enhances the expression of MFN2 and suppressed OXPHOS and PDAC growth. We repurposed leflunomide in stringent preclinical models of PDAC and observed potent single-agent activity with few observable side effects. This excellent tolerability of the oral drug reflects the experiences of rheumatologic patients who are dosed at levels similar to the animals in this study (29).
We recognize that leflunomide is known to affect several biological pathways, including signaling through β-catenin (30), EGF (31), PDGF (32), and VEGF (33). A small phase I study in solid tumors based on this panoply of growth factor inhibition showed that intravenous leflunomide was well tolerated (34) but never progressed past phase II/III trials due to marginal improvements in response compared with standard of care in prostate cancers and variations in pharmacokinetic responses in glioblastomas (35, 36). Leflunomide is used to treat rheumatoid arthritis through its effects as a weak inhibitor of DHODH, a key enzymatic step in pyrimidine biosynthesis (37). Recent studies have suggested that altering pyrimidine biosynthesis affects OXPHOS in pancreatic tumors (38, 39), which may partially explain gemcitabine efficacy and subsequent resistance in pancreatic cancer (25). Thus, while it is likely that leflunomide may also suppress PDAC progression through other mechanisms in addition to mitochondrial fusion, we believe that these multiple effects may contribute to the drug’s overall efficacy. In fact, the weak DHODH inhibition by leflunomide would likely synergize with standard-of-care regimens, such as gemcitabine/nab-paclitaxel (40), which target PDAC metabolism at different branch points in the pyrimidine biosynthesis pathway in addition to directly targeting mitochondrial function.
Our data predict that mitochondrial fusion should be specific to cancer cells with abnormal fragmented mitochondria, such as pancreatic cancer (10). This effect may be limited to cancers with KRAS driver mutations, as we demonstrated that leflunomide did not inhibit OXPHOS in KRAS WT cancer lines. Future experiments should determine whether this approach is useful in other cancers with KRAS driver mutations, such as adenocarcinomas of the colon (41), lung (42), and biliary tract (43). Importantly, this treatment strategy would not be expected to cause significant off-target effects, unlike direct complex I inhibitors (44), which often cause unwanted dose-limiting side effects in normal tissues that also require mitochondrial function (45). Mitochondrial fusion would not be expected to affect normal tissues, such as the heart (46) and muscle, since mitochondrial dynamics in those tissues tend to favor fusion (47).
Methods
Animal studies.
Mice were between 8 and 12 weeks of age at the start of the study and were observed until tumor burden required their euthanasia as per the Institutional Animal Care and Use Committee protocol. Mice were kept in 12-hour light/dark cycles and given free access to standard rodent chow (Prolab Isopro RMH 3000 Irradiated Feed) and sterilized water. C57BL/6J mice (JAX stock no. 000664) were purchased from The Jackson Laboratory. KrasLSL/+ and Trp53FL/FL; Ptf1aCre/+ animals were backcrossed to a pure C57BL/6 background as previously described (48).
Orthotopic pancreas injection.
Orthotopic pancreatic injections were performed as previously described (49). Before surgery, animals were shaved around the surgical site to remove excess fur; 0.1 mg/kg extended release buprenorphine was given as an analgesic. Mice were anesthetized with 2% isoflurane and given artificial eye drops during surgery. Tet-On-Mfn2 KPC cells were inoculated into the pancreata of recipient C57BL/6 mice in DMEM (catalog D5796, MilliporeSigma) mixed 1:1 in ice-cold Matrigel (catalog 354234, Corning Inc.) for a total of 20 μl. The day after innoculation, mice were given water with 200 μg/ml doxycycline or PBS and water was changed weekly. Tumors were imaged twice per week with an ultrasound (Vevo 2100 system, FujiFilm VisualSonics) to assess growth. KPC sgGFP/sgDrp1 cells expressing luciferase were implanted in recipient C57BL/6 mice in the same way. Tumors were imaged by luminescence twice a week.
Mdivi-1 administration.
For in vivo Mdivi-1 treatment, mice were heterotopically transplanted with KPC cells in the flank, and when tumors reached 7 mm, mice were assigned to receive intraperitoneal vehicle control (10% DMSO; catalog D2650, MilliporeSigma) or intraperitoneal Mdivi-1 (10 mg/kg) (CAS no. 338967-87-6, MilliporeSigma) daily for 1 week, as previously published (50). Tumor sizes were measured twice per week until any dimension of the tumor reached 15 mm unless there were any obvious signs of morbidity that required euthanasia per the Institutional Animal Care and Use Committee protocol.
