Abstract
A revolution in cellular measurement technology is under way. While prior studies have only been able to analyze the averaged outputs from renal tissue, now we can accurately monitor genome-wide gene expression, regulation, function, cellular history, and cellular interactions in thousands of individual cells in a single experiment. These methods are key drivers to changing our morphotype-based organ and disease descriptions to a new era of unbiased genomic definitions and to improving our understanding of kidney development, homeostasis and disease.
Keywords: single cell, kidney disease, epigenome, RNA-seq
The cell is the fundamental unit of the living organism. For centuries, cell types have been defined phenotypically by their structure and function, also referred to as morphotypes. Cells are named after their discoverer (Loop of Henle), their location (proximal tubule), or their phenotypic feature on microscopical examination. Downstream of cellular classification lies disease taxonomy, which are also based on microscopical changes observed in kidney cell morphotypes. There are several key deficiencies in our current cell and downstream disease categorization. For example, the current morphological classification does not account for the developmental origin and evolution of cells and diseases. Cells can originate from different lineages, but they take on similar and sometimes indistinguishable phenotypes - a phenomenon called phenotypic convergence. For example, cells in glomerular crescents look very similar, yet they likely have could different developmental origins. Morphology-based disease terminology is highly operator dependent and does not capture the most important disease discriminative and prognostic features. Most currently used kidney disease descriptors are unable to capture molecular mechanisms that underlie disease driving molecular pathways and are therefore not sufficient for prognostication, target identification and drug development.
The desire for a more rigorous and unbiased cell-type classification has recently been met with an explosion of development in new and feasible methods for systematic cell characterization. These methods mostly rely on genomic profiling approaches such as large-scale profiling RNA, epigenome, chromatin, protein, and metabolites of single cells. In this review, we briefly touch on some of the recent methods and discuss new results from recent publications to illustrate how this information improved our understanding of kidney health and disease.
METHODOLOGIES
Single-cell RNA-Seq (scRNA-seq)
scRNAseq refers to methods for unbiased genome wide RNA profiling of individual cells. Transcriptome profiling of individual cells can be split into four major components: RNA molecule capture, reverse transcription and transcriptome amplification, library preparation, and sequencing and quantification (Figure 1). One of the key developments leading to the current popularity of single cell transcriptomics is the PCR-based amplification[1], which has made the amplification step far less complicated compared to the prior in vitro transcription method.[2] The in vitro transcription method still has certain advantages, such as the lower amplification distortion and the ability to amplify certain materials that the PCR reaction cannot. Most commercial platforms currently rely on PCR amplification. Once amplification is achieved, library and sequencing follow the standard RNAsequencing pipeline. After sequencing, informatics remains challenges to analyze single cell data, such as alignments from 3’-biased amplicons and expression quantification that takes on the unique noise structure of scRNAseq. Informatics approaches to analyze single cell datasets are still in flux and due to space limitations, we do not include an overview here.[3-6]
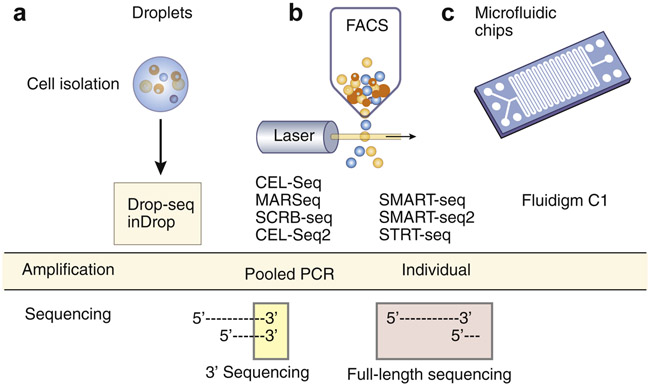
Figure 1: Various single cell RNAseq methods and technologies.
Many methods have been developed to separate cells systematically. a. Microfluidic flow technology encapsulates single cells into aqueous droplets in oil. b. Fluorescent Activated Cell Sorting (FACS) can be used for single cell plating in a highly specific manner. FACS allows for concurrent quantitative and qualitative multi-parametric analyses of single cells c. Commercially available microfluidic chips can provide programmed single-cell lysis, RNA extraction, and cDNA synthesis for numerous cells on a single chip at the same time. RNA amplification could be done either in a pooled PCR reaction or individually, and sequencing can be performed for length transcripts or only for the 3’ end of the transcripts. The green indicates the use of individual amplification methods, whereas the red indicates the use of pooled amplification methods.
