Abstract
Idiopathic pulmonary fibrosis (IPF) is a chronic and fatal lung disease. A maladaptive epithelium due to chronic injury is a prominent feature and contributor to pathogenic cellular communication in IPF. Recent data highlight the concept of a “reprogrammed” lung epithelium as critical in the development of lung fibrosis. Extracellular vesicles (EVs) are potent mediators of cellular crosstalk, and recent evidence supports their role in lung pathologies, such as IPF. Here, we demonstrate that syndecan-1 is overexpressed by the epithelium in the lungs of patients with IPF and in murine models after bleomycin injury. Moreover, we find that syndecan-1 is a profibrotic signal that alters alveolar type II cell phenotypes by augmenting TGF-β and Wnt signaling among other profibrotic pathways. Importantly, we demonstrate that syndecan-1 controls the packaging of several antifibrotic microRNAs into EVs that have broad effects over several fibrogenic signaling networks as a mechanism of regulating epithelial plasticity and pulmonary fibrosis. Collectively, our work reveals new insight into how EVs orchestrate cellular signals that promote lung fibrosis and demonstrate the importance of syndecan-1 in coordinating these programs.
Keywords: Pulmonology
Keywords: Fibrosis
Syndecan-1 modulates packaging of anti-fibrotic microRNA into extracellular vesicles to regulate epithelial reprogramming as a mechanism to promote lung fibrosis.
Introduction
Idiopathic pulmonary fibrosis (IPF), a progressive fibrotic disease of the lungs, has a substantial impact on patient mortality, with a median survival of 2–3 years (1, 2). Although the abnormal fibroproliferation that occurs in IPF involves a complex interplay between parenchymal and immune cells, the epithelial barrier has a fundamental role, particularly in early events that lead to lung fibrosis (3). Recently, senescence or depletion of alveolar epithelial cells has been implicated in the pathogenesis of lung fibrosis (4–9). In addition, several epithelial-expressed genes have been found to confer a risk for the development of pulmonary fibrosis (e.g., MUC5B, SFTPC, SFTPA2, ABCA), further supporting dysfunctional alveolar epithelium as a sentinel event in the development of fibrosis (10).
Within the distal lung, alveolar type II (ATII) cells can undergo phenotypic changes and exhibit abnormalities referred to as “reprogramming” during fibrogenesis (11, 12). These alterations to the alveolar epithelium shift their normal reparative response to injury toward pathogenic consequences, such as loss of epithelial integrity and excessive matrix deposition (13). The process of alveolar epithelial reprogramming is clearly multifactorial, requiring alterations in several signaling pathways. One prominent example is TGF-β, a cytokine that is central to the fibrogenic process and can induce changes to the epithelial phenotype (14–17). Moreover, reactivated developmental pathways, such as Wnt signaling, can also affect alveolar epithelial plasticity in facilitating lung fibrosis (18–22).
Extracellular vesicles (EVs) are released from the cell and have the capacity to alter signaling networks in distal cells through intercellular delivery of various functional cargo (e.g., nucleic acids, proteins, lipids) (23, 24). Several subsets of EVs have been well described, with exosomes (40–150 nm in diameter) and microvesicles (50–1000 nm in diameter) being the predominant forms. Collectively, EVs play an important role in tissue homeostasis, and alterations to their production (quantity and quality) occur during and contribute to various diseases. In pathological states, dysregulation of EV cargo can have profound effects in disease by fundamentally altering the microenvironment and cellular phenotypes through disruption of normal homeostatic pathways while augmenting pathological signals. To this end, several studies reported EVs having a functional role in pulmonary diseases, such as acute lung injury, chronic obstructive pulmonary disease, and lung cancer (25–31). Only recently have studies begun unraveling the role that EVs play in mediating fibroproliferation (32–37).
Syndecan-1 is a type I transmembrane protein with glycosaminoglycan side chains that is localized to the epithelium, placing it in an ideal location to govern lung mucosal immune responses (38–43). The mechanisms by which syndecan-1 mediates its effects have revolved predominantly around its ability to act as a cell surface coreceptor or for the shed ectodomain to facilitate cell signals through presentation or sequestration of ligands with cognate receptors (44). Recently, Baetti et al. made a novel observation that syndecan-1 is found in exosomes and has a distinct role in biogenesis (45). Subsequent studies not only confirmed this seminal work but also demonstrated the ability of syndecan-1 to regulate the packaging of specific cargo within EVs (25, 46, 47).
Here, we identify a potentially novel phenotype where syndecan-1 overexpression by the epithelium during lung fibrosis promotes fibroproliferation. Our studies demonstrate that syndecan-1 supports reprogramming of the alveolar epithelium toward a profibrotic phenotype. Furthermore, we reveal that syndecan-1 controls alveolar epithelial plasticity by altering microRNA cargo within EVs that can govern networks of fibrogenic pathways, such as TGF-β and Wnt signaling.
Results
Syndecan-1 is overexpressed by the alveolar epithelium in fibrotic lungs.
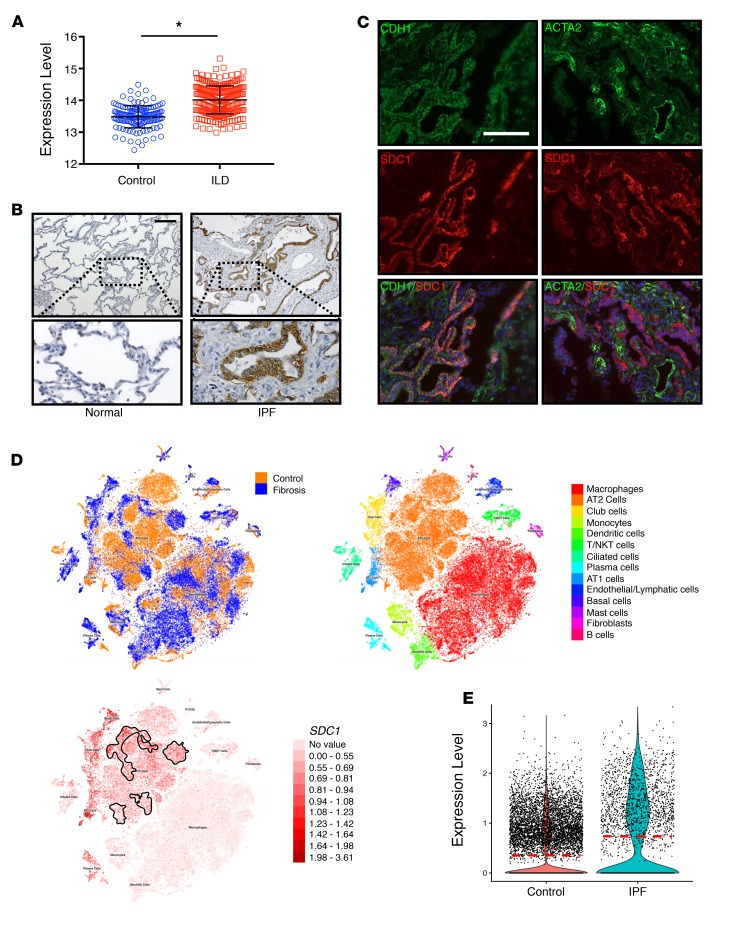
To discern the importance of syndecan-1 in pulmonary fibrosis, we first evaluated the expression of syndecan-1 in interstitial lung disease (ILD) populations by mining the Lung Genomics Research Consortium (LGRC) transcriptomic database (http://www.lung-genomics.org) and found that syndecan-1 expression was significantly elevated in the lungs of patients with ILD (Figure 1A). We then determined that syndecan-1 was overexpressed in IPF lungs and localized to the alveolar epithelium (Figure 1, B and C). Next, we used a recently published single-cell RNA-sequencing (scRNA-Seq) data set (48) to further localize syndecan-1 expression in IPF versus normal lungs and found that syndecan-1 was significantly upregulated by ATII cells in fibrotic lungs (Figure 1, D and E).
Figure 1. Syndecan-1 is overexpressed by alveolar type II epithelial cells in IPF lungs.
(A) Transcriptomic data was derived from the Lung Genomic Research Consortium (LGRC) database (http://www.lung-genomics.org) and analyzed for syndecan-1 expression in the lungs from normal patients and those with interstitial lung disease. Bars represent mean ± SEM. *P < 0.05 by 2-tailed Student’s t test. (B and C) Lung explants from normal and IPF patients were processed for (B) syndecan-1 immunohistochemical staining (brown; scale bar: 200 μm) (original magnification ×20) and (C) immunofluorescent staining for epithelial cells (CDH1; green), myofibroblasts (ACTA2; green), and syndecan-1 (SDC1; red); scale bar: 100 μm. (D and E) We analyzed a published whole-lung scRNA-Seq data set of control (n = 8) and IPF (n = 8) lungs (48). (D) Using the University of California Santa Cruz cell browser (http://www.nupulmonary.org/resources), cell populations were displayed in t-distributed stochastic neighbor embedding (t-SNE) plots. The IPF alveolar type II (ATII) cell cluster is outlined by a solid line (bottom). (E) Syndecan-1 expression was determined in ATII cells in control and IPF lungs and displayed in a violin plot. Horizontal red dotted line is the mean value.
