Abstract
The development of new advances in the understanding of neutrophil biochemistry requires effective procedures for isolating purified neutrophil populations. Although methods for human neutrophil isolation are now standard, similar procedures for isolating neutrophils from many of the nonhuman species used to model human diseases are not as well developed. Since neutrophils are reactive cells, the method of isolation is extremely important to avoid isolation technique-induced alterations in cell function. We present methods here for reproducibly isolating highly purified neutrophils from large animals (bovine, equine, ovine), small animals (murine and rabbit), and nonhuman primates (cynomolgus macaques), and describe optimized details for obtaining the highest cell purity, yield, and viability. We also describe methods to verify phagocytic capacity in the purified cell populations using a flow cytometry-based phagocytosis assay.
Keywords: Inflammation, Phagocytosis, Large animal model, Granulocyte, Polymorphonuclear leukocyte, Cell isolation, Flow cytometry, Blood, Bone marrow
1. Introduction
Over the years, various animal models have been developed for investigation of the pathogenesis of human inflammation and infectious diseases (reviewed in ref. 1). Although small animal models, such as rodents, are easier to handle, breed easily, require much less in the way of housing facilities, and are generally less expensive, they often do not provide an accurate reflection of human physiology [2]. Thus, nonhuman primates and larger animal models, such as bovine and sheep, are often desirable as models for human disease pathogenesis [2]. In these models, it is important to characterize neutrophil function, which requires efficient methods for purification of these phagocytic cells.
Currently, much of our understanding of neutrophil biology is based on studies using human cells, whereas much less is known regarding the biology of these cells in nonhuman species. It is clear, however, that neutrophils from other species differ from their human counterpart in a number of important functional characteristics [3], and one cannot assume that the basic features of human neutrophil biochemistry and function are representative of neutrophils in all other species. Many species are used as models to understand human pathophysiology; thus, it is essential that we gain a thorough understanding of neutrophil functions in these models. It is also important to characterize neutrophil biology in various nonhuman species in order to determine immune status and host defense mechanisms in these organisms. Additionally, comparison of conserved features between neutrophils from different species can contribute to our understanding of neutrophil biochemistry in general.
One of the major challenges associated with neutrophil studies is the isolation of highly purified cell preparations that are morphologically and functionally similar to cells found in the blood in vivo. Neutrophils are temperamental cells that can be easily altered by improper handling [4]. Thus, the method of cell isolation is extremely important to avoid isolation technique-induced alterations in neutrophil function [4]. For instance, neutrophils can be primed during isolation, resulting in altered neutrophil responses to subsequent stimuli and changes in surface antigen expression [5 – 7]. Furthermore, some neutrophil function, such as chemotaxis, can actually be inhibited by isolation procedures [4]. To completely eliminate any isolation artifacts, methods have been developed for analysis of neutrophils in whole blood (e.g., see refs. 5, 8). Conversely, the use of whole blood can be impractical for many biochemical studies, which require purified cells in the absence of other contaminating cells and serum proteins.
Although effective methods for isolation of human neutrophils have been extensively developed (see Chapter 2 in this volume), these methods do not work in many nonhuman species. We present methods here for neutrophil isolation from a range of large (bovine, equine, nonhuman primate, ovine) and small (murine, rabbit) animals and describe optimized details for obtaining the highest cell purity, yield, and viability. We also verify functional phagocytic capacity in the purified cell populations using a flow cytometry-based phagocytosis assay. Importantly, all of these isolation methods are relatively inexpensive, utilize commonly available reagents, and do not require the acquisition of specialized equipment. Thus, they can be easily implemented in any lab.
2. Materials
Fifteen-mL Vacutainer tubes (Becton Dickinson) containing 150 μL of 500-mM disodium ethylenediaminetetraacetic acid (EDTA), pH 7.4, prepared in sterile H2O and filtered (see Note 1).
Sterile endotoxin-free disposable plastic pipettes, polypropylene centrifuge tubes, and disposable polyethylene transfer pipettes (Fisher Scientific) (see Note 2).
CD66abce MicroBead Kit (Miltenyi Biotec).
MACS LS separation columns and MACS magnetic stand (Miltenyi Biotec) (see Note 3).
Sodium heparin 1,000 U/mL.
2.1. Buffers
Sterile injection-grade H2O and 0.9 % NaCl solution (Baxter Healthcare Corporation)(see Note 4).
Phosphate-buffered saline (PBS): 140-mM NaCl, 2.7-mM KCl, 8-mM Na2 HPO4, and 1.5-mM KH2 PO4 dissolved in sterile water. Adjust pH to 7.2 and sterile filtered. Store at 4 °C.
Dulbecco’s Modified Eagle Medium (DMEM), Dulbecco’s Phosphate Buffered Saline (DPBS), 10× Hanks’ balanced salt solution (10× HBSS) (without Ca2+, Mg2+, and phenol red), RPMI Medium 1640 without phenol red (RPMI) all from Gibco/Life Technologies.
HBSS: Dilute 10× HBSS in sterile H2O, adjust pH to 7.4, and sterile filter. Store at 4 °C (see Notes 4 and 5).
RPMI/HEPES: RPMI supplemented with 10-mM HEPES buffer.
Hetastarch solution containing 6 % hetastarch in 0.9 % NaCl (Abbott Laboratories).
Solutions of 9 % (w/v) and 10 % (w/v) NaCl in sterile H2O. Prepare fresh and sterile filter (see Notes 4 and 5).