The biggest and most unique challenge for scRNAseq involves the isolation of cells and the individualized RNA capture (Figure 1). The most common microtiter-plate-based and microfluidic scRNA-seq techniques and their applications have recently been reviewed by Wu et al.[7] RNA capture requires separating individual cells for the reverse transcription reaction. The separation of the cells can be done manually into standard microtubes or microwell plates, by sorting into microwell plates, or by the original FuidigmC1 into micro-fabricated reaction chambers [1],[2],[8]. Microfluidics allows higher-throughput scRNA-seq workflows, and more uniform reactions, along with cost saving from reaction volume reduction. Another advantage of the microfluidics based high-throughput method is the droplet generation methods used, which use an oil-water mixed flow to create a water-in-oil droplet, at high rate that encapsulates individual cells. Two droplet-based methods, inDrops [9] and Drop-seq [10], were developed in parallel with related commercial systems, allowing straightforward implementation. In particular, inDrops encapsulates cells by using hydrogel beads bearing poly(T) primers with defined barcodes, after which the photo-releasable primers are detached from the beads to improve molecule-capture efficiency and initiate in-drop reverse transcription reactions. The inDrops system is licensed to 1CellBio, and a variant protocol has been commercialized as the Chromium Single Cell 3′ Solution (10x Genomics)[11]. The Chromium system currently has considerably higher library-preparation costs but has gained incredible popularity over the last year, and a large portion of single cell papers now use this method [12]. The droplet-based high throughput methods have revolutionized the sampling depth, but current platforms still have relatively low efficiency in RNA capture and the costs are still challenging for deep profiling of individual cells (Figure 1). We note that despite the tremendous rate of technology development, much of the challenges stem from the distinct nature of each tissue and disassociation of individual cells from that particular tissue, especially with respect to sampling biases that are unique to each tissue and experiment. There is no automated approach to this component of the problem, and full sampling of the kidney from various organisms still remains an important challenge.
Single nuclear sequencing (snRNASeq) is becoming a popular alternative to single cell sequencing (scRNAseq). Here nuclei are isolated from cells and used for droplet-based sequencing. The main advantages of nuclear sequencing include portability as reasonably good quality nuclei can be isolated from snap frozen samples. In addition, certain cell types such as fibroblasts, which are difficult to digest off of the basement membrane, can be captured more completely. On the other hand, nuclear sequencing seems to capture other cells, such as immune cells, with lower efficiency. Furthermore, as most of the RNA in the nucleus is preRNA (unspliced), slightly different methods are needed for data alignment and downstream analysis [13]. Further studies are needed to quantitatively analyze and compare “cell drop-outs” by scRNAseq and snRNAseq.
Single-cell epigenome analysis
The epigenome is broadly believed to capture the gene expression logic and cellular history of individual cells (Figure 2). Therefore, it could potentially be a more robust feature for defining cell types. Single cell epigenome mapping can identify gene regulatory elements such as promoters, enhancers and insulators. Methods for single cell histone chromatin immunoprecipitation (ChIP-Seq)[14], open chromatin area (ATAC-Seq; Assay for Transposase-Accessible Chromatin using sequencing)[15],[16], DNA methylation[17],[18] and 3-D chromatin organization (such single cell Hi-C)[19],[20], have been developed. Of these methods, the chromatin accessibility analysis (ATAC-Seq) seems to be the most established. ATAC-Seq employs transposase Tn5 to cut accessible (non-histone bound) DNA and ligate sequencing adaptors simultaneously, which efficiently map the open chromatin regions in the whole genome. At the time of writing this review, scATAC-Seq is still less robust than the scRNAseq due to the multiple problems posed by ATAC-Seq, however, there is considerable excitement around the recently commercialized ATAC-Seq method by 10x Genomics. The first problem is that it is difficult to saturate the transposase cleavage and insertion to target every accessible site at the single cell level, creating a high false negative rate. In addition, recovering the “tagged” regions and amplifying the appropriate DNA are still a challenge, as PCR duplicates can interfere with the biological signal of interest. Lastly, given that epigenome capturing methods analyze the entire genome, not just the ~2% transcribed region, the cost associated with epigenome analysis is significant.
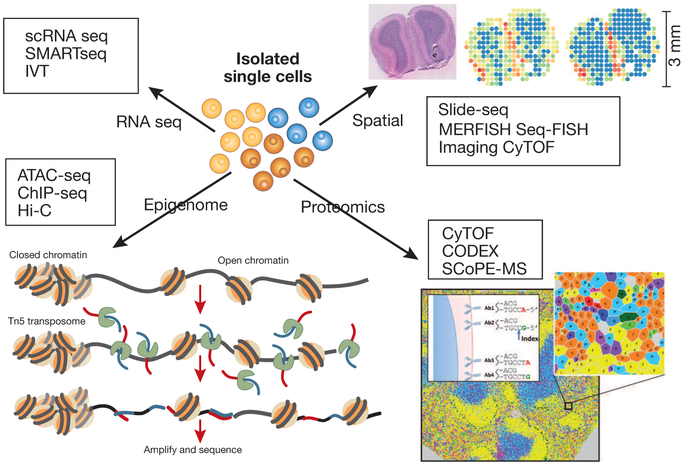
Figure 2: Single cells omics analysis Various methods exist to analyze single cell data to provide transcriptomic, epigenomic, spatial, and proteomic information.
Borrowed/Adapted from Goltsev Y, Samusik N, Kennedy-Darling J et al. Deep Profiling of Mouse Splenic Architecture with CODEX Multiplexed Imaging Cell vol 174, Issue 4, Pages 968-981.e15; https://doi.org/10.1016/j.cell.2018.07.010
[29] and Borrowed/Adapted from Ståhl P, Salmén F, Vickovic S et al Visualization and analysis of gene expression in tissue sections by spatial transcriptomics Science 01 Jul 2016: Vol. 353, Issue 6294, pp. 78-82 DOI: 10.1126/science.aaf2403
[59] with permission from AAAS .