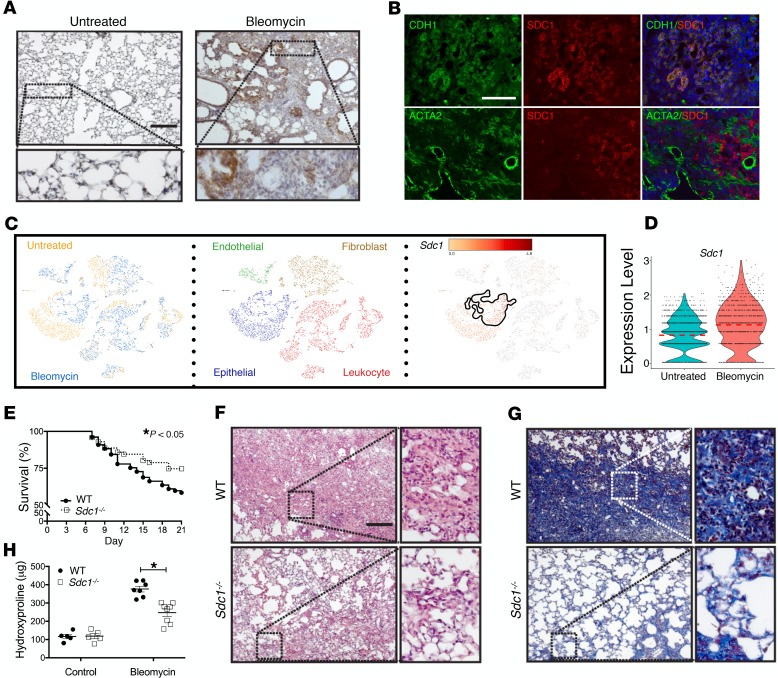
Bleomycin injury is a common murine model of lung fibrosis, so we evaluated syndecan-1 expression in mouse lungs after treatment. Similar to the findings in IPF lungs, syndecan-1 was overexpressed by the ATII cells in the fibrotic region of bleomycin-injured mice (Figure 2, A and B). We also evaluated syndecan-1 expression by scRNA-Seq analysis of bleomycin-injured mice and found augmented syndecan-1 expression by ATII cells in fibrotic lungs versus control (Figure 2, C and D). Altogether, our data reveal that syndecan-1 is overexpressed by ATII cells in both human IPF disease and murine bleomycin-induced lung fibrosis.
Figure 2. Syndecan-1 overexpression by the lung epithelium promotes fibrosis.
WT and Sdc1–/– mice were injured with bleomycin (0.75 unit/kg) and sacrificed 21 days after injury. Lungs from WT mice were processed for (A) syndecan-1 immunohistochemical staining (brown; scale bar: 200 μm) and (B) immunofluorescent staining for epithelial cells (CDH1; green), myofibroblasts (ACTA2; green), and syndecan-1 (SDC1; red); scale bar: 100 μm. (C) Lungs from WT control (n = 3) and bleomycin-injured (n = 3) mice were processed for scRNA-Seq, and t-SNE plots were generated by the Loupe Cell Browser (10× Genomics). Syndecan-1 expression in ATII cells in the bleomycin-injured condition is outlined by a solid line. (D) Syndecan-1 expression was determined in ATII cells in control and bleomycin-injured lungs and displayed in a violin plot. Horizontal red dotted line is the mean value. (E) Survival of bleomycin-injured WT and Sdc1–/– mice (n = 37 in each group); *P < 0.05 by log-rank (Mantel-Cox) survival analysis. (F) H&E staining and (G) Masson’s trichrome staining of bleomycin-injured WT and Sdc1–/– mouse lungs; scale bar: 200 μm. (H) Hydroxyproline content in lungs from bleomycin-injured WT and Sdc1–/– mouse lungs (n = 8 in each group). *P < 0.0001 by 2-way ANOVA.
Syndecan-1 overexpression is profibrotic.
The similarities in syndecan-1 overexpression between murine and human lung fibrosis suggest a conserved effect in lung fibrosis across species. Therefore, we injured wild-type (WT) and syndecan-1–deficient (Sdc1–/–) mice with bleomycin to determine the functional role of syndecan-1 during lung fibrosis. No appreciable difference was found between the uninjured lungs of WT and Sdc1–/– mice (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.129359DS1; ref. 38). WT mice exhibited significantly higher mortality (Figure 2E) and histologically developed more fibrosis in comparison with Sdc1–/– mice (Figure 2, F and G). Moreover, WT mice had higher lung collagen content (Figure 2H) and profibrotic gene expression (Supplemental Figure 2). Finally, we demonstrated syndecan-1 promotes fibroproliferation and expansion of the fibroblast compartment with more immunostaining of α–smooth muscle actin in lung histology from bleomycin-injured WT compared with Sdc1–/– mice (Supplemental Figure 3). Additionally, scRNA-Seq analysis of fibroblasts in WT and Sdc1–/– bleomycin-injured lungs demonstrated a 1.81-fold increased expression of Mki67, suggesting more proliferation of fibroblasts in WT compared with Sdc1–/– conditions (FDR < 0.05). These findings clarify the significance of syndecan-1 overexpression and implicate it in the pathogenesis of fibrosis in mice and humans.
Syndecan-1 promotes alveolar epithelial reprogramming toward a fibroproliferative phenotype.
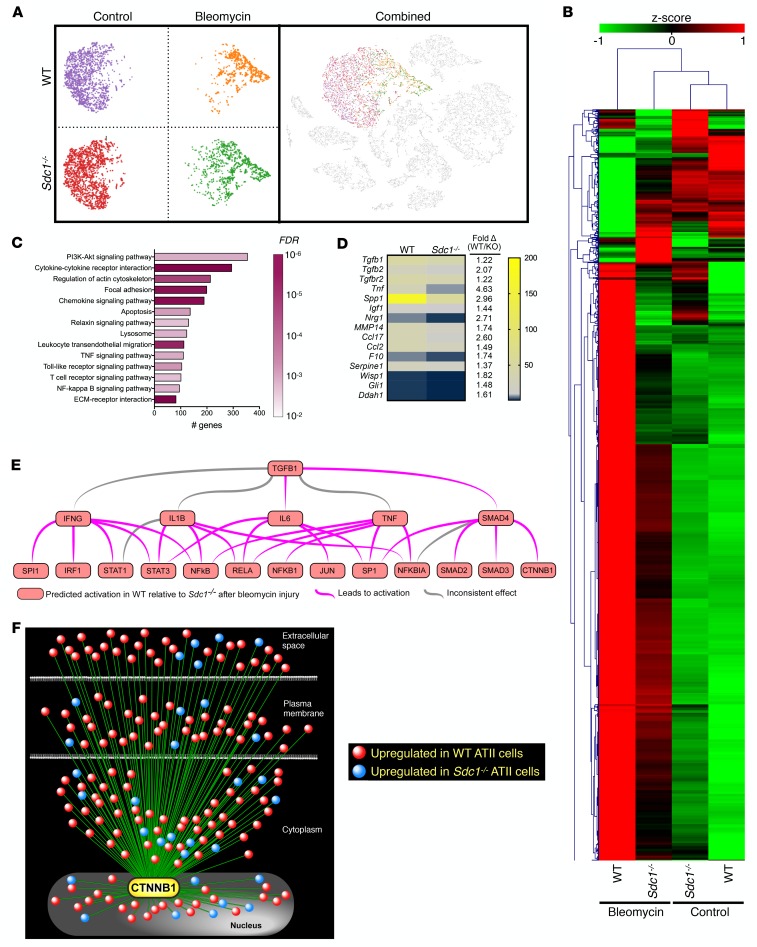
ATII cells, and more importantly their dysfunction, are an important driver of lung fibrosis (11–13). We postulated that aberrant syndecan-1 expression by ATII cells facilitates alterations to the phenotypes of these critical lung progenitors during lung fibrosis. Cultured cells can have altered behavior, so we used scRNA-Seq transcriptomic analysis of cells isolated from the lungs of control and bleomycin-injured WT and Sdc1–/– mice to understand how syndecan-1 controlled ATII behavior in vivo. Visualization by t-SNE plots revealed WT and Sdc1–/– ATII cells clustered based on exposure condition (Figure 3A). Moreover, a shift of ATII cell populations was seen following bleomycin treatment in both genotypes, indicating a strong transcriptional signal elicited by injury. However, when we evaluated the profile of differentially expressed genes based on the interaction between genotype and treatment, a different picture emerged wherein Sdc1–/– ATII cells from bleomycin-injured mice displayed transcriptional patterns more similar to untreated conditions (Figure 3B).
Figure 3. Syndecan-1 regulates alveolar epithelial reprogramming by facilitating TGF-β and Wnt signaling.
Uninjured or bleomycin-injured (0.75 unit/kg; 21 days) WT and Sdc1–/– mice (n = 3 per condition) were sacrificed and processed for scRNA-Seq of cells isolated from dissociated lungs. (A) t-SNE plots of ATII cells were generated by the Loupe Cell Browser (10× Genomics). (B) Hierarchical cluster analysis of differentially expressed genes (genotype × condition) in ATII cells between control and bleomycin-injured WT and Sdc1–/– mice. (C) Using genes significantly upregulated (FDR < 0.01) in ATII cells of bleomycin-injured WT lungs relative to Sdc1–/– lungs, a KEGG pathway analysis was performed to determine the significantly enriched functional pathways represented by the gene set. Bars indicate the number of genes in the pathway, and color indicates FDR level of the KEGG analysis. (D) Heatmap of the expression (transcript counts) of epithelial-derived profibrotic mediators that were significantly different in scRNA-Seq analysis of ATII cells from WT compared with Sdc1–/– mice. (E) Cluster network analysis using Ingenuity Pathway Analysis (QIAGEN Inc.; www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/) of differentially expressed genes between ATII cells in WT and Sdc1–/– mice identified TGF-β signaling as a highly significant upstream regulator (P < 10–60). (F) Wnt signaling interaction network using β-catenin (Ctnnb1) as a seed to visualize the connectivity of differentially expressed genes in ATII cells between WT and Sdc1–/– mice. The majority of genes interacting with Wnt signaling were upregulated in WT conditions. Please refer to Supplemental Figure 5 for an annotated version with the gene symbols.
