Abstract
Purpose of review:
The influence of environmental factors on Type 2 diabetes (T2D) risk is now well recognized and highlights the contribution of epigenetic mechanisms. This review will focus on the role of epigenetic factors in the risk and pathogenesis of T2D.
Recent findings:
Epigenetic dysregulation has emerged as a key mechanism underpinning the pathogenesis of T2D and its complications. Environmental variations, including alterations in lifestyle, nutrition, and metabolic demands during prenatal and postnatal life, can induce epigenetic changes that may impact glucose homeostasis and the function of different metabolic organs. Accumulating data continues to uncover the specific pathways that are epigenetically dysregulated in T2D, providing an opportunity for therapeutic targeting.
Summary:
Environmental changes can disrupt specific epigenetic mechanisms underlying metabolic homeostasis, thus contributing to T2D pathogenesis. Such epigenetic changes can be transmitted to the next generation, contributing to the inheritance of T2D risk. Recent advances in epigenome wide association studies and epigenetic editing tools presents the attractive possibility of identifying epimutations associated with T2D, correcting specific epigenetic alterations, and designing novel epigenetic biomarkers and interventions for T2D.
Keywords: Epigenetics, type 2 diabetes, glucose homeostasis, diabetes complications, biomarkers, epigenetic therapies
Introduction
Type 2 diabetes (T2D) is defined by hyperglycemia, and results from metabolic syndrome and inadequate insulin availability in response to relative insulin resistance. T2D is fast turning into a global pandemic [1]. T2D and chronic hyperglycemia can also lead to significantly increased rates of multiple micro- and macrovascular complications such as retinopathy, nephropathy and neuropathy, as well as atherosclerosis [2, 3].
T2D pathogenesis has a strong hereditary component, such that family history of disease confers a much higher risk of developing T2D [4, 5]. This recognition has led to an intense search for genetic factors responsible for T2D pathogenesis. While genome-wide association studies have identified multiple loci associated with T2D risk [6], genetic factors account for only a small fraction of diabetes associated with family history [7]. Furthermore, adult-onset diabetes was recently recognized to be a heterogeneous disease with five subgroups differing in disease progression and complications risk [8]. The incidence of T2D has increased drastically in the past few decades, coincident with the increase in food availability, sedentary lifestyle, and obesity [9]. However, this time span is unlikely to cause significant changes in the human genome. Altogether, this points to a strong influence of environmental factors and gene-environment interactions on obesity and T2D risk [10].
The effect of environmental factors including diet, physical activity, circadian rhythms, stress, temperature etc. on gene expression can be mediated by epigenetic mechanisms, which dictate how cells respond and adapt to their environment [11]. Epigenetic changes refer to mitotically or meiotically heritable changes in gene function without alterations in the underlying DNA sequence. Epigenetic regulation of gene expression occurs through changes in chromatin accessibility, mediated by the individual or combinatorial involvement of multiple mechanisms such as DNA cytosine methylation, histone post-translational modifications, and noncoding RNAs [3, 12]. Environmental changes can drive transient or persistent changes in the epigenome, which may alter gene expression and cellular phenotypes. For example, metabolic variations can directly alter the epigenome, given that many enzymatic regulators of epigenetic modifications require metabolic intermediates as cofactors [13]. Epigenetic mechanisms play a critical role in governing the expression of key genes involved in the development and homeostasis of metabolic organs, such as the pancreatic insulin-producing beta cells [14]. An altered metabolic state can thus affect the epigenome and phenotype of different organs, and contribute to the development of T2D and its multiple peripheral complications [3]. The present review focuses on the epigenetic basis of glucose homeostasis in health and diabetes, and potential implications for epigenetic biomarkers and therapies.
Developmental origins of T2D risk: the contribution of epigenetics
A strong case for the involvement of epigenetic factors in T2D is made by studies on the effect of maternal and intrauterine nutrition and growth retardation on diabetes development in multiple species [15]. Studies on the Dutch Hunger Winter famine have shown that intrauterine malnutrition and low birth weight leads to an increased likelihood for developing diabetes in subsequent generations [16]. This phenomenon, referred to as the “thrifty phenotype hypothesis”, proposes that under-nutrition during development leads to permanent changes in glucose homeostasis [17]. Similarly, maternal over-nutrition (such as a high-fat diet) and gestational diabetes can also adversely affect the metabolic health of the offspring [15]. Impaired glucose homeostasis in the parent has been shown to alter the metabolic program in the offspring coincident with very specific epigenetic changes, suggesting an epigenetic basis for the transmission of metabolic disease risk [16, 18, 19]. Thus, factors such as poor maternal health, as well as over- and under-nutrition during the fetal and postnatal growth phase can impact the development and function of key metabolic organs, and predispose the offspring to metabolic syndrome and diabetes in early or later life [11, 15]. Environmentally induced epigenetic alterations can also occur in the germline, and may therefore be potentially transmitted to subsequent generations, contributing to the (epigenetic) inheritance of diabetes risk (reviewed in [20]).
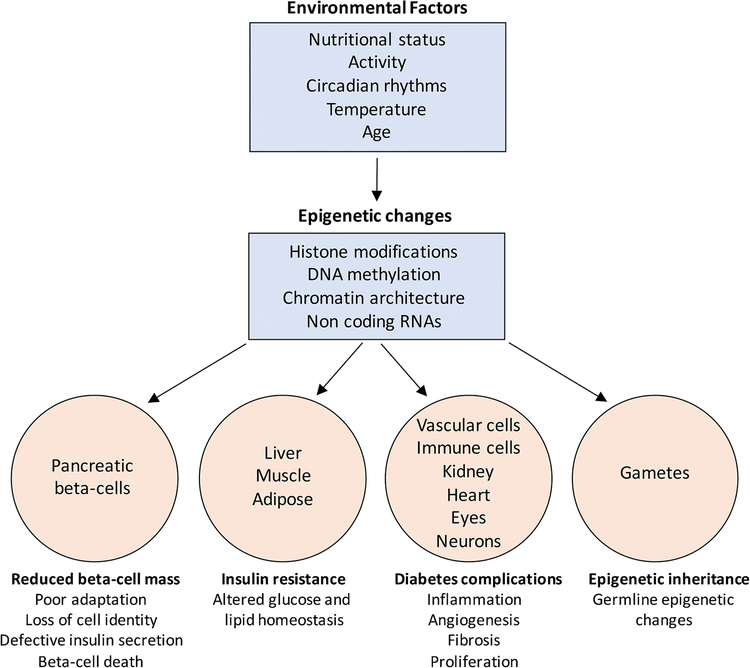
While the contribution of maternal health to disease risk in the offspring is well recognized (reviewed in [15]), recent data suggest that the epigenome of male germ cells is also altered by nutritional imbalance during intrauterine life [21], and can influence gene regulation during the development of the offspring. Paternal diet has been shown to influence cholesterol and lipid metabolism in the offspring [18]. Over- and under-nutrition, as well obesity in the paternal generation leads to reprogramming of the sperm epigenome, resulting in a trans-generational influence on metabolic homeostasis in the offspring [22, 23]. These studies suggest that parental metabolic environment and lifestyle can induce transgenerational changes in epigenome and metabolic fitness. Disturbances in the epigenetic regulation of imprinted genes (genes with differential allelic regulation based on parental origin) can further dictate the pattern of inheritance of diabetes risk [24]. Epigenetic factors may therefore not only mediate the effect of environmental factors on the development of T2D and its various complications, but also contribute to the transmission of disease risk to subsequent generations (Figure 1).
Figure 1.