Spatial techniques
The complex 3-dimensional organization of organs is important for cell-cell interactions and therefore their functions. Several methods have been or are being developed to detect spatial organization. The most popular technique is multiplex in situ RNA detection, as exemplified by MERFISH[21],[22] and Seq-FISH[23], which combine multiplex hybridization with microscopy- based quantification to assess distributions at both the cellular and subcellular level. The multiplexing strategies typically involve a scheme to sequentially hybridize combinatorial barcodes that, when combined, uniquely identify a transcript. FISSEQ and variations generate in situ cDNA that are amplified and sequenced in spatially localized reactions.[24] The various in situ methods, especially in situ sequencing, are limited in terms of spatial resolution that limit the number of molecules detected (typically 1% of total RNA molecules). Other approaches are based on protein detection, such as Imaging Mass Cytometry[25] and Multiplexed Ion Beam Imaging[26], and involve staining tissue specimen with antibodies (see later). These in situ results can then be used to map massive amounts of single-cell genomic information from dissociated cells onto the tissue samples by using statistical mapping of single cell profiles to spatial profiles, thus combining the depth of transcript coverage of individual scRNAseq and the breadth of the spatial information from in situ methods. Integration of high-throughput scRNAseq and spatial transcriptomics can provide important clues about spatial relationships and cell-cell communication. However, combining these assay modalities is a non-trivial task, requiring both novel analytical methods as well as careful tissue sampling.
Proteomics, metabolomics, multi-omics
One of the many limitations of single cell RNA-Seq is that for low or moderately expressed transcripts, large fractions of cells in a cluster show no expression, usually called a “transcript dropout phenomenon”. This is presumably due to transcriptional dynamics in the cell where transcription occurs in bursts that are punctuated by periods of up to several hours with no transcription. Thus, despite continuous expression of a particular protein, its transcript may not be detectable for a large fraction of the time. Proteomics profiling would offer the advantage of analyzing protein levels that are much closer linked to function.
Most single cell proteomics methods use antibody-based detection. CyTOF (flow Cytometer, which utilizes Time-Of-Flight mass spectrometer) is a variation of flow cytometry in which antibodies are labeled with heavy metal ion tags, allowing multiplexing [27],[28]. Readout is by time-of-flight mass spectrometry of the lanthanide isotopes that are labeling the antibodies. CyTOF can detect 40 or more proteins in millions of cells. While CyTOF requires a significant instrumentation investment, a newly-developed highly multiplexed cytometric imaging approach, termed co-detection by indexing (CODEX), uses regular motorized stage three-color fluorescence microscopes, available in most labs for multidimensional tissue rendering [29]. CODEX iteratively visualizes multiplexed antibody binding events using DNA barcodes, fluorescent dNTP analogs, and an in situ polymerization-based indexing procedure. In a recent publication, CODEX was able to detect 66 antigens by the “activation primer”-based extension system and the results were comparable to CyTOF.[29] While antibody-based detection of proteomics has been popular, recently, mass spectrometry methods have also been developed. Single Cell ProtEomics by Mass Spectrometry (SCoPE-MS) is an unbiased mass spectrometry-based proteomics method that quantifies over a thousand proteins at single cell resolution.[30] A bottleneck for mass spectrometry is not so much in the sensitivity of the detection instrument but in the sample handling and in maximizing yield. For example, in standard MALDI procedure, 90% of the macromolecules are left behind. SCoPE-MS attempts to overcome these problems through specific processes to minimize front-end loss[30]. Another important challenge in single cell proteomics is the multi-modal omics detection at the single cell level. Many new methods have been recently developed to address this challenge: Sci-CAR (combinatorial indexing-based co-assay of chromatin accessibility and mRNA)[31] can simultaneously detect gene expression and open chromatin information for thousands of cells. Recently, with improved indexing, this method has been applied to survey 2 million cells derived from 61 embryos staged between 9.5 and 13.5 days of gestation.[32] ScTrio-seq[33] (single-cell triple omics sequencing) technique can assess somatic copy number alterations (SCNAs), DNA methylation, and transcriptome information simultaneously from an individual cell. An improved version of this scTrio-seq2 method has just been published [34]. Multi-modal assays remain challenging and the sensitivity and accuracy of these methods still need to be validated under wider use cases. Still, the pace of technological development is extremely promising and our ability to make cell-type informative measurements will continue to expand.
RECENT ADVANCES IN KIDNEY SINGLE CELL STUDIES
Single Cell Transcriptome based new taxonomy for the mouse kidneys
The most fundamental level of analysis is atlasing for the unbiased identification of cell types. The morphotype-based kidney cell atlas is several centuries old. In our initial paper, we generated a transcriptomic map for 70,000 cells sequenced from healthy mouse kidney samples using the droplet-based method established by the 10x Chromium system [12]. The key aspect of atlasing efforts is the use of dimension reduction methods to group cells based on their transcript abundance. Several such methods have been developed [35-37]. It is important to note that these methods can produce significant differences in the number of cell clusters and therefore these cell clusters should not be immediately interpreted as cell types. Cell type identification is based on mapping the unbiased transcript data onto prior function, structure, or morphotype based datasets. Validation studies are critical in case such prior datasets are not available for cell type identification.