Leflunomide administration.
Leflunomide was administered by oral gavage at a dose of 20 mg/kg in 10 (w/v)% ethanol (E7023, MilliporeSigma), 30% Poly (ethylene glycol) (catalog 202398, MilliporeSigma), and 60% UltraPure Distilled Water (catalog 10977015, Thermo Fisher). Drug or vehicle was given 5 days a week.
Lung metastatic model.
Tail vein injection was used to establish the lung metastasis model. C57BL/6J mice were ordered from The Jackson Laboratory. Tet-On-Mfn2 KPC cells were prepared with 107 cells/ml in PBS. Prior to injection, animals were warmed gently with a heating pad, and tail veins were injected with a 100 μl bolus of cells. Any mice that died within 48 hours of injection were excluded from analysis. After 3 weeks, we euthanized all the mice and fixed the lungs in order to count the nodules and perform H&E staining.
Cell culture.
Patient-derived MDA-PATC118 and human normal epithelial HPNE were a gift from Jason Fleming from the H. Lee Moffitt Center (16). Patient-derived MDA-PATC50, MDA-PATC102, MDA-PATC153, MDA-PATC53, MDA-PATC124, and MDA-PATC148 were a gift from the TRACTION Platform of The University of Texas MD Anderson Cancer Center. Cells were grown in RPMI-1640 supplemented with 2 mM L-glutamine and 10% FBS. Murine KPC cells syngeneic with C57BL/6 (K8484) were a gift from Anirban Maitra from the MD Anderson Cancer Center. The KPC cells were grown in RPMI-1640 (catalog R8768) with 10% FBS (catalog F4135) (both from MilliporeSigma), 2 mM GlutaMAX (catalog 35050-061), 1 mM sodium pyruvate (catalog 1130-070), and 7 μg/ml insulin (catalog 12585-014) (all from Gibco Life Technologies). For Tet-On KPC cell lines, the same media was used, with the exception of substitution of Tet system with FBS (catalog 631101, Clontech). KPC lines with iKras were a gift from Haoqiang Ying from MD Anderson Cancer Center. To induce gene overexpression, cells were seeded in 6-well plates in low density and treated with 1 μg/ml doxycycline (D9891, MilliporeSigma) for 48 hours. Lenti-X 293T cells (catalog 632180, Clontech) for lentivirus production were maintained in DMEM high glucose with 10% FBS and 2 mM GlutaMAX. For MEKi PD0325901 (catalog 41-9210, Thermo Fisher Scientific) treatment, KPC cells were treated with 10 μM inhibitor or vehicle for 24 hours. All cell lines were cultured in a humidified atmosphere containing 5% CO2 at 37°C.
Cell proliferation assay.
A total of 105 cells were seeded at day 0 in multiple 10-cm dishes. From day 1 to day 5, 1 dish was taken out per day and total cell number was counted. Total cell numbers were used to plot a cell growth curve.
Cell cycle and TUNEL assay.
Cells were harvested and fixed in cold 70% ethanol at 4°C overnight. They were then washed twice with PBS and centrifuged to discard the supernatant. Cells were treated with ribonuclease for 30 minutes to get rid of RNA. 1 mg/ml Propidium Iodide (P1304MP, Thermo Fisher Scientific) was used to stain cells for 30 minutes at 4°C. After staining, cell cycle was measured by flow cytometry (Gallios, Beckman Coulter), and data were analyzed by FlowJo.
TUNEL assay was performed with the In Situ Cell Death Detection Kit, TMR red (MilliporeSigma). KPC cells were seeded in chamber slides and followed by leflunomide or Mdivi-1 treatment for 24 hours. Air-dried cell samples were fixed with freshly prepared 4% paraformaldehyde for 1 hour at room temperature. Slides were rinsed and incubated in 0.1% Triton X-100 in 0.1% sodium citrate for 2 minutes on ice. Afterwards, slides were rinsed twice with PBS and dried at room temperature. We then added 50 μl TUNEL reaction mixture and incubated the slides in a humidified box for 1 hour at 37°C in the dark. Slides were rinsed 3 times with PBS and mounted with DAPI mounting medium (VectorLabs H-1200) before confocal imaging with an Olympus FV1000 laser scanning confocal microscope (Olympus).