Variations in environmental factors such as nutritional status (diet), activity (sedentary lifestyle), circadian rhythms (sleep disruption), seasonal changes in temperature, and even aging can alter the cellular epigenome. These changes may occur in the histone modifications, DNA methylation patterns, chromatin accessibility, as well as the expression of non-coding RNA species such as lncRNAs and miRNAs. The epigenetic dysregulation in response to adverse environmental exposure in turn drives transcriptional changes across several tissues such as the insulin producing beta-cells and insulin sensitive organs including liver, muscle, and adipose. This can eventually induce a deficit of functional beta-cell mass and impaired insulin secretion, as well as drive insulin resistance, thus disrupting glucose homeostasis towards the pathogenesis of T2D. In addition, epigenetic alterations in vascular cells, kidney, retina, neurons, and immune cells can lead to multiple micro- and macro-vascular complications of diabetes. Finally, epigenetic changes in response to adverse environment can also occur in the germline and be potentially transmitted to the offspring, contributing to the inheritance of T2D risk.
Epigenetic mechanisms of beta cell homeostasis and failure
Failure of beta cells to compensate for insulin resistance is central to the pathogenesis of T2D, and involves the progressive impairment of beta-cell identity, function, and survival. These aspects of beta cell homeostasis are governed by epigenetic mechanisms (reviewed in [14]), suggesting that epigenetic changes driven by adverse metabolic environment can potentially induce beta cell failure. A large body of evidence shows that stage-specific patterning of DNA methylation, histone modifications, and chromatin architecture is essential for pancreas lineage specification and endocrine differentiation [25–28]. Studies using human embryonic stem-cell differentiation have shown that epigenetic priming of lineage-specific enhancers dictates the stage-specific developmental competence and response to inductive signals throughout pancreatic differentiation [29].
Epigenetic regulation also plays a pivotal role in the establishment and maintenance of cellular identity and functional maturity of beta cells. DNA methylation patterning regulates the alpha-versus beta-cell fate choice by repressing the expression of the alpha-cell lineage determining transcription factor Arx in beta cells [30]. The DNA methyltransferase Dnmt1 maintains the methylated and repressed state of Arx locus during beta cell replication. Accordingly, loss of Dnmt1 in beta cells leads to induction of Arx expression due to promoter de-methylation, driving the trans-differentiation of beta-to alpha-cells [31]. DNA methylation also serves to establish the metabolic program that allows the establishment of glucose-stimulated insulin secretion (GSIS) in postnatal beta cells towards a functionally mature beta-cell phenotype [32]. A comparison of human alpha- and beta-cell DNA methylation profiles shows that differential methylation patterns are largely concentrated in enhancer regions, indicating putative roles of these regions in regulating cell identity [33]. Epigenetic regulation via micro RNAs (miRNAs) and long noncoding RNAs (lncRNAs) has also been implicated in islet development and functional maturation [34–36]. Mice lacking the miRNA processing enzyme Dicer in the pancreatic, endocrine, or beta cell lineages display severe beta cell deficits [37, 38]. In addition, changes in the beta-cell miRNA landscape in response to postnatal nutrient shifts are essential for beta cell functional maturation [39]. Similarly, the lncRNA blinc1 regulates beta-cell differentiation and function through its effect on specific islet transcription factors located in its genomic neighborhood [40]. Epigenetic mechanisms also control beta cell replication and expansion during postnatal growth, adaptation, and aging via the regulation of cell-cycle inhibitors such as p27Kip1 and p16Ink4a, and pro-replication imprinted genes such as the maternally imprinted lncRNA H19 [41–44]. The replicative and adaptive capacity of beta cells declines with age. Epigenetic regulation of p16Ink4a expression is also central to the Platelet Derived Growth Factor (PDGF) and Transforming Growth Factor-beta (TGF-beta) dependent control of age-related changes in beta cell replication [45, 46]. Furthermore, aging induces profound beta cell specific changes in the epigenetic states of genes involved in beta cell replication and function, such as Cdkn1a, Ccnd3, Plk1, Abcc8, and Kcnj11 [47]. Aging is a well-known risk factor for T2D, and it is likely that the age-dependent epigenetic changes in beta cell homeostasis play an instrumental role in this process.
The significance of epigenetic regulation of islet homeostasis is further highlighted by imprinting disorders such as the Beckwith-Wiedemann Syndrome (BWS) and Transient Neonatal Diabetes Mellitus (TNDM) (reviewed in [48]). In BWS, imprinting defects lead to lack of cell-cycle inhibitor CDKN1C (p57Kip2), leading to unrestrained beta cell proliferation, and consequent excessive beta cell mass, hyperinsulinemia, and hypoglycemia. Similarly, in TNDM, imprinting defects lead to the overexpression of two genes, namely ZAC and HYMAI, leading to hypoinsulinemia in neonatal life, which resolves subsequently [48]. Variants of imprinted genes GRB10 (regulates insulin signaling) and KCNQ1 (K+channel subunit, regulates insulin secretion) are also associated with increased T2D risk [49, 50], and islets from human subjects with T2D display differential methylation of KCNQ1 [51]. Human islets from donors with T2D display altered imprinting of the DLK1-MEG3 locus, which has important pathophysiological consequences. Hypermethylation of the MEG3 promoter in T2D islets leads to downregulation of a cluster of miRNAs which regulate genes involved in beta cell function and survival [52]. Locus-specific changes in histone modifications in T2D islets de-repress Neuropeptide Y (NPY) in beta cells, leading to impaired function. NPY is abundant in neonatal beta-cells, and is epigenetically repressed in beta cells during their functional maturation. Epigenetic dysregulation of NPY in diabetic beta cells leads them to resemble the functionally immature fetal beta cells [53]. These data, combined with the role of epigenetic mechanisms in beta cell identity, suggest that epigenetic dysregulation plays an important role in the loss of mature beta cell identity in diabetes, a phenomenon referred to as de-differentiation [54]. Recent work demonstrating the role of polycomb repressive complex 2 (PRC2)-dependent epigenetic regulation in beta cell identity, and the loss of PRC2-dependent gene repression in T2D islets further supports this idea [55].
A combination of sophisticated high-throughput sequencing techniques and powerful integrative data analysis approaches has led to a surge of epigenome-wide association studies (EWAS) in T2D cohorts to gain more insights into disease pathology [56–58]. Studies focusing on genome-wide profiling of DNA methylation in human islets from control and T2D donors show large-scale, but specific changes in the islet methylome in diabetes, translating into differential expression of loci critical for insulin secretion, adaptation, and survival [51, 59, 60]. Importantly, motifs for key islet transcription factors such as MAFA, PDX1, and RFX6, are enriched within the differentially methylated regions in T2D islets, suggesting dysregulation of islet transcriptional networks. These data indicate that epigenetic changes in diabetic islets lead are at least in part responsible for defects in beta cell function and survival in T2D (Table 1). Furthermore, environmentally induced epigenetic changes in the islets can be perpetuated trans-generationally, leading to increased T2D risk (reviewed in [15]). For example, the epigenetic landscape of genes related to beta-cell replication, function, and survival undergo profound changes in the progeny exposed to intrauterine growth retardation (IUGR) [61]. Altogether, these studies support the view that epigenetic alterations underlie beta cell defects in T2D, can be triggered by environmental factors, and transmitted to subsequent generations, contributing to T2D risk.
Table 1.
