Abstract
Duchenne Muscular Dystrophy (DMD) is a fatal X-linked genetic disorder. In DMD, the absence of the dystrophin protein causes decreased sarcolemmal integrity resulting in progressive replacement of muscle with fibrofatty tissue. The effects of lacking dystrophin on muscle and systemic metabolism are still unclear. Therefore, to determine the impact of the absence of dystrophin on metabolism, we investigated the metabolic and lipid profile at two different, well-defined stages of muscle damage and stabilization in mdx mice. We measured NMR-detectable metabolite and lipid profiles in the serum and muscles of mdx mice at 6 and 24 weeks of age. Metabolites were determined in muscle in vivo using 1H MRI/MRS, in isolated muscles using 1H-HR-MAS NMR, and in serum using high resolution 1H/13C NMR. Dystrophic mice were found to have a unique lipid saturation profile compared with control mice, revealing an age-related metabolic change. In the 6-week-old mdx mice, serum lipids were increased and the degree of lipid saturation changed between 6 and 24 weeks. The serum taurine-creatine ratio increased over the life span of mdx, but not in control mice. Furthermore, the saturation index of lipids increased in the serum but decreased in the tissue over time. Finally, we demonstrated associations between MRI-T2, a strong indicator of inflammation/edema, with tissue and serum lipid profiles. These results indicate the complex temporal changes of metabolites in the tissue and serum during repetitive bouts of muscle damage and regeneration that occur in dystrophic muscle.
Keywords: Duchenne Muscular Dystrophy (DMD), lipids, metabolism, metabolomics, muscle, neuromuscular disease
Graphical Abstract
This is the first study to investigate metabolite and lipid profiles at two different stages of disease progression in mdx mice using NMR of serum, HR-MAS NMR in tissues, and in vivo MRI T2. Lipid saturation and metabolite alterations suggest a complex, temporal interplay between metabolism in the tissue and serum at different stages of disease in mdx mice.
INTRODUCTION
Duchenne muscular dystrophy (DMD) is an X-linked recessive disorder with a prevalence of 1 in 5000 live male births1. DMD is caused by mutations in the dystrophin gene, which leads to progressive muscle deterioration, loss of functional abilities, and eventually death in the third decade of life2. Currently there is no cure for DMD, but there have been many promising clinical trials in the past five years3–5.
It is clear that the loss of dystrophin has both structural and metabolic consequences. Dystrophin acts to transmit lateral forces and helps to stabilize the muscle membrane through muscle contraction6–8. Without dystrophin to stabilize the sarcolemma, eccentric contractions lead to membrane damage initiating chronic bouts of muscle repair and damage6. This cycle continues until the healthy muscle is replaced by noncontractile tissue such as lipids and collagen/fibronectin and the cell environment is no longer conducive for muscle regeneration9. Additionally, dystrophin also acts to anchor neuronal nitric oxide synthase (nNOS) to the subsarcolemmal space, where localization is important for vasodilation1 and metabolism2.
Many studies have investigated the changes of metabolism secondary to the loss of dystrophin10–23. When dystrophin is not functional, there are significant alterations in nitric oxide synthesis10, energy metabolism11–16, and lipid synthesis17–21. Interestingly, investigations of serum17,18 and muscle biopsies19–21 have shown alterations in lipid metabolism in both DMD patients and in dystrophic mice, but, to date, there is no clear understanding of the mechanism by which lipids infiltrate the muscle and what corresponding metabolic changes occur systemically. It has been suggested that the muscle lipid infiltration is a consequence of an accumulation of lipids necessary for required membrane repair24. Milad et al found that a high-fat diet in mdx-ApoE dramatically worsened fat infiltration and replacement of muscle. One hypothesis was that the circulating molecules are more readily able to diffuse into the muscle cells through the semi-permeable membrane of dystrophin-deficient muscle25. Furthermore, they suggest that circulating lipids are potentially significant contributors to the mechanism and progression of DMD pathology. They reason that murine models might exhibit mild phenotypes due to the low plasma lipid levels generally seen in mice. Others have argued that lipid accumulation occurs through mitochondrial dysfunction, which causes the cell to lose its ability to maintain lipid vacuoles in the membrane, and they diffuse into the intercellular space26. Although mdx mice accumulate less fat than patients with DMD 27,28, using tissue NMR, we found that there are still significant changes in lipid composition in the muscles of mdx mice12. To our knowledge, the relationship between serum and muscle lipids in the mdx mouse at different stages of disease has not been investigated. Therefore, it is important to obtain a comprehensive set of measurements to improve our understanding of the metabolite equilibrium that exists between tissue and serum in control and dystrophin-deficient muscle at different stages of disease. In addition, it is of great importance to characterize disease-related changes that can be quantified and used to track disease progression through an affordable, easy to acquire blood test.
The mdx mouse model, which lacks functional dystrophin, is the most commonly studied mouse model in DMD; however, the phenotype is less severe than in humans lacking dystrophin, in part to the upregulation of utrophin in mice29 and the tremendous regenerative capacity of mice. Despite this, mdx mice demonstrate well-characterized stages, including an initial stage characterized by muscle damage and inflammation at four to six weeks. This is followed by a “recovery” phase in which regeneration appears to match degeneration from approximately 24 to 65 weeks. During the later stages of life, lipid infiltration, fibrosis, and necrosis become more predominant30.
The objective of this study was to investigate changes in muscle and serum metabolites and lipids from mdx mice at both the initial muscle damage phase (6 weeks) and during the recovery phase (24 weeks of age). We discovered abnormalities in lipid and glucose metabolism of the mdx mouse which may prove useful for mechanistic insights for translational investigations in DMD.
EXPERIMENTAL
Animals
C57BL/10ScSn-DMD mdx mice (mdx) and age-matched C57BL/10ScSn (controls) were purchased from Jackson Laboratories (Bar Harbor, ME, USA). The University of Florida’s institutional animal care and use committee (IACUC) approved the experimental protocol for this study. All animals were maintained with a 12-hour light-dark cycle, at 22.2 °C and 42% humidity at the University of Florida Animal Facilities, which are approved by the Accreditation of Laboratory Animal Care.
Study design
Both mdx and control (C57BL10) mice were obtained at 4 weeks of age and raised to either 6 [mdx (n = 6), control (n = 5)] or 24 weeks old [mdx (n = 10), control (n = 8)]. At each time point (6 or 24 weeks), in vivo mouse lower leg hindlimb data were collected using T2-magnetic resonance imaging (MRI), followed by blood and tissue collection when the animals were sacrificed. At sacrifice, while administering 2% isoflurane for anesthesia, 200 μL of blood were collected by submandibular puncture from randomized animals to reduce any potential order effect and bias. Blood was allowed to sit at room temperature for 30 minutes to clot, then was centrifuged to extract serum. The gastrocnemius muscles were extracted, flash-frozen and stored at –80 °C until further analysis. The mice were then euthanized as per approved IACUC protocols. All experiments were performed in the morning (from 8–10 AM; food was removed from cages one hour before euthanizing).