Plasmids, lentivirus production, and clone selection.
Human MFN2 were synthesized and codon optimized by GenScript and then subcloned into pLVX-TetOne-Puro vector (catalog 631849, Clontech). CRISPR gRNA–targeting mouse Dnm1l was designed at the Zhang laboratory at the Broad Institute (https://zlab.bio/guide-design-resources) and subcloned into LentiCRISPRv2 plasmid using a standard protocol; this plasmid was a gift from Feng Zhang (Addgene plasmid 52961) (51). The forward sequence is 5′-CACCGGGTCATGGAGGCGCTGATCC-3′, reverse 5′-AAACGGATCAGCGCCTCCATGACCC-3′. Lentiviruses were generated using the Lenti-X system from Clontech according to the manufacturer’s instructions. Lentiviruses with the gRNA-targeting GFP or mouse Dnm1l were further used to transfect KPC cells with luciferase expression.
KPC sgGFP/sgDrp1 and KPC Tet-on Mfn2 clones were generated by monoclonal selection. After lentivirus transfection, cells were diluted to low densities and seeded in a 10-cm dish to obtain well-isolated single-cell colonies. When the colonies reached appropriate size, we picked multiple clones for each line and tested those clones for target gene expression. At least 2 clones were used for further experiments.
Immunoblotting.
Cells were lysed in T-PER Tissue protein extraction reagent (catalog 78501, Thermo Fisher Scientific). Cell lysates were resolved on SDS-PAGE gel (catalog 456-8034, Bio-Rad Laboratories Inc.) and transferred onto PVDF (catalog 162-0239, Bio-Rad Laboratories Inc.) using Bio-Rad Trans-Blot Turbo transfer system. Primary antibodies were used to identify the relevant protein and loading control (β-actin) and then probed with HRP-conjugated secondary antibodies. The detection of bands was carried out on a ChemiDoc imaging system (Bio-Rad Laboratories Inc.). β-Tubulin (9F3), Tom20 (D8T4N), LC3 (E5Q2K), β-actin (13E5), Drp1 (D8H5), Mfn2 (D2D10), Mfn1 (D6E2S), and p-Drp1 Ser616 (D9A1) antibodies are from Cell Signaling Technology. OXPHOS antibody (ab110413) is from Abcam (52). DHODH antibody (14877-1-AP) is from Proteintech.
Immunofluorescence and mitochondrial morphology analysis.
Cells were seeded in coverslips with low density, and live cells were loaded with 25 nM MitoTracker Red CMXRos (M7512, Thermo Fisher Scientific) for 30 minutes. After several washes, cells were fixed in 4% paraformaldehyde in growth medium. Coverslips were mounted in mounting medium with DAPI (D3571, Thermo Fisher Scientific) (6); cells were visualized under a confocal microscope (model FV1000, Olympus Corp.) and processed using Fluoview software (Olympus Corp.). Mitochondrial morphology was scored in at least 100 cells per group by using 3 different categories of mitochondrial morphology (tubular, fragmented, and intermediate). Cells with >50% fragmented mitochondria were scored as mitochondrial fragmentation phenotype. Cells with over 80% elongated mitochondria were scored as tubular, and cells with over 50% short-rod like mitochondria were scored as intermediate (53).
For the mitophagy immunofluorescence, cells were permeabilized by 0.2% Triton X-100 (X100, MilliporeSigma) at room temperature for 10 minutes and then blocked using blocking buffer for 30 minutes. Primary antibodies, Tom20 (D8T4N) and LC3 (E5Q2K), were diluted in PBS to the proper concentration, together with 1% FBS. Slides were then incubated in a humidified chamber at 4°C overnight and then rinsed 3 times with 1× PBS for 5 minutes each. After washing, the slides were incubated in secondary antibody for 2 hours at room temperature: Goat anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 (catalog A28175, Thermo Fisher Scientific) and Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 (catalog A-11011, Thermo Fisher Scientific). Afterwards, we washed the slides 3 times with 1× PBS for 5 minutes each and mounted them for confocal imaging.
DNA and mtDNA copy number.