Tissue specific epigenetic regulation, EWAS, and T2D risk in humans
| Tissue | Context | Epigenetic mechanism | Key Finding | Reference |
|---|---|---|---|---|
| Islets | T2D | DNA methylation | Disrupted regulation of loci critical for islet function, adaptation, and survival. | [51, 59, 60] |
| Lymphocytes | Obesity | DNA methylation | Association of BMI with large scale DNA methylation changes. | [63] |
| Blood | T2D | DNA methylation | Identification of key DNA methylation sites associated with T2D, fasting blood glucose, and HbA1c levels. | [64] |
| Adipose | Obesity | DNA methylation | Obesity related changes in DNA methylation patterns may predict future development of T2D. | [65–67] |
| Adipose | Bariatric surgery, obesity | DNA methylation | Identification of key obesity related differentially methylated regions that overlap with specific genetic T2D risk loci. | [68] |
| Skeletal Muscle | T2D diagnosis and family history | DNA methylation | Epigenetic changes at loci related to insulin signaling, and association of some of these regions with T2D risk SNPs. | [71, 72] |
| Liver | Obesity, T2D | DNA methylation | DNA methylation changes at regions associated with T2D risk in the context of Obesity and T2D. | [74] |
| Adipose, skeletal muscle | Diet | DNA methylation | Epigenetic dysregulation of regions associated with metabolic pathways upon exposure to short-term high fat diet. | [75, 76] |
| Adipose, skeletal muscle | Exercise | DNA methylation | Both short- and long-term exercise reprograms the epigenetic landscape of genes involved in adipogenesis, and muscle contraction. | [71, 79, 80] |
Epigenetics of insulin resistance: the effect of obesity and metabolic health
The postprandial release of insulin ensures metabolic homeostasis by promoting nutrient uptake and storage in several tissues. Insulin promotes muscle glucose uptake, hepatic glycogen synthesis, and triglyceride synthesis, and suppresses lipolysis in adipose tissue. Insulin resistance refers to the impairment of such peripheral cellular responses to insulin, and can result from obesity, metabolic syndrome and chronic over-nutrition [62]. Epigenome-wide profiling has been very informative in elucidating novel epigenetic mechanisms underlying insulin resistance across metabolic tissues, especially in the context of obesity (Table 1). Recent EWAS data show that body mass index (BMI; a key measure of adiposity), is associated with large-scale changes in DNA methylation patterns in lymphocytes [63]. Additional EWAS studies using blood genomic DNA from various cohorts have demonstrated key DNA methylated sites associated with T2D, fasting blood glucose and HbA1c levels [64].
DNA methylation profiling of adipose tissue shows that the differentially methylated regions associated with obesity mark genes involved in lipid and lipoprotein metabolism, nutrient transport, inflammation, and T2D risk, and such alterations in the DNA methylation patterns are predictive of future development of T2D [65–67]. High throughput analysis of DNA methylation in adipose samples from patients pre- and post-gastric bypass surgery identified obesity-related differentially methylated regions that overlapped with 27 genetic T2D risk loci, implicating a cross-talk between genetics and epigenetic risk factors [68]. These data suggest that the epigenome is highly sensitive to body weight changes in either direction, and such epigenetic changes may be predictive of T2D risk. The importance of DNA methylation in metabolic homeostasis is further underscored by recent data implicating the DNA methyltransferase Dnmt3a in regulating insulin sensitivity in adipose tissue [69]. Epigenomic profiling of multiple histone modifications has also been instrumental in the identification of key enhancer elements and nuclear receptor pathways (glucocorticoid and vitamin D receptor) that drive insulin resistance in adipocytes, in response to cues such as steroid exposure and inflammation [70].
Studies using epigenetic and transcriptomic analysis of skeletal muscle in the context of newly diagnosed T2D and a family history of T2D show key differences in the muscle transcriptional program and insulin signaling, with some of the differentially regulated regions associated with T2D risk SNPs [71, 72]. Diet-induced obesity in the grand-paternal generation can lead to the transgenerational reprogramming of unfolded protein response (UPR) in skeletal muscle in the F2 (grand-child) generation [73].
The liver epigenome is also sensitive to obesity and hyperglycemia, as shown by large-scale epigenetic profiling. Obesity and T2D are associated with methylation changes at regions associated with T2D risk, and reprogram the liver epigenome towards increased glycolysis and lipolysis, which may promote the development of insulin resistance [74].
Collectively, these studies point to epigenetic dysregulation across multiple tissues as an underlying phenomenon in insulin resistance and T2D. Furthermore, they suggest that loci affected by both genetic and epigenetic changes may have a higher association with disease risk, or that epigenetics may confer functionality/causality to disease-related SNPs.
Epigenetics as a mediator of environmental influences on T2D risk
Changes in diet, including the fat content and composition have a strong impact on the adipose and muscle epigenome, especially at regions associated with metabolism [75, 76]. Besides diet, other environmental factors such as seasonal variation, exercise, and sleep can also shape the epigenome and metabolic homeostasis. Variations in temperature, such as heat or coldexposure, have been shown to change the epigenome and phenotype of beige adipocytes, to allow metabolic adaptation to temperature changes [77]. Cold exposure also induces epigenetic reprogramming in the sperm, with the offspring showing improved adaptation to over-nutrition and hypothermia [78]. Lifestyle interventions such as acute and chronic exercise lead to reprogramming of the DNA methylome in subcutaneous white adipose tissue (sWAT) and skeletal muscle in sedentary humans, affecting several genes involved in regulating adipogenesis, mitochondrial function, contraction, and inflammation [71, 79, 80]. Circadian rhythm is another critical environmental factor that directly affects the epigenome, as exemplified by the inherent histone acetyltransferase (HAT) activity of CLOCK (a core molecular component of the circadian clock). There is a strong link between metabolic and nutrient shifts, circadian clock, and epigenome, such that the feeding-fasting behavior regulates circadian gene-expression patterns to adapt to the diurnal variations in nutrient availability (reviewed in [81]). For example, an RNA-binding protein NONO serves as a novel epigenetic regulator of genes involved in glucose and lipid metabolism in the liver in response to nutrient availability [82]. The link between circadian clock and metabolism is further strengthened by data showing that circadian disruption is a major risk factor for T2D [83]. In line with this, time restricted feeding has been shown to prevent metabolic syndrome in mice harboring disruptions in the clock machinery [84]. Thus, circadian disruption can alter the cellular metabolic and epigenetic landscape, and consequently impair adaptation to nutrient availability, predisposing to an increased risk of T2D. Together, these studies show that adverse environmental and lifestyle changes can contribute to T2D pathogenesis as well as the inheritance of T2D risk [85] (Figure 1).
Epigenetic dysregulation as a mediator of diabetes complications
A significant number of patients with T2D develop serious secondary health problems that can severely impair quality of life, and increase morbidity and mortality. These include microvascular complications such as retinopathy, nephropathy and neuropathy, and macrovascular diseases such as atherosclerosis and hypertension [3]. Hyperglycemia and consequent metabolic dysregulation is one of the major triggers for vascular complications of diabetes, and can lead to vascular damage through multiple pathways, such as increased cellular stress, accumulation of advanced glycation end-products (AGEs), dysregulation of profibrotic and inflammatory pathways downstream of Transforming growth factor-beta (TGF-β, NF-κB, and angiotensin-II (AngII) [2]. These cellular alterations lead to upregulation of genes involved in growth, inflammation, apoptosis, and fibrosis resulting in endothelial dysfunction, vascular smooth muscle and renal cell growth and fibrosis, macrophage infiltration, inflammation and ultimately to multiple complications across different organs [2].