Our unbiased mouse kidney transcript-based analysis identified >19 distinct cell types in control (healthy) mouse kidneys. These cell types can be distinguished based on their unbiased genome-wide gene expression patterns. Using prior segment-based datasets, we were able to annotate all but three cell clusters in the kidney.[38] All major kidney cell types, such as endothelial cells, podocytes, proximal, distal and collecting duct cells and loop of Henle cells were identified. We found that for resident kidney cells, transcripts with the highest abundance and best discriminative characteristics in our single cell analysis showed high concordance with previously identified cell-type identifiers. For example, Nephrin was one of most abundant transcripts in podocytes and aquaporin 2 was such in principal cells. However, single markers do not seem to be sufficient in identifying cell type, and our dataset provides a comprehensive multi-marker (20-100) based genomic cell-type definition. In addition to the identification of kidney cell types, we have identified a large number of diverse immune cells even in mice that have been kept in a pathogen free environment. There are several cell types in the kidney that are purely morphotype defined without clear cell type specific markers such as mesangial cells. Identification of these cell types in single cell transcript datasets remains challenging due to the lack of anchoring gene expression data. Reanalysis of the existing datasets coupled with careful functional validation experiments will be essential for the molecular definition of these and other cells.
The kidney collecting duct represents the last segment of the tubule system. Developmentally, this segment originates from the ureteric bud, while the rest of the epithelial cells in the kidney come from the metanephric mesenchyme. Furthermore, this segment contains two different cell types that are located next to each-other: Principal cells (PC) express high levels of aquaporin 2 and vasopressin receptor as their cell discriminative genes, and intercalated cells (IC) express high levels of the specific vacuolar ATPase subunits. Our single cell cell-clustering algorithm identified a third distinct cell type in the collecting duct. We called this distinct cell type “transitional cells”, as these cells express both the ATPase and aquaporin 2. Several scientists have proposed that these cells could be progenitor or stem cells that differentiate into PC or IC cells. Our cell trajectory analysis indicated, however, that IC cells turn into PC cells via the transitional cell type (PC/IC cells). We experimentally confirmed these results using lineage-tagging analysis. We used V-ATPaseCre mated with Tomato GFP, which labeled all cells of IC cell origin with GFP. While the majority of these cells express the V-ATPase by immunostaining, there were some cells that no longer expressed the V-ATPase by staining (i.e. they are not IC cells) but instead expressed aquaporin 2 (they are PC cells). These cells are PC cells that converted from IC cells, as they were originally ATPase positive. Cell trajectory analysis indicated that the IC vs PC cell state is determined by Notch ligand vs receptor expression. Transgenic expression of the Notch receptor was sufficient to change the fate of these cells to take on the principal cell fate[12]. Recent studies corroborate the plasticity of the collecting duct cells in the mouse kidney. The Zhang lab showed that AQP2+ cells convert to IC cells, specifically following deletion of an epigenome editing enzyme Dot1 [39]. In addition, Mukherjee et al performed a series of experiments including genetically inactivating Notch1 and Notch2 or deleting the Notch target Hes1, or inducing expression of a Notch inhibitor in the collecting ducts in mice after kidney development. They show that Notch signaling was required for maintenance of Aqp2-expressing cells in distal nephron and collecting duct segments in adult kidneys. Loss of Notch signaling or use of lithium triggered transdifferentiation of mature principal cells to intercalated cells in adult kidneys[40].
Single cell transcriptome datasets for mouse kidneys have been generated as part of large whole-animal scale efforts, such as the Tabula Muris[41], which used 10x Genomics and SMARTSeq methods, or the Mouse Cell Atlas (MCA)[42], which used microwell-based sequencing to analyze more than 400,000 cells from more than 40 organs. While the resolution of these datasets for the kidney is not particularly high, these datasets allow for the comparison of kidney cell types with other organs and the identification of shared and specific signatures. These datasets could be particularly informative for organ-shared cell types such as immune cells, fibroblasts, or endothelial cells.
A limitation of shot-gun (or whole organ) scRNA-Seq for kidneys is that most of the sequencing is done for dominant cell types, viz a viz proximal tubule and thick ascending limb of Henle's loop, whereas nonabundant cell types with important functions may be "under profiled". This problem can be resolved through prudent use of biochemical separation methods, cell sorting methods, affinity methods, etc. to enrich certain cell types prior to scRNA-Seq. Several groups have focused on developing high resolution single cell transcriptomic maps for specific kidney segments, such as the glomerulus [43] or the collecting duct system by sorting cells based on known markers.[44] Another limitation of all single cell methods is the possible distortion of the transcriptomic profile from metabolically vulnerable cell types (i.e. thick ascending limb cells and proximal tubule cells) undergoing stress responses. Medullary thick ascending limb cells begin to die within 60 minutes, even at cold temperatures. While early publications reported significant stress signature issues, newer studies do not seem to show these changes. Several studies have offered improved methods that address this problem, such as the use of cold resistant enzymes or nuclear sequencing (snRNAseq, see above). It is clear that we are only at the beginning of this journey and that larger and more standardized datasets both at low and high resolution will be needed to detect sex-, strain- and age- specific differences.