To further understand how these changes correspond to differences in cellular phenotype, we performed functional enrichment analysis using the KEGG database on differentially expressed genes between WT and Sdc1–/– ATII cells. In contrast with downregulated genes, a distinctive enrichment pattern emerged with a preponderance of profibrotic signaling pathways among genes significantly upregulated after bleomycin injury in WT ATII cells (Figure 3C and Supplemental Table 1). Additionally, a number of profibrotic mediators are released by the lung epithelium in IPF (12), and we found that most of these genes were significantly upregulated in WT ATII cells after bleomycin injury, suggesting a dependence on syndecan-1 in their expression during fibrosis (Figure 3D).
These data suggest that syndecan-1 promotes ATII plasticity toward a fibrotic phenotype. To identify the regulatory signals modulated by syndecan-1 in mediating this effect, we performed causal network analysis using the set of differentially expressed genes following bleomycin injury between genotypes (49). This unbiased approach allows for statistical identification of upstream regulators as well as downstream consequences inferred from the overall changes found in the transcriptional responses. Interestingly, TGF-β was one of the most significant upstream regulators identified by this analysis (P < 10–60; Figure 3E), further supporting a role for syndecan-1 in promoting fibrogenesis. In addition, gene product network analysis using TGFB as a seed constructed an expansive interactome with relationships to many differentially expressed genes, which had a predominance with higher expression in the WT condition (Supplemental Figure 4). These results implicate TGF-β signaling as a dominant player in syndecan-1–dependent reshaping of ATII behavior during lung fibrosis.
We noted that β-catenin (Ctnnb1) was a major downstream consequence identified in the causal network analysis (Figure 3E). Reactivation of the developmental Wnt/β-catenin pathway contributes to ATII cell reprogramming and development of lung fibrosis (18–22, 48, 50). Therefore, we performed a network analysis using β-catenin as a seed and identified an extensive array of differentially expressed genes that interacted with the Wnt/β-catenin signaling pathway (Figure 3F and Supplemental Figure 5). Additionally, most of these genes had higher expression in WT compared with Sdc1–/– ATII cells, indicating that syndecan-1 overexpression augments Wnt/β-catenin signaling in ATII cells during lung fibrosis. Cumulatively, our data support the concept that syndecan-1 overexpression promotes lung fibrosis by facilitating a pathological reprogramming of ATII cells in vivo through modulation of fibrogenic pathways, such as TGF-β and Wnt/β-catenin.
EVs isolated from fibrotic lungs promote lung fibrosis.
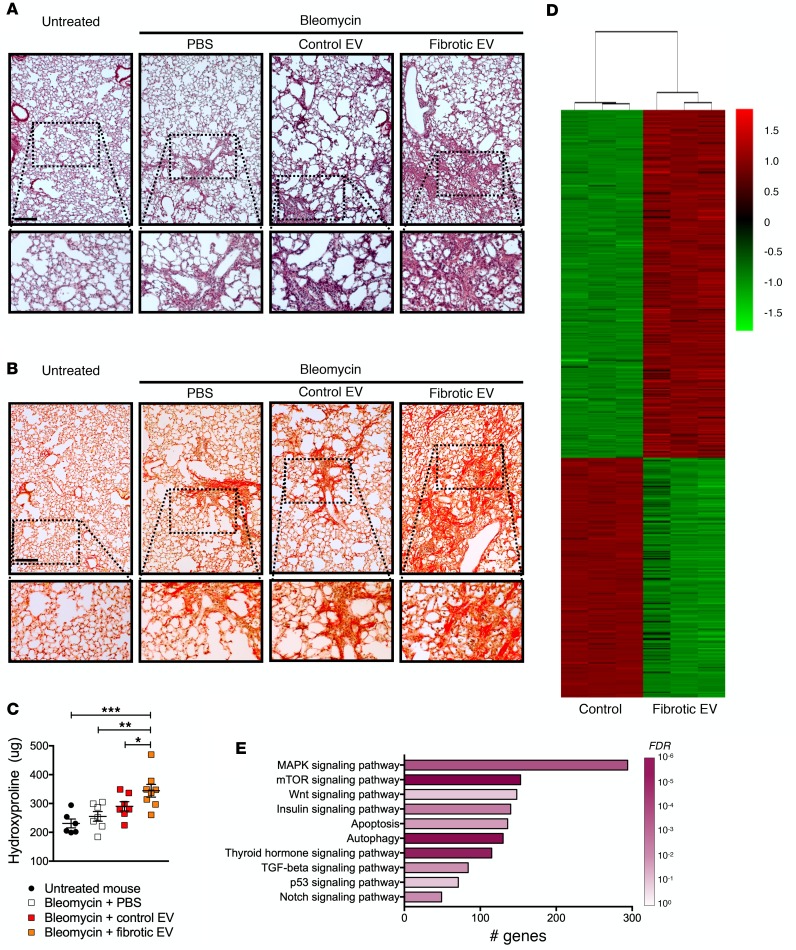
Recent studies have found that EVs carry profibrotic mediators that have fibrogenic consequences in vitro (34–37). However, no studies to date to our knowledge have determined the in vivo effects of EVs in lung fibrosis. Because cultured cells lose the context of a diseased microenvironment (e.g., matrix, other cells, microbiome, etc.), we chose to isolate EVs from the airspaces of fibrotic lungs (henceforth described as fibrotic EVs). EVs isolated from the airspaces originated primarily from epithelial cells, which is consistent with published results (Supplemental Figure 6 and refs. 29, 51). These fibrotic EVs were reinstilled into mice to test the functional consequences in lung fibrosis in vivo. No obvious effect on lung fibrosis was observed when fibrotic EVs isolated from bleomycin-injured mice were given to healthy mice (unpublished observations). In contrast, when WT mice were injured with a low dose of bleomycin that induced only mild fibrosis, fibrotic EVs augmented lung fibrosis (Figure 4, A–C). Similarly, mice treated with low-dose bleomycin and instilled with fibrotic EVs isolated from IPF lungs had less survival and trended toward more lung fibrosis in comparison with control EVs (Supplemental Figure 7). These results demonstrate that EVs in fibrotic lungs have the capacity to augment fibroproliferative signals.
Figure 4. EVs within fibrotic lungs have profibrotic properties.
(A–C) WT mice were injured with low-dose bleomycin (0.25 units/kg) and then given the respective treatments as indicated. Representative images of (A) H&E staining and (B) Picrosirius red staining (scale bar: 200 μm) on day 21 after bleomycin. Original magnification ×20. (C) Hydroxyproline content (n = 6–8 in each group) in the lungs after bleomycin treatment. *P < 0.05; **P < 0.005; ***P < 0.0005 by 1-way ANOVA analysis. (D and E) Mouse lung epithelial (MLE-12) cells were treated with control and fibrotic EVs and processed for RNA-Seq. (D) Heatmap of differentially expressed genes in MLE-12 cells treated with fibrotic EVs compared with control. (E) Profibrotic pathways identified by KEGG pathway analysis of upregulated MLE-12 cells in fibrotic EV–treated condition compared with control. Bars indicate the number of genes in the pathway and color indicates FDR level. Refer to Supplemental Table 2 for the entire gene list and KEGG pathway analysis.
To examine how fibrotic EVs functionally alter cellular signals, we evaluated murine lung epithelial cells treated with fibrotic EVs from bleomycin-injured lungs by RNA-Seq to identify the transcriptomic consequences on the cellular phenotype. A distinct change in the transcriptional architecture occurred with fibrotic EV treatment (Figure 4D and Supplemental Table 2). To understand the meaning of these changes, KEGG pathway enrichment analysis was performed using highly significantly upregulated and downregulated genes in fibrotic EV–treated cells compared with control (Supplemental Table 2). Many of the significant pathways identified in the KEGG analysis of upregulated genes mediate fibroproliferation (Figure 4E). Notably, TGF-β and Wnt signaling were identified, indicating fibrotic EVs can augment the signaling adapters of these important fibrogenic pathways. Furthermore, we treated cells with fibrotic EVs from IPF lungs and demonstrated an upregulation of Wnt/β-catenin signaling, which provides functional data supporting the concept that EVs promote profibrotic signaling (Supplemental Figure 8).
Syndecan-1 alters EV cargo to promote profibrotic reprogramming of epithelial cells.
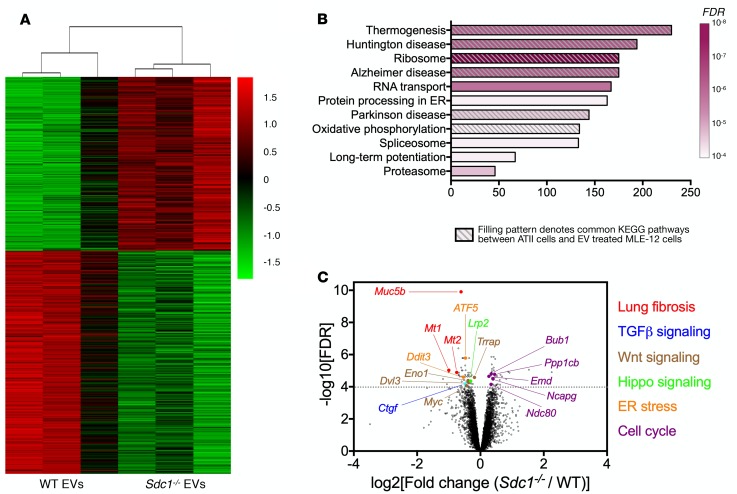
After establishing that fibrotic EVs can modulate fibroproliferation, our next step was to determine whether syndecan-1 overexpression facilitated the lung fibrotic phenotype through alterations in EV function. We incubated lung epithelial cells with fibrotic EVs from bleomycin-injured WT and Sdc1–/– mice to identify the syndecan-1–dependent transcriptomic changes in response to treatment (Figure 5A and Supplemental Table 3). KEGG pathway analysis of the differentially expressed genes between WT and Sdc1–/– fibrotic EV–treated cells revealed several significantly enriched pathways (Figure 5B and Supplemental Table 3). We noted a high degree of similarity in these pathways when compared to the significant KEGG pathways determined from the transcriptomic differences in ATII cells from IPF and normal lungs (using the scRNA-Seq data set from ref. 48; Supplemental Table 3). Indeed, 6 of 11 KEGG pathways (FDR < 0.0001) were identical (Figure 5B), suggesting that syndecan-1 regulates the ability of fibrotic EVs to alter epithelial cell plasticity similar to the changes found in IPF.