Epigenetic profiling studies have enhanced our understanding of the mechanisms underlying diabetes-related complications (reviewed in [3, 86], summarized in Table 2). A comparison of genome-wide DNA methylation data from renal tubules in humans with chronic kidney disease including diabetic nephropathy and control subjects shows significant differences in DNA methylation at loci involved in fibrosis [87], highlighting the significance of epigenetic dysregulation in diabetic nephropathy. Furthermore, EWAS of DNA methylation in human peripheral blood samples show specific and predictive changes in DNA methylation associated with the decline of renal function in diabetic nephropathy [88, 89]. TGF-β signaling plays a crucial pathologic role in diabetic nephropathy, and both DNA methylation and key histone modifications have been implicated in driving TGF-β dependent activation of genes associated with renal fibrosis (reviewed in [3, 86]). Enrichment of activating histone modifications at promoters of fibrotic genes associated with diabetic nephropathy are also observed in vivo in rodent models of diabetes [3]. A high glucose milieu has been shown to disrupt the DNA methylation patterns of key loci involved in endothelial and neuronal complications, in primary vascular cells and Schwann cells, respectively [90, 91]. Epigenetic dysregulation is also implicated in the disruption of redox homeostasis, extracellular matrix, and inflammation in retinal endothelial cells (RECs), in a model of diabetic retinopathy [92]. Genome-wide comparison of activating and repressive histone marks in monocytes cultured under high glucose conditions as well as monocytes from diabetic patients with controls further highlights large-scale changes in the epigenome in diabetes [93]. Similarly, in vascular smooth muscle cells (VSMCs), epigenetic changes in key histone modifications mediate the upregulation of inflammatory gene expression in response to hyperglycemic conditions in vitro and in mouse models of T2D [94].
Table 2.
EWAS and diabetes complications in humans
| Tissue | Context | Epigenetic mechanism | Key Finding | Reference |
|---|---|---|---|---|
| Human Renal Tubuli | Nephropathy | DNA methylation | Altered DNA methylation at loci involved in fibrosis in tubuli from humans with diabetic nephropathy and renal dysfunction. | [87] |
| Peripheral blood samples | Nephropathy | DNA methylation | Specific DNA methylation changes associated with eGFR identified, and a distinct subset of these also associated with kidney fibrosis and showed concordant DNA methylation changes in the kidney cortex biopsies from patients with chronic kidney disease. | [88] |
| Peripheral blood leukocytes | Nephropathy | DNA methylation | Key DNA methylation changes associated with decline of renal function (estimated glomerular filtration rate (eGFR)) identified in the context of diabetic nephropathy, in a cohort of Pima Indians with T2D. | [89] |
| Primary vascular endothelial cells | Vascular complications | Histone acetylation (activating), DNA methylation | Hyperglycemia mediated induction of genes associated with endothelial dysfunction occurs via histone acetylation, and is inversely correlated with DNA methylation. | [90] |
| Human monocytes (Primary and THP-1 cells) | Effect of hyperglycemia | Activating And repressive histone modifications | Chronic hyperglycemia can alter the chromatin states to drive changes in expression of key genes associated with inflammation. | [93] |
| Vascular smooth muscle cells from diabetic mice | Vascular complications, metabolic memory | Histone methylation (repressive) | Dysregulation of epigenetic states is a key mechanism underlying metabolic memory, as well as inflammation in vascular cells. | [94] |
| Human blood monocytes and lymphocytes | Metabolic memory | Histone modifications | Monocyte histone acetylation was associated with HbA1c level during the DCCT phase and the long-term (EDIC) follow-up, pointing to an epigenetic basis for metabolic memory. | [103] |
| Human whole blood and blood monocytes | Metabolic memory | DNA methylation | Several key genes associated with complications display sustained differential DNA methylation patterns in the same diabetic subjects over 16 years in association with HbA1c and an adverse diabetes complications outcome. | [104] |
Non-coding RNAs (miRNAs and lncRNAs) have also been identified as key epigenetic players in the development of diabetes complications (reviewed in [86, 95–97]). For example, the miR-216/miR-217 cluster promotes TGF-β dependent activation of Akt kinase and subsequent changes in extracellular matrix (ECM) gene expression and hypertrophy in mesangial cells by targeting PTEN (an inhibitor of Akt) [98]. Endoplasmic reticulum (ER)-stress induces lnc-MGC in mesangial cells treated with high glucose or TGF-β, as well as in the glomeruli of diabetic mice to mediate early events in diabetic nephropathy [99]. Similarly, upregulation of lncRNA Dnm3os in the macrophages of diabetic mice, as well as in monocytes from patients with T2D promotes inflammatory gene expression. Dnm3os interacts with the nucleolar protein, nucleolin, in macrophages and disruption of this interaction under diabetic conditions allows Dnm3os to enhance histone H3K9 acetylation at promoters of target inflammatory genes [100]. In VSMC, AngII, which is associated with numerous diabetic vascular complications, activates enhancers and super-enhancers associated with target genes, including lncRNAs, related to VSMC dysfunction [101]. In rat and human VSMCs, AngII also upregulates a novel lncRNA Giver which induces VSMC growth and oxidant stress [102]. Together these studies illustrate the emerging importance of lncRNAs in diabetic complications, as well the epigenetic cross talk between the non-coding RNA and chromatin layers.
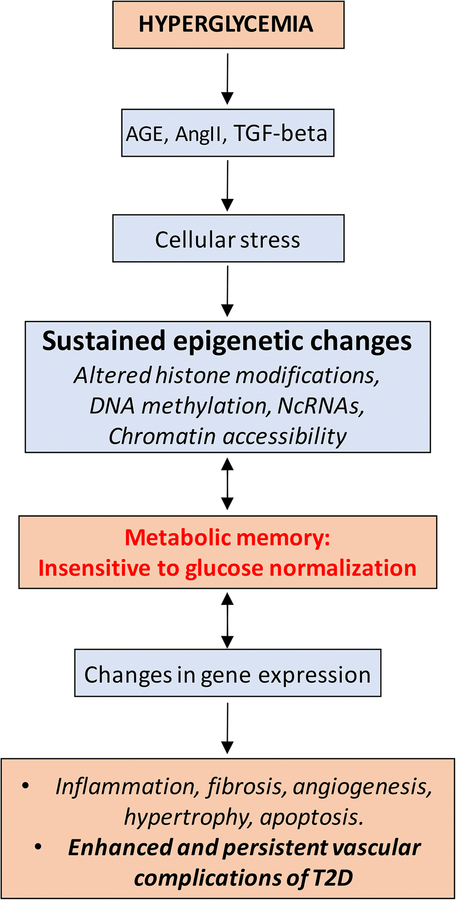
The importance of epigenetic regulation in the pathogenesis of diabetes complications is also evident from the phenomenon of metabolic memory, which underlies the long-term protection from intensive glycemic control, or conversely the continued progression of diabetes complications even upon achieving glycemic control. Metabolic memory refers to the observation that cells somehow retain the memory of prior exposure to hyperglycemic milieu, even after normoglycemia is attained. This phenomenon has been observed in experimental models as well as in clinical trials such as the Diabetes Control and Complications Trial (DCCT), and the long-term follow-up observational Epidemiology of Diabetes Interventions and Complications (EDIC) study [3, 103]. Even though all subjects maintained similar intensive glycemic control (HbA1c) in the EDIC phase, those with prior history of conventional glycemic control during the DCCT phase had higher risk of developing diabetes complications compared to subjects who received intensive glycemic control throughout [103]. Studies in T2D patients have similarly demonstrated that the benefits of intensive glycemic control lasted long after the completion of such a regimen [104]. These data suggest that epigenetic alterations conferred by prolonged exposure to hyperglycemic milieu may be responsible for the metabolic memory of dysfunction in target tissues. Accordingly, epigenetic profiling of multiple histone modifications and DNA methylation in blood monocytes from the DCCT/EDIC cohorts demonstrates clear epigenetic differences at key genes involved in inflammation between subjects on conventional control vs. intensive control [3, 105]. Notably, DNA methylation profiling of whole blood genomic DNA collected at the end of DCCT (~1993) and monocyte DNA collected ~17 years later during EDIC from the same patient demonstrated a persistence of DNA methylation at key loci, including those associated with complications, supporting a close connection between epigenetics and metabolic memory [106]. In summary, changes in the metabolic environment of a cell can drive changes in the epigenome possibly as an adaptive mechanism. However, such changes can epigenetically program the cells to sustain and continue to dictate the cellular response, even after the initial metabolic assault has ceased (Figure 2). A clearer understanding of epigenetic adaptive responses and the mechanisms that serve to maintain them will be essential to design therapeutic protocols that address the issue of metabolic memory.