Due to its well annotated cellular diversity, the mouse kidney is now increasingly used as a model organ for single cell methodology development. The Shendure lab has developed a combinatorial indexing assay called sci-ATAC-seq.[16] Using this assay, they profiled genome- wide chromatin accessibility in ~100,000 single cells from 13 adult mouse tissues including the mouse kidney. Later last year, the Shendure lab also reported on developing sci-CAR, a combinatorial indexing-based co-assay that jointly profiles chromatin accessibility and mRNA in each of the thousands of single cells.[31] As a proof-of-concept, the group has analyzed 11,296 cells from the adult mouse kidney. With the resulting data, the group was able to establish the dynamics of chromatin accessibility and gene expression, reconstruct the chromatin accessibility profiles of cell types defined by RNA profiles, and link cis-regulatory sites to their target genes on the basis of the covariance of chromatin accessibility and transcription across large numbers of single cells. While the initial steps are encouraging, more work is needed both in the technical development and analytical methods.
Cataloging cells in the human kidney
Single cell analysis of human kidney tissue samples is clearly of high importance for the understanding of human kidney physiology and disease states. Unfortunately, the generation of high-quality human kidney single cell data has been challenging mostly due to sample procurement and sample preparation issues. One of the first human kidney single cell data was published by Der et al and contained 899 cells from the skins and kidneys of lupus nephritis patients [45]. The authors were able to identify cell types based on cell-type specific marker expression for different immune cells and for different kidney tubule cells.
Young et al sought to define the identities of normal and cancerous human kidney cells.[46] The group has catalogued 72,501 single kidney cell transcriptomes including Wilms tumor, clear cell renal cell carcinoma, and papillary renal cell carcinoma in relation to healthy fetal, pediatric, adolescent, and adult kidneys, as well as to ureters. From these data, they determined that Wilms tumor originates from aberrant fetal cells, whereas adult kidney cancers are likely derived from a specific subtype of proximal convoluted tubular cell. By mapping the differentiation of fetal cells, they observed the abnormal differentiation that occurs in pediatric kidney cancer.
The Humphreys group has also generated adult human kidney single cell data by sequencing nuclei from a 62-year-old white male with a serum creatinine level of 1.03 mg/dL using the 10X Chromium platform.[47] They profiled 4,524 nuclei and identified 12 distinct epithelial cell clusters, including podocytes, proximal tubule cells (S1–S3), loop of Henle cells (descending and ascending), distal tubule cells, connecting segment cells, principal cells, and intercalated cells. It is clear that early results from human kidney single cell analysis shows exciting results; however, given the genetic and environmental diversity of the human population, larger higher resolution datasets and integrated analysis will be essential for future studies.
Mapping kidney development in mice, in humans and in a dish
Single cell studies for developing kidneys can provide unprecedented insight into cell diversification and differentiation. Newly developed pseudotemporal analytical methods seem to perform well for defining cellular transitions during the differentiation process (Figure 3). Pseudotime is a newly developed concept to measure the progress an individual cell has made through a specific cellular process such as differentiation. In many biological processes, cells do not change their state in perfect synchrony. Rather, cells in different stages of the process are present in the organ. In a population of cells captured at exactly the same time, some cells might be far along, while others might not yet even have begun the process. New methods enable cells to be ordered as they progress along a learned trajectory, so instead of tracking changes in expression as a function of time, the analysis tracks changes as a function of progress along the trajectory, which is termed “pseudotime". Pseudotime is an abstract unit of progress: it is simply the distance between a cell and the start of the trajectory, measured along the shortest path. The trajectory's total length is defined in terms of the total amount of transcriptional change that a cell undergoes as it moves from the starting state to the end state.
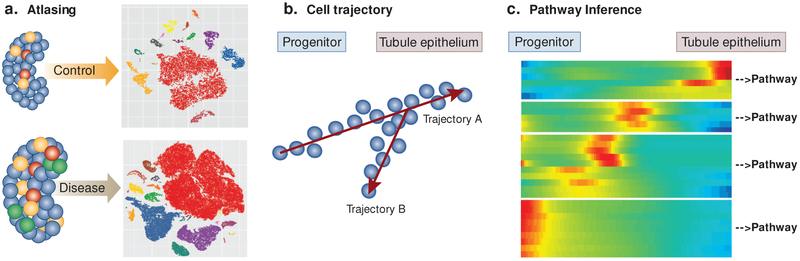
Figure 3: Application of Single Cell RNA Seq information to understand kidney health and disease.
a. Single-cell transcriptomic clustering profiles can reveal and identify cell clusters that represent specific cell populations changes in cell population in control and diseased patients’ populations. b. Using pseudotime analysis, single-cell RNA sequencing data can help elucidate cell developmental trajectories during differentiation. c. Single-cell RNA sequencing data can be used to determine expression of genes in specific cells. These genes can be clustered by expression to determine gene regulatory pathways.