Figure 5. Syndecan-1 alters EV cargo to promote reprogramming of epithelial cells.
MLE-12 cells were treated with fibrotic EVs isolated from WT and Sdc1–/– mice lungs injured with bleomycin. Cells were subsequently processed for RNA-Seq. (A) Heatmap of differentially expressed genes between MLE-12 cells treated with fibrotic EVs from bleomycin-injured WT and Sdc1–/– mice. (B) KEGG pathway analysis of differentially expressed genes in WT and Sdc1–/– fibrotic EV–treated cells. Bars indicate the number of genes in the pathway, and color indicates FDR level (only highly significant pathways with FDR < 0.0001 are presented). Furthermore, KEGG pathway analysis was performed for differentially expressed genes in ATII cells using a published data set from an scRNA-Seq evaluation of IPF versus control lungs (48). The hashed filling pattern indicates the identical KEGG pathway identified between analysis of the 2 scRNA-Seq data sets (EV-treated lung epithelial cells and ATII cells from patients with IPF). Please refer to Supplemental Table 3 for the complete KEGG analysis and gene lists. (C) Volcano plot of the FDR level versus magnitude of differentially expressed genes in WT and Sdc1–/– fibrotic EV–treated cells. Highly significant genes (FDR > 0.0001) pertinent to lung fibrosis were identified and notated in the graph and color-coded into groups consistent with their effect.
To further investigate syndecan-1–dependent effects on the ability of fibrotic EVs to control cellular behavior, we evaluated individual genes that were highly significant in their differential expression between cells treated with WT and Sdc1–/– fibrotic EVs (Figure 5C and Supplemental Table 3). Interestingly, Muc5b was the most significant gene identified in our analysis, with more expression in WT fibrotic EV–treated conditions. Considering MUC5B is highly expressed in IPF lungs, and a promoter variant is the most predictive risk factor for IPF (52), identification of this gene provides additional support for the importance of syndecan-1 in modulating the fibrogenic properties of fibrotic EVs. Furthermore, several other genes were identified to be upregulated in the WT fibrotic EV condition that aligned with important fibrogenic pathways (e.g., TGF-β and Wnt) whereas cell cycle genes decreased, suggesting impairment of cell proliferation. Together, these results indicate syndecan-1 facilitates the ability of fibrotic EVs to reprogram lung epithelial cells toward a profibrotic phenotype.
Syndecan-1 controls packaging of antifibrotic microRNAs into EVs.
MicroRNAs (miRNAs) are important mediators of cellular signaling networks that can have distal effects via EV delivery (53). We have previously shown that syndecan-1 regulates miRNA packaging in EVs in lung cancer (25). Therefore, we postulated that syndecan-1 modulates cellular phenotypes during lung fibrogenesis by altering miRNA profiles in EVs. To test this hypothesis, we first performed miRNA-Seq of fibrotic EVs isolated from bleomycin-injured WT and Sdc1–/– mice. We found that in comparison with the Sdc1–/– condition, fibrotic EVs from WT mice had significantly lower amounts of 5 miRNAs (miR-144-3p, miR-142a-3p, miR-142b, miR-503-3p, and miR-34b-5p) (Figure 6A). Because miR-142b does not have a human homolog, we excluded this miRNA from further analysis. Moreover, we found that miR-144-3p and miR-142-3p (the human homolog of murine miR-142a-3p) had a similar trend of decreased EV levels in IPF versus control (which is analogous to WT versus Sdc1–/– conditions, respectively) (Supplemental Figure 9).
Figure 6. Syndecan-1 controls packaging of antifibrotic miRNAs into EVs.
WT and Sdc1–/– mice were injured with bleomycin (0.75 unit/kg) and sacrificed 21 days after injury. Fibrotic EVs were collected from the airspaces, and miRNAs were isolated for RNA-Seq. (A) Analysis of the miRNA levels within fibrotic EVs from bleomycin-injured WT and Sdc1–/– mice identified significant differences in 5 miRNAs. (B) Predicted miR-34b-5p binding site in the Muc5b 3′ UTR region was identified with http://www.microrna.org/microrna/home.do (C) Muc5b dot blot of MLE-12 cells transfected with control and miR-34b-5p mimics. (D) Luciferase activity of HEK cells transfected with a reporter plasmid containing a chimeric firefly luciferase with the Muc5b 3′ UTR and either a control or miR-34b-5p. *P < 0.05; Fluc/Rluc (firefly luciferase/renilla luciferase). (E) Predicted miR-144-3p binding site in the Tgfbr1 3′ UTR region was identified with http://www.microrna.org/microrna/home.do (F) Tgfbr1 immunoblot of MLE-12 cells transfected with the control or miR-144-3p mimics. (G) Luciferase activity of HEK cells transfected with a reporter plasmid containing a chimeric firefly luciferase with the Tgfbr1 3′ UTR and either a control or miR-144-3p. *P < 0.05. (H) TGF-β signaling regulatory network of experimentally identified targets of miR-34b-5p (blue), miR-503-5p (purple), miR-144-3p (brick red), and miR-142-3p (orange). These curated targets were experimentally validated either by UTR analysis (closed circle) or with pull-down methods (dotted circle). (I) Immunoblot for phosphorylated SMAD2 and SMAD2 using MLE-12 cells transfected with control or miR-144-3p mimics followed by TGF-β stimulation. (J) Wnt signaling regulatory network of experimentally identified targets (labeling identical to H). (K) Luciferase activity of HEK cells cotransfected with a TopFlash reporter plasmid and miRNAs as labeled. *P < 0.05; **P < 0.005; ***P < 0.0005 by 1-way ANOVA analysis.
We proceeded to identify the targets of these miRNAs to understand the effect of their changes in EV levels. Muc5b was the most significantly upregulated gene in lung epithelial cells treated with WT fibrotic EVs (Figure 5C and Supplemental Table 3). We found that Muc5b contained a miR-34-5p seed sequence in its 3′ UTR region (Figure 6B). Furthermore, miR-34b-5p mimics reduced MUC5B protein expression (Figure 6C) and targeted the Muc5b 3′ UTR in a dual-luciferase reporter assay (Figure 6D), supporting Muc5b as a target of miR-34b-5p. We evaluated Muc5b mRNA levels and did not detect its presence in WT and Sdc1–/– EVs (unpublished observations). Altogether, these findings indicate that the difference seen in Muc5b expression after EV treatment (Figure 5C and Supplemental Table 3) was due to miRNA suppression and not direct transfer of the Muc5b transcript.
Our data consistently pointed to syndecan-1–dependent augmentation of TGF-β signaling, so we determined potential miRNA targets in this critical profibrotic pathway. We found Tgfbr1 contained several miR-144-3p seed sequences in the 3′ UTR region (Figure 6E). Transfection of cells with miR-144-3p mimics significantly downregulated TGFBR1 (Figure 6F) and could suppress the Tgfbr1 3′ UTR reporter (Figure 6G). For the differentially packaged miRNAs in EVs (Figure 6A), we mined available literature and curated all the miRNA targets that were identified experimentally by either 3′ UTR suppression assays or pull-down of miRNA/mRNA complexes (e.g., HITS-CLIP; ref. 54). These miRNAs suppressed numerous TGF-β signaling adapters that spanned across both canonical and noncanonical pathways (Figure 6H and Supplemental Table 4). In addition to finding an extensive network of suppression, several miRNAs had identical targets (e.g., TGFBR1, SMAD4), which would allow synergistic regulation of these specific adapters. Finally, we found that miR-144-3p and miR-142-3p could suppress TGF-β signal transduction in cells stimulated with TGF-β1 (Figure 6I and Supplemental Figure 10). These results functionally demonstrate that specific EV cargo, the loading of which is syndecan-1 dependent, can dampen TGF-β signaling and serve as a mechanism of regulating lung fibrosis.
In addition to TGF-β signaling, our results consistently identified alterations in Wnt signaling. Therefore, we constructed a miRNA regulatory network of experimentally validated targets that encompassed key components of the Wnt signaling pathway (Figure 6J and Supplemental Table 4). Further, we evaluated the ability of these miRNAs to suppress β-catenin signaling and observed that they effectively governed activation of the Wnt/β-catenin canonical pathway (Figure 6K). Moreover, we also found these miRNAs regulated other processes that mediate profibrotic signals, such as cellular senescence, ER stress, and Notch signaling (Supplemental Figure 11 and Supplemental Table 4). These findings support a general model by which EVs, through miRNA cargo, exert an overarching regulatory control of multiple profibrotic pathways in fibrogenic diseases, such as IPF.
Syndecan-1 regulation of EV cargo promotes lung fibrosis.