Acid citrate dextrose (ACD): 65-mM citric acid, 85-mM sodium citrate, and 2 % dextrose dissolved in sterile H2O and sterile filtered. Store at 4 °C (see Notes 4 and 5).
Murine neutrophil buffer: HBSS containing 0.1 % (w/v) bovine serum albumin, 1 % (w/v) glucose. Prepare fresh and sterile filter (see Notes 4 and 5).
Rabbit neutrophil buffer: 138-mM NaCl, 27-mM KCl, 8.1-mM Na2HPO4, 1.5-mM KH2PO4, and 5.5-mM glucose dissolved in sterile H2O and sterile filtered. Store at 4 °C (see Notes 4 and 5).
Dextran solution: 6 % (w/v) non-pyrogenic dextran 500 (Amersham Biosciences) dissolved in sterile 0.9 % NaCl solution and sterile filtered. Store at 4 °C (see Notes 4 – 6).
10× PBS/EDTA buffer: Dissolve 100-mM KH2PO4, 9 % (w/v) NaCl, 2-mg/L EDTA in sterile H2O, adjust pH to 7.4, and sterile filter (see Notes 4 and 5).
PBS/EDTA buffer: Dilute 10× PBS/EDTA 1:10 in sterile H2O, readjust pH to 7.4 if needed, and sterile filter (see Notes 4 and 5).
MACS erythrocyte lysis buffer: 155-mM NH4Cl, 10-mM KHCO3, and 0.1-mM EDTA dissolved in sterile water. Adjust pH to 7.3 and sterile filtered. Store at 4 °C (NHP protocol).
MACS neutrophil buffer: PBS (pH 7.2), 0.5 % human serum albumin, 2-mM EDTA. Sterile filtered and stored at 4 °C (see Note 7).
2.2. Density Gradient Solutions
Percoll, Histopaque 1077, and Histopaque 1119 (Sigma-Aldrich).
Histopaque 1077/1119 solution: Mix equal volumes of Histopaque 1077 and Histopaque 1119.
Percoll stock solution (100 % Percoll): Percoll and 10× HBSS, pH 7.4 mixed at a ratio of 9:1 (v/v) (see Note 8).
Percoll solutions: Mix 100 % Percoll stock with the appropriate volumes of 1× HBSS to obtain 85, 81, 70, 65, 62, 55, 50, and 45 % (v/v) Percoll solutions (see Note 8).
Percoll/EDTA stock solution (100 % Percoll/EDTA): Percoll and 10× PBS/EDTA, pH 7.4 mixed at a ratio of 9:1 (v/v) (see Note 8).
Percoll/EDTA solution: Mix 100 % Percoll/EDTA stock with the appropriate volume of 1× PBS/EDTA to obtain 65 % (v/v) Percoll/EDTA solution (see Note 8).
2.3. Cell Analysis Reagents
Acetic acid (2 %) prepared in sterile H2O.
Trypan blue solution (0.4 %) (Sigma-Aldrich).
Vybrant phagocytosis assay kit (Molecular Probes).
3. Methods
In the methods described below, we outline the steps to obtain highly purified and functionally active neutrophils from bovine, equine, nonhuman primates, ovine, and rabbit blood, as well as murine bone marrow. Note that some of the methods detailed here have been adapted with modifications from previously published methods for isolation of equine [9], murine [10], ovine [11], and rabbit neutrophils [12, 13]. Additionally, the method for isolation of neutrophils from nonhuman primates utilizes positive selection, which requires labeling of neutrophils with primary and secondary antibodies conjugated to MicroBeads and was adapted from the Miltenyi MicroBead Kit protocol (Miltenyi Biotec Inc., Auburn, CA, USA). Importantly, the presence of these antibodies on the surface of nonhuman primate neutrophils should be considered in the experimental design.
In addition to the basic neutrophil isolation procedures, we also provide details for quantifying cell yield, evaluating cell purity and viability, and measuring phagocytic function of the purified cells from several species. These parameters are compared with human neutrophils purified by a standard method. Although neutrophil isolation from all species except nonhuman primates is based on density gradient separation techniques, the gradient media and composition vary widely because of slight differences in neutrophil density between species. In addition, differences in red blood cell reactivity to aggregating reagents between species are reflected in the methods described below. Overall, these methods are efficient, easy to perform, and reproducibly generate high-quality neutrophil populations for biochemical and functional studies.
3.1. Bovine Neutrophil Isolation
Collect bovine blood into Vacutainer tubes containing EDTA. For the method outlined here, we collected 50 mL of blood. If different volumes of blood are required, adjust the indicated volumes proportionally.
Pool 50 mL of blood into a conical 50-mL polypropylene centrifuge tube and centrifuge at 740 × g for 10 min at room temperature with low brake.
Remove the upper plasma layer and buffy coat found at the plasma–red blood cell interface with a plastic transfer pipette. Transfer the remaining red blood cell layer into a conical 250-mL polypropylene tube.
Lyse red blood cells by adding 50 mL of sterile H2O. Mix by gently inverting the tube for 20 s at room temperature (see Note 9).
Immediately add 5 mL of 10 % NaCl solution, and mix well by gently inverting the tube.
Centrifuge at 585 × g for 10 min at room temperature with low brake.
Remove the supernatant using a plastic transfer pipette, and resuspend the cell pellet in 50 mL of HBSS.
Lyse any remaining red blood cells by repeating steps 4 through 6.