Figure 2.
Hyperglycemia in T2D can activate multiple pathways such as signaling via AGEs, AngII and TGF-beta, as well as induce a milieu of cellular-stress. This can lead to dysregulation of different epigenetic mechanisms such as histone modifications, DNA methylation, and ncRNAs, and consequently alter chromatin accessibility and gene expression profiles in multiple tissues, resulting in the development of diabetes complications. Such aberrant epigenetic patterns can persist and lead to metabolic memory, such that there is increased risk of developing diabetes complications even after achieving glycemic control.
Epigenetic biomarkers and therapies
Development of biomarkers for T2D is a challenging task, and demands practical, non-invasive methods, such as those using peripheral blood samples, that have minimal adverse impact on the patient. An ideal blood-based biomarker candidate would be a stable molecular species that can be reliably detected, and accurately reflect disease initiation/progression related molecular alterations in the affected tissue(s). Cell-free DNA is released into the bloodstream through cell death, necrosis, or active secretion in the body, and can mirror tissue changes during disease pathogenesis. Cell-free DNA is highly stable, and the epigenetic signatures of these DNA fragments faithfully mirror their tissue-of-origin [107, 108]. The epigenetic profiles of cell-free DNA in the peripheral blood can therefore be potentially used as biomarkers to detect tissue specific epigenetic changes in disease conditions. Several studies have demonstrated that beta-cell death can be detected by assaying for beta-cell specific DNA methylation patterns of genes such as INS in circulating DNA [109, 110]. While these approaches have primarily focused on type 1 diabetes (T1D), they may be useful in T2D, as well as highlighted by recent data demonstrating that the DNA methylation changes associated with T2D in beta cells and peripheral insulin sensitive tissues are reliably captured in circulating DNA [111, 112].
miRNAs represent another molecular species that can be found stably circulating in the serum, and their profiles undergo changes in response to pathological conditions, including T2D [97]. For example, the serum levels of miR-192 and miR-193b are increased in pre-diabetic human subjects [113], while levels of miR-155 are downregulated in T2D [114]. Circulating miRNAs are often present in exosomes, and shown to be involved in cell-cell communication in metabolic homeostasis, insulin sensitivity, and T2D pathogenesis [115]. Exosomes containing obesity-associated miRNAs can induce glucose intolerance in lean mice, highlighting their relevance to T2D [116]. Thus, disease-specific epigenetic mechanisms not only serve as a highly promising avenue for biomarker development, but also as potential therapeutic targets for T2D.
Approaches that target epigenetic marks such as DNA methylation and chromatin modifications systemically have been successfully used for cancer therapeutics, and are now beginning to be considered for diabetes. Among these, inhibitors of the bromodomain proteins (BRDs) have shown much promise for cancer and inflammatory disease therapeutics, and have been used in the context of autoimmune diabetes in mice [117]. Given the importance of BRD proteins in metabolic homeostasis [118], BRD inhibitors also hold promise for T2D therapy. Of relevance to beta-cell replacement strategies, BRD inhibitors have been shown to promote pancreatic endocrine differentiation from stem cells [119]. However, drugs such as BRD inhibitors which target a whole class of epigenetic regulators, may not be the most optimal avenue for therapeutic use, given the potential for side effects. The development of more selective BRD inhibitors is warranted to address these concerns, but will have to await clearer understanding of how individual BRD proteins regulate different aspects of metabolic homeostasis.
Recent advances in gene-editing using CRISPR/Cas9 and TALEN systems have now made it possible to tailor the epigenetic patterns at specific genomic regions, and thus potentially correct disease-specific epigenetic changes. Such locus-specific epigenetic tailoring can be used to target DNA methylation or demethylation, as well as alter chromatin structure [120]. A recent study used this approach to drive human beta-cell proliferation by tailoring the DNA methylation pattern of an imprinted cell-cycle inhibitor gene CDKN1C [121]. As discussed earlier, hypo-methylation at the CDKN1C locus in patients with BWS leads to reduced levels of p57Kip2, resulting in beta-cell hyperproliferation. By targeting the DNA demethylase TET1 to CDKN1C to tailor a locus-specific epigenetic milieu reminiscent of the BWS beta-cells, this study successfully induced replication of adult human beta-cells. Similarly, CRISPR/Cas9 based targeting of DNA methyltransferase Dnmt3a to drive the DNA methylation and repression of alpha-cell fate determinant gene Arx in the developing pancreatic progenitors has recently been used to promote beta-cell lineage [122]. In a slightly different approach, a CRISPR/Cas9 trans epigenetic remodeling system was employed to transcriptionally activate target genes in vivo by recruiting specific transcriptional machinery and modulating histone marks, rather than editing DNA sequences. This strategy was used to alter cell fates by inducing trans-differentiation factors, e.g. alter liver cells to an insulin expressing, “beta-cell like” phenotype by ectopically expressing Pdx1 [123].
Non-coding RNAs such as miRNAs and lncRNAs have also been widely studied as potential epigenetic therapeutic targets in T2D and its complications [86, 96]. For example, locked nucleic acid (LNA) modified oligonucleotide mediated inhibition of miR-192 or lnc-MGC attenuates features of early diabetic nephropathy in mice [96, 99]. Similarly, CRISPR/Cas9 based targeting of key enhancers regulated by AngII has been shown to ameliorate angiotensin-dependent gene expression (including lncRNAs) in VSMCs related to hypertensive phenotypes [101]. These studies collectively illustrate the potential therapeutic benefits of targeting the epigenetic landscape of specific loci involved in metabolic tissues homeostasis or T2D pathogenesis.
Conclusions
Together these reports illustrate the contribution of epigenetic factors to the pathogenesis and complications of T2D, as well as the inheritance of T2D risk across generations. Comprehensive epigenetic profiling and EWAS show that T2D pathogenesis is marked by highly specific epigenetic changes in distinct gene categories involved in cell identity, function, inflammation etc. across target organs. Combining EWAS with GWAS candidates for T2D can significantly enhance the identification of putative causal variants for further experimental validation. It is likely that variations in the macro and micro-environmental factors such as light/dark cycle, temperature, diet, activity, metabolism, cellular-stress etc. initially induce epigenetic changes as a means of adaptation. How sustained exposure to “adverse” environmental milieu leads to a failure of epigenetic regulation (Figure 1) and whether lifestyle changes such as exercise and improved diet can reverse pathological changes remains to be determined. Are there some regions of the genome that are more vulnerable to environmental changes? If so, what determines the epigenetic plasticity of any genomic region, and are such regions amenable to therapeutic interventions? Elucidation of the molecular basis of epigenetic dysregulation in T2D will not only inform our understanding of adaptive mechanisms in metabolic tissues, disease pathogenesis, and inheritance of disease risk, but will also guide the development of innovative epigenetic biomarkers and therapies. Modulation of the enzymatic regulators of epigenetic marks using small molecule drugs is being pursued with great interest for addressing different aspects of T2D pathology. Such approaches, however, often suffer from off-target effects, and require the development of more specific small molecule agents and tissue specific delivery methods to become therapeutically successful. Targeted epigenetic engineering of key genes that are dysregulated in T2D has emerged as a promising alternative avenue, and is likely to improve the efficacy of approaches such as beta-cell replacement. However, detailed studies are required to identify specific epigenetic regulators and changes that are cell/tissue-type specific in T2D, to develop novel and targeted therapeutic strategies that address diabetes pathogenesis and complications.