DNA was extracted using DNeasy Blood and Tissue Kit (catalog 69506, QIAGEN), using 10 ng genomic DNA as a template for quantitative PCR reaction. Two sets of primers were used for both mtDNA and nDNA amplification. Nuclear-encoded genes were used as a reference for relative quantification of the mtDNA copy number. The primers for mtDNA genes are as follows: mt-Nd1 forward 5′-CAGCCGGCCCATTCGCGT TA-3′, reverse 5′-AGCGGAAGCGTGGATAGGATGC-3′; mt-Nd2 forward 5′-TCCTCCTGGCCATCGTACTCAACT-3′, reverse 5′-AGAAGTGGAATGGGGCGAGGC-3′. The primers for nuclear encoded genes in this study are as follows: nGADPH forward 5′-ACAGCCGCATCTTCTTGTGCAGTG-3′, reverse 5′-GGCCTTGACTGTGCCGTTGAATTT-3′; nNDUFV1 forward 5′-CTTCCCCACTGGCCTCAAG-3′, reverse 5′-CCAAAACCCAGTGATCCAGC-3′.
Quantitative PCR.
Total cellular RNA was extracted using the RNeasy Plus Mini Kit (catalog 74136, QIAGEN) according to technical specifications. A mix of random primers and oligos (53) were applied for cDNA synthesis using the QuantiTect Reverse Transcription kit (catalog 205310, QIAGEN). Quantitative reverse transcription PCR was performed on a CFX384 Real-time System (Bio-Rad Laboratories Inc.), using SYBR Green Master mix (catalog 172-5124, Bio-Rad Laboratories Inc.) and gene-specific primers per the manufacturer’s advice. All PCR primers used to amplify mitochondrial-encoded proteins were synthesized from Integrated DNA Technologies Inc. (54).
TEM.
Cells were grown and processed in Permanox Petri plates as follows: 1× PBS (catalog SH30256.01, GE Healthcare Life Sciences), rinsed, and then fixed at 4°C for 2 days in 2% formaldehyde plus 2.5% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.4. After several 0.1 M cacodylate buffer rinses, cells were stained in 0.1% tannic acid followed by post-fixation in 1% OsO4 in 0.1 M cacodylate buffer plus 0.8% potassium ferricyanide. The cells were en bloc stained in 1% aqueous uranyl acetate followed by dehydration through a gradient series of ethanols (50%, 70%, 80%, 90%, 95%, 100%, 100%). Cells were then infiltrated with a progressively higher ratio of embedding resin to ethanol and given 3 changes of pure resin before embedded in a Petri plate with a fresh change of Spurr’s Low Viscosity resin 14. Embedded samples were polymerized at 60°C for 3 days and then separated manually from the plate, leaving cells on the embedding resin. Ultrathin sections (55–60 nm) were cut on a DiATOME Ultra 45° knife, using a Leica EM UC7 ultramicrotome. Sections were collected on 150 hex-mesh copper grids and viewed on a Hitachi H-7500 transmission electron microscope (Chiyoda) using an accelerating voltage of 80 kV. Images were captured using an AMT XR-16 digital camera and AMT Image Capture, v602.600.51 software. Mitochondrial length and number were measured using ImageJ version 1.8.0_112 (NIH).
Oxygen consumption and glycolytic capacity.
OCR and extracellular acidification rate were determined using the Seahorse XFe24 Extracellular Flux Analyzer (Seahorse Bioscience). Cells were seeded at 20,000-40,000 per well for measurement. We used the Seahorse XF Cell Mito Stress Test Kit (103015-100, Seahorse Bioscience), which contained electron transport chain inhibitors that were sequentially injected to each well at specific time points: oligomycin (2 μM), followed by FCCP (1 μM), followed by the addition of a combination of rotenone (0.5 μM) and antimycin (2 μM). To measure extracellular acidification rate, cells were plated at the same number as that used for OCR measurement. We also used the Seahorse XF Glycolysis Stress Test Kit (103020-100, Seahorse Bioscience), which included glucose (10 mM final concentration), oligomycin (2 μM final concentration), and 2-deoxy-glucose (100 mM final concentration), which were injected sequentially as per the manufacturer’s instructions. For assays with drug treatment, all raw readings were normalized to protein levels in the cells, as determined by an in-cell Bradford assay after the flux experiments were completed. For assays without drug treatment, cells were seeded and the assays were performed on the same day and cell number was used for quantification.