The Brunskill group was the first to profile mouse kidney samples at embryonic days 11.5, 12.5 and postnatal day 4 at single cell resolution [48]. They provided important evidence that initial kidney organogenesis involves a multilineage priming process. Cebrian et al. profiled 3,000 human fetal renal cells spanning 4 months of development in utero and identified transcriptional regulators and signaling pathways involved in segmentation of nephron tubules [49]. One of the most comprehensive renal development profiling studies was performed by Lindstom et al and published in a series of papers.[50],[51] They found that in humans, each progenitor population adopts a stereotypical arrangement: nephron progenitors capped outgrowing ureteric branch tips, whereas interstitial progenitors were located between the nephron progenitors and the renal capsule. An interesting finding was that transcriptional profile of human nephron and interstitial progenitors substantially overlapped. For example, FOXD1, a key interstitial progenitor marker in mice, was readily detected in human nephron progenitors. Comparative gene expression profiling in human and mouse nephron progenitors showed broad agreement between the species but also identified species-biased expression of some genes. Importantly, human nephron progenitors were enriched for human renal disease genes, such as DAPL1 and COL9A2. These findings could have implications for the evolutionary processes that drive the diversity of mammalian organ systems.
Wang et al performed modified single-cell tagged reverse-transcription sequencing (STRT-seq) with excellent sensitivity and precision for single-cell transcriptome detection, analyzing a total of 3,543 renal cells from 11 human embryos spanning gestational weeks 7–25 [52]. They evaluated the proportion of each cell type and found that mesenchymal cells account for a large fraction of the fetal human kidney. They also identified the persistent expression of SIX1 throughout human nephrogenesis. Meanwhile, they identified two subtypes of cells within the cap mesenchyme, one with self-renewal potential and the other exhibiting epithelial features through the mesenchymal epithelial transition. The cap mesenchyme cell cluster exhibited clear cell proliferation features in the human fetal stage, indicating its progenitor characteristics. This indicates that during the development the cap mesenchyme may sustain its proliferative capacity to constantly provide progenitor cells, ensuring the formation of large numbers of cells for the entire nephron. The co-expression of proliferation- and differentiation- related genes in the cap mesenchyme suggests a tightly controlled transcriptional program to ensure the accurate spatiotemporal expression of different regulators to fulfill the function of the cap mesenchyme. The expression of HES1 and NOTCH2 indicated an important role for Notch signaling pathway activation in cap mesenchyme morphogenesis.
In addition to mapping the development and differentiation of mouse and human kidneys, single cell sequencing efforts have proven to be highly informative for evaluating the differentiation of stem cells in kidney organoid systems. In these experiments, a cocktail of factors is applied sequentially to pluripotent stem cells to induce their differentiation into kidney cell types and structure, called organoids. The Humphreys group recently very elegantly compared multiple different kidney organoid differentiation methods and profiled gene expression at the single cell level for 83,130 cells isolated from 65 organoids [47]. The authors have found that the differentiation protocol generates a broad range of cells, including multiple early kidney epithelial cells, podocytes, and proximal tubules, in addition to non-renal cells that could be as abundant as 20% of the population. They have performed a systematic comparison of cells from the organoids to cells from fetal and adult kidneys. They reconstructed differentiation by pseudotemporal ordering and identified ligands, receptors and transcription factor networks associated with fate decisions. They found that inhibition of the brain-derived neurotrophic factor and its receptor neurotrophic tyrosine kinase, receptor, type 2 markedly decreased neuronal differentiation in the culture system without altering kidney differentiation. Studies from the Freedman group reported an automated organoid differentiation protocol coupled with immunofluorescence and single-cell RNA sequencing methods.[53] They identified previously undetected parietal, interstitial, and partially differentiated cells within organoids and defined conditions that can be used to expand the vascular endothelium of the organoids. Both groups showed similarities between organoids and developing fetal human kidneys. However, most cells did not differentiate into terminally differentiated renal epithelial cells, and multiple aberrant cell types were observed in the culture indicating the need for improved organoid differentiation methods.
Kidney disease at single cell resolution
One key purpose of the single cell analysis efforts to move morphotype-based disease definitions into a molecular era is to identify disease-causing cell types and disease-causing pathways [54]. As a first step, we mapped the expression of genes causing monogenic genetic diseases. For example, mutations in close to 30 genes have been associated with nephrotic range proteinuria.[55] Upon mapping the expression of these genes in our mouse kidney single cell atlas, we found that almost all of the nephrotic syndrome causing genes were expressed in only a single cell type in the kidney: the podocytes (or glomerular epithelial cells) [12]. While earlier studies had implicated defects in endothelial cells and proximal tubules in the development of proteinuria, and indeed functional and structural changes in these cell types can be seen in patients with proteinuria, our results unequivocally show that podocyte dysfunction is the principal cause of inherited nephrotic syndrome. As another example, we found that the mouse homologs of genes associated with renal tubule acidosis in humans were expressed only by intercalated cells of the collecting duct, and genes implicated in blood pressure regulation were enriched for distal convoluted tubule expression [12].
Following the same logic, we annotated the expression of putative complex trait disease genes that have been associated with blood pressure and serum metabolite levels [12]. The expression of genes associated with blood pressure in genome wide association studies was also mostly enriched in collecting duct cells, indicating the role of the same cell type both for monogenic and complex disease and a conserved cell-endophenotype correlation. Genes associated with serum metabolite levels in people showed very strong enrichment for proximal tubule expression. Finally, we mapped the expression of genes association with eGFR in large genome wide association studies [56]. We found that loci associated with kidney function in the human population regulate genes that are mainly expressed in the kidney proximal tubule [57]. Similar observations have been made by the Humphreys group by analyzing human organoids. The major challenge when annotating complex disease genes with single cell gene expression data is actually knowing the causal gene(s) in an associated GWAS locus, which requires genotype and gene expression association analysis by eQTL (expression quantitative trait)[47]. In addition to the genetic mapping, the Humphreys group has generated single-cell transcriptome maps from human kidney transplant biopsies.[58] Unsupervised clustering analysis of the biopsy specimen was performed to identify 16 distinct cell types, including all of the major immune cell types and most native kidney cell types, in this biopsy specimen, for which the histologic read was mixed rejection [47]. While initial studies are encouraging, larger datasets are needed both from healthy and disease samples to identify key disease driving cell types.