In vivo imaging and spectroscopy
Magnetic resonance imaging and spectroscopy (MRS) were performed on an 11.1-T horizontal bore magnet (Magnex) equipped with an Agilent spectrometer. Mice were anesthetized with 3% isoflurane mixed with oxygen followed by maintenance at 0.5–1% isoflurane throughout the scanning. Temperature (maintained with a circulated warm water heating pad) and respiration were monitored throughout the entire scanning period using a fiber optic small animal physiological monitoring device (SAI; Stoney Brook). The lower hindlimbs of the mdx (6-week, n = 6; 24-week, n = 10) and control (6-week, n = 5; 24-week, n = 8) mice were inserted into a 2.0-cm internal-diameter, custom-built 1H solenoid coil, up to the knee. For T2-weighted MR images, multiple slice, single spin-echo images were acquired with a repetition time (TR) of 2000 ms, echo times (TE) of 14 ms and 40 ms, a field of view of 10–20 mm2, slice thickness of 0.5–1 mm, and with an acquisition matrix of 128 × 256 for two signal averages. The T2 decay was fit to a single-exponential curve. A 1 × 2 × 2 mm3 voxel was positioned for spectroscopy in the gastrocnemius located in the posterior compartment of the lower leg, where care was taken to avoid other muscles. These voxels were placed as precisely as possible between mice. Spectroscopy was performed using a localization by adiabatic selective refocusing (LASER)31 pulse sequence with a TE of 14 ms, TR of two seconds, 1024 spectral data points, a spectral width (SW) of 15 ppm, and using the WET32 water suppression sequence.
Solution NMR of serum
Serum was separated from the red blood cells by allowing samples to sit at room temperature for 30 minutes before centrifugation for 15 minutes at 10 000 g at 4 °C. Samples were then transferred to an Eppendorf tube, flash-frozen in liquid nitrogen and stored at –80 °C. At the time of analysis, samples were thawed and approximately 100 μL serum was lyophilized and re-suspended in 40 μL 99% D2O (Cambridge Isotope Laboratories, Tewksbury, MA, USA) and transferred into 1.5 mm NMR tubes (Norell). One-dimensional 1H and 13C spectra were collected at a controlled temperature of 25° C on an Agilent VNMRS-600 spectrometer using a custom 1.5 mm 13C high temperature superconducting (HTS) probe33.
One-dimensional 1H data were acquired in approximately 18 minutes using a Carr-Purcell-Meiboom-Gill (CPMG) sequence, which was utilized to remove the macromolecule and lipoprotein signal contribution due to T2 decay, resulting in a flat baseline34,35. Data were recorded with the following parameters: SW of 9615.4 Hz, a 2.0 s relax delay, a 90-degree initial pulse, a train of 124 × 180-degree CPMG pulses with a 1 ms inter-pulse delay, and a 2.0 s acquisition time and 128 scans.
1D 13C spectra were collected in approximately two hours under conditions that favor nuclei with short T1 relaxation times in order to maximize overall sensitivity and minimize measurement time, as described previously36. Protein contribution was found to affect the baseline in 13C spectra and therefore a 13C CPMG sequence with a 90-degree excitation pulse followed by a train of 18 individual 180-degree pulses with a 1 ms inter-pulse delay was used. The 13C CPMG sequence used a 60° excitation pulse with a very short 0.1 s relaxation delay (D1) at the start of each experiment, a 0.8 s acquisition time, a 212 ppm (32 051.3 Hz) spectral window, and a carrier frequency of 98.0 ppm at a frequency of 150.79 MHz. 1H was decoupled and allowed to build up an NOE at 599.68 MHz with a power of 37 dB using a continuous WALTZ-16 sequence.
HR-MAS NMR on ex vivo gastrocnemius muscle
The gastrocnemius muscle of the control and mdx mice at 6 (mdx = 6, control = 5) and 24 weeks (mdx = 10, control = 8) was analyzed using high-resolution magic-angle spinning (HR-MAS) NMR. Data were acquired using a 4 mm HR-MAS probe on a Bruker 600 MHz spectrometer (AVIII) running Topspin 3.2 software. Sample preparation and data acquisition followed the protocols from Beckonert et al37. In addition, the wet weight of the muscle was measured and used for normalization. The magic angle was optimized and set for all samples using potassium bromide (KBr). Pulse calibration, tune, match and manual shimming were performed for each muscle sample. One-dimensional NOESY with water presaturation was acquired with a total of 128 scans, a SW of 12 ppm, 16 000 complex data points, a relaxation delay of two seconds and a NOESY mixing time of 90 ms. All samples were acquired at 4 °C and in under 30 minutes as per protocol.
Data processing
In vivo imaging and spectroscopy was used to measure the muscle T2 and muscle metabolites, respectively, in the gastrocnemius. An average of six slices of in vivo MRI images were used to measure T2 values using echo times of 14 and 40 ms 38. In vivo spectra were processed and analyzed using Mnova NMR software [MestReNova (Mnova) NMR, version 11.0.3, Mestrelab Research S. L., Santiago de Compostela, Spain; www.mestrelab.com, 2017]. Spectra were apodized, zero-filled twice, Fourier-transformed, phased, baseline-corrected, referenced using the in vivo creatine peak at 3.02 ppm, aligned and normalized to water. Each metabolite was quantified by integration of resolved peaks.
HR-MAS and solution NMR spectra were processed using NMRPipe39. Spectra were apodized (cosine squared for 1H and exponential with 2 Hz line broadening for 13C), zero-filled twice, Fourier-transformed, phased, and baseline-corrected. 1H spectra were referenced to the lactate peak at 4.1 ppm. 13C spectra were referenced to the anomeric carbon glucose peak at 98.64 ppm. Further processing and analysis of NMR spectra were performed using an in-house MATLAB metabolomics toolbox40. Spectra were referenced, aligned using the peak alignment fast Fourier transform (PAFFT) algorithm41 and normalized using Probabilistic Quotient Normalization (PQN)42. PQN is a method used to normalize spectra by finding the most probable dilution factor using the distribution of quotients of amplitudes of each test spectrum compared with a reference spectrum (average spectrum). Metabolites were quantified using integration of resolved 1H resonances with chemical shift ranges defined in Table S1 (tissue) and Table S2 (serum). The polyunsaturation index (pi) and unsaturation index (ui) were calculated according to Mosconi et al43 using the following equations:
| (1) |
| (2) |
where Lipid A (CH3)is the peak at 0.9 ppm, Lipid B (HC=CH-CH2-HC=CH) is the peak at 2.75 ppm, and Lipid C (HC=CH) is the peak at 5.4 ppm.