Resuspend the leukocyte pellet in 10 mL of HBSS.
Prepare Histopaque gradients by first pipetting 15 mL of Histopaque 1077 into the bottom of a conical 50-mL centrifuge tube. Place a borosilicate glass Pasteur pipette into the tube so the pipette tip rests on the bottom of the tube. Use this pipette as a funnel to carefully underlay 15 mL of Histopaque 1077/1119 solution.
Layer the 10-mL leukocyte suspension on top of the Histopaque gradient using a plastic transfer pipette. This must be done carefully to avoid mixing the cell suspension with the Histopaque.
Centrifuge the gradient at 440 × g for 25 min at room temperature with no brake.
Remove the supernatant with a plastic transfer pipette and discard (see Note 10).
Wash the neutrophil pellet by resuspending the cells in 50 mL of HBSS and centrifuging at 585 × g for 10 min at room temperature.
Resuspend purified cells in the desired assay buffer.
3.2. Equine Neutrophil Isolation
Collect equine blood into Vacutainer tubes containing EDTA. For the method outlined here, we collected 24 mL of blood. If different volumes of blood are required, adjust the indicated volumes proportionally.
Prepare Percoll gradients by underlaying 2.5 mL of 85 % Percoll solution below 2.5 mL of 70 % Percoll solution in a conical 15-mL polypropylene centrifuge tube. Use a borosilicate glass Pasteur pipette as a funnel to underlay the Percoll solution (see step 11 of Subheading 3.1).
Carefully layer 3 mL of blood on top of each gradient using a plastic transfer pipette.
Centrifuge the gradients for 20 min at 400 × g with no brake at room temperature.
The neutrophil band sediments at the interface between 70 and 85 % Percoll solutions. Carefully remove all of the supernatant above the neutrophil band with a plastic transfer pipette and discard (see Note 10).
Collect the neutrophil band with a clean plastic transfer pipette.
Wash the cells by resuspending them in 50 mL of HBSS and centrifuging at 200 × g for 10 min at room temperature.
Wash the cells twice more by repeating step 7 above.
Resuspend purified cells in the desired assay buffer.
3.3. Human Neutrophil Isolation
Collect human blood into Vacutainer tubes containing EDTA. For the method outlined here, we collected 30 mL of blood. If different volumes of blood are required, adjust the indicated volumes proportionally.
Combine 6.7 mL of dextran solution with 30 mL of blood in a conical 50-mL polypropylene centrifuge tube. Mix by gently inverting the tube.
Allow the blood–dextran mixture to sediment for 45 min at room temperature. Dextran causes the red blood cells to form aggregates, which sediment to the bottom of the tube. This leaves a clear, red blood cell-depleted layer above the red blood cell-rich lower layer.
Transfer the upper cell layer to a clean conical 50-mL polypropylene centrifuge tube with a plastic transfer pipette.
Centrifuge the tube at 740 × g for 10 min with low brake at room temperature.
Remove the supernatant using a plastic transfer pipette and discard.
Resuspend the white blood cell pellet in 7 mL of sterile 0.9 % NaCl solution.
Place 7 mL of Histopaque 1077 into a conical 50-mL polypropylene centrifuge, and carefully layer the white blood cell suspension on top of the Histopaque. This must be done carefully to avoid mixing the cell suspension with the Histopaque.
Centrifuge at 700 × g for 15 min with no brake at room temperature.
Remove the supernatant using a plastic transfer pipette and discard (see Note 10).
Resuspend the neutrophil pellet in 6 mL of sterile 0.9 % NaCl solution.
Lyse contaminating red blood cells by adding 20 mL of sterile H2O. Mix by gently inverting tubes for 20 s at room temperature (see Note 9).
Immediately add 1.8 mL of 10 % NaCl solution, and mix well by gently inverting tubes.
Centrifuge at 740 × g for 10 min at room temperature with low brake.
Remove the supernatant using a plastic transfer pipette and discard.
Resuspend the neutrophil pellet in 6 mL of sterile 0.9 % NaCl solution.
Lyse any remaining red blood cells by repeating steps 12 through 15 above.
Remove the supernatant with a plastic transfer pipette.
Wash the neutrophil pellet by resuspending the cells in 50 mL of 0.9 % NaCl solution and centrifuging at 740 × g for 10 min at room temperature.
Resuspend purified cells in the desired assay buffer.
3.4. Murine Neutrophil Isolation
Dissect femurs and tibias from 8- to 12-week-old male mice. BALB/c mice were used here, but this procedure should also work for other strains of mice.
Clip the ends of each tibia and femur with dissecting scissors to expose the marrow.
Flush bone marrow cells from the tibias and femurs with murine neutrophil buffer using a syringe with 27-G needle. Use two 1-mL volumes of buffer for tibias and three 1-mL volumes of buffer for femurs.
Resuspend the pooled bone marrow eluates by gentle pipetting, followed by filtration through a 70-μm nylon cell strainer (Becton Dickinson) to remove cell clumps and bone particles.
Centrifuge pooled bone marrow cells at 600 × g for 10 min at 4 °C with low brake.
Remove the supernatant with a plastic transfer pipette and discard.
Resuspend the cell pellet in 3 mL of 45 % Percoll solution.
Prepare Percoll gradients by layering 2 mL each of the 62, 55, and 50 % Percoll solutions successively on top of 3 mL of 81 % Percoll solution in a conical 15-mL polypropylene tube.