Acknowledgments
The authors gratefully acknowledge research funding from the National Institutes of Health (NIDDK and NHLBI), the Wanek Family Project to Cure Type 1 Diabetes at City of Hope, and the Juvenile Diabetes Research Foundation (to RN), and from the Wanek Family Project to Cure Type1 Diabetes at City of Hope, Human Islet Research Network (NIH) UC4 DK104162, and the National Institutes of Health (NIDDK; R01 grant DK120523) (to SD).
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Conflict of Interest
Sangeeta Dhawan declares no conflict of interest.
Rama Natarajan reports a pending patent on inhibitors of epigenetically modified targets.
Human and Animal Rights and Informed Consent
All the studies noted in this review that were performed by the authors involving animals were done in compliance with appropriate institutional committees for animal research (IACUC) and all institutional guidelines for the care and use of animals were followed. Studies quoted in this article undertaken by the authors involving human subjects were either carried out on retrospective de-identified samples obtained from appropriate repositories, in compliance with institutional review boards (IRB), or all the procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional review boards (IRB), and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1.Zimmet PZ, Magliano DJ, Herman WH, Shaw JE. Diabetes: a 21st century challenge. Lancet Diabetes Endocrinol. 2014;2(1):56–64. doi: 10.1016/S2213-8587(13)70112-8. [DOI] [PubMed] [Google Scholar]
- 2.Forbes JM, Cooper ME. Mechanisms of diabetic complications. Physiol Rev. 2013;93(1):137–88. doi: 10.1152/physrev.00045.2011. [DOI] [PubMed] [Google Scholar]
- 3.Reddy MA, Zhang E, Natarajan R. Epigenetic mechanisms in diabetic complications and metabolic memory. Diabetologia. 2015;58(3):443–55. doi: 10.1007/s00125-014-3462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Almgren P, Lehtovirta M, Isomaa B, Sarelin L, Taskinen MR, Lyssenko V et al. Heritability and familiality of type 2 diabetes and related quantitative traits in the Botnia Study. Diabetologia. 2011;54(11):2811–9. doi: 10.1007/s00125-011-2267-5. [DOI] [PubMed] [Google Scholar]
- 5.Meigs JB, Cupples LA, Wilson PW. Parental transmission of type 2 diabetes: the Framingham Offspring Study. Diabetes. 2000;49(12):2201–7. [DOI] [PubMed] [Google Scholar]
- 6. •.Fuchsberger C, Flannick J, Teslovich TM, Mahajan A, Agarwala V, Gaulton KJ et al. The genetic architecture of type 2 diabetes. Nature. 2016;536(7614):41–7. doi: 10.1038/nature18642. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study provides a comprehensive analysis of the contribution of genetic factors to T2D risk.
- 7.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. •.Ahlqvist E, Storm P, Karajamaki A, Martinell M, Dorkhan M, Carlsson A et al. Novel subgroups of adult-onset diabetes and their association with outcomes: a data-driven cluster analysis of six variables. Lancet Diabetes Endocrinol. 2018;6(5):361–9. doi: 10.1016/S2213-8587(18)30051-2. [DOI] [PubMed] [Google Scholar]; This study highlights the heterogeneity and complexity of adult-onset diabetes.
- 9.Unnikrishnan R, Pradeepa R, Joshi SR, Mohan V. Type 2 Diabetes: Demystifying the Global Epidemic. Diabetes. 2017;66(6):1432–42. doi: 10.2337/db16-0766. [DOI] [PubMed] [Google Scholar]
- 10.Rosen ED, Kaestner KH, Natarajan R, Patti ME, Sallari R, Sander M et al. Epigenetics and Epigenomics: Implications for Diabetes and Obesity. Diabetes. 2018;67(10):1923–31. doi: 10.2337/db18-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ling C, Groop L. Epigenetics: a molecular link between environmental factors and type 2 diabetes. Diabetes. 2009;58(12):2718–25. doi: 10.2337/db09-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17(8):487–500. doi: 10.1038/nrg.2016.59. [DOI] [PubMed] [Google Scholar]
- 13.Sassone-Corsi P. Physiology. When metabolism and epigenetics converge. Science. 2013;339(6116):148–50. doi: 10.1126/science.1233423 339/6116/148 [pii]. [DOI] [PubMed] [Google Scholar]
- 14.Arnes L, Sussel L. Epigenetic modifications and long noncoding RNAs influence pancreas development and function. Trends Genet. 2015;31(6):290–9. doi: 10.1016/j.tig.2015.02.008 S0168–9525(15)00036–0 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bansal A, Simmons RA. Epigenetics and developmental origins of diabetes: correlation or causation? Am J Physiol Endocrinol Metab. 2018;315(1):E15–E28. doi: 10.1152/ajpendo.00424.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A. 2008;105(44):17046–9. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hales CN, Barker DJ. The thrifty phenotype hypothesis. Br Med Bull. 2001;60:5–20. [DOI] [PubMed] [Google Scholar]
- 18.Carone BR, Fauquier L, Habib N, Shea JM, Hart CE, Li R et al. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell. 2010;143(7):1084–96. doi: 10.1016/j.cell.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng SF, Lin RC, Laybutt DR, Barres R, Owens JA, Morris MJ. Chronic high-fat diet in fathers programs beta-cell dysfunction in female rat offspring. Nature. 2010;467(7318):963–6. doi: 10.1038/nature09491. [DOI] [PubMed] [Google Scholar]
- 20.Sales VM, Ferguson-Smith AC, Patti ME. Epigenetic Mechanisms of Transmission of Metabolic Disease across Generations. Cell Metab. 2017;25(3):559–71. doi: 10.1016/j.cmet.2017.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez D, Pentinat T, Ribo S, Daviaud C, Bloks VW, Cebria J et al. In utero undernutrition in male mice programs liver lipid metabolism in the second-generation offspring involving altered Lxra DNA methylation. Cell Metab. 2014;19(6):941–51. doi: 10.1016/j.cmet.2014.03.026. [DOI] [PubMed] [Google Scholar]
- 22.de Castro Barbosa T, Ingerslev LR, Alm PS, Versteyhe S, Massart J, Rasmussen M et al. High-fat diet reprograms the epigenome of rat spermatozoa and transgenerationally affects metabolism of the offspring. Mol Metab. 2016;5(3):184–97. doi: 10.1016/j.molmet.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. ••.Donkin I, Versteyhe S, Ingerslev LR, Qian K, Mechta M, Nordkap L et al. Obesity and Bariatric Surgery Drive Epigenetic Variation of Spermatozoa in Humans. Cell Metab. 2016;23(2):369–78. doi: 10.1016/j.cmet.2015.11.004. [DOI] [PubMed] [Google Scholar]; This study demonstrates that variations in the body mass index can reprogram the epigenome of spermatozoa, providing novel insights into the inheritance of metabolic disease risk.