Data analysis
Multivariate analyses were conducted using nonlinear iterative partial least squares (NIPALS) principle component analysis (PCA)44 and partial least squares discriminant analysis (PLS-DA)45 using our in-house MATLAB metabolomics toolbox40. Targeted comparisons were analyzed using Graphpad Prism version 6. Quantified metabolites were tested for normality using the Shapiro-Wilk test46. Comparisons between controls and mdx at both ages determined to be normal were analyzed using a one-way ANOVA and a Bonferonni correction. Comparisons between mdx and controls at both ages that were not determined as normal using the Shapiro-Wilk normality test were compared using a Mann-Whitney test that was corrected for multiple comparisons. Correlations of metabolite and MR parameters were determined using a linear correlation matrix, in MATLAB (corrcoef), which calculated both the correlation coefficients and P-values and used a false discovery rate (FDR) of 0.05 to correct for the multiple comparisons.
RESULTS
In vivo MR characterization of muscle
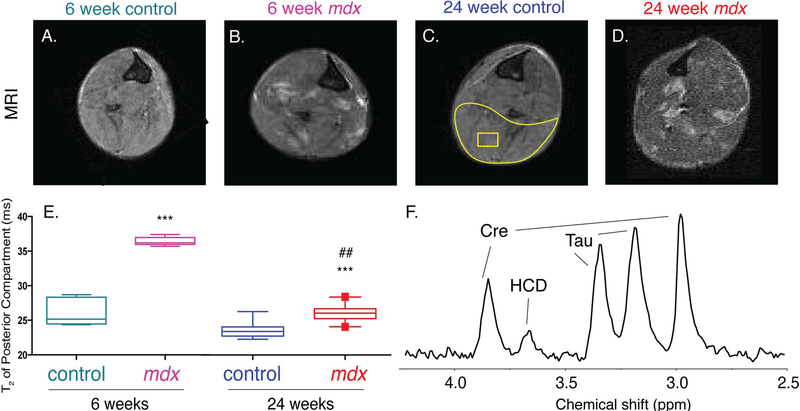
We used MRI T2–weighted imaging to visualize and to quantify the state of disease in vivo in mdx mice at 6 and 24 weeks of age. Figure 1 shows representative MR images of hind limbs from 6-week control (A; n = 5) and mdx (B; n = 6) mice, and 24-week control (C; n = 8) and mdx (D; n = 10) mice. Control mice demonstrated homogenous signal throughout the muscle, indicating healthy muscle tissue. The mdx mice showed regions of hyperintense signal, symptomatic of muscle damage and/or inflammation. The MRI T2 was calculated from the posterior compartment (outlined in yellow) of the hind limb of each mouse to quantify the amount of damage/inflammation (Figure 1E). We found that the posterior hindlimb muscle T2 of 6-week-old mdx mice were significantly (P = 0.0003) higher (37.4 ± 1.5 ms) than that of 6-week-old controls (27.3 ± 1.8 ms), 24-week-old controls (23.8 ± 0.3 ms) and 24-week-old mdx (26 ± 1.3 ms) (Figure 1E). The muscle T2 from the 24-week-old mdx mice was significantly (P = 0.01) higher than that of 24-week-old control mice. There was no significant difference between 6- and 24-week controls (Figure 1E). Figure 1F shows a representative localized 1H spectrum from a voxel (represented as a yellow square) in the posterior compartment of the hind limb, from which creatine (Cr), taurine (Tau) and a histidine-containing derivative (HCD) were quantified for all mice. An overlay of all spectra is included as Figure S1. There were no significant changes between groups in Cr, Tau or HCD. Consistent with previous reports in mdx mice47,48, the in vivo muscle lipid content was low and below the level necessary to reliably calculate an in vivo pi and ui.
FIGURE 1.
Characterization of disease state with MRI T2. MRI images of the hind limb (A)-(D) of 6-week control, 6-week mdx, 24-week control and 24-week mdx, respectively. MRI T2 was taken from an area of interest (yellow area circling muscle) in the posterior compartment of the hind limb (E). A voxel (yellow square) was placed in the posterior compartment of the lower hind limb for quantification of in vivo metabolites using MRS (F). Significance was determined by corrected P ≤ 0.05 or lower. Comparisons to 6-week controls with P < 0.001 are denoted with ***. Comparisons to 6-week mdx with P < 0.01 are denoted with ##. Abbreviations: Cre, creatine; Tau, taurine peaks; HCD, histidine-containing derivatives
Multivariate global analysis of control and dystrophic mice using 13C spectra
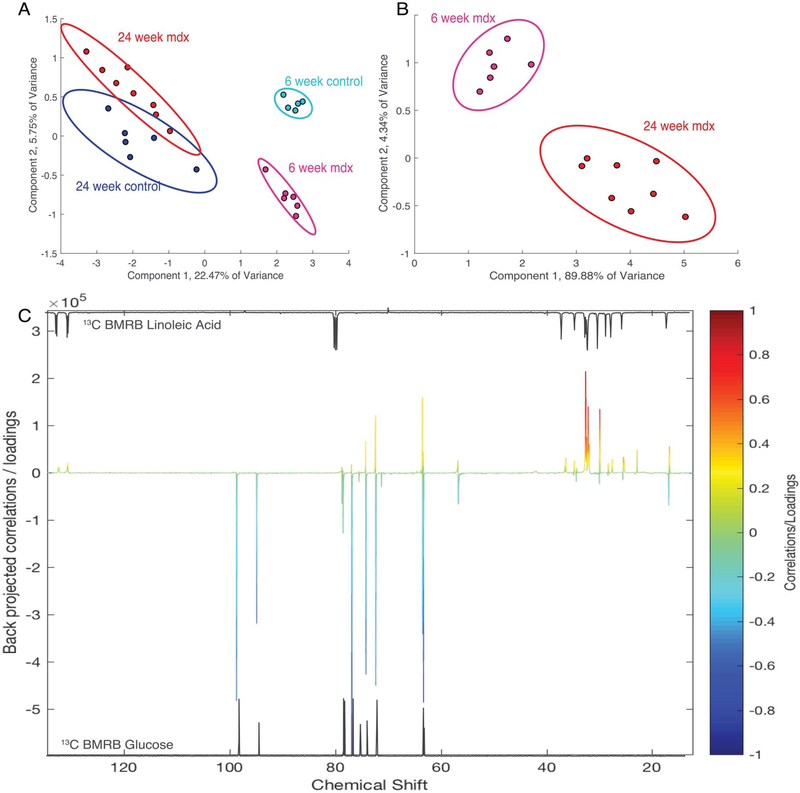
13C spectra of control and dystrophic serum at both 6 and 24 weeks were used to perform PCA to identify the largest variation between the cohorts. Figure 2A shows a PCA plot where there is a strong separation of mdx from controls at 6 weeks but less so at 24 weeks. PCA of the 13C spectra of the mdx mouse serum (Figure 2B) separated 6-week mdx from the 24-week mdx serum spectra using PCA (verified with PLS-DA with a Q2 = 0.82, R2 = 0.98). The loadings plot for principle component (PC) 2 reveals that peaks related to linoleic acid (fatty acids) showed a greater association with 6-week mdx and glucose was more associated with the 24-week-old mice (Figure 2C). To guide the eye, the BioMagResBank49 reference spectra of linoleic acid and glucose are displayed above and below the loadings spectrum, respectively. Peaks for both linoleic acid and glucose were also verified using 2D heteronuclear single quantum correlation (HSQC) spectroscopy (Figure S2).