Carefully layer the bone marrow cell suspension on top of the gradient.
Centrifuge at 1,600 × g for 30 min with no brake at 10 °C.
Remove the supernatant down to the 62 % Percoll layer using a plastic transfer pipette and discard (see Note 10).
Collect the cell band located between the 81 and 62 % Percoll layer.
Wash the collected cells by resuspending them in 10 mL of murine neutrophil buffer and centrifuging at 600 × g for 10 min at 10 °C.
Wash the cells again by repeating step 13 above, and resuspend the final pellet in 3 mL of murine neutrophil buffer.
Carefully layer the cell suspension on top of 3 mL of Histopaque 1119 in conical 15-mL polypropylene tubes.
Centrifuge the gradients at 1,600 × g for 30 min at 10 °C and no brake to remove contaminating red blood cells.
Remove the supernatant using a plastic transfer pipette and discard (see Note 10).
Collect the cell layer between the Histopaque and buffer layers with a plastic transfer pipette.
Wash the cells by resuspending them in 10 mL of murine neutrophil buffer and centrifuging at 600 × g for 10 min at 10 °C.
Wash the cells again by repeating step 19 above.
Resuspend purified cells in the desired assay buffer.
3.5. Nonhuman Primate Neutrophil Isolation
Collect blood from cynomolgus macaques (Macaca fascicularis) into syringes containing 100–150 μL of heparin (1,000 U/mL) per 10 mL of blood (see Note 11).
Transfer blood to a fresh tube and mix gently with 5–10 volumes of MACS erythrocyte lysis buffer (see Note 12).
Rotate tube continuously at low settings on a rotation mixer for slow and gentle rotation of cells (e.g., Rotamix, ATR, Laurel, MD) for 10 min at room temperature or rotate tubes by hand several times during the incubation.
Pellet cells by centrifugation at 300 × g for 10 min at room temperature.
Aspirate supernatant and wash cells once with 10 mL of MACS neutrophil buffer (see Note 13).
Pellet cells by centrifugation at 200 × g for 10 min at room temperature.
Remove supernatant and resuspend cell in 0.5 mL of MACS neutrophil buffer.
3.5.1. Magnetic Labeling
Count cells using a hemacytometer or estimate based on the known number of neutrophils per mL of blood.
If it is necessary to concentrate cells, centrifuge the cell suspension at 200 × g for 10 min at room temperature.
Aspirate supernatant and resuspend cell pellet in MACS neutrophil buffer (40-μL buffer per 107 cells).
Add CD66abce-Biotin-labeled antibody to cell suspension (10-μL antibody solution per 107 cells). Mix and incubate for 10 min at 2–8 °C.
Add MACS neutrophil buffer (30-μL buffer per 107 cells).
Add 20 μL of Anti-Biotin MicroBeads to the cell suspension. Mix and incubate for 15 min at 2–8 °C.
Wash cells by adding 1–2 mL of MACS neutrophil buffer per 107 cells and centrifuge at 300 × g for 10 min at 4 °C. Aspirate supernatant.
Resuspend cells in 500-μL MACS neutrophil buffer (up to 108 cells for every 500 μL of buffer).
3.5.2. Magnetic Separation
Place an LS column in the magnetic field of the MACS separator (see Note 3).
Rinse the LS column with 3 mL of MACS neutrophil buffer.
Transfer cell suspension to the LS column.
Collect cells that pass through the column (these will be unlabeled cells) and wash column three times with 3 mL of MACS neutrophil buffer. The total effluent should be collected (i.e., the unlabeled cell fraction) (see Note 14).
Transfer the column from the MACS separator to the top of an appropriate collection tube.
Pipette 5-mL MACS buffer onto the LS column. Flush out the magnetically labeled cells by depressing the plunger into the column.
Pellet cells by centrifugation at 300 × g for 10 min at 4 °C.
Resuspend cell pellet in 500-μL RPMI/HEPES.
Count cells and dilute to desired concentration.
3.6. Ovine Neutrophil Isolation
Collect ovine blood into Vacutainer tubes containing EDTA. For the method outlined here, we collected 50 mL of blood. If different volumes of blood are required, adjust the indicated volumes proportionally.
Transfer 50 mL of blood into a conical 50-mL polypropylene tube and centrifuge at 400 × g for 20 min with low brake at room temperature.
Remove the upper plasma layer and buffy coat found at the plasma–red blood cell interface with a plastic transfer pipette.
Dilute the red blood cell layer up to the starting blood volume (50 mL in this case) with PBS/EDTA buffer.
Pipette 25 mL of the diluted cells into each of two conical 250-mL polypropylene tubes.
Lyse red blood cells by adding 150 mL of sterile H2O into each tube. Mix by gently inverting tubes for 20 s at room temperature (see Note 9).
Immediately add 15 mL of 9 % NaCl solution, and mix well by gently inverting tubes.
Centrifuge at 250 × g for 5 min at room temperature with low brake.
Remove the supernatant using a plastic transfer pipette, and resuspend the cell pellet in 50 mL of PBS/EDTA buffer.
Centrifuge at 250 × g for 5 min at room temperature.
Resuspend the leukocyte pellet in 9 mL of PBS/EDTA buffer.
Carefully layer 3 mL of the white blood cell suspension on top of 5 mL of 65 % Percoll/EDTA solution using a plastic transfer pipette.
Centrifuge the gradients at 400 × g for 20 min at room temperature with no brake.