- 24.Kong A, Steinthorsdottir V, Masson G, Thorleifsson G, Sulem P, Besenbacher S et al. Parental origin of sequence variants associated with complex diseases. Nature. 2009;462(7275):868–74. doi: 10.1038/nature08625 nature08625 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu CR, Cole PA, Meyers DJ, Kormish J, Dent S, Zaret KS. Chromatin “prepattern” and histone modifiers in a fate choice for liver and pancreas. Science. 2011;332(6032):963–6. doi: 10.1126/science.1202845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKenna B, Guo M, Reynolds A, Hara M, Stein R. Dynamic recruitment of functionally distinct Swi/Snf chromatin remodeling complexes modulates Pdx1 activity in islet beta cells. Cell Rep. 2015;10(12):2032–42. doi: 10.1016/j.celrep.2015.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Georgia S, Kanji M, Bhushan A. DNMT1 represses p53 to maintain progenitor cell survival during pancreatic organogenesis. Genes Dev. 2013;27(4):372–7. doi: 10.1101/gad.207001.112 27/4/372 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lenoir O, Flosseau K, Ma FX, Blondeau B, Mai A, Bassel-Duby R et al. Specific control of pancreatic endocrine beta- and delta-cell mass by class IIa histone deacetylases HDAC4, HDAC5, and HDAC9. Diabetes. 2011;60(11):2861–71. doi: 10.2337/db11-0440 db11–0440 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang A, Yue F, Li Y, Xie R, Harper T, Patel NA et al. Epigenetic priming of enhancers predicts developmental competence of hESC-derived endodermal lineage intermediates. Cell Stem Cell. 2015;16(4):386–99. doi: 10.1016/j.stem.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papizan JB, Singer RA, Tschen SI, Dhawan S, Friel JM, Hipkens SB et al. Nkx2.2 repressor complex regulates islet beta-cell specification and prevents beta-to-alpha-cell reprogramming. Genes Dev. 2011;25(21):2291–305. doi: 10.1101/gad.173039.11125/21/2291 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dhawan S, Georgia S, Tschen SI, Fan G, Bhushan A. Pancreatic beta cell identity is maintained by DNA methylation-mediated repression of Arx. Dev Cell. 2011;20(4):419–29. doi: 10.1016/j.devcel.2011.03.012 S1534–5807(11)00118–3 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dhawan S, Tschen SI, Zeng C, Guo T, Hebrok M, Matveyenko A et al. DNA methylation directs functional maturation of pancreatic beta cells. J Clin Invest. 2015;125(7):2851–60. doi: 10.1172/JCI79956 79956 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neiman D, Moss J, Hecht M, Magenheim J, Piyanzin S, Shapiro AMJ et al. Islet cells share promoter hypomethylation independently of expression, but exhibit cell-type-specific methylation in enhancers. Proc Natl Acad Sci U S A. 2017;114(51):13525–30. doi: 10.1073/pnas.1713736114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinez-Sanchez A, Rutter GA, Latreille M. MiRNAs in beta-Cell Development, Identity, and Disease. Front Genet. 2016;7:226. doi: 10.3389/fgene.2016.00226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singer RA, Sussel L. Islet Long Noncoding RNAs: A Playbook for Discovery and Characterization. Diabetes. 2018;67(8):1461–70. doi: 10.2337/dbi18-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.LaPierre MP, Stoffel M. MicroRNAs as stress regulators in pancreatic beta cells and diabetes. Mol Metab. 2017;6(9):1010–23. doi: 10.1016/j.molmet.2017.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanji MS, Martin MG, Bhushan A. Dicer1 is required to repress neuronal fate during endocrine cell maturation. Diabetes. 2013;62(5):1602–11. doi: 10.2337/db12-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lynn FC, Skewes-Cox P, Kosaka Y, McManus MT, Harfe BD, German MS. MicroRNA expression is required for pancreatic islet cell genesis in the mouse. Diabetes. 2007;56(12):2938–45. doi: 10.2337/db07-0175. [DOI] [PubMed] [Google Scholar]
- 39.Jacovetti C, Matkovich SJ, Rodriguez-Trejo A, Guay C, Regazzi R. Postnatal beta-cell maturation is associated with islet-specific microRNA changes induced by nutrient shifts at weaning. Nat Commun. 2015;6:8084. doi: 10.1038/ncomms9084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. ••.Arnes L, Akerman I, Balderes DA, Ferrer J, Sussel L. betalinc1 encodes a long noncoding RNA that regulates islet beta-cell formation and function. Genes Dev. 2016;30(5):502–7. doi: 10.1101/gad.273821.115. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study discovered a novel role for lncRNAs in the regulation of beta cell homeostasis.
- 41.Karnik SK, Hughes CM, Gu X, Rozenblatt-Rosen O, McLean GW, Xiong Y et al. Menin regulates pancreatic islet growth by promoting histone methylation and expression of genes encoding p27Kip1 and p18INK4c. Proc Natl Acad Sci U S A. 2005;102(41):14659–64. doi:0503484102 [pii] 10.1073/pnas.0503484102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dhawan S, Tschen SI, Bhushan A. Bmi-1 regulates the Ink4a/Arf locus to control pancreatic beta-cell proliferation. Genes Dev. 2009;23(8):906–11. doi:23/8/906 [pii] 10.1101/gad.1742609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen H, Gu X, Su IH, Bottino R, Contreras JL, Tarakhovsky A et al. Polycomb protein Ezh2 regulates pancreatic beta-cell Ink4a/Arf expression and regeneration in diabetes mellitus. Genes Dev. 2009;23(8):975–85. doi: 10.1101/gad.1742509 23/8/975 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. •.Sanchez-Parra C, Jacovetti C, Dumortier O, Lee K, Peyot ML, Guay C et al. Contribution of the Long Noncoding RNA H19 to beta-Cell Mass Expansion in Neonatal and Adult Rodents. Diabetes. 2018;67(11):2254–67. doi: 10.2337/db18-0201. [DOI] [PubMed] [Google Scholar]; This study identified a role for combinatorial epigenetic regulation via genomic imprinting and lncRNAs in beta cell homeostasis.
- 45.Chen H, Gu X, Liu Y, Wang J, Wirt SE, Bottino R et al. PDGF signalling controls age-dependent proliferation in pancreatic beta-cells. Nature. 2011;478(7369):349–55. doi: 10.1038/nature10502 nature10502 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. •.Dhawan S, Dirice E, Kulkarni RN, Bhushan A. Inhibition of TGF-beta signaling promotes human pancreatic beta cell replication. Diabetes. 2016. doi:db151331 [pii] db15–1331 [pii] 10.2337/db15-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study highlights how cellular signals regulate beta cell replication via epiegentic remodeling of cell-cycle machinery, with implications for therapeutic targeting.