Mutational profiling of patient-derived pancreatic cell lines.
Putative biologically relevant mutations were selected for using an annotation filtering strategy. Called mutations were annotated with Oncotator (55). Mutations observed at 0.001% or higher in the population were filtered out based on population allele frequencies observed in the ExAC (56) and 1000 genomes (57) variant data sets. Further, only those mutations that were recurrent (observed 2 or more times) in COSMIC (58) were retained. These select mutations were then visualized in a heatmap using the R package ComplexHeatmap (59).
Statistics.
Data acquisition and analysis was not blinded. All grouped data are presented as mean ± SEM. Differences between groups were assessed by 2-tailed Student’s t test or 1-way and 2-way ANOVA using GraphPad Prism version 7.0c for macOS. Kaplan-Meier curves were generated, and log-rank and Gehan Breslow Wilcoxon analyses were performed using GraphPad. A P value less than 0.05 was considered significant.
Study approval.
Animal studies were performed in accordance with the animal protocol procedures approved by the MD Anderson Cancer Center Institutional Review Board and Institutional Animal Care and Use Committee.
Author contributions
MY and CMT designed the experiments. MY executed most of the experiments with assistance from NDN. YH created the KPC luciferase cell lines used in in vivo study and performed the orthotopic experiments. TNF, AD, JMM, and DL directly contributed to leflunomide experiments, tumor growth, and maintenance of our KPC colony. YK and FASL performed analysis of the MDA-PATC lines. CJF generated reagents and performed preliminary experiments. EJK, JMH, AM, and ACK provided critical input, and SG, HY, JBF, and AM provided the KPC and MDA-PATC cell lines in the study. The manuscript was written by MY, NDN, and CMT.
Supplementary Material
Acknowledgments
CMT was supported by funding from NIH under award R01CA227517-01A1, Cancer Prevention & Research Institute of Texas (CPRIT) grant RR140012, V Foundation (V2015-22), Kimmel Foundation, Sabin Family Foundation Fellowship, and the McNair Foundation. We thank The University of Texas MD Anderson Cancer Center TRACTION Platform for their gift of the MDA-PATC cell lines. This research was performed in the Flow Cytometry & Cellular Imaging Facility, which is supported in part by the NIH through an MD Anderson’s Cancer Center support grant (CA016672). TEM imaging was supported by the Integrated Microscopy Core at Baylor College of Medicine with funding from NIH (DK56338 and CA125123), CPRIT (RP150578), Dan L. Duncan Comprehensive Cancer Center, and John S. Dunn Gulf Coast Consortium for Chemical Genomics. Additional funding sources that supported this work include the National Cancer Institute of the NIH under award CA214793-01A1 (to HY). The graphical abstract created with BioRender.
Version 1. 07/23/2019
In-Press Preview
Version 2. 08/22/2019
Electronic publication
Footnotes
Conflict of interest: JMH receives research support from Oncosil, Galera, Adura, Augmenix, and RaySearch and is a consult for BTG, AbbVie, Celgene, AstraZeneca, Bristol-Myers Squibb, Varian, and Sirtex.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(16):e126915.https://doi.org/10.1172/jci.insight.126915.
Contributor Information
Meifang Yu, Email: MYu2@mdanderson.org.
Nicholas D. Nguyen, Email: NDNguyen1@mdanderson.org.
Yanqing Huang, Email: YHuang10@mdanderson.org.
Daniel Lin, Email: DLin4@mdanderson.org.
Tara N. Fujimoto, Email: tnfujimoto@mdanderson.org.
Jessica M. Molkentine, Email: molkentinej@upmc.edu.
Amit Deorukhkar, Email: adeorukh@mdanderson.org.
F. Anthony San Lucas, Email: FASaan@mdanderson.org.
Conrad J. Fernandes, Email: Conrad.Fernandes@bcm.edu.
Eugene J. Koay, Email: ekoay@mdanderson.org.
Sonal Gupta, Email: SGupta8@mdanderson.org.
Haoqiang Ying, Email: hying@mdanderson.org.
Albert C. Koong, Email: ackoong@mdanderson.org.
Joseph M. Herman, Email: JMHerman@mdanderson.org.
Jason B. Fleming, Email: Jason.Fleming@moffitt.org.