Remove supernatant using a plastic transfer pipette and discard (see Note 10).
Wash the cells by resuspending them in 50 mL of PBS/EDTA buffer and centrifuging at 400 × g for 10 min at room temperature.
Wash the cells again by repeating step 15.
Resuspend purified cells in the desired assay buffer.
3.7. Rabbit Neutrophil Isolation
Collect rabbit blood into a conical 50-mL polypropylene tube containing ACD so that a 4:1 (v/v) ratio of blood:ACD is achieved. For the method outlined here, we collected 24 mL of blood into a tube containing 6 mL of ACD. If different volumes of blood are required, adjust the indicated volumes proportionally.
Transfer 30 mL of blood into a conical 250-mL conical centrifuge tube and add 5 volumes of hetastarch to each tube (150 mL in this case). Mix by gently inverting the tube.
Allow the blood–hetastarch mixture to sediment for 40 min at room temperature. Hetastarch causes the red blood cells to form aggregates, which sediment to the bottom of the tube. This leaves a clear, red blood cell-depleted layer above the red blood cell-rich lower layer (see Note 15).
Transfer the upper red blood cell-depleted layer to a clean conical 250-mL polypropylene tube with a plastic transfer pipette.
Centrifuge the solutions at 585 × g for 10 min with low brake at room temperature.
Remove the supernatant using a plastic transfer pipette and discard.
Resuspend the white blood cell pellet in 10 mL of rabbit neutrophil buffer.
Lyse red blood cells by adding 100 mL of sterile H2O. Mix by gently inverting tubes for 20 s at room temperature (see Note 9).
Immediately add 10 mL of 10 % NaCl solution, and mix well by gently inverting tubes.
Centrifuge at 585 × g for 10 min with low brake at room temperature.
Remove the supernatant using a plastic transfer pipette and discard.
Resuspend the cell pellet in 10 mL of rabbit neutrophil buffer.
Lyse any remaining red blood cells by repeating steps 8 – 10 above.
Resuspend the leukocyte pellet in 5 mL of rabbit neutrophil buffer.
Carefully layer cell suspension on top of 7 mL of Histopaque 1077 in a conical 50-mL polypropylene tube.
Centrifuge the gradients at 475 × g for 25 min with no brake at room temperature.
Remove the supernatant using a plastic transfer pipette and discard (see Note 10).
Wash the neutrophil pellet by resuspending the cells in 50 mL of rabbit neutrophil buffer and centrifuging at 585 × g for 10 min at room temperature.
Resuspend purified cells in the desired assay buffer.
3.8. Quantifying Cell Number and Viability
Resuspend the final neutrophil pellet into the desired volume of assay buffer to achieve the appropriate cell concentration (usually 2 to 5 mL) and remove an aliquot for counting.
To quantify cell number, dilute 10 μL of the final cell suspension in 190 μL of 2 % acetic acid. Pipette a few microliters onto a hemacytometer, and count the cells contained in the 25 squares inside the central double lines. Count only neutrophils, which are easily identified by their characteristic multilobed nuclei. Divide the neutrophil count by 25 to obtain the average per square. Multiply the average per square by 5 × 106 and then by the volume (in ml) of the final cell suspension to determine the total number of isolated neutrophils. A summary of the neutrophil recovery data determined for all species is shown in Table 1.
Cell viability is determined by mixing equal aliquots of neutrophil suspension and trypan blue, pipetting the mixture onto microscope slides, and viewing the cells under a microscope. Cells that exclude the trypan blue and appear transparent are counted as viable, whereas cells that turn blue are counted as dead cells. A summary of the cell viability data determined for all species is shown in Table 1.
Table 1.
Average neutrophil purity, yield, and cell viability using the described methods
| Species | Total neutrophilsa | Neutrophil purity (%) | Yield (per mL blood)b | Viability (%) |
|---|---|---|---|---|
| Bovine | 9.79 × 107 | 93.7 | 6.94 × 105 | >99 |
| Equine | 3.9 × 107 | 97.8 | 1.63 × 106 | >99 |
| Human | 2.53 × 107 | 99.1 | 8.42 × 105 | >99 |
| Murine | 5.12 × 106 | 85.9 | 1.71 × 106 | >99 |
| Nonhuman primatec | 1.74 × 107 | 97.4 | 2.4 × 106 | ≥98 |
| Ovine | 1.8 × 107 | 93.6 | 4.89 × 105 | >99 |
| Rabbit | 1.83 × 106 | 90.7 | 7.62 × 104 | >99 |
Number of neutrophils obtained from the described method
Murine neutrophil yield is presented as cells per mouse
Cynomolgus macaques. The data represent the average from at least three separate neutrophil preparations per species
3.9. Analysis of Cell Purity
Purity can be evaluated with the hemacytometer (see step 2 of Subheading 3.8) by differential counting of neutrophils versus non-neutrophils.
Analysis of cell purity can also be performed by flow cytometric analysis, which provides an effective approach to evaluate the cells present and their level of activation.
Collect 10,000 events for each sample using a flow cytometer with linear amplification of forward- and side-scatter channels.