FIGURE 2.
PCA of serum 13C NMR spectra can differentiate control and mdx at different ages. (A) PCA plot of all groups; (B) PCA of mdx at 6 weeks and 24 weeks of age demonstrate the separation on the second component. In (C), the loadings plot of PC 2 showing the 6-week (positive) separate based on lipids and the 24-week (negative) separate based on glucose. (D) and (E) are BMRB standard spectra that reveal the similar peaks to the compounds separated out in the loadings plot.
Quantification of serum glucose and lipids
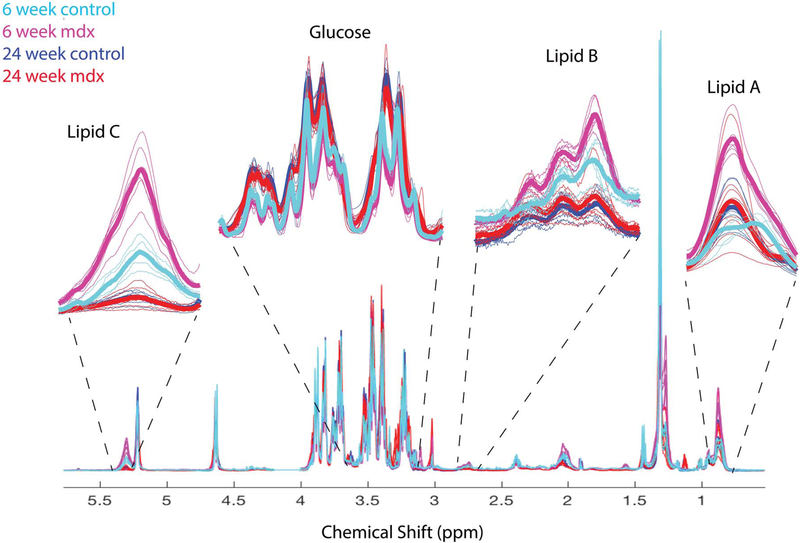
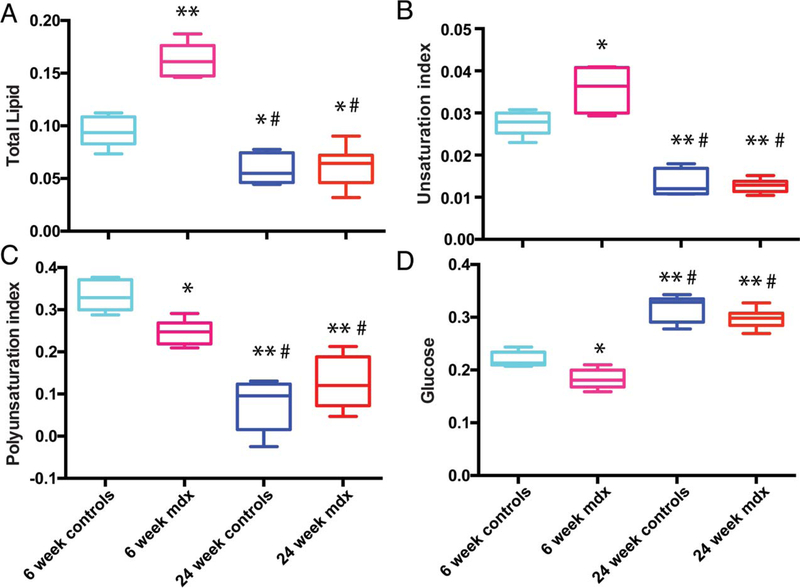
Figure 3 shows the overlay of all 1H NMR serum spectra from dystrophic and control mice at both 6 and 24 weeks of age with inset expansions of lipid and glucose peaks. Integration of A (0.9 ppm), B (2.75 ppm) and C (5.4 ppm) lipid peaks were used to calculate the pi (Equation 1) and ui (Equation 2) in the lipids43. Total lipids (Figure 4A) and ui (Figure 4B) were significantly (P < 0.0001, P < 0.01, respectively) higher in the 6-week-old mdx mice compared with age-matched controls, but this was not seen at 24 weeks. The pi in the 6-week-old mdx mice were lower (P < 0.01) than that of the 6-week-old controls, but still higher than both the 24-week-old mdx and controls (P < 0.0001 for both) (Figure 4C). Glucose levels in the 6-week-old mdx mice were significantly (P < 0.01) lower than 6-week-old controls (Figure 4D). Conversely, 24-week-old control and mdx mice had significantly higher glucose than controls and mdx at 6 weeks (Figure 4D).
FIGURE 3.
Serum 1H spectra indicating peaks of interest. Spectra from control (teal and blue) and mdx (pink and red) mice at 6 and 24 weeks. Large changes found in the 1H are magnified to demonstrate the differences. Thin lines are from individual mice, and the bold lines are the mean for each group.
FIGURE 4.
Quantification of serum metabolites. Relative abundance quantification from the 1H spectra of lipids and glucose in the serum of control and mdx mice at both 6 and 24 weeks of age. Significance was determined by P = 0.05. Comparisons with 6-week controls with P < 0.01 are denoted with *. Comparisons with 6-week controls with P < 0.001 are denoted with **. Comparisons with 6-week mdx with P < 0.001 are denoted with #. Comparisons with 24-week controls with P < 0.01 are denoted with †
Tissue metabolite comparison at different ages
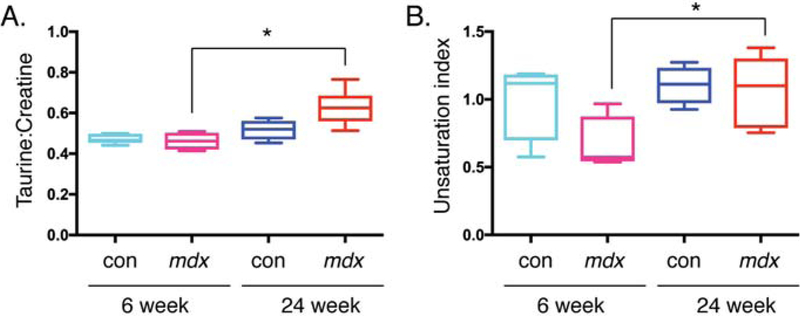
The metabolic profile of gastrocnemius muscle from both 6- and 24-week-old control and mdx mice was determined using 1H HR-MAS NMR. Table S1 summarizes the metabolites, the ppm range used to integrate each metabolite and normalized quantifications (with standard deviations) of both the mdx and controls at each age. The taurine-creatine ratio (Figure 5A, P = 0.009), and ui (Equation 2) (Figure 5B, P = 0.035) in the lipids significantly changed between the two ages in mdx mice only, but remained unchanged within controls at different ages. This suggests that these metabolic changes are related to the state of the disease in the mdx and not merely an effect of age.
