Abstract
`NK cell–mediated regulation of antigen-specific T cells can contribute to and exacerbate chronic viral infection, but the protective mechanisms against NK cell–mediated attack on T cell immunity are poorly understood. Here, we show that progranulin (PGRN) can reduce NK cell cytotoxicity through reduction of NK cell expansion, granzyme B transcription, and NK cell–mediated lysis of target cells. Following infection with the lymphocytic choriomeningitis virus (LCMV), PGRN levels increased — a phenomenon dependent on the presence of macrophages and type I IFN signaling. Absence of PGRN in mice (Grn–/–) resulted in enhanced NK cell activity, increased NK cell–mediated killing of antiviral T cells, reduced antiviral T cell immunity, and increased viral burden, culminating in increased liver immunopathology. Depletion of NK cells restored antiviral immunity and alleviated pathology during infection in Grn–/– mice. In turn, PGRN treatment improved antiviral T cell immunity. Taken together, we identified PGRN as a critical factor capable of reducing NK cell–mediated attack of antiviral T cells.
Keywords: Immunology, Infectious disease
Keywords: Adaptive immunity, Innate immunity, NK cells
Progranulin is a critical factor capable of protecting anti-viral CD8+ T cells from NK cell cytotoxicity.
Introduction
Progranulin (PGRN) is a secreted protein associated with cancer and inflammation (1–3). PGRN is expressed and secreted during wound healing and can accelerate the healing process (4) and alleviate TNF-mediated rheumatoid arthritis (5). PGRN deficiency triggers increased expression of proinflammatory cytokines, including TNF and IL-6, by macrophages and microglia (6, 7). Elastase cleaves PGRN into smaller granulin peptides, which have been shown to augment TLR9 signaling by enhancing delivery of CpG-oligodeoxynucleotides (CpG-ODNs) (8).
The functional relevance of PGRN is demonstrated through its direct contribution to several diseases. Inactivating mutations in the GRN gene are one underlying cause of frontotemporal dementia, a neurodegenerative disorder resulting in language, personality, and behavioral changes (9, 10). PGRN haploinsufficiency results in the accumulation of pathologic inclusions in neurons and glia cells containing phosphorylated and ubiquitinated TDP-43 (11, 12). TDP-43 inclusions are also present in the motor neuron disease amyotrophic lateral sclerosis, indicating overlapping molecular etiologies (11). As PGRN can be transported into lysosomes through the binding receptor sortilin-1 (SORT1) (13) defects in PGRN are also associated with reduced autophagic pathways, which also possibly contribute to the formation of the pathologic TDP-43 inclusions (14, 15).
In addition to PGRN, SORT1 can bind and direct other ligands into the lysosome, including PGRN’s heterodimeric binding partner prosaposin (16, 17). Prosaposin can also facilitate SORT1-independent trafficking of PGRN into lysosomes (18). As defects in PGRN cause reduced lysosomal trafficking of prosaposin, a strong functional interaction between PGRN, prosaposin, and SORT1 is plausible (19). SORT1 deficiency can result in reduced cytotoxicity but enhanced cytokine production in T and NK cells (20). Interestingly, PGRN can bind to Lamp-1, a membrane protein, which is critical in protecting cells from NK cell–mediated cytotoxicity (14, 21, 22). Taken together, PGRN’s importance in several physiological and cellular processes is highlighted by its role in human disease. While there is a strong implication of PGRN’s involvement in modulation of the immune system, whether and how PGRN modulates NK cell–dependent, antigen-specific, or antiviral immunity is not well studied.
It is well established that NK cells have antiviral activity and that NK cell–mediated killing of virus infected cells can contribute to elimination of viral infections (23). However, during chronic viral infections, NK cell–mediated attack of antiviral T cells can limit T cell immunity (24, 25), and NK cell depletion can result in clearance of an otherwise chronic viral infection (26–28). NK cell activity is orchestrated by a balanced expression of activating and inhibitory receptors on NK cells — as well as ligands expressed on other immune cells, including T cells — that trigger them (29). Type I IFN (IFN-I), induced during the course of a viral infection, protects antiviral T cells from NK cell–mediated attack (30, 31). IFN-I reduces expression of NKp46 activating ligands on the surface of antiviral T cells and increases expression of the inhibitory NK cell receptor ligands MHC-I and MHC-Ib, thus shifting the equilibrium toward NK cell inhibition (30, 31). On the other hand, lack of inhibitory molecules during viral infection can result in increased NK cell activation and consequent deletion of antiviral T cells (32). NK cell–mediated regulation of antigen-specific T cells is also observed in human disease. Specifically, expression of NK cell inhibitory receptors and their ligands correlates with clearance of hepatitis C virus infection (33, 34). NK cells can target anti-HBV–specific T cells, contributing to T cell dysfunction (35). Notably, an ILC population induced suppression of antitumor-specific T cells in ovarian cancer (36). Taken together, NK cell–mediated T cell inhibition during infection is a complex and finely tuned process, and there are many unknown factors regulating T cell escape from NK cell–mediated regulation.
In the current study, we have identified PGRN to inhibit NK cell–mediated cytotoxicity and to promote T cell immunity during lymphocytic choriomeningitis virus (LCMV) infection. Absence of PGRN resulted in enhanced regulatory NK cell function and reduced antiviral T cell immunity. In turn, NK cell depletion restored T cell immunity and prevented viral persistence in Grn–/– mice.
Results
NK cell–mediated cytotoxicity is limited by PGRN.
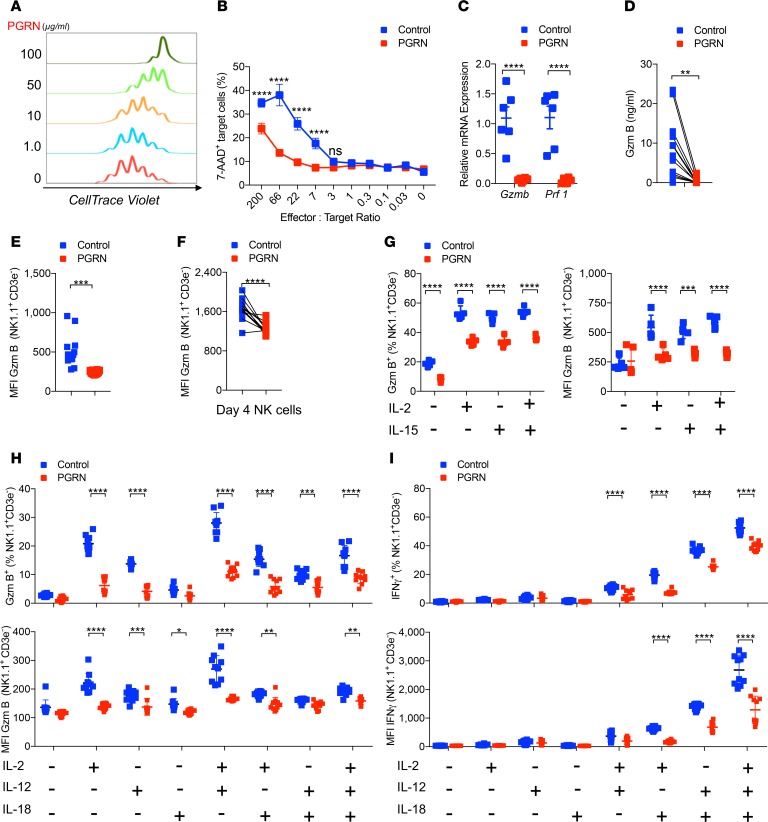
In order to test whether PGRN directly affects NK cells, we exposed murine NK cells to different doses of recombinant human PGRN. Interestingly, the presence of PGRN reduced the IL-2–mediated expansion of NK cells in a dose-dependent manner (Figure 1A and Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.129856DS1). Flow cytometry analysis, by staining these NK cells with 7-AAD/Annexin V, suggested that PGRN inhibits NK cell expansion on a mechanism that is independent of proapoptotic effects (Supplemental Figure 1B). In turn, we found an increased presence of NK cells in the G0/G1 stage and, consequently, a reduced proportion of NK cells in the S phase (Supplemental Figure 1C). Notably, PGRN-treated and untreated NK cells were characterized by comparable expression levels of activating and inhibitory receptors (Supplemental Figure 1D). Furthermore, PGRN reduced NK cell–mediated cytotoxicity toward RMA/S cells (Figure 1B). NK cell–mediated cytotoxicity is triggered by perforin and granzymes (37). Indeed, granzyme B and perforin mRNA expression levels in NK cells were reduced following exposure to PGRN (Figure 1C). Consistently, granzyme B protein levels were lower in PGRN-treated cells than control cells (Figure 1, D and E). We also observed reduced protein levels of granzyme B at later time points during NK cell tissue culture following PGRN addition (Figure 1F). We speculated that PGRN interferes with NK cell activation and expansion. On the basis of our in vitro observations, we stimulated NK cells harvested from spleen tissue with cytokines, IL-2, IL-12, IL-15, and/or IL-18 and found an increase in granzyme B expression, which was blunted in the presence of PGRN (Figure 1, G and H). Consistently, IFN-γ expression in NK cells was reduced when incubated with PGRN (Figure 1I). Proteolytic cleavage of the PGRN by elastase gives rise to smaller peptide fragments, termed granulin (38), and we wondered whether granulin would have a similar effect on NK cells (Supplemental Figure 1E). Interestingly, following exposure of NK cells with elastase-treated PGRN, no granzyme B–suppressing effects were observed (Supplemental Figure 1, E and F). These data suggest that PGRN mediated NK cell suppression.