Anirban Maitra, Email: amaitra@mdanderson.org.
Cullen M. Taniguchi, Email: ctaniguchi@mdanderson.org.
References
- 1.Viale A, et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514(7524):628–632. doi: 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen H, Chan DC. Mitochondrial Dynamics in Regulating the Unique Phenotypes of Cancer and Stem Cells. Cell Metab. 2017;26(1):39–48. doi: 10.1016/j.cmet.2017.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337(6098):1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boland ML, Chourasia AH, Macleod KF. Mitochondrial dysfunction in cancer. Front Oncol. 2013;3:292. doi: 10.3389/fonc.2013.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen H, McCaffery JM, Chan DC. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell. 2007;130(3):548–562. doi: 10.1016/j.cell.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 6.Zhao J, et al. Mitochondrial dynamics regulates migration and invasion of breast cancer cells. Oncogene. 2013;32(40):4814–4824. doi: 10.1038/onc.2012.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson GR, et al. Dysregulation of mitochondrial dynamics proteins are a targetable feature of human tumors. Nat Commun. 2018;9(1):1677. doi: 10.1038/s41467-018-04033-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Serasinghe MN, et al. Mitochondrial division is requisite to RAS-induced transformation and targeted by oncogenic MAPK pathway inhibitors. Mol Cell. 2015;57(3):521–536. doi: 10.1016/j.molcel.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luchsinger LL, de Almeida MJ, Corrigan DJ, Mumau M, Snoeck HW. Mitofusin 2 maintains haematopoietic stem cells with extensive lymphoid potential. Nature. 2016;529(7587):528–531. doi: 10.1038/nature16500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kashatus JA, et al. Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Mol Cell. 2015;57(3):537–551. doi: 10.1016/j.molcel.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manczak M, Kandimalla R, Yin X, Reddy PH. Mitochondrial division inhibitor 1 reduces dynamin-related protein 1 and mitochondrial fission activity. Hum Mol Genet. 2019;28(2):177–199. doi: 10.1093/hmg/ddy335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith G, Gallo G. To mdivi-1 or not to mdivi-1: Is that the question? Dev Neurobiol. 2017;77(11):1260–1268. doi: 10.1002/dneu.22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miret-Casals L, et al. Identification of New Activators of Mitochondrial Fusion Reveals a Link between Mitochondrial Morphology and Pyrimidine Metabolism. Cell Chem Biol. 2018;25(3):268–278.e4. doi: 10.1016/j.chembiol.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 14.Sykes DB, et al. Inhibition of Dihydroorotate Dehydrogenase Overcomes Differentiation Blockade in Acute Myeloid Leukemia. Cell. 2016;167(1):171–186.e15. doi: 10.1016/j.cell.2016.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ying H, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149(3):656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang Y, et al. Two-dimensional culture of human pancreatic adenocarcinoma cells results in an irreversible transition from epithelial to mesenchymal phenotype. Lab Invest. 2015;95(2):207–222. doi: 10.1038/labinvest.2014.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim MP, Evans DB, Wang H, Abbruzzese JL, Fleming JB, Gallick GE. Generation of orthotopic and heterotopic human pancreatic cancer xenografts in immunodeficient mice. Nat Protoc. 2009;4(11):1670–1680. doi: 10.1038/nprot.2009.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koay EJ, et al. A Visually Apparent and Quantifiable CT Imaging Feature Identifies Biophysical Subtypes of Pancreatic Ductal Adenocarcinoma. Clin Cancer Res. 2018;24(23):5883–5894. doi: 10.1158/1078-0432.CCR-17-3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D’Erchia AM, et al. Tissue-specific mtDNA abundance from exome data and its correlation with mitochondrial transcription, mass and respiratory activity. Mitochondrion. 2015;20:13–21. doi: 10.1016/j.mito.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 20.LeBleu VS, et al. PGC-1α mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat Cell Biol. 2014;16(10):992–1003. doi: 10.1038/ncb3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abeliovich H, Zarei M, Rigbolt KT, Youle RJ, Dengjel J. Involvement of mitochondrial dynamics in the segregation of mitochondrial matrix proteins during stationary phase mitophagy. Nat Commun. 2013;4:2789. doi: 10.1038/ncomms3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rahib L, Fleshman JM, Matrisian LM, Berlin JD. Evaluation of Pancreatic Cancer Clinical Trials and Benchmarks for Clinically Meaningful Future Trials: A Systematic Review. JAMA Oncol. 2016;2(9):1209–1216. doi: 10.1001/jamaoncol.2016.0585. [DOI] [PubMed] [Google Scholar]