FIGURE 5.
Tissue metabolites that change in mdx at different ages. Quantification from the 1H HR-MAS spectra of tissue from control and mdx mice at both 6 and 24 weeks of age. Significance was determined by P = 0.05. Comparisons with 6-week controls with P < 0.05 are denoted with *
Correlations of metabolites between serum and tissue
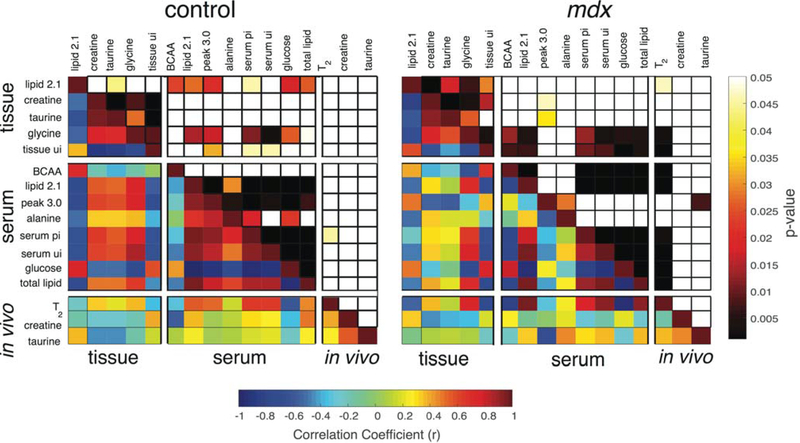
A correlation matrix of pairwise comparisons was made between five tissue metabolites, eight serum metabolite/metabolite ratios, two in vivo metabolites and in vivo T2 in both controls (left) and mdx (right) (Figure 6). The control matrix showed zones of strong and significant correlations (r > 0.50, P < 0.05) within the serum metabolite group and within tissue metabolites separately. In the tissue, the lipid peak at 2.1 ppm (HC=CH-CH2) and glycine were the only two metabolites that demonstrate a relationship (r > 0.6, P < 0.05) with serum metabolites.
FIGURE 6.
Correlations between serum and tissue measurements. The lower left of each correlation matrix are the correlation coefficients for each comparison which are labeled on the left and at the top. The upper-right portion of each correlation matrix are the P-values for the paired comparison. The scale bar at the bottom of the figure corresponds to the correlations and the one on the right represents the corresponding P-values. Each metabolite is grouped into tissue, serum or in vivo, denoted on the left-hand side
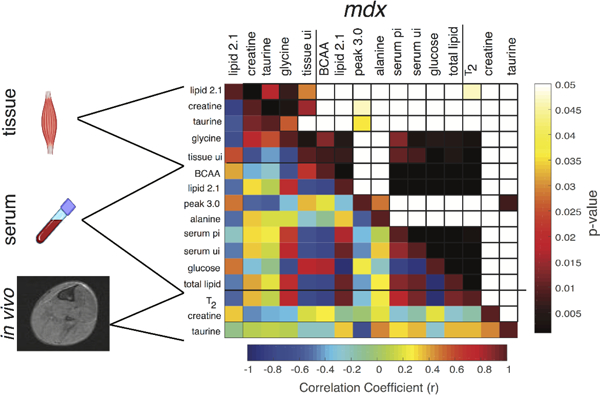
Similarly, in mdx mice there was also a strong (r > 0.6) and significant (P < 0.001) positive relationship between glycine and serum metabolites but this was not seen for the lipid peak at 2.1 ppm. Compared with controls, the mdx showed no relationship between the peak at 3.0 ppm (most likely an amino acid derivative but this could not be definitively identified by 2D NMR36) and alanine with the rest of the serum metabolites. Conversely, there were stronger relationships (r > 0.6; P < 0.01) between the serum branched chain amino acids (BCAA) and glycine with serum lipids, than was observed in controls. The mdx mice showed a strong negative correlation (r > 0.6, P < 0.005) between the serum peak at 3.0 ppm and the in vivo taurine, whereas no such relationship was seen in controls. However, the most striking contrast was that mdx in vivo T2 strongly correlated (r > 0.5; P < 0.001) to both tissue and serum lipid measurements, but almost no relationship was observed between control T2 and lipid measurements. Since MRI T2 has proven to be a reliable marker for inflammation/damage in mdx mice 38, these data suggest serum and tissue lipid saturation alterations have a relationship to MRI T2 and, potentially, disease progression or state.
DISCUSSION
In this study, tissue and serum NMR were used to investigate the metabolite and lipid profile in mdx mice during both the initial muscle damage phase (6 weeks) and during the recovery phase (24 weeks of age). We found strong associations between the lipid profiles in serum with both tissue metabolites and MRI T2. MRI T2 can be elevated by increased inflammation and lipid composition, both of which are present in dystrophic mice38 and individuals with DMD50–55. As mdx mice age, upregulation of utrophin partially compensates for the loss of dystrophin and ameliorates disease phenotypes56,57, shown previously by both histology and MRI38. Using MRI T2, ex vivo tissue HR-MAS and solution NMR of the serum, we are able to show that systemic lipid changes of mdx mice correlate with state of disease.
Correlations between serum metabolites, tissue metabolites and in vivo measurements were paramount in determining the systematic alterations occurring in lipids throughout disease. Measured lipids between both the tissue and serum had strong and significant correlation to in vivo MRI T2. However, it is important to point out that the tissue ui is negatively correlated with MRI T2, whereas serum lipids (pi, ui and total lipids) all positively correlate with MRI T2. Altered lipid saturation in the mdx mice returned to control levels by 24 weeks, the same time that utrophin is upregulated and disease symptoms are ameliorated. The metabolic profile we observed suggests that the initial bout of damage includes elevated levels of lipids in the serum as damage and repair are occurring within the muscle. As lipids in the serum reduce by 24 weeks, the increase of lipid in the tissue may be coming from the overproduction of lipids to repair cell membrane after damage, as previously suggested24. Infiltration of lipids from the circulating blood has previously been suggested by Milad et al25 as a means to infiltrate leaky muscles. However, the exact mechanism by which fatty tissue infiltrates and replaces muscle tissue is still unclear and needs to be further investigated.
We found that the amount of total lipid increased during the initial wave of muscle damage in 6-week-old mice. However, not all types of lipids increased equally. Others have shown that lipid composition in the muscle and blood of patients with DMD is altered12,17–20,58. Following repeated bouts of muscle damage, the contractile tissue of the muscle is replaced by fibrofatty tissue23,59,60. We found that the ui increased but the pi decreased in 6-week-old mdx mice compared with controls. These findings are consistent with Gillet et al19, who found similar changes in lipid saturation between 3-week-old mdx and control mice using 2D NMR. Linoleic acid was higher in controls than in mdx mice. In DMD muscle, it has been recently shown by Srivastava et al23 that linoleic acids are significantly reduced in DMD muscle compared with normal/control individuals. Alterations in polyunsaturated fatty acids in erythrocytes from DMD patients61 have also been reported. These data demonstrate that in addition to changes in total muscle lipids, there are also systemic lipid changes in people with DMD. Changes in muscle and membrane lipid composition during muscle damage and recovery have been documented both in vivo19,48 and in vitro19. For example, Gillet et al19 observed that changes in linoleic acid track with membrane fusion and skeletal muscle differentiation and recovery19,24. Differences in lipid composition have also been observed in areas of muscle damage in mdx muscle using an imaging mass spectrometry technique62.