Figure 1. Progranulin limits NK cell–mediated cytotoxicity.
(A) Isolated NK cells were cultured with 1000 IU/ml IL-2 and indicated doses of progranulin (PGRN) for 4 days. The division of these NK cells were determined by flow cytometry (n = 3). (B) The cytotoxic activity of PGRN (100 μg/ml) treated NK cells to RMA/S cell was measured at indicated effector/target ratios (n = 4–5). (C) Granzyme B (Gzmb) and perforin (Prf1) mRNA levels from PGRN-treated (100 μg/ml) NK cells and controls were determined by qPCR (n = 6). (D) Granzyme B protein concentrations were measured in the supernatants (n = 12). (E) Mean fluorescence intensity (MFI) of granzyme B was determined in NK cells in presence of RMA/S cells with or without PGRN treatment (n = 16). (F) The expanded NK cells were treated with PGRN overnight. Gzm B MFI in NK cell was measured by flow cytometry (n = 12). (G) Splenocytes from naive WT mice were incubated with 1000 IU/ml IL-2 and/or 100 IU/ml IL-15 with or without PGRN (25 μg/ml) for 16 hours. Gzm B expression was measured by flow cytometry. The left panel represents the Gzm B+ NK cell frequency, and the right panel represents the MFI of Gzm B (n = 5). (H and I) Splenocytes from naive WT mice were incubated with 1000 U/ml IL-2, 20 ng/ml IL-12, and 5 ng/ml IL-18 with or without PGRN (25 μg/ml) for 6 hours. Gzm B (H, n = 10) and IFN-γ (I, n = 10) was determined in NK cells by flow cytometry. The upper panels represent the frequency, whereas the lower panels indicate the MFI. Data in B–I show mean ± SEM. P values calculated by 2-way ANOVA (B, C, and G–I); D, E, and F by Student’s t test; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Next, we wondered whether PGRN would suppress human NK cells. We cultured whole healthy donor PBMCs and analyzed NK cell activity. We found a significant reduction in degranulation of total and CD56dim NK cells in response to HLA-devoid cell lines K562 and 721.221 in the presence of PGRN (Figure 2, A and B). Furthermore, IFN-γ levels in human NK cells were also reduced following exposure to PGRN (Figure 2, C and D). Taken together, these data indicate that PGRN can reduce NK cell cytotoxicity by limiting expression of effector molecules.
Figure 2. PGRN supresses human NK cell activation.
PBMCs from healthy donors were pretreated with 100 μg/ml PGRN overnight and stimulated with K562 or 721.221 cells for 5 hours at an effector/target ratio of 10:1. The frequency of IFN-γ–producing and CD107a-expressing NK cells was determined by intracellular cytokine staining. (A and B) CD107a levels were measured in total NK cells (A) and CD56dim NK cells (B) by flow cytometry (n = 15). (C and D) IFN-γ levels in total NK cells (C) and CD56dim NK cells (D) were measured by flow cytometry (n = 15). Data in A–D show mean ± SEM. Each symbol represents experimental data from an individual healthy donor. P values calculated by paired Student’s t test; *P < 0.05; ***P < 0.001; ****P < 0.0001.
PGRN decreases cyclin T1 and CDK9 expression in NK cells.
Next, we explored the mechanism by which PGRN might limit NK cell cytotoxicity. NK cells can be self-regulated through cytotoxicity-mediated fratricide, which itself is triggered by the critical effector molecule perforin (37, 39). When we cultured perforin-deficient (Prf1–/–) NK cells with PGRN, we still found inhibition of NK cell expansion in tissue culture (Supplemental Figure 2A), suggesting that perforin is not functionally contributing to the phenotype. PGRN has been shown to reduce TNF receptor 1 (TNFR1) signaling (5). However, Tnfrsf1a–/– and Tnfrsf1b–/– NK cells were characterized by similarly reduced levels of granzyme B compared with control NK cells following PGRN treatment (Supplemental Figure 2B). These results suggest that PGRN-mediated NK suppression is not TNFR dependent. We therefore wondered whether transcription factors regulating granzyme B expression could be affected following PGRN treatment. It has been previously shown that PGRN can bind to the positive transcription elongation factor b (P-TEFb), which consists of cyclin T1 and CDK9 (40). Knockdown of CDK9 and cyclin T1 results in the inhibition of T cell effector function attributable to the reduced expression of perforin and granzyme B (41). We wondered whether the same connection linking PGRN and CDK9/Cylclin T1 in T cells would also be applicable to NK cells.
Firstly, we treated NK cell cultures with SNS-032, a cyclin T1/CDK9 inhibitor, and found that granzyme B expression in NK cell cultures was blocked (Figure 3A). We wondered whether PGRN was taken up by the NK cells, and we confirmed this using immunoblot analysis of NK cell lysates (Figure 3B). Furthermore, in the presence of PGRN, there was a significant reduction of Cdk9 mRNA expression levels in NK cells compared with the control conditions (Figure 3C), which was corroborated on a protein level using immunoblot analysis (Figure 3D). CDK9 cooperates with cyclin T1 to induce transcription (42), and we also found reduced protein levels of cyclin T1 following treatment of NK cells with PGRN (Figure 3D). The reduction of cyclin T1 protein levels following incubation with PGRN was further confirmed by flow cytometry (Figure 3E) and immunofluorescence analyses (Figure 3F). It has been previously reported that IL-2 promotes RNA polymerase II recruitment to the promoter of IL-2–induced genes (43). A histidine-rich domain in cyclin T1 promotes phosphorylation of the C-terminal domain of RNA polymerase II, which can be prevented by PGRN (40, 44). Reduced phosphorylation (pSer2) of RNA polymerase II was indeed detected in NK cells during PGRN treatment (Figure 3G). The above data indicate that PGRN limits the expression of key transcription factors CDK9 and cyclin T1, which are, in turn, critical for the transcription of NK cell effector molecules.
Figure 3. PGRN decreases cyclin T1 and CDK9 expression in NK cells.
(A) Splenocytes were treated with 1000 U/ml IL-2 and the cyclin T1/CDK9 inhibitor SNS-032 for 6 hours (concentration as indicated). Gzm B levels were measured by flow cytometry (n = 4). The left panel represents the frequency of Gzm B in NK cells, and the right panel represents the Gzm B MFI in NK cells. (B–F) Freshly isolated NK cells were treated with IL-2 and PGRN for 4 days. (B) The uptake of PGRN into NK cells was measured by Western blotting (n = 4). The right panel indicates quantification of the data. (C) Cyclin T1 and CDK9 mRNA levels were examined by qPCR (n = 6). Relative mRNA levels were normalized to Actin. (D) Cyclin T1 and CDK9 protein levels were examined by Western blotting (n = 7). The middle panel represents the quantification of cyclin T1; the right panel represents CDK9 quantification. (E) Cyclin T1 protein levels were examined by flow cytometry at day3 after IL-2 treatment (n = 6). The right panel represents the quantification of cyclin T1 MFI. (F) Cyclin T1 protein levels were examined by immunofluorescence (n = 3). The right panel represents the quantification of cyclin T1 by cyclin T1 MFI/DAPI MFI by ImageJ. Scale bars: 20 μm. (G) Isolated NK cells were treated by IL-2 with or without PGRN. The phospho-Ser2 RNA polymerase II frequency (pSer2-RII, upper panel) and MFI (lower panel) were measured by flow cytometry at the indicated time points (n = 6). Data in A–G show mean ± SEM. Each symbol represents an individual mouse. P values calculated by 2-way ANOVA (except for B, D, E, and F by Student’s t test); *P < 0.05; **P < 0.001; ***P < 0.001; ****P < 0.0001.
IFN-I triggers PGRN secretion following infection with LCMV.