Conclusion
The kidney is composed of more than 20 distinct cell types that work in concert to regulate organismal electrolyte and small molecular homeostasis and respond to environmental changes to maintain health. The new single cell methods can change our morphotype-based cell type definition to transcript, epigenome and cell-of-origin classifications. These methods can improve our understanding of developmental biology and define key pathways of cell diversification and differentiation. Using these tools, we can identify transcriptional changes in kidney disease and understand pathways that drive these changes and define new cell-cell communication alterations. Combining single-cell genomics, emerging spatial approaches, and multiplex phenotyping, together with functional analysis, could help to develop the next generation of diagnostics and therapies.
Acknowledgement
Work in the Susztaklab is supported by Juvenile Diabetes Research organization, by the National Institute of Health R01 DK076077, R01 DK087635 and DP3 DK108220. Dr. Park is supported by the American Diabetes Association Training grant #1-17-PDF-036. Dr. Kim is supported by the NIDDK 3U01DK107350-02S1 as a subcontract from USC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Picelli S, et al. , Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc, 2014. 9(1): p. 171–81. [DOI] [PubMed] [Google Scholar]
- 2.Hashimshony T, et al. , CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep, 2012. 2(3): p. 666–73. [DOI] [PubMed] [Google Scholar]
- 3.Butler A, et al. , Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol, 2018. 36(5): p. 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang M, et al. , SAVER: gene expression recovery for single-cell RNA sequencing. Nat Methods, 2018. 15(7): p. 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang J, et al. , Gene expression distribution deconvolution in single-cell RNA sequencing. Proc Natl Acad Sci U S A, 2018. 115(28): p. E6437–E6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, et al. , Bulk Tissue Cell Type Deconvolution with Multi-Subject Single-Cell Expression Reference. bioRxiv, 2018: p. 354944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu H and Humphreys BD, The promise of single-cell RNA sequencing for kidney disease investigation. Kidney Int, 2017. 92(6): p. 1334–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Islam S, et al. , Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res, 2011. 21(7): p. 1160–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klein AM, et al. , Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell, 2015. 161(5): p. 1187–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Macosko EZ, et al. , Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell, 2015. 161(5): p. 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng GX, et al. , Massively parallel digital transcriptional profiling of single cells. Nat Commun, 2017. 8: p. 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park J, et al. , Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science, 2018. 360(6390): p. 758–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu H, et al. , Advantages of Single-Nucleus over Single-Cell RNA Sequencing of Adult Kidney: Rare Cell Types and Novel Cell States Revealed in Fibrosis. J Am Soc Nephrol, 2019. 30(1): p. 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rotem A, et al. , Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat Biotechnol, 2015. 33(11): p. 1165–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buenrostro JD, et al. , Single-cell chromatin accessibility reveals principles of regulatory variation. Nature, 2015. 523(7561): p. 486–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cusanovich DA, et al. , A Single-Cell Atlas of In Vivo Mammalian Chromatin Accessibility. Cell, 2018. 174(5): p. 1309–1324 e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smallwood SA, et al. , Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat Methods, 2014. 11(8): p. 817–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo H, et al. , Single-cell methylome landscapes of mouse embryonic stem cells and early embryos analyzed using reduced representation bisulfite sequencing. Genome Res, 23(12): p. 2126–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagano T, et al. , Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature, 2013. 502(7469): p. 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stevens TJ, et al. , 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature, 2017. 544(7648): p. 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen KH, et al. , RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science, 2015. 348(6233): p. aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moffitt JR, et al. , High-throughput single-cell gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization. Proc Natl Acad Sci U S A, 2016. 113(39): p. 11046–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shah S, et al. , seqFISH Accurately Detects Transcripts in Single Cells and Reveals Robust Spatial Organization in the Hippocampus. Neuron, 2017. 94(4): p. 752–758 e1. [DOI] [PubMed] [Google Scholar]
- 24.Lee JH, et al. , Highly multiplexed subcellular RNA sequencing in situ. Science, 2014. 343(6177): p. 1360–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giesen C, et al. , Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods, 2014. 11(4): p. 417–22. [DOI] [PubMed] [Google Scholar]
- 26.Angelo M, et al. , Multiplexed ion beam imaging of human breast tumors. Nat Med, 20(4): p. 436–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baranov VI, et al. , A sensitive and quantitative element-tagged immunoassay with ICPMS detection. Anal Chem, 2002. 74(7): p. 1629–36. [DOI] [PubMed] [Google Scholar]