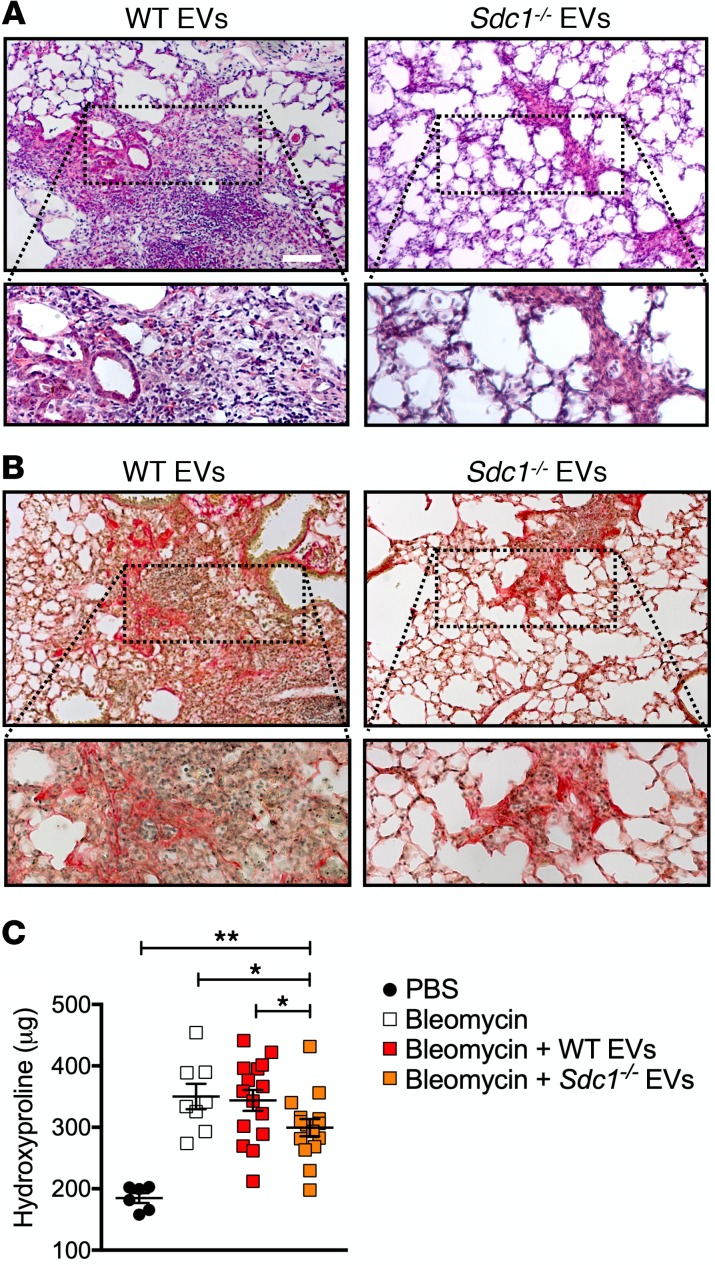
To elucidate the syndecan-1–dependent effects on the ability of fibrotic EVs to modulate lung fibrosis in vivo, we performed EV transfer experiments. Following bleomycin lung injury of WT mice, fibrotic EVs from the bronchoalveolar lavage of WT or Sdc1–/– conditions were delivered intratracheally. We found that mice treated with Sdc1–/– EVs had histological evidence of reduced lung fibrosis compared with those receiving WT EVs (Figure 7, A and B). Whole-lung collagen content was also significantly reduced in Sdc1–/– EV-treated mice as measured by hydroxyproline assay (Figure 7C). In contrast with the results with low-dose bleomycin–injured mice (Figure 4, A–C), fibrotic WT EVs did not augment fibrosis compared to saline control, which we attribute to the near maximal achievable fibrosis from the higher dose of bleomycin used in these experiments. These studies demonstrate that syndecan-1 has the capacity to drive lung fibrosis in vivo through the differential regulation of EV cargo.
Figure 7. Syndecan-1 dependence of EVs in modulating lung fibrosis.
WT mice were injured with bleomycin (0.75 unit/kg) and then intratracheally instilled with fibrotic EVs isolated from bleomycin-injured WT and Sdc1–/– mice. Representative images of (A) H&E and (B) Picrosirius red staining and (C) hydroxyproline content (n = 12–15 per group) of lungs recovered from WT versus Sdc1–/– fibrotic EV–treated mice; scale bar: 200 μm. Original magnification ×20.*P < 0.05; **P < 0.005 by 1-way ANOVA analysis.
Discussion
Reprogramming of the alveolar epithelium is an early event in a multistep process that culminates in the end-stage fibrosis of IPF (13). Our study provides important, potentially new information by revealing a syndecan-1–dependent mechanism where EVs modulate multiple signaling pathways that control epithelial plasticity and the development of lung fibrosis. Indeed, we demonstrate EVs from fibrotic lungs can augment profibrotic pathways and lung fibrosis. Moreover, syndecan-1 participates in this process by regulating antifibrotic miRNA cargo within the EVs. Interestingly, our data consistently showed a strong predilection for syndecan-1 in governing TGF-β and Wnt signaling among other important developmental pathways that are reactivated during fibrosis. Changes in the cellular response to biological signals is the basis of epithelial reprogramming, and our work builds upon this framework by providing evidence for syndecan-1–dependent EV regulation of profibrotic signals that govern epithelial plasticity in lung fibrosis.
Having pleiotropic effects, TGF-β is a key profibrotic cytokine that has well-known effects for stimulating extracellular matrix deposition (55, 56). Although activation of TGF-β is a major checkpoint for its function, our data suggest that EVs can modulate this pathway through delivery of miRNAs that suppress signaling adapters. Not only could the constellation of miRNA targets affect the overall magnitude of response to the receptor/ligand interaction, but a shift in the overall cellular response between canonical (i.e., SMAD) and noncanonical (i.e., PI3K, p38, ERK) signaling could occur depending on the preferential suppression of signaling adapters within each downstream signaling pathway. The same concept can be true for any of the other pathways (e.g., Wnt) that are regulated by the altered miRNAs and brings into focus how changes to the EV cargo could have dramatic effects on cellular phenotypes and lead to the epithelial reprogramming seen in lung fibrosis.
Syndecan-1 can regulate Wnt/β-catenin signaling in development and tumorigenesis by presenting Wnt ligands to the cognate receptor (57–59). This study provides a new level of understanding of how syndecan-1 controls Wnt signaling and the pathological consequences associated with disturbances in this pathway. Wnt/β-catenin signaling, an important developmental pathway, is reactivated and profibrotic in lung fibrosis (19–21). Furthermore, recent studies using scRNA-Seq approaches have reinforced the importance of aberrant Wnt activation in the ATII cells within the fibrotic lungs (48, 50). Wnt/β-catenin signaling is dysregulated in the aged lung and promotes senescence, and a pathogenic feature of reprogrammed fibrotic lung epithelial cells is cellular senescence (8, 9, 48, 60, 61). Altogether, these findings suggest that aberrant signaling through this pathway can promote lung fibrosis via initiation of cellular senescence. Our work further adds to this concept by demonstrating that EVs have regulatory roles by directly targeting cellular senescence and/or indirectly through Wnt/β-catenin signaling.
Although our results indicate syndecan-1 has the strongest effect on TGF and Wnt signaling, we also found effects on the unfolded protein response and developmental signals, such as Notch and hedgehog, which are functional participants in pathogenic lung fibroproliferation (22). Moreover, Muc5b was identified as the most significantly upregulated gene by an unbiased transcriptomic evaluation of cells treated with WT compared with Sdc1–/– EVs. This result is intriguing when considering a common promoter variant of MUC5B (rs35705950) is the strongest risk factor for the development of IPF (52). In fact, rs35705950 also is a risk factor for the development of lung fibrosis in patients with interstitial lung abnormalities and rheumatoid arthritis (62, 63). Patients with IPF have increased MUC5B expression independent of genetic risks (52), and increased Muc5b in the distal airways appears to impair mucociliary clearance as the mechanism of promoting fibrosis (64). Our findings uncover another regulatory step (i.e., EVs) governing its expression in fibrotic lungs.
The effects on profibrotic signaling pathways are mediated by miRNAs that are specifically packaged by syndecan-1–dependent mechanisms within EVs. By compiling the targets of these miRNAs (miR-144-3p, miR-142-3p, miR-34b-5p, miR-503-5p), we were able to demonstrate broad effects on multiple elements within each signaling pathway. The power of miRNAs to control signaling networks is their ability to simultaneously suppress multiple targets (65). Indeed, most studies, ours included, typically find modest changes of any given miRNA. In contrast, target identification studies typically transfect mimetics in vitro, resulting in hundreds-fold overexpression of the miRNA and demonstrating regulation of a signaling pathway. These studies are necessary for validation of miRNA targets, but this level of overexpression is rarely seen in vivo and likely does not reflect the true mechanism of action (i.e., through profound suppression of a singular target). Instead, our findings build a model by which miRNAs govern cellular signaling through simultaneous suppression of multiple components of a given pathway (66–69). As such, modest changes in multiple signaling adapters can synergize to modulate signal transduction. Moreover, pathological alterations in several miRNAs that have overlapping targets will further magnify this effect (70). The implication of our findings suggests that in contrast with a single therapeutic miRNA approach, which may have significant off-target effects, a better strategy may be to use combinatory miRNAs that have overlapping targets, thus allowing cooperative suppression of pathogenic pathways with lower dosing of any given miRNA.
The method by which miRNA cargo is packaged into EVs is still not fully elucidated. However, it is now evident that sorting of cargo into EVs is not a stochastic process, but instead, cellular mechanisms specifically facilitate loading (24). Indeed, certain miRNAs are found only in EVs but not in the cell of origin and vice versa, indicating cellular processes specifically shuttle cargo into the EV (71–73). In fact, 2 independent studies have identified miR-144-3p and miR-142-3p, both of which were identified in our study, to be preferentially loaded into EVs (73, 74). A general concept is that pathways mediating EV biogenesis also facilitate cargo loading (24). However, miRNAs may also be shuttled by RNA binding proteins into the EVs during biogenesis through specific binding motifs (71–73, 75–78). In this vein, our findings demonstrate that aberrant overexpression of syndecan-1 in lung fibrosis mediates its detrimental effects through the alteration of miRNA cargo loaded into EVs. It remains to be determined whether this is from the ability of syndecan-1 to regulate EV biogenesis or through interactions with RNA binding proteins. To date, efforts to target a single pathway in the pathogenesis of IPF have failed to yield successful therapies. In particular, targeting TGF-β and Wnt has been elusive, and both pirfenidone and nintedanib, which are approved for IPF, have pleiotropic targets (2). These data support the concept that targeting upstream mechanisms that mediate TGF-β and Wnt effector functions may be a novel approach to drug development for IPF.