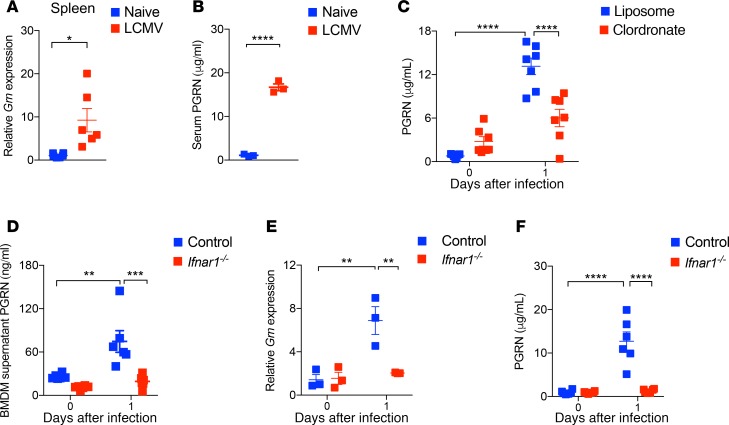
PGRN can be produced by macrophages, microglia, and endothelial cells in response to inflammatory stimuli and/or tissue damage (1). We wondered about the effects of PGRN during viral infection. To address this, we used the noncytopathic lymphocytic choriomeningitis virus (LCMV), which is commonly used to study CD8+ effector T cell–mediated immunity. We found increased mRNA expression levels of Grn following infection with LCMV (Figure 4A). As expected, serum PGRN protein levels were elevated compared with uninfected controls (Figure 4B). To investigate which cell types might be a source of PGRN, we analyzed serum PGRN levels in mice deficient for different immune subsets after LCMV infection. We did not find a reduction of PGRN levels in the absence of B cells and T cells (Rag1–/– mice), CD169+ cells (CD169 diphtheria toxin receptor mice; CD169-DTR), or CD11c+ (CD11c-DTR) cells after LCMV infection (Supplemental Figure 3, A–C). However, serum PGRN levels were decreased following treatment with the phagocyte-depleting compound Clodronate during LCMV infection (Figure 4C). As the reduction of PGRN was not complete compared with basal preinfection levels, this indicates that cell types other than macrophages also contribute to PGRN secretion. Consistently, we observed comparable PGRN levels in lethally irradiated WT mice reconstituted with either control or Grn–/– BM cells (Supplemental Figure 3D). This result indicates that cell subsets other than immune cells can produce PGRN during LCMV infection.
Figure 4. IFN-I triggers PGRN expression following viral infection.
(A) WT mice were infected with 2 × 106 pfu LCMV (WE strain). Organs were collected at day 1 after viral infection and subjected to qPCR analysis. The relative mRNA levels were normalized to Gapdh (n = 6). (B) Serum PGRN protein concentrations were determined by ELISA at indicated time points after LCMV infection (2 × 106 pfu, n = 3). (C) Clodronate treated (day –1) WT mice were infected with 2 × 106 pfu LCMV-WE. Serum PGRN levels (day 1) were measured by ELISA (n = 7). (D) WT and Ifnar1–/– BMDMs were infected with LCMV-WE (MOI = 1) for 48 hours. PGRN protein levels in cell supernatants were examined by ELISA (n = 6). (E and F) Ifnar1–/– and WT mice were infected with 2 × 106 pfu LCMV-WE. The Grn mRNA levels in the spleen were measured at day 1 following infection by qPCR (E, n = 3), and serum PGRN levels (F, n = 6) were determined by ELISA. Data in A–G show mean ± SEM. Each symbol represents an individual mouse. P values calculated by 2-way ANOVA (C–F) and Student’s t test (A and B); *P < 0.05; **P < 0.001; ***P < 0.001; ****P < 0.0001.
Next, we wondered how PGRN secretion was triggered following infection. Early antiviral defenses highly depend on IFN-I (45). Therefore, we speculated that innate IFN-I might trigger PGRN secretion. When BM-derived macrophages (BMDMs) were infected with LCMV in vitro, we observed increased secretion of PGRN in the supernatants (Figure 4D). However, in IFN-I binding receptor–deficient (Ifnar1–/–) BMDMs, PGRN secretion was not elevated and supernatant PGRN levels were similar to the naive control conditions (Figure 4D). When Ifnar1–/– mice were infected with LCMV, we measured a reduction of Grn mRNA levels in spleen tissue when compared with WT control animals (Figure 4E). Furthermore, we observed no serum PGRN increase in Ifnar1–/– mice following infection when compared with serum samples from WT control animals (Figure 4F). Taken together, these data demonstrate a striking increase in PGRN secretion during LCMV infection, which was dependent on IFN-I signaling.
T cell immunity following LCMV infection is blunted by PGRN deficiency.
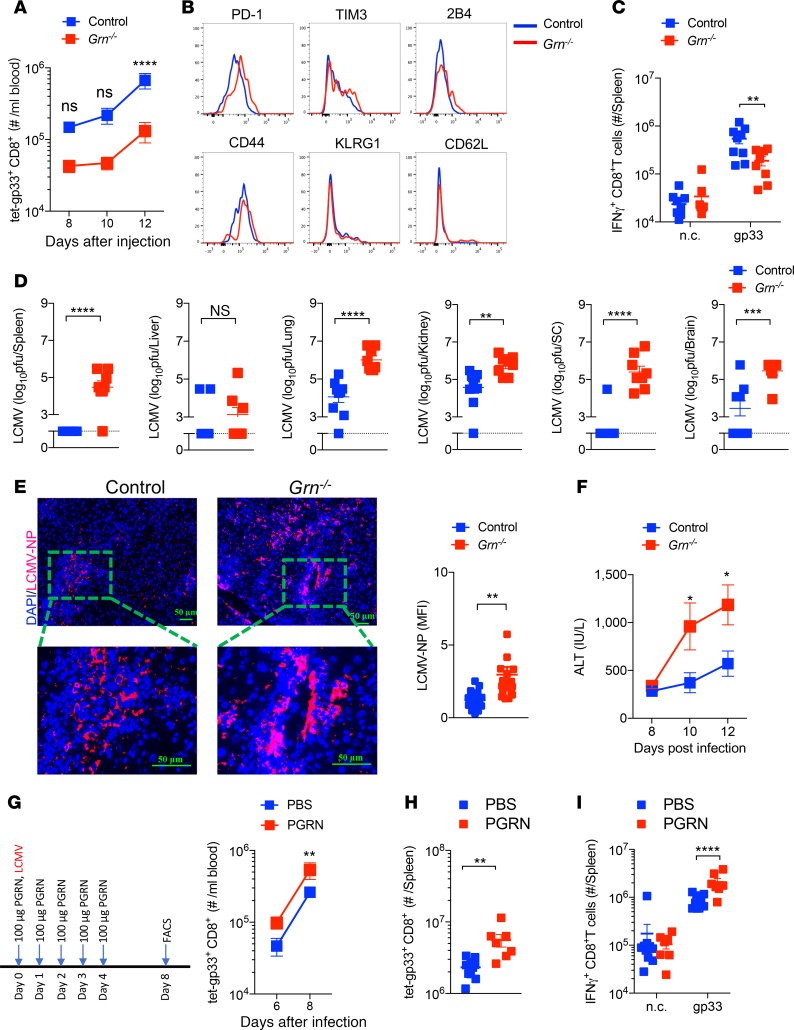
Next, we wondered whether there were functional consequences affecting antiviral immunity of the PGRN secretion triggered by LCMV infection. Naive PGRN-deficient mice (Grn–/–) exhibited comparable CD8+ and CD4+ T cells compared with WT animals (Supplemental Figure 4, A and B). However, following infection with LCMV, Grn–/– mice showed a reduced presence of antiviral CD8+ T cells when compared with corresponding control animals (Figure 5A and Supplemental Figure 5A). Consistently, we found increased expression of PD-1 on antiviral (gp33-H2-Db-tetramer+) T cells from Grn–/– mice, which is associated with T cell exhaustion (Figure 5B and Supplemental Figure 5B). Furthermore, when we restimulated splenocytes with LCMV epitope gp33 peptides, we observed reduced IFN-γ production in CD8+ T cells from Grn–/– mice compared with controls (Figure 5C and Supplemental Figure 5C). Control and clearance of LCMV depends on antiviral CD8+ T cell immunity, and as expected, increased LCMV titers were detected in Grn–/– mice when compared with controls (Figure 5D). While control animals eliminated LCMV at later time points following LCMV infections (day 20), we still observed LCMV titers in Grn–/– mice, indicating that absence of PGRN can result in a prolonged viral infection (Supplemental Figure 5D). As we observed increased LCMV nuclear protein–positive (LCMV-NP+) cells in sections of snap-frozen liver tissue harvested from Grn–/– mice when compared with control animals (Figure 5E), we speculated that PGRN might exert direct antiviral effects. However, when N2a neuroblastoma, Vero, or L929 cells were exposed to PGRN in tissue culture experiments, we did not observe any changes in LCMV replication (Supplemental Figure 6, A–C). Hepatic replication of LCMV can result in T cell–mediated liver damage (46, 47). The enzyme alanine aminotransferase (ALT) activity in the serum of LCMV-infected Grn–/– animals was markedly increased compared with control animals, suggesting that liver pathology was increased in the absence of PGRN (Figure 5F). In addition, when we treated WT mice with recombinant PGRN throughout LCMV infection, we observed enhanced antiviral CD8+ T cell immunity (Figure 5, G–I).