Our observation that tissue taurine levels were associated with lipid composition and MRI T2 is consistent with other reports of changes in taurine during muscle regeneration63 and renormalization following steroid treatment65. Changes in choline and taurine levels following acute muscle damage in mdx mice have been observed in vivo using in vivo spectroscopy and validated by histology48. In a comparison of injured and uninjured muscles from mdx mice, taurine decreased by 17% and choline increased by 25% in injured muscles. In wildtype mice, intramyocellular lipids and total muscle lipid levels increase significantly with injury48. Taurine has consistently been shown to be altered in dystrophic muscles11–13,63,64. Overall, the altered taurine-creatine levels in mdx vs control muscle suggests that taurine could be a useful biomarker for dystrophic muscle damage11–13,63,64.
We also observed a negative correlation between serum glucose and MRI T2 and serum lipids. Glucose levels were lower during the period of severe muscle damage in mdx compared with controls but were not different at 24 weeks of age. Previous studies in the literature66–68 have shown alterations in blood glucose and insulin resistance in preclinical models of muscular dystrophy and in DMD patients. However, it should be noted that our mice were not fasted for 12 hours prior to sacrifice and therefore this could interfere with interpretation of the data. In addition, we did not examine mice at ages older than 24 weeks and it is possible that blood glucose levels may differ at those ages. Future studies examining mdx mice at multiple time points postfasting at different ages could provide further mechanistic insight into glucose metabolism in dystrophic mice.
In conclusion, we found a complex, temporal interplay between metabolism in the tissue and serum during periods of elevated muscle damage/inflammation and recovery in dystrophic muscle. Specifically, metabolic profile changes and systemic lipid saturation in mdx mice were altered at different ages vs controls. Correlations between serum lipids to tissue lipids and in vivo MRI T2 suggest that lipid saturation in serum is a potential means to monitor disease state in dystrophic muscle, and this is obtainable in an uncomplicated, minimally invasive, and translational manner.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by UF Wellstone Muscular Dystrophy Cooperative Center (U54 AR052646). Support for student training include the T32 Neuromuscular Training Program (HD043730) and the TL1 Clinical and Translational Training Program (TR000066). Data were collected by the Southeast Center for Integrated Metabolomics (NIH/NIDDK 1U24DK097209-01A1) using the National High Magnetic Field Laboratory’s AMRIS Facility, which is supported by National Science Foundation Cooperative Agreement No. DMR-1157490 and the State of Florida. ASE is supported by the Georgia Research Alliance. This work was supported in part by an NIH award, S10RR025671 & S10RR031637 for MRI/S instrumentation.
Abbreviations used:
- BCAA
branched chain amino acids
- COSY
correlation spectroscopy
- CPMG
Carr-Purcell-Meiboom-Gill
- DMD
Duchenne Muscular Dystrophy
- TE
echo time
- GRMD
golden retriever muscular dystrophy model
- HCD
histidine-containing derivative
- HR-MAS
high resolution magic angle spinning
- HSQC
heteronuclear single quantum correlation
- HTS
high temperature superconducting
- mdx
DMD mouse model
- MRI
magnetic resonance imaging
- MRS
magnetic resonance spectroscopy
- NIPALS
nonlinear iterative partial least squares
- nNOS
neuronal nitric oxide synthase
- PLS-DA
partial least squares discriminant analysis
- PAFFT
peak alignment fast Fourier transform
- pi
polyunsaturation index
- PCA
principle component analysis
- PQN
probabilistic quotient normalization
- SW
spectral width
- TR
repetition time
- ui
unsaturation index
REFERENCES
- 1.Mendell JR, Lloyd-Puryear M. Report of MDA muscle disease symposium on newborn screening for Duchenne muscular dystrophy. Muscle Nerve 2013;48:21–26. [DOI] [PubMed] [Google Scholar]
- 2.Bushby K, Finkel R, Birnkrant DJ, et al. Diagnosis and management of Duchenne muscular dystrophy, part 2: implementation of multidisciplinary care. Lancet Neurol 2010;9:177–189. [DOI] [PubMed] [Google Scholar]
- 3.Bushby K, Finkel R, Wong B, et al. Ataluren treatment of patients with nonsense mutation dystrophinopathy. Muscle Nerve 2014;50:477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fairclough RJ, Wood MJ, Davies KE. Therapy for Duchenne muscular dystrophy: renewed optimism from genetic approaches. Nat Rev Genet 2013;14:373–378. [DOI] [PubMed] [Google Scholar]
- 5.Konieczny P, Swiderski K, Chamberlain JS. Gene and cell-mediated therapies for muscular dystrophy. Muscle Nerve 2013;47:649–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ervasti JM, Campbell KP. A role for the dystrophin-glycoprotein complex as a transmembrane linker between laminin and actin. J Cell Biol 1993;122:809–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ervasti JM, Ervasti JM, Ohlendieck K, et al. Deficiency of a glycoprotein component of the dystrophin complex in dystrophic muscle. Nature 1990;345:315–319. [DOI] [PubMed] [Google Scholar]
- 8.Ervasti JM, Campbell KP. Membrane organization of the dystrophin-glycoprotein complex. Cell 1991;66:1121–1131. [DOI] [PubMed] [Google Scholar]
- 9.Boldrin L, Zammit PS, Morgan JE. Satellite cells from dystrophic muscle retain regenerative capacity. Stem Cell Res 2015;14:20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tidball JG, Wehling-Henricks M. Nitric oxide synthase deficiency and the pathophysiology of muscular dystrophy. J Physiol 2014;592:4627–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffin JL, Rosiers Des C. Applications of metabolomics and proteomics to the mdx mouse model of Duchenne muscular dystrophy: lessons from downstream of the transcriptome. Genome Med 2009;1:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffin JL, Williams HJ, Sang E, Nicholson JK. Abnormal lipid profile of dystrophic cardiac tissue as demonstrated by one- and two-dimensional magic-angle spinning (1)H NMR spectroscopy. Magn Reson Med 2001;46:249–255. [DOI] [PubMed] [Google Scholar]