There are numerous other possible cargoes that could also be adding to the fibrotic EV function (24). In fact, our data suggest that other unidentified factors exist. The miRNAs we identified had an overwhelming antifibrotic profile in the identified targets and do not explain the ability of fibrotic EVs to augment lung fibrosis in mice injured with low-dose bleomycin. It is possible that miRNA targets that have yet to be identified could have profibrotic consequences. However, the more plausible explanation is that fibrotic EVs also carry profibrotic factors. In support of this, we recently reported that fibrotic EVs isolated from IPF lungs carried functional WNT5A (34), but additional protein cargoes likely exist that drive fibrogenesis. Perhaps, other noncoding RNAs, such as long noncoding RNAs, may be present that antagonize antifibrotic miRNAs, resulting in a profibrotic state. To further complicate things, lipids, which are EV constituents, can also mediate lung fibrosis (79). Future studies will require multiomics approaches with integrated bioinformatic analyses to model the complicated interaction network of profibrotic and antifibrotic signals to fully appreciate the complex means by which EVs moderate lung fibrosis.
Our results reveal a role by which EVs orchestrate the multiple profibrotic pathways that lead to pulmonary fibrosis and provide new insight into how these fibrogenic signals are modulated. Furthermore, we demonstrate that syndecan-1 has a key role in controlling the EV loading of antifibrotic miRNAs. Our findings support an integrated model by which EVs coordinate system-wide networks of cellular signals whose disruption leads to pathological states. Discovering mechanisms that control these global changes in biological networks could provide novel therapeutic approaches to reverse pathological changes found in complex diseases, such as lung fibrosis.
Methods
Cell line cultures and transfections.
MLE-12 (originally provided by William Altemeier, University of Washington), human bronchial epithelial cells (16HBE14o-; originally provided by Steve White, University of Chicago, Chicago, Illinois, USA), and human embryonic kidney cells (HEK293; ATCC) were propagated and maintained in 1× DMEM (Corning) supplemented with 10% fetal bovine serum (Hyclone) and 1% penicillin-streptomycin (Corning) and cultured in 5% CO2 at 37°C. For EV treatment, EVs of a ratio of 100 particles per cell were used for all cell culture assays. Transfection of miRNAs and cotransfection of miRNAs and DNA plasmids were performed using lipofectamine RNAiMAX and lipofectamine 2000 (Life Technologies), respectively, per the manufacturer’s protocol.
Animals.
C57BL/6 WT and syndecan-1–null (Sdc1–/–) mice, age- and sex-matched and 8–12 weeks old, were maintained in a specific pathogen–free environment. All mice were from breeding of in-house colonies. For the bleomycin lung injury experiment, mice were anesthetized by isoflurane inhalation, and bleomycin (0.25–0.75 units/kg, APP Pharmaceuticals) or sterile Dulbecco’s phosphate-buffered saline (DPBS) was intratracheally administered once. At day 21 after injury, bronchoalveolar lavage fluid was collected for EV isolation, and lungs were harvested for histology, protein, RNA, and hydroxyproline assays. For all EV treatment experiments, EVs at 1 × 1010 particles diluted in 50 μl of sterile DPBS were administered intratracheally every other day beginning at day 7 following bleomycin injury for a total of 6 doses.
Protein assays, histology, and immunostaining.
Proteins were quantitated using bicinchoninic acid protein assay (Pierce, Thermo Fisher Scientific) per the manufacturer’s instructions. Western blot, dot blot, and hydroxyproline assays were performed as previously published (43, 66).
Paraffin-embedded, 10% normal formalin buffer–fixed human and mouse lung tissues were sectioned at 5-μm thickness and processed for histology studies. H&E, Masson’s trichrome, and Picrosirius red staining and immunostaining were performed according to laboratory protocols as previously described (66, 80). All antibody clones are listed in Supplemental Table 5.
RNA isolation, quantitative reverse transcriptase, and polymerase chain reactions.
TRIzol reagent (Thermo Fisher Scientific) was used to isolate RNA from animal lung tissue and mammalian cells as previously described (66). QIAGEN Inc. miRNeasy Micro Kit was used for small RNA isolation and purification. All mRNA primers were purchased from Integrated DNA Technology with detailed sequences provided in Supplemental Table 5. All miRNA primers were purchased from QIAGEN Inc. Quantitative PCR was performed using Applied Biosystems ViiA 7 Real-Time PCR (Thermo Fisher Scientific).
EV isolation and quantitation.
EVs were isolated by ultrafiltration centrifugation, quantified by Nanoparticle Tracking Analysis (Nanosight 300, Malvern Panalytical; Supplemental Figure 4), and further characterized by flow cytometry and Western blotting as previously described (81), which demonstrated the EVs to comprise largely exosomes. In most instances, EVs were immediately processed for in vivo and in vitro applications. Excess EVs were aliquoted and stored in –80°C for future use.
scRNA-Seq.
In scRNA-Seq experiments, freshly harvested WT and Sdc1–/– mouse lungs (control and bleomycin treated) were enzymatically digested with elastase (Worthington Biochemical Corporation) and mechanically disrupted to generate single-cell suspensions per previously published protocol (50). Mouse lung cells were hashed with mouse cell hashing antibodies (Totalseq, BioLegend) before pooling and sorting. The viable lung cell suspension (DAPI–) was flow sorted for lung immune cells (CD45+), endothelial cells (CD31+), epithelial cells (CD326+), and stromal cells (CD45–CD31–CD326–). Equal proportions of all 4 cell types were pooled and subjected to single-cell capture. Single-cell capture and library construction using 10× Genomics Chromium system and 3′ library and gel bead kit v2 (10× Genomics) followed standard vendor-guided protocol (82). Sequencing was performed using Illumina Novaseq 6000 (S2) system at UCLA Technology Center for Genomics and Bioinformatics.
RNA-Seq data analysis.
The detailed information for miRNA-Seq and data analysis was previously described by our group (25). The data set of EV miRNAs can be viewed through the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) database with accession number GSE134948. In bulk mRNA-Seq data, TopHat2 (2.0.7) was applied to align sequencing reads to the reference mouse genome (GRCm38/mm10) (83). The relative gene expression reads per kilobase of transcript, per million mapped reads (RPKM) and read counts were estimated using SAMMate (2.7.4) and Ensembl database (Mus Musculus.GRCm38.82) (84). Protein coding genes with at least 2 RPKM on average were used to perform the differential gene expression analysis using DESeq2 (85). To control for multiple testing, the P values of multiple tests were adjusted using Benjamini-Hochberg method (86), and the significance level was designated as FDR < 0.01. The mRNA data set can be viewed through GEO database with accession number GSE134741.
In scRNA-Seq analysis, CellRanger v3.0.2 software was used with the default settings for demultiplexing, aligning reads with STAR software to mm10 mouse reference genome, and counting unique molecular identifiers. Cell hashing data were demultiplexed using CITE-seq Count 1.4.1 (87). Downstream analysis was performed using the Seurat v2.3 R package (88) and Partek Flow software. Canonical correlation analysis was applied to integrate and combine data sets for unsupervised clustering. The t-SNE method was used for visualization of unsupervised clustering. Differentially expressed genes between treatment group and control groups of each cell type were identified using Seurat package, and significant differences were designated as an adjusted P < 0.01. The data set can be viewed through GEO database with accession number GSE131800.
For the bleomycin injury model in WT and Sdc1–/– ATII cells, one-way ANOVA analysis was performed to identify genes differentially expressed because of genotype by exposure (G × E) interaction. Adjusted P value (Benjamini-Hochberg) < 0.01 was used for the significance threshold. Upstream regulator and causal network analysis (Ingenuity Pathway Analysis software, QIAGEN Inc.; www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/) was applied to identify known transcriptional regulators by comparing the direction of change observed in our differentially expressed genes (G × E interaction) and expected effects based on prior knowledge (49). Functional enrichment analysis using KEGG pathways was performed using Webgestaldt as previously described (89).
Luciferase reporter assay.
The 3′ UTR binding site of Tgfbr1 and Muc5b was functionally assessed using methods as previously described (66).
Statistics.
One-way and 2-way ANOVA analyses were used for multiple comparisons. Student’s t test (2 tailed) was used for 2-group analysis. Survival comparison was performed with the log-rank and Gehan-Wilcoxon tests. All analyses were performed in GraphPad Prism 7.0. Results were presented as the mean ± SEM. P value < 0.05 was considered statistically significant.
Study approval.
Human lung tissue was collected at Cedars-Sinai Medical Center (CSMC) in accordance with guidelines of and with approval from the CSMC Institutional Board Review Committee. Animal experiments were conducted in accordance with and approved by the Institution Animal Care and Use Committee at CSMC.
Author contributions
TP, CY, LG, SAB, RB, DJ, PWN, CMH, TJA, MK, BRS, and PC contributed to conceptual design of all experiments. TP, CY, LG, RB, FAV, and SAB performed experiments. TP, CY, LG, RB, SAB, ND, MK, SAG, and PC analyzed data. DMH, CME, TX, DJ, TJA, CMH, OB, MK, LKMK, and BRS provided material and technical assistance. TP, CY, ND, SAB, PWN, MK, SAG, and PC were involved in manuscript writing.