Figure 5. Grn deficiency limits CD8+ T cell immunity and triggers LCMV-induced immunopathology.
WT and Grn–/– mice were infected with 2 × 106 pfu LCMV-WE. (A) Absolute numbers of antiviral CD8+ T cells in the blood were determined at the indicated time points after infection (n = 7–10). (B) The expression of surface molecules on spleen antiviral CD8+ T cells was examined by flow cytometry. (C) At day 12 after infection, splenocytes were restimulated with the LCMV-specific epitope gp33, followed by staining for IFN-γ (n = 7–10). n.c. indicates negative control. (D) Virus titers were determined from spleen, liver, lung, kidney, spinal cord (SC), and brain tissues at day 12 after infection (n = 7–10). (E) Sections of snap-frozen liver tissue from WT and Grn–/– mice (day 12 after infection) were analyzed for LCMV nucleoprotein (LCMV-NP) expression by IHC. The bar graph depicts the fluorescence intensity of LCMV-NP (n = 6). Scale bars: 50 μm. (F) ALT activity in the serum of control and Grn–/– mice was determined at the indicated time points after infection (n = 6). (G–I) WT mice were injected with PGRN at the indicated dose and time points (G, left panel); the absolute number of antiviral T cells in blood (G, right panel) were measured by flow cytometry (n = 7–10). (H) The absolute number of antiviral T cells in the spleen were measured by flow cytometry at day 8 (n = 7–10). (I) Splenocytes were restimulated with gp33 peptides and IFN-γ+CD8+ T cells were measured by flow cytometry (n = 7–10). Data show mean ± SEM. Each symbol represents an individual mouse. P values calculated by 2-way ANOVA (A, C, F, G, I) and Student’s t test (D, E, and H), *P < 0.05; **P < 0.001; ***P < 0.001; ****P < 0.0001.
Next, we examined whether the observed defective T cell immunity following LCMV infection in Grn–/– mice was caused by T cell intrinsic effects. We adoptively transferred negatively sorted CD8+ T cells, from a mouse carrying a TCR recognizing the LCMV antigen gp33 (P14 cells) (48), to Grn–/– and control mice. We found reduced expansion of P14 cells in Grn–/– hosts following LCMV infection when compared with control hosts (Figure 6A). Moreover, we found persistence of LCMV in Grn–/– mice in the presence of P14 cells when compared with PGRN-expressing control animals (Figure 6B). ALT and aspartate aminotransferase (AST) activities were increased in Grn–/– hosts when compared with control animals (Figure 6C). Furthermore, when we analyzed the liver fibrosis marker α-smooth muscle actin (α-SMA), we found increased presence of α-SMA in Grn–/– liver tissue sections, when compared with control mice (Figure 6D). Next, we wondered whether PGRN affected T cell activation. Stimulation of WT CD8+ T cells with anti-CD3/anti-CD28 antibodies or PMA/ionomycin in the presence or absence of PGRN did not affect proliferation or activation of CD8+ T cells (Supplemental Figure 7, A and B). To further investigate the effects of PGRN on T cells, purified CD8+ T cells were activated with anti-CD3/anti-CD28 antibodies and recombinant PGRN for 24 hours. Flow cytometry analysis showed that PGRN had no effects on apoptosis of CD8+ T cells in this setting (Supplemental Figure 7C). Additionally, PGRN also had no effects on IFN-γ expression of CD8+ T cells in the absence or presence of PGRN, when splenocytes from LCMV-infected mice were rechallenged with gp33 peptides (Supplemental Figure 7D).
Figure 6. PGRN competent CD8+ T cells cannot restore antiviral immunity in Grn–/– mice.
A total of 3000 negatively sorted P14+ (TCR transgenic CD8+ T cells recognizing the LCMV epitope gp33) T cells were transferred into WT and Grn–/– mice followed by infection with 2 × 106 pfu LCMV-WE. (A) The absolute numbers of P14+ T cells were determined at the indicated time points after infection (n = 6). (B) Virus titers from spleen, liver, lung, kidney, spinal cord (SC), and brain tissue were determined at day 12 after infection (n = 6). (C) Serum ALT (left panel) and AST (right panel) activities were examined at the indicated time points after infection (n = 6). (D) Sections of snap-frozen liver tissue (day 12) were stained with α-smooth muscle actin (α-SMA) antibodies (n = 3). Right panel indicates quantification of fluorescence intensity. Scale bars: 100 μm. Data show mean ± SEM. Each symbol represents an individual mouse. P values calculated by 2-way ANOVA (A and C) and Student’s t test (B and D), *P < 0.05; **P < 0.001; ****P < 0.0001.
Taken together, these data indicate that PGRN deficiency resulted in decreased antiviral T cell immunity and increased pathology during LCMV infection and these effects were independent of T cell intrinsic effects.
PGRN deficiency results in excessive NK cell–mediated regulation of antiviral T cells.
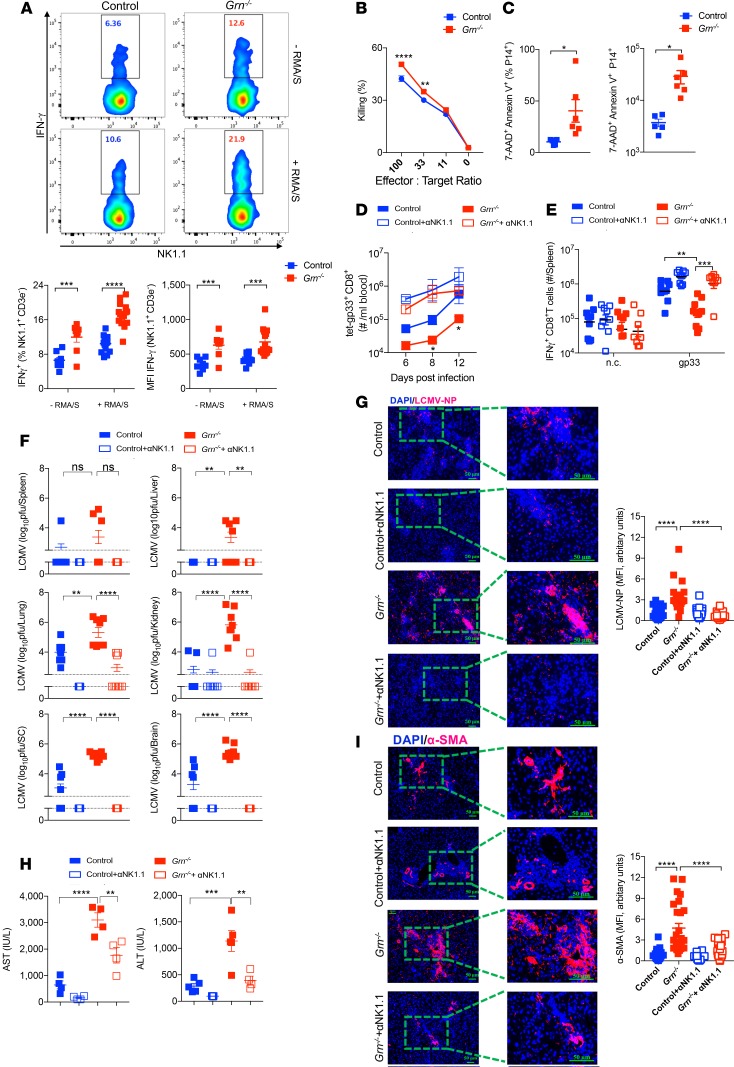
While PGRN had a direct NK cell inhibitory activity in vitro, absence of PGRN in vivo resulted into reduced T cell activity following infection. We therefore speculated that NK cells in Grn–/– mice limited antiviral T cells during LCMV infection, which could contribute to establishment of a persistent viral infection (26–28). We first examined the development of NK cell and innate lymphoid cells (ILCs) in naive animals. Grn–/– mice had the same amount of the NK cell precursors as control mice (Supplemental Figure 8A). Absolute numbers of both NK cell and ILC subsets were comparable between naive control and Grn–/– mice (Supplemental Figure 8, B and C). The surface receptor expression on NK cells from different organs was also similar in both groups before infection (Supplemental Figure 8D). In addition, NK cell numbers and surface NK cell receptor expression changed in a similar way in control and Grn–/– mice following LCMV infection (Supplemental Figure 9, A and B). NK cells from Grn–/– and control splenocytes showed comparable granzyme B and IFN-γ production following NK cell stimulation (Supplemental Figure 9, C and D). These data suggest that NK cell development was not dependent on PGRN. As PGRN can be produced in vitro by macrophages following LCMV infection, we mixed naive WT NK cells with LCMV-infected BMDMs from control and Grn–/– mice. CD27+CD11b+ NK cells have been shown to exhibit highly cytotoxic effects to RMA/S cells, whereas CD27–CD11b+ NK cells only displayed background levels of cytotoxicity in this setting (49). Following exposure of LCMV-infected Grn–/– BMDMs, we observed an increased presence of CD27+CD11b+ NK cells compared with control BMDMs (Supplemental Figure 9E).