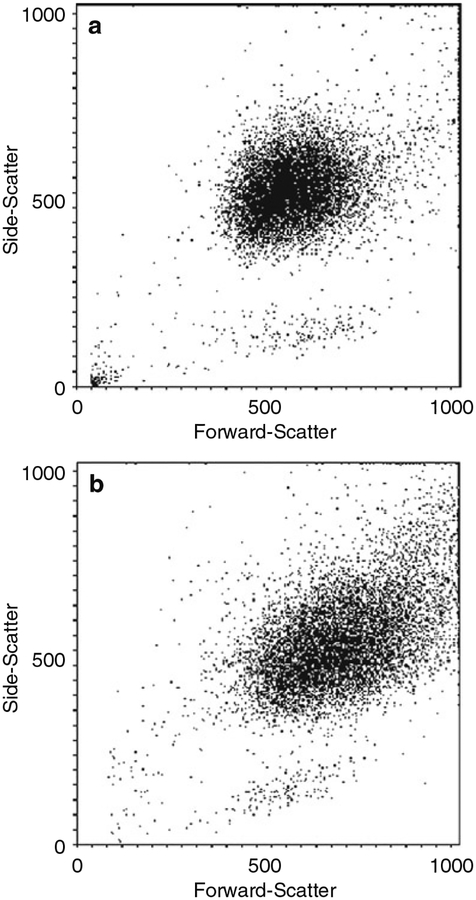
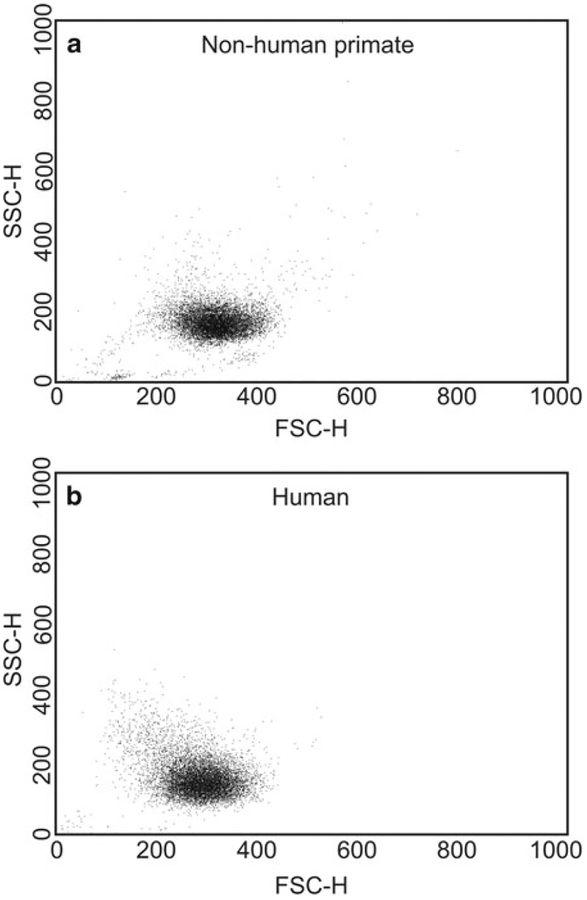
Create a forward-scatter versus side-scatter dot plot, and gate out any cellular debris. Set a gate around the neutrophil population to obtain gate statistics, such as percent of total events (a measure of purity) and relative size and granularity. Figure 1a shows a representative dot plot, where the neutrophils form a relatively uniform profile (see Note 16). Likewise, Fig. 2 shows a comparison of representative dot plot of neutrophils isolated from human and nonhuman primate blood using positive selection on MicroBeads.
A summary of the neutrophil purity data obtained for all species is shown in Table 1.
Fig. 1.
Analysis of neutrophil Purity by flow cytometry. A forward-scatter versus side-scatter dot plot was used to evaluate neutrophil purity (equine cells shown here as an example). Resting (panel a) and mildly activated neutrophils (panel b) are shown (see Note 17). Neutrophils from all species showed similar forward-scatter versus side-scatter profi les. Reproduced by permission of Humana Press©2007 [15]
Fig. 2.
Analysis of neutrophils from nonhuman primates (Macaca fascicularis) and humans by flow cytometry. Neutrophils were obtained from venous blood of nonhuman primates (a) or humans (b) using the positive selection methods described and then analyzed by flow cytometry to determine forward (FSC-H)-and side (SSC-H)-angle light scatter
3.10. Phagocytosis Assay
Phagocytosis assays were performed using a Vybrant phagocytosis assay kit with modifications for use with flow cytometry.
Thaw one vial each of fluorescent Escherichia coli K-12 bioparticles and concentrated HBSS.
To prepare stock bioparticles, pipette concentrated HBSS into the vial containing the bioparticles and sonicate. Transfer the solution into a clean glass test tube, add 4.5 mL of sterile H2O, and sonicate again until the beads are completely dispersed.
Thaw one vial of trypan blue solution, transfer the trypan blue solution to a polycarbonate test tube, dilute with 4 mL of sterile H2O, and sonicate.
Dilute isolated neutrophils from any species to a final concentration of 1 × 106 cells/mL in DMEM.
Dilute stock bioparticles by mixing 1 mL of bioparticles and 0.5 mL of DMEM.
Aliquot 150-μL DMEM, 100-μL neutrophils, and 10 μL of diluted bioparticles into 1.5-mL microcentrifuge tubes. For control samples, substitute 10-μL DMEM instead of bioparticles. This dilution gives a final bioparticle to neutrophil ratio of 20:1 (see Note 17).
Incubate triplicate samples for 2, 5, 10, and 15 min at 37 °C. We normally incubate controls samples of neutrophils alone and neutrophils with trypan blue quench controls for 15 min to evaluate any effects resulting from the extended incubation time.