Supplementary Material
Acknowledgments
The work is supported by NIH grants HL120947 (to PC), HL103868 (to PC), HL137076 (to PC), HL134637 (PWN), and AI137111 (to SAG); American Heart Association Grant-in-Aid (to PC); and the Samuel Oschin Comprehensive Cancer Institute Lung Cancer Research Award (to PC). We would like to thank the Smidt Heart Institute at CSMC for sharing a Nanosight 300 instrument for EV quantification.
Version 1. 08/08/2019
In-Press Preview
Version 2. 09/05/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(17):e129359.https://doi.org/10.1172/jci.insight.129359.
Contributor Information
Tanyalak Parimon, Email: tanyalak.parimon@cshs.org.
Changfu Yao, Email: Changfu.Yao@CSMC.edu.
David M. Habiel, Email: david.habiel@gmail.com.
Lingyin Ge, Email: lingyin.ge@wolfgreenfield.com.
Stephanie A. Bora, Email: stephanie.bora@cshs.org.
Rena Brauer, Email: rena.brauer@gmail.com.
Christopher M. Evans, Email: christopher.evans@ucdenver.edu.
Ting Xie, Email: ting.xie@cshs.org.
Felix Alonso-Valenteen, Email: Felix.alonso-valenteen@cshs.org.
Lali K. Medina-Kauwe, Email: Lali.Medina-Kauwe@cshs.org.
Dianhua Jiang, Email: Dianhua.Jiang@cshs.org.
Paul W. Noble, Email: paul.noble@cshs.org.
Cory M. Hogaboam, Email: Cory.Hogaboam@cshs.org.
Nan Deng, Email: Nan.Deng@cshs.org.
Olivier Burgy, Email: olivier.burgy@ucdenver.edu.
Travis J. Antes, Email: travantes@gmail.com.
Barry R. Stripp, Email: barry.stripp@cshs.org.
Sina A. Gharib, Email: sagharib@u.washington.edu.
Peter Chen, Email: peter.chen@cshs.org.
References
- 1.Ley B, Collard HR, King TE. Clinical course and prediction of survival in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2011;183(4):431–440. doi: 10.1164/rccm.201006-0894CI. [DOI] [PubMed] [Google Scholar]
- 2.Lederer DJ, Martinez FJ. Idiopathic pulmonary fibrosis. N Engl J Med. 2018;378(19):1811–1823. doi: 10.1056/NEJMra1705751. [DOI] [PubMed] [Google Scholar]
- 3.Wynn TA, Ramalingam TR. Mechanisms of fibrosis: therapeutic translation for fibrotic disease. Nat Med. 2012;18(7):1028–1040. doi: 10.1038/nm.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sisson TH, et al. Targeted injury of type II alveolar epithelial cells induces pulmonary fibrosis. Am J Respir Crit Care Med. 2010;181(3):254–263. doi: 10.1164/rccm.200810-1615OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naikawadi RP, et al. Telomere dysfunction in alveolar epithelial cells causes lung remodeling and fibrosis. JCI Insight. 2016;1(14):e86704. doi: 10.1172/jci.insight.86704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Povedano JM, et al. Therapeutic effects of telomerase in mice with pulmonary fibrosis induced by damage to the lungs and short telomeres. Elife. 2018;7:e31299. doi: 10.7554/eLife.31299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Povedano JM, Martinez P, Flores JM, Mulero F, Blasco MA. Mice with pulmonary fibrosis driven by telomere dysfunction. Cell Rep. 2015;12(2):286–299. doi: 10.1016/j.celrep.2015.06.028. [DOI] [PubMed] [Google Scholar]
- 8.Lehmann M, et al. Senolytic drugs target alveolar epithelial cell function and attenuate experimental lung fibrosis ex vivo. Eur Respir J. 2017;50(2):1602367. doi: 10.1183/13993003.02367-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang C, et al. Serpine 1 induces alveolar type II cell senescence through activating p53-p21-Rb pathway in fibrotic lung disease. Aging Cell. 2017;16(5):1114–1124. doi: 10.1111/acel.12643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kropski JA, Blackwell TS, Loyd JE. The genetic basis of idiopathic pulmonary fibrosis. Eur Respir J. 2015;45(6):1717–1727. doi: 10.1183/09031936.00163814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blackwell TS, et al. Future directions in idiopathic pulmonary fibrosis research. An NHLBI workshop report. Am J Respir Crit Care Med. 2014;189(2):214–222. doi: 10.1164/rccm.201306-1141WS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borok Z, et al. Cell plasticity in lung injury and repair: report from an NHLBI workshop, April 19–20, 2010. Proc Am Thorac Soc. 2011;8(3):215–222. doi: 10.1513/pats.201012-067CB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Selman M, Pardo A. Revealing the pathogenic and aging-related mechanisms of the enigmatic idiopathic pulmonary fibrosis. an integral model. Am J Respir Crit Care Med. 2014;189(10):1161–1172. doi: 10.1164/rccm.201312-2221PP. [DOI] [PubMed] [Google Scholar]
- 14.Kim KK, et al. Alveolar epithelial cell mesenchymal transition develops in vivo during pulmonary fibrosis and is regulated by the extracellular matrix. Proc Natl Acad Sci U S A. 2006;103(35):13180–13185. doi: 10.1073/pnas.0605669103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou B, et al. Interactions between β-catenin and transforming growth factor-β signaling pathways mediate epithelial-mesenchymal transition and are dependent on the transcriptional co-activator cAMP-response element-binding protein (CREB)-binding protein (CBP) J Biol Chem. 2012;287(10):7026–7038. doi: 10.1074/jbc.M111.276311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willis BC, et al. Induction of epithelial-mesenchymal transition in alveolar epithelial cells by transforming growth factor-beta1: potential role in idiopathic pulmonary fibrosis. Am J Pathol. 2005;166(5):1321–1332. doi: 10.1016/S0002-9440(10)62351-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim KK, et al. Epithelial cell alpha3beta1 integrin links beta-catenin and Smad signaling to promote myofibroblast formation and pulmonary fibrosis. J Clin Invest. 2009;119(1):213–224. doi: 10.1172/JCI36940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mutze K, Vierkotten S, Milosevic J, Eickelberg O, Königshoff M. Enolase 1 (ENO1) and protein disulfide-isomerase associated 3 (PDIA3) regulate Wnt/β-catenin-driven trans-differentiation of murine alveolar epithelial cells. Dis Model Mech. 2015;8(8):877–890. doi: 10.1242/dmm.019117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Königshoff M, et al. WNT1-inducible signaling protein-1 mediates pulmonary fibrosis in mice and is upregulated in humans with idiopathic pulmonary fibrosis. J Clin Invest. 2009;119(4):772–787. doi: 10.1172/JCI33950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Königshoff M, et al. Functional Wnt signaling is increased in idiopathic pulmonary fibrosis. PLoS One. 2008;3(5):e2142. doi: 10.1371/journal.pone.0002142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chilosi M, et al. Aberrant Wnt/β-catenin pathway activation in idiopathic pulmonary fibrosis. Am J Pathol. 2003;162(5):1495–1502. doi: 10.1016/S0002-9440(10)64282-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chanda D, Otoupalova E, Smith SR, Volckaert T, De Langhe SP, Thannickal VJ. Developmental pathways in the pathogenesis of lung fibrosis. Mol Aspects Med. 2019;65:56–69. doi: 10.1016/j.mam.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19(4):213–228. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 25.Parimon T, et al. Syndecan-1 controls lung tumorigenesis by regulating miRNAs packaged in exosomes. Am J Pathol. 2018;188(4):1094–1103. doi: 10.1016/j.ajpath.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bourdonnay E, et al. Transcellular delivery of vesicular SOCS proteins from macrophages to epithelial cells blunts inflammatory signaling. J Exp Med. 2015;212(5):729–742. doi: 10.1084/jem.20141675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider DJ, Speth JM, Penke LR, Wettlaufer SH, Swanson JA, Peters-Golden M. Mechanisms and modulation of microvesicle uptake in a model of alveolar cell communication. J Biol Chem. 2017;292(51):20897–20910. doi: 10.1074/jbc.M117.792416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moon HG, Cao Y, Yang J, Lee JH, Choi HS, Jin Y. Lung epithelial cell-derived extracellular vesicles activate macrophage-mediated inflammatory responses via ROCK1 pathway. Cell Death Dis. 2015;6:e2016. doi: 10.1038/cddis.2015.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee H, Zhang D, Zhu Z, Dela Cruz CS, Jin Y. Epithelial cell-derived microvesicles activate macrophages and promote inflammation via microvesicle-containing microRNAs. Sci Rep. 2016;6:35250. doi: 10.1038/srep35250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoshino A, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527(7578):329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Genschmer KR, et al. Activated PMN exosomes: pathogenic entities causing matrix destruction and disease in the lung. Cell. 2019;176(1–2):113–126.e15. doi: 10.1016/j.cell.2018.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Njock MS, et al. Sputum exosomes: promising biomarkers for idiopathic pulmonary fibrosis. Thorax. 2019;74(3):309–312. doi: 10.1136/thoraxjnl-2018-211897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Makiguchi T, et al. Serum extracellular vesicular miR-21-5p is a predictor of the prognosis in idiopathic pulmonary fibrosis. Respir Res. 2016;17(1):110. doi: 10.1186/s12931-016-0427-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin-Medina A, et al. Increased extracellular vesicles mediate WNT-5A signaling in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. doi: 10.1164/rccm.201708-1580OC. doi: 10.1164/rccm.201708-1580OC. [published online ahead of print July 25, 2018]. [DOI] [PubMed] [Google Scholar]