NK cell–mediated IFN-γ production was increased in Grn–/– mice when compared with PGRN-competent mice during LCMV infection (Figure 7A). Notably, we monitored, that NK cell–mediated killing was slightly enhanced in ex vivo assays using splenocytes harvested from LCMV-infected Grn–/– mice compared with controls (Figure 7B). Consistently, when we transferred LCMV-specific P14 T cells, we observed increased cell death of transferred cells following LCMV infection in Grn–/– mice when compared with controls (Figure 7C). Notably, MHC-I expression levels on overall CD8+ T cells were not affected in Grn–/– mice during LCMV infection (Supplemental Figure 9F). We reasoned that antiviral T cells were being attacked by NK cells in the absence of PGRN. As expected, when we depleted NK cells in Grn–/– and control mice prior to infection with LCMV, antiviral CD8+ T cell immunity was restored (Figure 7D). Furthermore, we found highly increased CD8+ T cell–mediated IFN-γ production following restimulation with LCMV gp33 peptides in NK cell–depleted Grn–/– mice (Figure 7E). Notably, NK cell–depleted Grn–/– mice exhibited disease symptoms over the course of infection and were sacrificed, which is consistent with increased T cell immunity during LCMV infection (Supplemental Figure 9G and ref. 26). Consistently, LCMV was eliminated at day 12 after infection following NK cell depletion in Grn–/– mice, in sharp contrast with NK cell–competent Grn–/– mice, which showed high LCMV titers (Figure 7F). Furthermore, when we analyzed LCMV-NP expression in liver tissue, we found enhanced LCMV-NP expression in Grn–/– mice but reduced expression in NK cell–depleted Grn–/– mice (Figure 7G). At this time point, liver enzymes (AST/ALT; Figure 7H), as well as α-SMA expression (Figure 7I), were markedly reduced in the absence of NK cells in Grn–/– animals. Taken together, we conclude that PGRN reduces cytotoxic NK cell activity and, as a consequence, increases antiviral T cell immunity, resulting in clearance of LCMV.
Figure 7. NK cell depletion restores defective T Cell immunity and prevents pathology in Grn–/– mice.
(A and B) WT and Grn–/– mice were infected with 2 × 106 LCMV. Splenocytes were collected and incubated with RMA/S cells for 16 hours. (A) IFN-γ+ NK cells were measured by flow cytometry (n = 8–15). (B) NK cell–mediated killing to RMA/S cells were measured (n = 12–14). (C) WT and Grn–/– mice were transferred with 1 × 106 P14 T cells, followed by infection with 2 × 106 pfu LCMV. The frequency (left panel) and cell numbers (right panel) of apoptotic P14 cells were determined at day 2 after infection. (n = 5–6). (D–I) WT and Grn–/– mice were treated with anti-NK1.1 (αNK1.1) antibody, followed by infection with 2 × 106 pfu WE. (D) Gp33-specific CD8+ T cells were determined by flow cytometry in blood samples from all groups, as indicated at the specified time points after infection (n = 5–8. *P < 0.05 between Grn–/– and WT group). (E) IFN-γ+CD8+ T cells were measured by flow cytometry following gp33 peptide stimulation (n = 8–10). n.c. represents negative control. (F) Virus titers were measured in indicated organs at day 12 after infection (n =8–9). (G) Sections of snap-frozen liver tissue (day 12 after infection) were analyzed for LCMV-NP expression (n = 6–8). Right panel indicates quantification of fluorescence intensity. Scale bars: 50 μm. (H) Serum AST (left panel) and ALT (right panel) activities were measured at day 8 after infection (n = 4–5). (I) Sections of snap-frozen liver tissue harvested from all groups as indicated 12 days after infection were stained with α-SMA (n = 6–8). Right panel indicates quantification of fluorescence intensity. Scale bars: 50μm. Data show mean ± SEM. Each symbol represents an individual mouse. P values calculated by 2-way ANOVA (except for C by Student’s t test), *P < 0.05; **P < 0.001; ***P < 0.001; ****P < 0.0001; ns, not statistically significant between the indicated groups.
Discussion
In this study, we discovered that PGRN limited NK cell cytotoxicity through suppressing cyclin T1 and CDK9 expression in NK cells. Grn–/– mice demonstrated increased NK cell–mediated regulation of antiviral T cells during LCMV infection, which was associated with prolonged LCMV infection. In turn, NK cell depletion rescued defective T cell immunity and prevented viral persistence in Grn–/– mice during infection with LCMV.
As has been previously demonstrated, PGRN plays a role during immune reactions and inflammation. Absence of PGRN in mice results in increased cytokine production by macrophages and microglia, which can contribute to increased neuroinflammation (6). PGRN also promotes autophagy and is important during infections with L. monocytogenes (6, 15). Considering our data, increased NK cell activity in Grn–/– mice might also trigger enhanced activation of macrophages. Furthermore, depletion of NK cells results in accelerated clearance of L. monocytogenes (50), while activation of adaptive T cell immunity is protective (51). It is tempting to speculate that increased NK cell activity in the absence of PGRN might also cause diminished T cell immunity during L. monocytogenes infection and contribute to the reported severe pathological effects (6, 15). In addition, PGRN can prevent establishment of rheumatoid arthritis, while PGRN-deficient mice are highly susceptible to arthritis (5). Grn–/– mice have decreased Tregs compared with control animals and, therefore, exhibit enhanced inflammation during arthritis (52). Consistently, T cell proliferation could be reduced during PGRN treatment by induction of Tregs in bulk cultures (53). Tregs rather limit T cell immunity during LCMV infection (54). Hence, we speculate that NK cells, rather than Tregs, affect T cell immunity in our settings. Although the role of NK cells during the establishment of collagen-induced arthritis is not entirely clear (55), NK cells might exert protective effects by limiting T cell immunity (56, 57). In turn, reports have described that NK cells contribute to bone erosions during rheumatoid arthritis (58). Our data showing that PGRN inhibits NK cell immunity might either point to a pathological role of NK cells or NK cell–independent effects of PGRN during rheumatoid arthritis. PGRN can promote tumor growth and metastasis (2, 3). Considering our data, PGRN might also inhibit cytotoxic NK cell immunity against tumor cells and, through this mechanism, promote their growth and dissemination. Taken together, we have discovered an additional potentially novel mechanism by which PGRN, through its modulation of NK cells, might contribute to the course of bacterial and viral infections, autoimmune disease, and cancer.
Our findings indicate that PGRN affects NK cell effector molecule expression. Previous studies have shown that the PGRN binding receptor SORT1 regulates trafficking of vesicles within T cells (20). Accordingly, T cell–mediated cytotoxicity was reduced in sharp contrast to IFN-γ secretion (20). In our experimental setup, we observed reduced antiviral CD8+ T cell–mediated IFN-γ production following infection with LCMV. However, following NK cell depletion, IFN-γ production of LCMV-specific T cells was restored. Hence, we speculate that the observed defects during LCMV infection mainly depend on NK cells. Notably, we did not find an intrinsic difference between cultured NK cells from WT and Grn–/– mice. Therefore, PGRN as a modulator of NK cells is produced by other cell types, including innate immune cells. SORT1 has been shown to recruit PGRN toward the lysosome (13). Absence of PGRN results in defective autophagy and accumulation of inclusions of TDP-43 (15). Moreover, PGRN can bind to Lamp-1, which itself protects against perforin-mediated killing (22). However, as we still identified an inhibitory role of PGRN in perforin-deficient NK cells, PGRN likely does not affect autoregulation by NK cells. In contrast, our data show that PGRN reduces RNA expression levels of genes encoding for NK cell effector proteins. Expression of effector molecules triggering cytotoxicity is promoted by the P-TEFb subunits cyclin T1 and CDK9 (41). PGRN can bind to cyclin T1 and inhibit phosphorylation of the RNA polymerase II, which results in inhibition of transcription (40). Hence, we speculate that PGRN can reduce the cytotoxicity of NK cells through transcriptional downregulation of critical effector proteins. It remains unclear, however, whether these effects would also be observed in cytotoxic T cells, as — during LCMV infection — NK cell–mediated regulation of antiviral T cells could have masked and outweighed potential direct effects of PGRN on T cells.
In conclusion, we show that PGRN inhibits NK cell function in vitro and in vivo and critically affects the outcome of antigen-specific T cell immunity.