After each incubation time, the neutrophils are pelleted by centrifuging at 3,000 × g for 30 s in a microfuge at room temperature.
The pellet is very small and easy to lose, so carefully aspirate the supernatant.
To quench free bioparticles and neutrophil surface-associated bioparticles, add 100 μL of the trypan blue solution and mix well.
Incubate for 1 min at room temperature, and centrifuge at 3,000 × g for 30 s in a microfuge at room temperature.
Carefully aspirate the supernatant. Again, use care because the neutrophil pellet is very small and easily lost.
Resuspend each pellet in 250-μL DPBS, and transfer the samples to flow cytometer tubes.
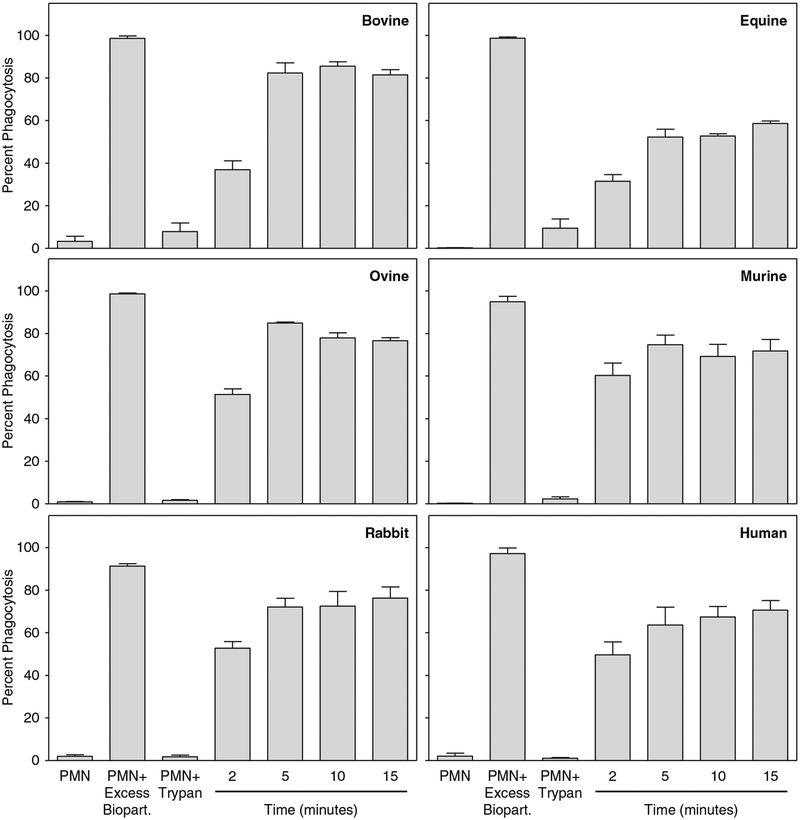
Collect 10,000 events for each sample using a flow cytometer with linear amplification of forward-scatter and side-scatter channels and logarithmic amplification for the FL1 channel (see Note 18). Analyze the data using flow cytometry software (e.g., CellQuest software) to determine the percent of neutrophils containing fluorescent bioparticles. A summary of the neutrophil phagocytosis data obtained for all species is shown in Fig. 3 (see Note 19).
Fig. 3.
Functional analysis of purified neutrophils. Neutrophils purified from the indicated species were analyzed for their ability to phagocytose fluorescent bioparticles, as described. Cells were incubated with bioparticles (1:20 ratio) for 2, 5, 10, and 15 min at 37 °C. Control samples include neutrophils alone (negative control), neutrophils incubated with trypan blue (trypan control), and neutrophils incubated with excess (1:200 ratio) bioparticles (positive control). In each panel, the results are presented as the mean ± SD of triplicate samples. Representative of at least three experiments for each panel. Reproduced from ref. 15 with permission from Humana Press
4. Notes
EDTA was added to Vacutainer tubes by injection with a 1-mL syringe and 27-G needle.
It is essential that the blood and subsequently isolated neutrophils do not ever come into contact with glass, which leads to cell activation. Thus, plasticware should be used throughout all procedures, with exception of the Vacutainer tubes, which are silicone coated.
An adequate-sized column should be selected based on its maximum cell capacity. The volumes provided here are for LS columns. Check the Miltenyi instructions on the package insert for appropriate volumes for use with other columns.
Neutrophils are highly susceptible to priming and/or activation by endotoxin or lipopolysaccharide (LPS) (e.g., see ref. 14), which is often a contaminant in biological reagents. Thus, all plasticware must be endotoxin-free. In addition, all buffers and reagents are prepared in sterile H2O or saline and sterile filtered to avoid endotoxin contamination.
All buffers should be sterile filtered through 0.2-μm filter units (Fisher Scientific).
To avoid possible contamination, which is a common problem with dextran, 30 g of dextran 500 is weighed directly into a sterile plastic 500-mL Nalgene container and dissolved in sterile 0.9 % NaCl solution, followed by sterile filtering.
As suggested by manufacturer, human serum albumin (HSA) can be substituted with bovine serum albumin, fetal bovine serum, or human serum.
Use extreme care and accuracy when preparing Percoll mixtures, as small variations in the final density of Percoll mixtures affects the purity and yield of neutrophil preparation.
Do not extend this incubation longer than 20 s, as longer incubation in hypotonic solution can alter and/or damage the neutrophils.