- 35.Chanda D, et al. Fibronectin on the surface of extracellular vesicles mediates fibroblast invasion. Am J Respir Cell Mol Biol. 2019;60(3):279–288. doi: 10.1165/rcmb.2018-0062OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lacy SH, et al. Activated human lung fibroblasts produce extracellular vesicles with antifibrotic prostaglandins. Am J Respir Cell Mol Biol. 2019;60(3):269–278. doi: 10.1165/rcmb.2017-0248OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang D, et al. Dysregulated lung commensal bacteria drive interleukin-17B production to promote pulmonary fibrosis through their outer membrane vesicles. Immunity. 2019;50(3):692–706.e7. doi: 10.1016/j.immuni.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 38.Brauer R, et al. Syndecan-1 attenuates lung injury during influenza infection by potentiating c-Met signaling to suppress epithelial apoptosis. Am J Respir Crit Care Med. 2016;194(3):333–344. doi: 10.1164/rccm.201509-1878OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Altemeier WA, et al. Transmembrane and extracellular domains of syndecan-1 have distinct functions in regulating lung epithelial migration and adhesion. J Biol Chem. 2012;287(42):34927–34935. doi: 10.1074/jbc.M112.376814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altemeier WA, Schlesinger SY, Buell CA, Parks WC, Chen P. Syndecan-1 controls cell migration by activating Rap1 to regulate focal adhesion disassembly. J Cell Sci. 2012;125(pt 21):5188–5195. doi: 10.1242/jcs.109884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen P, Abacherli LE, Nadler ST, Wang Y, Li Q, Parks WC. MMP7 shedding of syndecan-1 facilitates re-epithelialization by affecting alpha(2)beta(1) integrin activation. PLoS One. 2009;4(8):e6565. doi: 10.1371/journal.pone.0006565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gill SE, et al. Shedding of Syndecan-1/CXCL1 Complexes by matrix metalloproteinase 7 functions as an epithelial checkpoint of neutrophil activation. Am J Respir Cell Mol Biol. 2016;55(2):243–251. doi: 10.1165/rcmb.2015-0193OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Q, Park PW, Wilson CL, Parks WC. Matrilysin shedding of syndecan-1 regulates chemokine mobilization and transepithelial efflux of neutrophils in acute lung injury. Cell. 2002;111(5):635–646. doi: 10.1016/S0092-8674(02)01079-6. [DOI] [PubMed] [Google Scholar]
- 44.Teng YH, Aquino RS, Park PW. Molecular functions of syndecan-1 in disease. Matrix Biol. 2012;31(1):3–16. doi: 10.1016/j.matbio.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baietti MF, et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14(7):677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- 46.Roucourt B, Meeussen S, Bao J, Zimmermann P, David G. Heparanase activates the syndecan-syntenin-ALIX exosome pathway. Cell Res. 2015;25(4):412–428. doi: 10.1038/cr.2015.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Imjeti NS, et al. Syntenin mediates SRC function in exosomal cell-to-cell communication. Proc Natl Acad Sci U S A. 2017;114(47):12495–12500. doi: 10.1073/pnas.1713433114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reyfman PA, et al. Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am J Respir Crit Care Med. 2019;199(12):1517–1536. doi: 10.1164/rccm.201712-2410OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krämer A, Green J, Pollard J, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30(4):523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu Y, et al. Single-cell RNA sequencing identifies diverse roles of epithelial cells in idiopathic pulmonary fibrosis. JCI Insight. 2016;1(20):e90558. doi: 10.1172/jci.insight.90558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee H, Groot M, Pinilla-Vera M, Fredenburgh LE, Jin Y. Identification of miRNA-rich vesicles in bronchoalveolar lavage fluid: Insights into the function and heterogeneity of extracellular vesicles. J Control Release. 2019;294:43–52. doi: 10.1016/j.jconrel.2018.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seibold MA, et al. A common MUC5B promoter polymorphism and pulmonary fibrosis. N Engl J Med. 2011;364(16):1503–1512. doi: 10.1056/NEJMoa1013660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 54.Licatalosi DD, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456(7221):464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sime PJ, Xing Z, Graham FL, Csaky KG, Gauldie J. Adenovector-mediated gene transfer of active transforming growth factor-β1 induces prolonged severe fibrosis in rat lung. J Clin Invest. 1997;100(4):768–776. doi: 10.1172/JCI119590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Munger JS, et al. The integrin αvβ6 binds and activates latent TGF-β1: a mechanism for regulating pulmonary inflammation and fibrosis. Cell. 1999;96(3):319–328. doi: 10.1016/S0092-8674(00)80545-0. [DOI] [PubMed] [Google Scholar]
- 57.Alexander CM, et al. Syndecan-1 is required for Wnt-1-induced mammary tumorigenesis in mice. Nat Genet. 2000;25(3):329–332. doi: 10.1038/77108. [DOI] [PubMed] [Google Scholar]
- 58.Dejima K, Kang S, Mitani S, Cosman PC, Chisholm AD. Syndecan defines precise spindle orientation by modulating Wnt signaling in C. elegans. Development. 2014;141(22):4354–4365. doi: 10.1242/dev.113266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ren Z, et al. Syndecan-1 promotes Wnt/β-catenin signaling in multiple myeloma by presenting Wnts and R-spondins. Blood. 2018;131(9):982–994. doi: 10.1182/blood-2017-07-797050. [DOI] [PubMed] [Google Scholar]
- 60.Kovacs T, et al. Alteration in the Wnt microenvironment directly regulates molecular events leading to pulmonary senescence. Aging Cell. 2014;13(5):838–849. doi: 10.1111/acel.12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu H, et al. Augmented Wnt signaling in a mammalian model of accelerated aging. Science. 2007;317(5839):803–806. doi: 10.1126/science.1143578. [DOI] [PubMed] [Google Scholar]
- 62.Hunninghake GM, et al. MUC5B promoter polymorphism and interstitial lung abnormalities. N Engl J Med. 2013;368(23):2192–2200. doi: 10.1056/NEJMoa1216076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Juge PA, et al. MUC5B promoter variant and rheumatoid arthritis with interstitial lung disease. N Engl J Med. 2018;379(23):2209–2219. doi: 10.1056/NEJMoa1801562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hancock LA, et al. Muc5b overexpression causes mucociliary dysfunction and enhances lung fibrosis in mice. Nat Commun. 2018;9(1):5363. doi: 10.1038/s41467-018-07768-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ge L, et al. miR-323a-3p regulates lung fibrosis by targeting multiple profibrotic pathways. JCI Insight. 2016;1(20):e90301. doi: 10.1172/jci.insight.90301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu J, et al. MicroRNA-188 suppresses G1/S transition by targeting multiple cyclin/CDK complexes. Cell Commun Signal. 2014;12:66. doi: 10.1186/s12964-014-0066-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhu H, et al. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147(1):81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim NH, et al. A p53/miRNA-34 axis regulates Snail1-dependent cancer cell epithelial-mesenchymal transition. J Cell Biol. 2011;195(3):417–433. doi: 10.1083/jcb.201103097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pencheva N, et al. Convergent multi-miRNA targeting of ApoE drives LRP1/LRP8-dependent melanoma metastasis and angiogenesis. Cell. 2012;151(5):1068–1082. doi: 10.1016/j.cell.2012.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shurtleff MJ, et al. Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proc Natl Acad Sci U S A. 2017;114(43):E8987–E8995. doi: 10.1073/pnas.1712108114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Koppers-Lalic D, et al. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. 2014;8(6):1649–1658. doi: 10.1016/j.celrep.2014.08.027. [DOI] [PubMed] [Google Scholar]
- 73.Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, Ri S, Schekman R. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. Elife. 2016;5:e19276. doi: 10.7554/eLife.19276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guduric-Fuchs J, O’Connor A, Camp B, O’Neill CL, Medina RJ, Simpson DA. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genomics. 2012;13:357. doi: 10.1186/1471-2164-13-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Villarroya-Beltri C, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Santangelo L, et al. The RNA-binding protein SYNCRIP is a component of the hepatocyte exosomal machinery controlling microRNA sorting. Cell Rep. 2016;17(3):799–808. doi: 10.1016/j.celrep.2016.09.031. [DOI] [PubMed] [Google Scholar]
- 77.Teng Y, et al. MVP-mediated exosomal sorting of miR-193a promotes colon cancer progression. Nat Commun. 2017;8:14448. doi: 10.1038/ncomms14448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bolukbasi MF, et al. miR-1289 and “Zipcode”-like sequence enrich mRNAs in microvesicles. Mol Ther Nucleic Acids. 2012;1:e10. doi: 10.1038/mtna.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tager AM, et al. The lysophosphatidic acid receptor LPA1 links pulmonary fibrosis to lung injury by mediating fibroblast recruitment and vascular leak. Nat Med. 2008;14(1):45–54. doi: 10.1038/nm1685. [DOI] [PubMed] [Google Scholar]
- 80.Chen P, et al. Tissue inhibitor of metalloproteinase-1 moderates airway re-epithelialization by regulating matrilysin activity. Am J Pathol. 2008;172(5):1256–1270. doi: 10.2353/ajpath.2008.070891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Parimon T, Garrett NE, Chen P, Antes TJ. Isolation of extracellular vesicles from murine bronchoalveolar lavage fluid using an ultrafiltration centrifugation technique. J Vis Exp. 2018;(141):e58310. doi: 10.3791/58310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xie T, et al. Single-cell deconvolution of fibroblast heterogeneity in mouse pulmonary fibrosis. Cell Rep. 2018;22(13):3625–3640. doi: 10.1016/j.celrep.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xu G, Deng N, Zhao Z, Judeh T, Flemington E, Zhu D. SAMMate: a GUI tool for processing short read alignments in SAM/BAM format. Source Code Biol Med. 2011;6(1):2. doi: 10.1186/1751-0473-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodological) 1995;57(1):289–300. [Google Scholar]
- 87.Stoeckius M, et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods. 2017;14(9):865–868. doi: 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen P, Edelman JD, Gharib SA. Comparative evaluation of miRNA expression between in vitro and in vivo airway epithelium demonstrates widespread differences. Am J Pathol. 2013;183(5):1405–1410. doi: 10.1016/j.ajpath.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.