Methods
Mice.
Grn–/– (B6[Cg]-Grntm1.1Aidi/J) mice, which were previously described (from The Jackson Laboratories, stock no. 01375) (6), were used and compared with C57BL/6J mice. Rag1–/–, Ifnar1–/–, CD169-DTR, CD11c-DTR, and CD45.1+P14 mice were maintained under specific pathogen-free (SPF) conditions. For chimera experiments, WT mice were irradiated with 10.5 Gy. One day later, mice were reconstituted with BM cells from indicated donors. Six- to 8-week-old age-matched and sex-matched mice were used for all experiments. All mice used in this study were maintained in standard barrier facility at Heinrich-Heine-University Düsseldorf.
Virus.
LCMV strain WE was originally obtained from F. Lahmann-Grube (Heinrich Pette Institute, Hamburg, Germany). LCMV-WE was propagated in L929 cells as previously described (59). Virus titers were assessed using a plaque-forming assay as previously described (60).
Reagents.
Mouse-specific antibody to CD3ε (145-2C11, 11-0031-85), CD3e (17A2, 47-0032-82), NKp46 (29A1.4, 11-3351-82), CD11b (M1/70, 47-0112-82), IL-7R (A7R34, 11-1271-85), 2B4 (ebio244F4, 25-2441-82), CD5 (53-7.3, 47-0051-82), CD8 (53-6.7, 47-0081-82), CD19 (eBio1D3, 47-0193-82), Ly6G (RB6-8C5, 47-5931-82), TCR-β (H57-597, 47-5961-82), FcεR1 (MAR-1, 47-5898-82), Emoes (Dan11mag, 61-4875-82), CD4 (GK1.5, 48-0041-82), and GATA3 (TWAJ, 50-9966-42) from eBioscience were used. Mouse-specific antibody to NK1.1 (PK136, 25-5941-82), CD49b (DX5, 17-5971-81), granzyme B (NGZB, 12-8898-82), perforin (eBioOMAK-D, 11-9392-82), IFN-γ (XMG1.2, 17-7311-82), Tim3 (RMT3-23, 12-5870-82), CD16/32 (93, 11-0161-82), NKG2D (CX5, 25-5882-82), Ly49F/C/I/H (14B11, 12-5991-81), and Ter119 (TER119, 47-5921-82) were from Invitrogen. Mouse-specific antibody to CD27 (LG.3A10, 563365), CD44 (IM7, 563736), CD69 (H1.2F3, 561238), KLRG1 (2F1, 740553), CD62L (MEL-14, 563117), PD-1 (J43, 562523), CXCR5 (2G8, 563981), Lag3 (C9B7W, 563179), NKG2A/C/E (20d5, 740153), and RORγT (Q31-378, 562607) were from BD Biosciences. Human-specific antibody to CD56 (NCAM, 318328) was from BioLegend; IFN-γ (4S.B3, 11-7319-82), CD3e (OKT3, 45-0037-42), CD14 (61D3, 45-0149-42), CD19 (SJ25C1, 45-0198-42), and CD16 (eBioCB16, 47-0168-42) were from eBioscience; and CD107a (H4A3, 561348) was from BD Biosciences. Inhibitor (SNS-032) against cyclin T1/CDK9 was purchased from Selleck Chemicals (S1145). Mouse PGRN ELISA kit (EMGRN) and granzyme B (catalog 88-8022-22) were purchased from Invitrogen. Foxp3 mouse Treg Staining Buffer Set (eBioscience, 00-5523-00) was used. NK cell isolation kits (catalog 130-052-501) were from Miltenyi Biotec. Cell Proliferation Dye eFluor 450 (Invitrogen, 65-0842-85) was used for NK cell, Rma, and RMA/S cell labeling. Apoptotic cells or dead cells were stained with 7-AAD (Invitrogen, 00-6993-50) and Annexin V (BD Biosciences, 550474) in Annexin V staining buffer (BD Pharmingen, 51-66121E). Complete protease inhibitor cocktail (MilliporeSigma, 329-98-6) was used to lysate NK cells. RNeasy Mini Kits (250) (QIAGEN, 74106) were used for RNA extraction. Elastase was used for PGRN digestion to granulin (MilliporeSigma, E8140). CDK9 (Cell Signaling Technology, 2316S), cyclin T1 (Abcam, ab184703), phosphor-Ser2-RNA polymerase II (pSer2-R II, Abcam, ab193468) and α-SMA (Abcam, ab32575) antibodies were used. Anti–Rat IgG (Jackson ImmunoResearch, 112-116-072) were used for LCMV-NP staining; Cy3-conjugated goat anti–rabbit IgG (Jackson ImmunoResearch, 111-165-144) was used for cyclin T1 and phosphor-RNA polymerase II (pSer2-R II) flow cytometry staining. Horseradish peroxidase–conjugated (HRP-conjugated) goat anti–rat IgG (Jackson ImmunoResearch, 112-035-003) was used for plaque assasys. IRDye 800CW goat anti–rabbit IgG secondary antibody (LI-COR, 926-32211) was used for Western blotting (see complete unedited blots in the supplemental material). HRP-conjugated rabbit anti–β-actin antibody was used for Western blotting (Cell Signaling Technology, 5125S). PageRuler Prestained Protein Ladder (Thermo Fisher Scientific, 26617) was used. Recombinant mouse IL-2 (Miltenyi Biotec, 130-12-333); mouse IL-12 (BioLegend, 577002), mouse IL-15 (Peprotech, 210-15), and mouse IL-18 (BioLegend, 767002) were used. PMA (MilliporeSigma, P8139) and ionomycin calcium salt (MilliporeSigma, I0634) were used for T cell activation.
ELISA.
The serum from infected mice were collected for examining the PGRN levels at day 1 after infection (LCMV-WE). Granzyme B levels in the supernatant of NK cell cultures were measured by granzyme B ELISA kit. All experiments were performed according to the manufacturers’ instructions.
Recombinant human PGRN purification.
Stable PGRN-expressing HEK-293T cells were used. The supernatants from these cells had been collected and HiTrap TALON (GE Healthcare) crude columns, and AktaPrime were used for PGRN purification according to the manufacturers’ instructions.
Cell purification and culture.
Single-cell suspended splenocytes were enriched following the manufacturer’s instructions with the DX5 MACS kit. Isolated NK cells were expanded by 1,000 IU/ml IL-2 with or without indicated concentration of PGRN in RPMI-1640 (containing 10% FBS, 2 mM L-glutamin, and 100 U/ml penicillin-streptomycin) for 4 days in a humidified cell culture incubator at 37°C with 5% CO2. Vero, N2a neuroblastoma, and L929 cells were cultured in Eagle’s Minimum Essential Medium (Hyclone) supplemented with 10% FBS and 100 U/ml penicillin-streptomycin (Thermo Fisher Scientific) in a humidified cell culture incubator at 37°C with 5% CO2.
Flow cytometry.
Tetramer and intracellular cytokine staining were performed as described previously (59). For intracellular staining, Foxp3 mouse Treg staining buffer sets were used according to the manufacturer’s protocol. ILC and NK cell staining were performed as previously described (32). Experiments were performed using a FACS Fortessa (BD Bioscience) and analyzed with FlowJo software (Treestar).
NK cell cytotoxicity assays.
Freshly insolated NK cells were expanded by IL-2 for 4 days. Susceptible RMA/S and RMA cells were labeled by 10 μM Cell Proliferation Dye eFluor 450 and exposed to NK cells at indicated effectors/targets ratios. Eighteen hours later, 7-AAD was added, and the percentage of 7-AAD+ cells among eF450+ target cells was determined by flow cytometry. Control and Grn–/– mice were infected with LCMV-WE strain (2 × 106 pfu) for 24 hours. Splenocytes were collected and mixed with RMA/S cells at indicated effector/target ratio for 18 hours. 7-AAD was added, and the percentage of 7-AAD+ cells among eF450+ target cells was determined by flow cytometry.
Cell depletion.
NK cells were depleted with i.v. injections of anti-NK1.1 (clone PK136) as previously described (61). For depletion of macrophages, mice were treated with Clodronate liposomes (200 μl), and control mice were treated with empty control liposomes (200 μl) as previously described (62). For CD11c+ and CD169+ cell depletion, CD11c-DTR and CD169-DTR mice were injected with 0.01 mg/kg body weight diphtheria toxin (DT) for 24 hours.
IHC.
Histological analysis of snap-frozen tissue was performed as previously described by using anti–α-SMA and self-made anti–LCMV-NP monoclonal antibody (clone VL4) (32). NK cell cyclin T1 was stained using the Foxp3 staining kit, and these cells were cytospun onto slides. Cells were visualized with the Zeiss fluorescence microscope after staining with 4,6-diamidino-2-phenylindole.
Quantitative PCR.