- 23.Aguirre AJ, et al. Real-time Genomic Characterization of Advanced Pancreatic Cancer to Enable Precision Medicine. Cancer Discov. 2018;8(9):1096–1111. doi: 10.1158/2159-8290.CD-18-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cancer Genome Atlas Research Network Electronic address: andrew_aguirre@dfci.harvard.edu, Cancer Genome Atlas Research Network. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell. 2017;32(2):185–203.e13. doi: 10.1016/j.ccell.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shukla SK, et al. MUC1 and HIF-1alpha Signaling Crosstalk Induces Anabolic Glucose Metabolism to Impart Gemcitabine Resistance to Pancreatic Cancer. Cancer Cell. 2017;32(3):392. doi: 10.1016/j.ccell.2017.08.008. [DOI] [PubMed] [Google Scholar]
- 26.Bryant KL, et al. Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat Med. 2019;25(4):628–640. doi: 10.1038/s41591-019-0368-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bordt EA, et al. The Putative Drp1 Inhibitor mdivi-1 Is a Reversible Mitochondrial Complex I Inhibitor that Modulates Reactive Oxygen Species. Dev Cell. 2017;40(6):583–594.e6. doi: 10.1016/j.devcel.2017.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosdah AA, K Holien J, Delbridge LM, Dusting GJ, Lim SY. Mitochondrial fission - a drug target for cytoprotection or cytodestruction? Pharmacol Res Perspect. 2016;4(3):e00235. doi: 10.1002/prp2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kunkel G, W Cannon G. Leflunomide in the treatment of rheumatoid arthritis. Expert Rev Clin Immunol. 2006;2(1):17–31. doi: 10.1586/1744666X.2.1.17. [DOI] [PubMed] [Google Scholar]
- 30.Chen Y, Huang Q, Zhou H, Wang Y, Hu X, Li T. Inhibition of canonical WNT/β-catenin signaling is involved in leflunomide (LEF)-mediated cytotoxic effects on renal carcinoma cells. Oncotarget. 2016;7(31):50401–50416. doi: 10.18632/oncotarget.10409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mattar T, Kochhar K, Bartlett R, Bremer EG, Finnegan A. Inhibition of the epidermal growth factor receptor tyrosine kinase activity by leflunomide. FEBS Lett. 1993;334(2):161–164. doi: 10.1016/0014-5793(93)81704-4. [DOI] [PubMed] [Google Scholar]
- 32.Xu X, et al. In vitro and in vivo antitumor activity of a novel immunomodulatory drug, leflunomide: mechanisms of action. Biochem Pharmacol. 1999;58(9):1405–1413. doi: 10.1016/S0006-2952(99)00228-2. [DOI] [PubMed] [Google Scholar]
- 33.Casadio F, et al. Toward the definition of immunosuppressive regimens with antitumor activity. Transplant Proc. 2005;37(5):2144–2147. doi: 10.1016/j.transproceed.2005.03.089. [DOI] [PubMed] [Google Scholar]
- 34.Eckhardt SG, et al. Phase I and pharmacologic study of the tyrosine kinase inhibitor SU101 in patients with advanced solid tumors. J Clin Oncol. 1999;17(4):1095–1104. doi: 10.1200/JCO.1999.17.4.1095. [DOI] [PubMed] [Google Scholar]
- 35.Ko YJ, et al. A multi-institutional phase ii study of SU101, a platelet-derived growth factor receptor inhibitor, for patients with hormone-refractory prostate cancer. Clin Cancer Res. 2001;7(4):800–805. [PubMed] [Google Scholar]
- 36.Shawver LK, Slamon D, Ullrich A. Smart drugs: tyrosine kinase inhibitors in cancer therapy. Cancer Cell. 2002;1(2):117–123. doi: 10.1016/S1535-6108(02)00039-9. [DOI] [PubMed] [Google Scholar]
- 37.Lolli ML, Sainas S, Pippione AC, Giorgis M, Boschi D, Dosio F. Use of human Dihydroorotate Dehydrogenase (hDHODH) Inhibitors in Autoimmune Diseases and New Perspectives in Cancer Therapy. Recent Pat Anticancer Drug Discov. 2018;13(1):86–105. doi: 10.2174/1574892812666171108124218. [DOI] [PubMed] [Google Scholar]