- 28.Bendall SC, et al. , Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science, 2011. 332(6030): p. 687–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goltsev Y, et al. , Deep Profiling of Mouse Splenic Architecture with CODEX Multiplexed Imaging. Cell, 2018. 174(4): p. 968–981 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Budnik B, et al. , SCoPE-MS: mass spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation. Genome Biol, 2018. 19(1): p. 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao J, et al. , Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science, 2018. 361(6409): p. 1380–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao J, et al. , The single-cell transcriptional landscape of mammalian organogenesis. Nature, 2019. 566(7745): p. 496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hou Y, et al. , Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res, 2016. 26(3): p. 304–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bian S, et al. , Single-cell multiomics sequencing and analyses of human colorectal cancer. Science, 2018. 362(6418): p. 1060–1063. [DOI] [PubMed] [Google Scholar]
- 35.Becht E, et al. , Dimensionality reduction for visualizing single-cell data using UMAP. Nature Biotechnology, 2018. [DOI] [PubMed] [Google Scholar]
- 36.Haghverdi L, Buettner F, and Theis FJ, Diffusion maps for high-dimensional single-cell analysis of differentiation data. Bioinformatics, 2015. 31(18): p. 2989–98. [DOI] [PubMed] [Google Scholar]
- 37.Kobak D and Berens P, The art of using t-SNE for single-cell transcriptomics. bioRxiv, 2018: p. 453449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee JW, Chou CL, and Knepper MA, Deep Sequencing in Microdissected Renal Tubules Identifies Nephron Segment-Specific Transcriptomes. J Am Soc Nephrol, 2015. 26(11): p. 2669–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu H, et al. , Aqp2-expressing cells give rise to renal intercalated cells. J Am Soc Nephrol, 2013. 24(2): p. 243–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mukherjee M, et al. , Endogenous Notch Signaling in Adult Kidneys Maintains Segment-Specific Epithelial Cell Types of the Distal Tubules and Collecting Ducts to Ensure Water Homeostasis. J Am Soc Nephrol, 2019. 30(1): p. 110–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tabula Muris C, et al. , Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature, 2018. 562(7727): p. 367–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han X, et al. , Mapping the Mouse Cell Atlas by Microwell-Seq. Cell, 2018. 173(5): p. 1307. [DOI] [PubMed] [Google Scholar]
- 43.Karaiskos N, et al. , A Single-Cell Transcriptome Atlas of the Mouse Glomerulus. J Am Soc Nephrol, 2018. 29(8): p. 2060–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen L, et al. , Transcriptomes of major renal collecting duct cell types in mouse identified by single-cell RNA-seq. Proc Natl Acad Sci U S A, 2017. 114(46): p. E9989–E9998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Der E, et al. , Single cell RNA sequencing to dissect the molecular heterogeneity in lupus nephritis. JCI Insight, 2017. 2(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Young MD, et al. , Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science, 2018. 361(6402): p. 594–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu H, et al. , Comparative Analysis and Refinement of Human PSC-Derived Kidney Organoid Differentiation with Single-Cell Transcriptomics. Cell Stem Cell, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brunskill EW, et al. , Single cell dissection of early kidney development: multilineage priming. Development, 2014. 141(15): p. 3093–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Menon R, et al. , Single-cell analysis of progenitor cell dynamics and lineage specification in the human fetal kidney. Development, 2018. 145(16). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lindstrom NO, et al. , Progressive Recruitment of Mesenchymal Progenitors Reveals a Time-Dependent Process of Cell Fate Acquisition in Mouse and Human Nephrogenesis. Dev Cell, 2018. 45(5): p. 651–660 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lindstrom NO, et al. , Conserved and Divergent Features of Mesenchymal Progenitor Cell Types within the Cortical Nephrogenic Niche of the Human and Mouse Kidney. J Am Soc Nephrol, 2018. 29(3): p. 806–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang P, et al. , Dissecting the Global Dynamic Molecular Profiles of Human Fetal Kidney Development by Single-Cell RNA Sequencing. Cell Rep, 2018. 24(13): p. 3554–3567 e3. [DOI] [PubMed] [Google Scholar]
- 53.Czerniecki SM, et al. , High-Throughput Screening Enhances Kidney Organoid Differentiation from Human Pluripotent Stem Cells and Enables Automated Multidimensional Phenotyping. Cell Stem Cell, 2018. 22(6): p. 929–940 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gluck C, Ko YA, and Susztak K, Precision Medicine Approaches to Diabetic Kidney Disease: Tissue as an Issue. Curr Diab Rep, 2017. 17(5): p. 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Malaga-Dieguez L and Susztak K, ADCK4 "reenergizes" nephrotic syndrome. J Clin Invest, 2013. 123(12): p. 4996–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pattaro C, et al. , Genetic associations at 53 loci highlight cell types and biological pathways relevant for kidney function. Nat Commun, 2016. 7: p. 10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qiu C, et al. , Renal compartment-specific genetic variation analyses identify new pathways in chronic kidney disease. Nat Med, 2018. 24(11): p. 1721–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu H, et al. , Single-Cell Transcriptomics of a Human Kidney Allograft Biopsy Specimen Defines a Diverse Inflammatory Response. J Am Soc Nephrol, 2018. 29(8): p. 2069–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stahl PL, et al. , Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science, 2016. 353(6294): p. 78–82. [DOI] [PubMed] [Google Scholar]