RNA purification was performed according to the manufacturer’s instructions (QIAGEN). Gene expression analysis of Cdk9, Ccnt 1, Gzmb, Grn, and Prf1 were performed using kits from Applied Biosystems. All primers are listed in Supplemental Table 1. For analysis, the expression levels of all genes were normalized to GAPDH or Actin. Then, gene expression values were calculated based on the ΔΔCt method relative to controls.
Immunoblotting.
NK cells were lysed with lysis buffer (20 mM Tris-HCL, pH 7.5; 0.5% Nonidet P-40; 10 mM NaCl; and 3 mM EDTA) containing complete protease inhibitor cocktail for 30 minutes on ice. Cell lysates were separated by SDS-PAGE and analyzed with immunoblotting. Immunoblots were probed with primary antibody: cyclin T1, PGRN, CDK9, and β-actin, followed by secondary antibody or enhanced chemiluminescence detection of fluorescence secondary antibody, and detected by LI-COR imager (Odyssey Fc, LI-COR Biosciences).
BMDM culture.
BM cells were collected and cultured in DMEM supplemented with 10% FBS and 20% L929 cell culture supernatant. At day 7, differentiated BMDMs were treated with LCMV-WE (MOI, 1) for 48 hours. The levels of PGRN in these supernatants were measured by ELISA.
Statistics.
Data are expressed as mean ± SEM. For analysis of statistical significance between 2 groups, a Student’s t test (2-tailed) was used. For the analysis of human NK cell data, paired Student’s t test (2-tailed) was used. For the analysis of multiple time point experiments, 2-way ANOVA was used; a P value less than 0.05 was considered significant. Statistical analysis was performed using GraphPad Prism.
Study approval.
Cryopreserved PBMCs from 15 healthy donors were used in this study. Informed consent was obtained from each donor, and the study protocol was approved by the local ethics committee of the University Hospital Dusseldorf (Ethical number is NR: 2018-131-KFogU). Animal experiments were performed under the authorization of Landesamt für Natur, Umwelt, und Verbraucherschutz Nordrhein-Westfalen (LANUV) in accordance with German law for animal protection.
Author contributions
AH and PVS designed, performed, and analyzed experiments. TS and JT discussed and performed the human NK cell experiments. AH, JH, TS, HCX, CM, DH, TEW, JZ, AAP, JT, and SW performed and/or discussed experiments and analyzed data. KSL and PAL directed the study and wrote the manuscript.
Supplementary Material
Acknowledgments
This study was supported by the German Research Council (SFB974, LA2558/5-1, RTG1949), by the Jürgen Manchot Graduate School MOI III, by the NIH tetramer Facility, and by the Federal Ministry of Education and Research (Project 01GQ1422A to SW).
Version 1. 09/05/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019 American Society for Clinical Investigation
Reference information: JCI Insight. 2019;4(17):e129856. https://doi.org/10.1172/jci.insight.129856.
Contributor Information
Anfei Huang, Email: mm2huanganfei@gmail.com.
Prashant V. Shinde, Email: prashantshinde787@gmail.com.
Jun Huang, Email: erichuang2012@hotmail.com.
Tina Senff, Email: tina.senff@med.uni-duesseldorf.de.
Haifeng C. Xu, Email: chrishaifeng.xu@gmail.com.
Cassandra Margotta, Email: cassandra.margotta@gmail.com.
Dieter Häussinger, Email: haeussin@uni-duesseldorf.de.
Thomas E. Willnow, Email: willnow@mdc-berlin.de.
Jinping Zhang, Email: j_pzhang@suda.edu.cn.
Aleksandra A. Pandyra, Email: apandyra@gmail.com.
Jörg Timm, Email: Joerg.Timm@med.uni-duesseldorf.de.
Sascha Weggen, Email: sweggen@uni-duesseldorf.de.
Karl S. Lang, Email: KarlSebastian.Lang@uk-essen.de.
Philipp A. Lang, Email: philipp.lang@gmx.net.
References
- 1.Kao AW, McKay A, Singh PP, Brunet A, Huang EJ. Progranulin, lysosomal regulation and neurodegenerative disease. Nat Rev Neurosci. 2017;18(6):325–333. doi: 10.1038/nrn.2017.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He Z, Bateman A. Progranulin gene expression regulates epithelial cell growth and promotes tumor growth in vivo. Cancer Res. 1999;59(13):3222–3229. [PubMed] [Google Scholar]
- 3.He Z, Ismail A, Kriazhev L, Sadvakassova G, Bateman A. Progranulin (PC-cell-derived growth factor/acrogranin) regulates invasion and cell survival. Cancer Res. 2002;62(19):5590–5596. [PubMed] [Google Scholar]
- 4.He Z, Ong CH, Halper J, Bateman A. Progranulin is a mediator of the wound response. Nat Med. 2003;9(2):225–229. doi: 10.1038/nm816. [DOI] [PubMed] [Google Scholar]
- 5.Tang W, et al. The growth factor progranulin binds to TNF receptors and is therapeutic against inflammatory arthritis in mice. Science. 2011;332(6028):478–484. doi: 10.1126/science.1199214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yin F, et al. Exaggerated inflammation, impaired host defense, and neuropathology in progranulin-deficient mice. J Exp Med. 2010;207(1):117–128. doi: 10.1084/jem.20091568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martens LH, et al. Progranulin deficiency promotes neuroinflammation and neuron loss following toxin-induced injury. J Clin Invest. 2012;122(11):3955–3959. doi: 10.1172/JCI63113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park B, et al. Granulin is a soluble cofactor for toll-like receptor 9 signaling. Immunity. 2011;34(4):505–513. doi: 10.1016/j.immuni.2011.01.018. [DOI] [PubMed] [Google Scholar]
- 9.Baker M, et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442(7105):916–919. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- 10.Cruts M, et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442(7105):920–924. doi: 10.1038/nature05017. [DOI] [PubMed] [Google Scholar]
- 11.Neumann M, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314(5796):130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 12.Zhang YJ, et al. Progranulin mediates caspase-dependent cleavage of TAR DNA binding protein-43. J Neurosci. 2007;27(39):10530–10534. doi: 10.1523/JNEUROSCI.3421-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu F, et al. Sortilin-mediated endocytosis determines levels of the frontotemporal dementia protein, progranulin. Neuron. 2010;68(4):654–667. doi: 10.1016/j.neuron.2010.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka Y, Chambers JK, Matsuwaki T, Yamanouchi K, Nishihara M. Possible involvement of lysosomal dysfunction in pathological changes of the brain in aged progranulin-deficient mice. Acta Neuropathol Commun. 2014;2:78. doi: 10.1186/s40478-014-0078-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang MC, et al. Progranulin deficiency causes impairment of autophagy and TDP-43 accumulation. J Exp Med. 2017;214(9):2611–2628. doi: 10.1084/jem.20160999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lefrancois S, Zeng J, Hassan AJ, Canuel M, Morales CR. The lysosomal trafficking of sphingolipid activator proteins (SAPs) is mediated by sortilin. EMBO J. 2003;22(24):6430–6437. doi: 10.1093/emboj/cdg629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeng J, Racicott J, Morales CR. The inactivation of the sortilin gene leads to a partial disruption of prosaposin trafficking to the lysosomes. Exp Cell Res. 2009;315(18):3112–3124. doi: 10.1016/j.yexcr.2009.08.016. [DOI] [PubMed] [Google Scholar]
- 18.Zhou X, et al. Prosaposin facilitates sortilin-independent lysosomal trafficking of progranulin. J Cell Biol. 2015;210(6):991–1002. doi: 10.1083/jcb.201502029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou X, et al. Impaired prosaposin lysosomal trafficking in frontotemporal lobar degeneration due to progranulin mutations. Nat Commun. 2017;8:15277. doi: 10.1038/ncomms15277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herda S, et al. The sorting receptor Sortilin exhibits a dual function in exocytic trafficking of interferon-γ and granzyme A in T cells. Immunity. 2012;37(5):854–866. doi: 10.1016/j.immuni.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 21.Gowrishankar S, et al. Massive accumulation of luminal protease-deficient axonal lysosomes at Alzheimer’s disease amyloid plaques. Proc Natl Acad Sci USA. 2015;112(28):E3699–E3708. doi: 10.1073/pnas.1510329112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohnen A, et al. Surface CD107a/LAMP-1 protects natural killer cells from degranulation-associated damage. Blood. 2013;122(8):1411–1418. doi: 10.1182/blood-2012-07-441832. [DOI] [PubMed] [Google Scholar]
- 23.Cerwenka A, Lanier LL. Natural killer cell memory in infection, inflammation and cancer. Nat Rev Immunol. 2016;16(2):112–123. doi: 10.1038/nri.2015.9. [DOI] [PubMed] [Google Scholar]