- 38.Santana-Codina N, et al. Oncogenic KRAS supports pancreatic cancer through regulation of nucleotide synthesis. Nat Commun. 2018;9(1):4945. doi: 10.1038/s41467-018-07472-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bajzikova M, et al. Reactivation of Dihydroorotate Dehydrogenase-Driven Pyrimidine Biosynthesis Restores Tumor Growth of Respiration-Deficient Cancer Cells. Cell Metab. 2019;29(2):399–416.e10. doi: 10.1016/j.cmet.2018.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Von Hoff DD, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med. 2013;369(18):1691–1703. doi: 10.1056/NEJMoa1304369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arnold D, et al. Prognostic and predictive value of primary tumour side in patients with RAS wild-type metastatic colorectal cancer treated with chemotherapy and EGFR directed antibodies in six randomized trials. Ann Oncol. 2017;28(8):1713–1729. doi: 10.1093/annonc/mdx175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manchado E, et al. A combinatorial strategy for treating KRAS-mutant lung cancer. Nature. 2016;534(7609):647–651. doi: 10.1038/nature18600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Churi CR, et al. Mutation profiling in cholangiocarcinoma: prognostic and therapeutic implications. PLoS ONE. 2014;9(12):e115383. doi: 10.1371/journal.pone.0115383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Molina JR, et al. An inhibitor of oxidative phosphorylation exploits cancer vulnerability. Nat Med. 2018;24(7):1036–1046. doi: 10.1038/s41591-018-0052-4. [DOI] [PubMed] [Google Scholar]
- 45.Benej M, et al. Papaverine and its derivatives radiosensitize solid tumors by inhibiting mitochondrial metabolism. Proc Natl Acad Sci USA. 2018;115(42):10756–10761. doi: 10.1073/pnas.1808945115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eisner V, et al. Mitochondrial fusion dynamics is robust in the heart and depends on calcium oscillations and contractile activity. Proc Natl Acad Sci USA. 2017;114(5):E859–E868. doi: 10.1073/pnas.1617288114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen H, et al. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141(2):280–289. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Molkentine JM, et al. Enteral Activation of WR-2721 Mediates Radioprotection and Improved Survival from Lethal Fractionated Radiation. Sci Rep. 2019;9(1):1949. doi: 10.1038/s41598-018-37147-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fujimoto TN, et al. Selective EGLN Inhibition Enables Ablative Radiotherapy and Improves Survival in Unresectable Pancreatic Cancer. Cancer Res. 2019;79(9):2327–2338. doi: 10.1158/0008-5472.CAN-18-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rappold PM, et al. Drp1 inhibition attenuates neurotoxicity and dopamine release deficits in vivo. Nat Commun. 2014;5:5244. doi: 10.1038/ncomms6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11(8):783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Esteves P, et al. Mitochondrial retrograde signaling mediated by UCP2 inhibits cancer cell proliferation and tumorigenesis. Cancer Res. 2014;74(14):3971–3982. doi: 10.1158/0008-5472.CAN-13-3383. [DOI] [PubMed] [Google Scholar]
- 53.Rolland SG, Lu Y, David CN, Conradt B. The BCL-2-like protein CED-9 of C. elegans promotes FZO-1/Mfn1,2- and EAT-3/Opa1-dependent mitochondrial fusion. J Cell Biol. 2009;186(4):525–540. doi: 10.1083/jcb.200905070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tan AS, et al. Mitochondrial genome acquisition restores respiratory function and tumorigenic potential of cancer cells without mitochondrial DNA. Cell Metab. 2015;21(1):81–94. doi: 10.1016/j.cmet.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 55.Ramos AH, et al. Oncotator: cancer variant annotation tool. Hum Mutat. 2015;36(4):E2423–E2429. doi: 10.1002/humu.22771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walsh R, et al. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet Med. 2017;19(2):192–203. doi: 10.1038/gim.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.1000 Genomes Project Consortium. et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Forbes SA, et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45(D1):D777–D783. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32(18):2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.