- 13.Griffin JL, Sang E, Evens T, Davies K, Clarke K. Metabolic profiles of dystrophin and utrophin expression in mouse models of Duchenne muscular dystrophy. FEBS Lett 2002;530:109–116. [DOI] [PubMed] [Google Scholar]
- 14.Kuznetsov AV, Winkler K, Wiedemann FR, Bossanyi von P, Dietzmann K, Kunz WS. Impaired mitochondrial oxidative phosphorylation in skeletal muscle of the dystrophin-deficient mdx mouse. Mol Cell Biochem 1998;183:87–96. [DOI] [PubMed] [Google Scholar]
- 15.Percival JM, Siegel MP, Knowels G, Marcinek DJ. Defects in mitochondrial localization and ATP synthesis in the mdx mouse model of Duchenne muscular dystrophy are not alleviated by PDE5 inhibition. Hum Mol Genet 2012;22:153–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tarnopolsky MA, Mahoney DJ, Vajsar J, et al. Creatine monohydrate enhances strength and body composition in Duchenne muscular dystrophy. Neurology 2004;62:1771–1777. [DOI] [PubMed] [Google Scholar]
- 17.Srivastava NK, Pradhan S, Mittal B, Gowda GAN. High resolution NMR based analysis of serum lipids in Duchenne muscular dystrophy patients and its possible diagnostic significance. NMR Biomed 2010;23:13–22. [DOI] [PubMed] [Google Scholar]
- 18.Srivastava NK, Mukherjee S, Sinha N. Alteration of phospholipids in the blood of patients with Duchenne muscular dystrophy (DMD): in vitro, high resolution 31P NMR-based study. Acta Neurol Belg 2016:116:573–581. [DOI] [PubMed] [Google Scholar]
- 19.Gillet B, Doan BT, Verre-Serrie C, et al. In vivo 2D 1H NMR of mdx mouse muscle and myoblast cells during fusion: Evidence for a characteristic signal of long chain fatty acids. Neuromuscul Disord 1993;3:433–438. [DOI] [PubMed] [Google Scholar]
- 20.Touboul D, Piednoël H, Voisin V, et al. Changes of phospholipid composition within the dystrophic muscle by matrix-assisted laser desorption/ionization mass spectrometry and mass spectrometry imaging. Eur J Mass Spectrom 2004;10:657–659. [DOI] [PubMed] [Google Scholar]
- 21.Sharma U, Atri S, Sharma MC, Sarkar C, Jagannathan NR. Skeletal muscle metabolism in Duchenne muscular dystrophy (DMD): an in-vitro proton NMR spectroscopy study. Magn Reson Imaging 2003;21:145–153. [DOI] [PubMed] [Google Scholar]
- 22.Srivastava NK. Proton Nuclear Magnetic Resonance ((1)H NMR) Spectroscopy-Based Analysis of Lipid Components in Serum/Plasma of Patients with Duchenne Muscular Dystrophy (DMD). Methods Mol Biol 2018;1687:195–204. [DOI] [PubMed] [Google Scholar]
- 23.Srivastava NK, Yadav R, Mukherjee S, Pal L, Sinha N. Abnormal lipid metabolism in skeletal muscle tissue of patients with muscular dystrophy: In vitro, high-resolution NMR spectroscopy based observation in early phase of the disease. Magn Reson Imaging 2017;38:163–173. [DOI] [PubMed] [Google Scholar]
- 24.Sébrié C, Gillet B, Lefaucheur JP, Sebille A, Beloeil JC. Mouse muscle regeneration: an in vivo 2D 1H magnetic resonance spectroscopy (MRS) study. FEBS Lett 1998;423:71–74. [DOI] [PubMed] [Google Scholar]
- 25.Milad N, White Z, Tehrani AY, Sellers S, Rossi FMV, Bernatchez P. Increased plasma lipid levels exacerbate muscle pathology in the mdx mouse model of Duchenne muscular dystrophy. Skeletal Muscle 2017;7:809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Timpani CA, Hayes A, Rybalka E. Revisiting the dystrophin-ATP connection: How half a century of research still implicates mitochondrial dysfunction in Duchenne Muscular Dystrophy aetiology. Med Hypotheses 2015;85:1021–1033. [DOI] [PubMed] [Google Scholar]
- 27.McIntosh LM, Baker RE, Anderson JE. Magnetic resonance imaging of regenerating and dystrophic mouse muscle. Biochem Cell Bio 1998;76:532–541. [DOI] [PubMed] [Google Scholar]
- 28.Carnwath JW, Shotton DM. Muscular dystrophy in the mdx mouse: Histopathology of the soleus and extensor digitorum longus muscles. J Neurol Sci 1987;80:39–54. [DOI] [PubMed] [Google Scholar]
- 29.Martins-Bach AB, Bloise AC, Vainzof M, Rahnamaye Rabbani S, Rabbani SR. Metabolic profile of dystrophic mdx mouse muscles analyzed with in vitro magnetic resonance spectroscopy (MRS). Magn Reson Imaging 2012;30:1167–1176. [DOI] [PubMed] [Google Scholar]
- 30.Pastoret C, Sebille A. mdx mice show progressive weakness and muscle deterioration with age. J Neurol Sci 1995;129:97–105. [DOI] [PubMed] [Google Scholar]
- 31.Slotboom J, Bovée WMMJ. Adiabatic slice-selective rf pulses and a single-shot adiabatic localization pulse sequence. Concepts Magn Reson 1995;7:193–217. [Google Scholar]
- 32.Ogg RJ, Kingsley PB, Taylor JS. WET, a T1- and B1-insensitive water-suppression method for in vivo localized 1H NMR spectroscopy. J Magn Reson B 1994;104:1–10. [DOI] [PubMed] [Google Scholar]
- 33.Ramaswamy V, Hooker JW, Withers RS, Nast RE, Brey WW, Edison AS. Development of a 13C-optimized 1.5-mm high temperature superconducting NMR probe. J Magn Reson 2013;235:58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carr HY, Purcell EM. Effects of Diffusion on Free Precession in Nuclear Magnetic Resonance Experiments. Phys Rev 1954;94:630–638. [Google Scholar]
- 35.Meiboom S, Gill D. Modified Spin-Echo Method for Measuring Nuclear Relaxation Times. Rev Sci Instrum 1958;29:688–691. [Google Scholar]
- 36.Clendinen CS, Lee-McMullen B, Williams CM, et al. 13C NMR Metabolomics: Applications at Natural Abundance. Anal Chem 2014;86:9242–9250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beckonert O, Coen M, Keun HC, et al. High-resolution magic-angle-spinning NMR spectroscopy for metabolic profiling of intact tissues. Nat Protoc 2010;5:1019–1032. [DOI] [PubMed] [Google Scholar]
- 38.Vohra RS, Mathur S, Bryant ND, Forbes SC, Vandenborne K, Walter GA. Age-related T 2changes in hindlimb muscles of mdxmice. Muscle Nerve 2015;53:84–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 1995;6:277–293. [DOI] [PubMed] [Google Scholar]
- 40.Robinette SL, Ajredini R, Rasheed H, et al. Hierarchical Alignment and Full Resolution Pattern Recognition of 2D NMR Spectra: Application to Nematode Chemical Ecology. Anal Chem 2011;83:1649–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong JWH, Durante C, Cartwright HM. Application of Fast Fourier Transform Cross-Correlation for the Alignment of Large Chromatographic and Spectral Datasets. Anal Chem 2005;77:5655–5661. [DOI] [PubMed] [Google Scholar]