After the supernatant has been removed from the gradients, cotton applicators may be used to wipe the walls of the centrifuge tube to remove any adherent debris, which may contaminate the preparation. Be sure to avoid touching the neutrophil band or pellet with the applicator.
Sodium heparin can be replaced by other anticoagulants such as EDTA or sodium citrate. Alternatively, blood can be collected into Vacutainer tubes (Becton Dickinson) as described for the other neutrophil isolation methods.
Typically we use 35 mL of lysis buffer per 5 mL of heparinized blood (7:1 ratio).
Handle neutrophils as gently as possible, e.g., have a pipette on the lowest settings.
Both purified neutrophil fraction and effluent (unlabeled) fraction should be analyzed to determine purification efficiency.
As an alternative, rabbit red blood cells can also be aggregated with 6 % dextran (100,000–200,000 molecular weight) for 30–40 min [13]; however, hetastarch seems to be more efficient. For some reason, rabbit red blood cells do not lyse as readily as those from other species. Even after two rounds of H2O lysis, some red blood cells may still be present. If this is the case, remaining red blood cells may be removed by very gently washing the surface of the neutrophil pellet.
Note that neutrophil priming or activation causes an increase in cell size and granularity, which can also be evaluated with these dot plots (see Fig. 1b).
The relative amount of bioparticles and DMEM can be adjusted up or down to achieve different neutrophil:bioparticle ratios. However, we found that increased bioparticle concentrations resulted in close to 100 % phagocytosis at the 2-min time point, making it difficult to distinguish differences in rates of phagocytosis between samples.
Forward-scatter versus side-scatter plots yield different profiles due to varying lots of trypan blue used to quench external fluorescence.
Most phagocytosis experiments showed a decrease in the percent of positive cells after 5 min, which has been reported to be a result of acidification of the phagosomal compartments over time, resulting in quenching of the bioparticle fluorescence [12].
Acknowledgements
This work was supported in part by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences, National Institutes of Health under grant number GM103500 (D.S., I.A.S., L.K.N., B.L., M.Q.), the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health (N.M., A.R.W., F.R.D.), and the Montana State University Agricultural Experimental Station.
References
- 1.Wiles S, Hanage WP, Frankel G et al. (2006) Modelling infectious disease – time to think outside the box? Nat Rev Microbiol 4: 307–312 [DOI] [PubMed] [Google Scholar]
- 2.Casal M, Haskins M (2006) Large animal models and gene therapy. Eur J Hum Genet 14:266–272 [DOI] [PubMed] [Google Scholar]
- 3.Styrt B (1989) Species variation in neutrophil biochemistry and function. J Leukoc Biol 46: 63–74 [DOI] [PubMed] [Google Scholar]
- 4.Glasser L, Fiederlein RL (1990) The effect of various cell separation procedures on assays of neutrophil function. A critical appraisal. Am J Clin Pathol 93:662–669 [DOI] [PubMed] [Google Scholar]
- 5.Watson F, Robinson JJ, Edwards SW (1992) Neutrophil function in whole blood and after purification – changes in receptor expression, oxidase activity and responsiveness to cytokines. Biosci Rep 12:123–133 [DOI] [PubMed] [Google Scholar]
- 6.Forsyth KD, Levinsky RJ (1990) Preparative procedures of cooling and re-warming increase leukocyte integrin expression and function on neutrophils. J Immunol Methods 128: 159–163 [DOI] [PubMed] [Google Scholar]
- 7.Macey MG, Jiang XP, Veys P et al. (1992) Expression of functional antigens on neutrophils. Effects of preparation. J Immunol Methods 149:37–42 [DOI] [PubMed] [Google Scholar]
- 8.Alvarez-Larrán A, Toll T, Rives S et al. (2005) Assessment of neutrophil activation in whole blood by flow cytometry. Clin Lab Haematol 27:41–46 [DOI] [PubMed] [Google Scholar]
- 9.Pycock JF, Allen WE, Morris TH (1987) Rapid, single-step isolation of equine neutrophils on a discontinuous Percoll density gradient. Res Vet Sci 42:411–412 [PubMed] [Google Scholar]
- 10.Lowell CA, Fumagalli L, Berton G (1996) Deficiency of Src family kinases p59/61hck and p58c-fgr results in defective adhesion-dependent neutrophil functions. J Cell Biol 133:895–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woldehiwet Z, Scaife H, Hart CA et al. (2003) Purification of ovine neutrophils and eosinophils: anaplasma phagocytophilum affects neutrophil density. J Comp Pathol 128:277–282 [DOI] [PubMed] [Google Scholar]
- 12.White-Owen C, Alexander JW, Sramkoski RM et al. (1992) Rapid whole-blood microassay using flow cytometry for measuring neutrophil phagocytosis. J Clin Microbiol 30: 2071–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doerschuk CM, Allard MF, Martin BA et al. (1987) Marginated pool of neutrophils in rabbit lungs. J Appl Physiol 63:1806–1815 [DOI] [PubMed] [Google Scholar]
- 14.DeLeo FR, Renee J, Mccormick S et al. (1998) Neutrophils exposed to bacterial lipopolysaccharide upregulate NADPH oxidase assembly. J Clin Invest 101:455–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siemsen DW, Schepetkin IA, Kirpotina LN, Lei B, Quinn MT (2007) Neutrophil isolation from non-human species. Methods Mol Biol 412:21–34 [DOI] [PubMed] [Google Scholar]