- 24.Crouse J, Xu HC, Lang PA, Oxenius A. NK cells regulating T cell responses: mechanisms and outcome. Trends Immunol. 2015;36(1):49–58. doi: 10.1016/j.it.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 25.Crouse J, Kalinke U, Oxenius A. Regulation of antiviral T cell responses by type I interferons. Nat Rev Immunol. 2015;15(4):231–242. doi: 10.1038/nri3806. [DOI] [PubMed] [Google Scholar]
- 26.Waggoner SN, Cornberg M, Selin LK, Welsh RM. Natural killer cells act as rheostats modulating antiviral T cells. Nature. 2011;481(7381):394–398. doi: 10.1038/nature10624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lang PA, et al. Natural killer cell activation enhances immune pathology and promotes chronic infection by limiting CD8+ T-cell immunity. Proc Natl Acad Sci USA. 2012;109(4):1210–1215. doi: 10.1073/pnas.1118834109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cook KD, Whitmire JK. The depletion of NK cells prevents T cell exhaustion to efficiently control disseminating virus infection. J Immunol. 2013;190(2):641–649. doi: 10.4049/jimmunol.1202448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chiossone L, Dumas PY, Vienne M, Vivier E. Natural killer cells and other innate lymphoid cells in cancer. Nat Rev Immunol. 2018;18(11):671–688. doi: 10.1038/s41577-018-0061-z. [DOI] [PubMed] [Google Scholar]
- 30.Crouse J, et al. Type I interferons protect T cells against NK cell attack mediated by the activating receptor NCR1. Immunity. 2014;40(6):961–973. doi: 10.1016/j.immuni.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 31.Xu HC, et al. Type I interferon protects antiviral CD8+ T cells from NK cell cytotoxicity. Immunity. 2014;40(6):949–960. doi: 10.1016/j.immuni.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 32.Xu HC, et al. Lymphocytes Negatively Regulate NK Cell Activity via Qa-1b following Viral Infection. Cell Rep. 2017;21(9):2528–2540. doi: 10.1016/j.celrep.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 33.Khakoo SI, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305(5685):872–874. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 34.Thöns C, et al. HLA-Bw4 80(T) and multiple HLA-Bw4 copies combined with KIR3DL1 associate with spontaneous clearance of HCV infection in people who inject drugs. J Hepatol. 2017;67(3):462–470. doi: 10.1016/j.jhep.2017.03.040. [DOI] [PubMed] [Google Scholar]
- 35.Peppa D, et al. Up-regulation of a death receptor renders antiviral T cells susceptible to NK cell-mediated deletion. J Exp Med. 2013;210(1):99–114. doi: 10.1084/jem.20121172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crome SQ, et al. A distinct innate lymphoid cell population regulates tumor-associated T cells. Nat Med. 2017;23(3):368–375. doi: 10.1038/nm.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kägi D, et al. Cytotoxicity mediated by T cells and natural killer cells is greatly impaired in perforin-deficient mice. Nature. 1994;369(6475):31–37. doi: 10.1038/369031a0. [DOI] [PubMed] [Google Scholar]
- 38.Zhu J, et al. Conversion of proepithelin to epithelins: roles of SLPI and elastase in host defense and wound repair. Cell. 2002;111(6):867–878. doi: 10.1016/S0092-8674(02)01141-8. [DOI] [PubMed] [Google Scholar]
- 39.Madera S, Rapp M, Firth MA, Beilke JN, Lanier LL, Sun JC. Type I IFN promotes NK cell expansion during viral infection by protecting NK cells against fratricide. J Exp Med. 2016;213(2):225–233. doi: 10.1084/jem.20150712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoque M, Young TM, Lee CG, Serrero G, Mathews MB, Pe’ery T. The growth factor granulin interacts with cyclin T1 and modulates P-TEFb-dependent transcription. Mol Cell Biol. 2003;23(5):1688–1702. doi: 10.1128/MCB.23.5.1688-1702.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen R, et al. In vivo RNA interference screens identify regulators of antiviral CD4(+) and CD8(+) T cell differentiation. Immunity. 2014;41(2):325–338. doi: 10.1016/j.immuni.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Z, Zhu Q, Luo K, Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414(6861):317–322. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]
- 43.Pipkin ME, Sacks JA, Cruz-Guilloty F, Lichtenheld MG, Bevan MJ, Rao A. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity. 2010;32(1):79–90. doi: 10.1016/j.immuni.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu H, et al. Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature. 2018;558(7709):318–323. doi: 10.1038/s41586-018-0174-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Müller U, et al. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264(5167):1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 46.Lang PA, et al. Aggravation of viral hepatitis by platelet-derived serotonin. Nat Med. 2008;14(7):756–761. doi: 10.1038/nm1780. [DOI] [PubMed] [Google Scholar]
- 47.Zinkernagel RM, Haenseler E, Leist T, Cerny A, Hengartner H, Althage A. T cell-mediated hepatitis in mice infected with lymphocytic choriomeningitis virus. Liver cell destruction by H-2 class I-restricted virus-specific cytotoxic T cells as a physiological correlate of the 51Cr-release assay? J Exp Med. 1986;164(4):1075–1092. doi: 10.1084/jem.164.4.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pircher H, Bürki K, Lang R, Hengartner H, Zinkernagel RM. Tolerance induction in double specific T-cell receptor transgenic mice varies with antigen. Nature. 1989;342(6249):559–561. doi: 10.1038/342559a0. [DOI] [PubMed] [Google Scholar]
- 49.Hayakawa Y, Smyth MJ. CD27 dissects mature NK cells into two subsets with distinct responsiveness and migratory capacity. J Immunol. 2006;176(3):1517–1524. doi: 10.4049/jimmunol.176.3.1517. [DOI] [PubMed] [Google Scholar]
- 50.Takada H, Matsuzaki G, Hiromatsu K, Nomoto K. Analysis of the role of natural killer cells in Listeria monocytogenes infection: relation between natural killer cells and T-cell receptor gamma delta T cells in the host defence mechanism at the early stage of infection. Immunology. 1994;82(1):106–112. [PMC free article] [PubMed] [Google Scholar]
- 51.Lara-Tejero M, Pamer EG. T cell responses to Listeria monocytogenes. Curr Opin Microbiol. 2004;7(1):45–50. doi: 10.1016/j.mib.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 52.Wei F, Zhang Y, Zhao W, Yu X, Liu CJ. Progranulin facilitates conversion and function of regulatory T cells under inflammatory conditions. PLoS ONE. 2014;9(11):e112110. doi: 10.1371/journal.pone.0112110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kwack KH, Lee HW. Progranulin Inhibits Human T Lymphocyte Proliferation by Inducing the Formation of Regulatory T Lymphocytes. Mediators Inflamm. 2017;2017:7682083. doi: 10.1155/2017/7682083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Penaloza-MacMaster P, et al. Interplay between regulatory T cells and PD-1 in modulating T cell exhaustion and viral control during chronic LCMV infection. J Exp Med. 2014;211(9):1905–1918. doi: 10.1084/jem.20132577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ahern DJ, Brennan FM. The role of Natural Killer cells in the pathogenesis of rheumatoid arthritis: major contributors or essential homeostatic modulators? Immunol Lett. 2011;136(2):115–121. doi: 10.1016/j.imlet.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 56.Leavenworth JW, Wang X, Wenander CS, Spee P, Cantor H. Mobilization of natural killer cells inhibits development of collagen-induced arthritis. Proc Natl Acad Sci USA. 2011;108(35):14584–14589. doi: 10.1073/pnas.1112188108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lo CK, et al. Natural killer cell degeneration exacerbates experimental arthritis in mice via enhanced interleukin-17 production. Arthritis Rheum. 2008;58(9):2700–2711. doi: 10.1002/art.23760. [DOI] [PubMed] [Google Scholar]
- 58.Söderström K, et al. Natural killer cells trigger osteoclastogenesis and bone destruction in arthritis. Proc Natl Acad Sci USA. 2010;107(29):13028–13033. doi: 10.1073/pnas.1000546107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Welsh RM, Seedhom MO. Lymphocytic choriomeningitis virus (LCMV): propagation, quantitation, and storage. Curr Protoc Microbiol. 2008;Chapter 15:Unit 15A.1. doi: 10.1002/9780471729259.mc15a01s8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lang PA, et al. Reactive oxygen species delay control of lymphocytic choriomeningitis virus. Cell Death Differ. 2013;20(4):649–658. doi: 10.1038/cdd.2012.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koo GC, Peppard JR. Establishment of monoclonal anti-Nk-1.1 antibody. Hybridoma. 1984;3(3):301–303. doi: 10.1089/hyb.1984.3.301. [DOI] [PubMed] [Google Scholar]
- 62.Lang PA, et al. Tissue macrophages suppress viral replication and prevent severe immunopathology in an interferon-I-dependent manner in mice. Hepatology. 2010;52(1):25–32. doi: 10.1002/hep.23640. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.