- 42.Dieterle F, Ross A, Schlotterbeck G, Senn H. Probabilistic Quotient Normalization as Robust Method to Account for Dilution of Complex Biological Mixtures. Application in 1H NMR Metabonomics. Anal Chem 2006;78:4281–4290. [DOI] [PubMed] [Google Scholar]
- 43.Mosconi E, Mosconi E, Fontanella M, et al. Investigation of adipose tissues in Zucker rats using in vivo and ex vivo magnetic resonance spectroscopy. J Lipid Res 2011;52:330–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wold S, Esbensen K, Geladi P. Principal component analysis. Chemom Intell Lab Syst 1987;2:37–52. [Google Scholar]
- 45.Lindgren F, Geladi P, Wold S. The kernel algorithm for PLS. J Chemometrics 1993;7:45–59. [Google Scholar]
- 46.SHAPIRO SS, WILK MB. An analysis of variance test for normality (complete samples). Biometrika 1965;52:591–611. [Google Scholar]
- 47.Walter G, Cordier L, Bloy D, Lee Sweeney H. Noninvasive monitoring of gene correction in dystrophic muscle. Magn Reson Med 2005;54:1369–1376. [DOI] [PubMed] [Google Scholar]
- 48.Xu S, Pratt SJP, Spangenburg EE, Lovering RM. Early metabolic changes measured by 1H MRS in healthy and dystrophic muscle after injury. J Appl Physiol 2012;113:808–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ulrich EL, Ulrich EL, Akutsu H, et al. BioMagResBank. Nucleic Acids Res 2007;36:D402–D408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Willcocks RJ, Rooney WD, Triplett WT, et al. Multicenter prospective longitudinal study of magnetic resonance biomarkers in a large duchenne muscular dystrophy cohort. Ann Neurol 2016;79:535–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Forbes SC, Willcocks RJ, Triplett WT, et al. Magnetic Resonance Imaging and Spectroscopy Assessment of Lower Extremity Skeletal Muscles in Boys with Duchenne Muscular Dystrophy: A Multicenter Cross Sectional Study. Taneja R, ed. PLoS One 2014;9:e106435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arpan I, Forbes SC, Lott DJ, et al. T2mapping provides multiple approaches for the characterization of muscle involvement in neuromuscular diseases: a cross-sectional study of lower leg muscles in 5–15-year-old boys with Duchenne muscular dystrophy. NMR Biomed 2012;26:320–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnston JH, Kim HK, Merrow AC, et al. Quantitative Skeletal Muscle MRI: Part 1, Derived T2 Fat Map in Differentiation Between Boys With Duchenne Muscular Dystrophy and Healthy Boys. Am J Roentgenol 2015;205:W207–W215. [DOI] [PubMed] [Google Scholar]
- 54.Willcocks RJ, Arpan IA, Forbes SC, et al. Longitudinal measurements of MRI-T2 in boys with Duchenne muscular dystrophy: Effects of age and disease progression. Neuromuscul Disord 2014;24:393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hollingsworth KG, Garrood P, Eagle M, Bushby K, Straub V. Magnetic resonance imaging in duchenne muscular dystrophy: Longitudinal assessment of natural history over 18 months. Muscle Nerve 2013;48:586–588. [DOI] [PubMed] [Google Scholar]
- 56.Gramolini AO, Gramolini AO, Jasmin BJ, Jasmin BJ. Duchenne muscular dystrophy and the neuromuscular junction: The utrophin link. Bioessays 1997;19:747–750. [DOI] [PubMed] [Google Scholar]
- 57.Villalta SA, Rinaldi C, Deng B, Liu G, Fedor B, Tidball JG. Interleukin-10 reduces the pathology of mdx muscular dystrophy by deactivating M1 macrophages and modulating macrophage phenotype. Hum Mol Genet 2011;20:790–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Touboul D Lipid imaging by gold cluster time-of-flight secondary ion mass spectrometry: application to Duchenne muscular dystrophy. J Lipid Res 2005;46:1388–1395. [DOI] [PubMed] [Google Scholar]
- 59.Lott DJ, Forbes SC, Mathur S, et al. Assessment of intramuscular lipid and metabolites of the lower leg using magnetic resonance spectroscopy in boys with Duchenne muscular dystrophy. Neuromuscul Disord 2014;24:574–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tuazon MA, Henderson GC. Fatty acid profile of skeletal muscle phospholipid is altered in mdx mice and is predictive of disease markers. Metab Clin Exp 2012;61:801–811. [DOI] [PubMed] [Google Scholar]
- 61.Piperi C, Papapanagiotou A, Kalofoutis C, et al. Altered long chain fatty acids composition in Duchenne muscular dystrophy erythrocytes. In Vivo 2004;18:799–802. [PubMed] [Google Scholar]
- 62.Benabdellah F, Yu H, Brunelle A, Laprévote O, La Porte De S. MALDI reveals membrane lipid profile reversion in MDX mice. Neurobiol Dis 2009;36:252–258. [DOI] [PubMed] [Google Scholar]
- 63.Griffin JL, Williams HJ, Sang E, Clarke K, Rae C, Nicholson JK. Metabolic Profiling of Genetic Disorders: A Multitissue 1H Nuclear Magnetic Resonance Spectroscopic and Pattern Recognition Study into Dystrophic Tissue. Anal. Biochem 2001;293:16–21. [DOI] [PubMed] [Google Scholar]
- 64.McIntosh LM, Garrett KL, Megeney L, Rudnicki MA, Anderson JE. Regeneration and myogenic cell proliferation correlate with taurine levels in dystrophin- and MyoD-deficient muscles. Anat Rec 1998;252:311–324. [DOI] [PubMed] [Google Scholar]
- 65.McIntosh LM, Garrett KL, Megeney L, Rudnicki MA, Anderson JE. Regeneration and myogenic cell proliferation correlate with taurine levels indystrophin- and MyoD-deficient muscles. Anat Rec 1998;252:311–324. [DOI] [PubMed] [Google Scholar]
- 66.Rodríguez-Cruz M, Sanchez R, Escobar RE, et al. Evidence of insulin resistance and other metabolic alterations in boys with duchenne or becker muscular dystrophy. Int J Endocrinol 2015;2015(4):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Raith M, Valencia RG, Fischer I, et al. Linking cytoarchitecture to metabolism: sarcolemma-associated plectin affects glucose uptake by destabilizing microtubule networks in mdx myofibers. Skeletal Muscle 2013;3(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stapleton DI, Lau X, Flores M, et al. Dysfunctional Muscle and Liver Glycogen Metabolism in mdx Dystrophic Mice. Gaetano C, ed. PLoS ONE 2014;9(3): e91514 10.1371/journal.pone.0091514 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.