- 47.Avrahami D, Li C, Zhang J, Schug J, Avrahami R, Rao S et al. Aging-Dependent Demethylation of Regulatory Elements Correlates with Chromatin State and Improved beta Cell Function. Cell Metab. 2015;22(4):619–32. doi: 10.1016/j.cmet.2015.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Golson ML, Kaestner KH. Epigenetics in formation, function, and failure of the endocrine pancreas. Mol Metab. 2017;6(9):1066–76. doi: 10.1016/j.molmet.2017.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prokopenko I, Poon W, Magi R, Prasad BR, Salehi SA, Almgren P et al. A central role for GRB10 in regulation of islet function in man. PLoS Genet. 2014;10(4):e1004235. doi: 10.1371/journal.pgen.1004235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Travers ME, Mackay DJ, Dekker Nitert M, Morris AP, Lindgren CM, Berry A et al. Insights into the molecular mechanism for type 2 diabetes susceptibility at the KCNQ1 locus from temporal changes in imprinting status in human islets. Diabetes. 2013;62(3):987–92. doi: 10.2337/db12-0819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dayeh T, Volkov P, Salo S, Hall E, Nilsson E, Olsson AH et al. Genome-wide DNA methylation analysis of human pancreatic islets from type 2 diabetic and non-diabetic donors identifies candidate genes that influence insulin secretion. PLoS Genet. 2014;10(3):e1004160. doi: 10.1371/journal.pgen.1004160 PGENETICS-D-13–01899 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kameswaran V, Bramswig NC, McKenna LB, Penn M, Schug J, Hand NJ et al. Epigenetic regulation of the DLK1-MEG3 microRNA cluster in human type 2 diabetic islets. Cell Metab. 2014;19(1):135–45. doi: 10.1016/j.cmet.2013.11.016 S1550–4131(13)00487–7 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. •.Rodnoi P, Rajkumar M, Moin ASM, Georgia SK, Butler AE, Dhawan S. Neuropeptide Y expression marks partially differentiated beta cells in mice and humans. JCI Insight. 2017;2(12). doi: 10.1172/jci.insight.94005. [DOI] [PMC free article] [PubMed] [Google Scholar]; The data presented in this study point to epigenetic dysregulation as a key mechanism contrbuting to beta cell de-differentiation and dysfunction in T2D.
- 54.Talchai C, Xuan S, Lin HV, Sussel L, Accili D. Pancreatic beta cell dedifferentiation as a mechanism of diabetic beta cell failure. Cell. 2012;150(6):1223–34. doi: 10.1016/j.cell.2012.07.029 S0092–8674(12)00940–3 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. ••.Lu TT, Heyne S, Dror E, Casas E, Leonhardt L, Boenke T et al. The Polycomb-Dependent Epigenome Controls beta Cell Dysfunction, Dedifferentiation, and Diabetes. Cell Metab. 2018;27(6):1294–308 e7. doi: 10.1016/j.cmet.2018.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study uncovered a novel regulatory role for polycomb protein Eed in beta cell identity and function, and showed that polycomb disruption leads to beta cell de-differentiation and dysfunction in T2D.
- 56.Zhang H, Pollin TI. Epigenetics Variation and Pathogenesis in Diabetes. Curr Diab Rep. 2018;18(11):121. doi: 10.1007/s11892-018-1091-4. [DOI] [PubMed] [Google Scholar]
- 57.Dirks RA, Stunnenberg HG, Marks H. Genome-wide epigenomic profiling for biomarker discovery. Clin Epigenetics. 2016;8:122. doi: 10.1186/s13148-016-0284-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17(6):333–51. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Volkmar M, Dedeurwaerder S, Cunha DA, Ndlovu MN, Defrance M, Deplus R et al. DNA methylation profiling identifies epigenetic dysregulation in pancreatic islets from type 2 diabetic patients. EMBO J. 2012;31(6):1405–26. doi: 10.1038/emboj.2011.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. •.Volkov P, Bacos K, Ofori JK, Esguerra JL, Eliasson L, Ronn T et al. Whole-Genome Bisulfite Sequencing of Human Pancreatic Islets Reveals Novel Differentially Methylated Regions in Type 2 Diabetes Pathogenesis. Diabetes. 2017;66(4):1074–85. doi: 10.2337/db16-0996. [DOI] [PubMed] [Google Scholar]; The data presented in this article point to large-scale epigenetic dysregulation in pancreatic islets in the context of T2D, and provide mechanistic insights into how such epigenetic changes may alter islet function.
- 61.Thompson RF, Fazzari MJ, Niu H, Barzilai N, Simmons RA, Greally JM. Experimental intrauterine growth restriction induces alterations in DNA methylation and gene expression in pancreatic islets of rats. J Biol Chem. 2010;285(20):15111–8. doi: 10.1074/jbc.M109.095133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Samuel VT, Shulman GI. The pathogenesis of insulin resistance: integrating signaling pathways and substrate flux. J Clin Invest. 2016;126(1):12–22. doi: 10.1172/JCI77812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Feinberg AP, Irizarry RA, Fradin D, Aryee MJ, Murakami P, Aspelund T et al. Personalized epigenomic signatures that are stable over time and covary with body mass index. Sci Transl Med. 2010;2(49):49ra67. doi: 10.1126/scitranslmed.3001262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Walaszczyk E, Luijten M, Spijkerman AMW, Bonder MJ, Lutgers HL, Snieder H et al. DNA methylation markers associated with type 2 diabetes, fasting glucose and HbA1c levels: a systematic review and replication in a case-control sample of the Lifelines study. Diabetologia. 2018;61(2):354–68. doi: 10.1007/s00125-017-4497-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nilsson E, Jansson PA, Perfilyev A, Volkov P, Pedersen M, Svensson MK et al. Altered DNA methylation and differential expression of genes influencing metabolism and inflammation in adipose tissue from subjects with type 2 diabetes. Diabetes. 2014;63(9):2962–76. doi: 10.2337/db13-1459. [DOI] [PubMed] [Google Scholar]
- 66.Crujeiras AB, Diaz-Lagares A, Moreno-Navarrete JM, Sandoval J, Hervas D, Gomez A et al. Genome-wide DNA methylation pattern in visceral adipose tissue differentiates insulin-resistant from insulin-sensitive obese subjects. Transl Res. 2016;178:13–24 e5. doi: 10.1016/j.trsl.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 67. ••.Wahl S, Drong A, Lehne B, Loh M, Scott WR, Kunze S et al. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature. 2017;541(7635):81–6. doi: 10.1038/nature20784. [DOI] [PMC free article] [PubMed] [Google Scholar]; The data presented in this study provide a novel insight into the specific physiological pathways that are epigenetically dysregulated by adiposity.
- 68.Multhaup ML, Seldin MM, Jaffe AE, Lei X, Kirchner H, Mondal P et al. Mouse-human experimental epigenetic analysis unmasks dietary targets and genetic liability for diabetic phenotypes. Cell Metab. 2015;21(1):138–49. doi: 10.1016/j.cmet.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.You D, Nilsson E, Tenen DE, Lyubetskaya A, Lo JC, Jiang R et al. Dnmt3a is an epigenetic mediator of adipose insulin resistance. Elife. 2017;6. doi: 10.7554/eLife.30766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kang S, Tsai LT, Zhou Y, Evertts A, Xu S, Griffin MJ et al. Identification of nuclear hormone receptor pathways causing insulin resistance by transcriptional and epigenomic analysis. Nat Cell Biol. 2015;17(1):44–56. doi: 10.1038/ncb3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nitert MD, Dayeh T, Volkov P, Elgzyri T, Hall E, Nilsson E et al. Impact of an exercise intervention on DNA methylation in skeletal muscle from first-degree relatives of patients with type 2 diabetes. Diabetes. 2012;61(12):3322–32. doi: 10.2337/db11-1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. •.Scott LJ, Erdos MR, Huyghe JR, Welch RP, Beck AT, Wolford BN et al. The genetic regulatory signature of type 2 diabetes in human skeletal muscle. Nat Commun. 2016;7:11764. doi: 10.1038/ncomms11764. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study illustrates the power of combining genome- and epigenome-wide association data to identify the molecular underpinnings of type 2 diabetes pathogenesis.
- 73. •.Alm PS, de Castro Barbosa T, Barres R, Krook A, Zierath JR. Grandpaternal-induced transgenerational dietary reprogramming of the unfolded protein response in skeletal muscle. Mol Metab. 2017;6(7):621–30. doi: 10.1016/j.molmet.2017.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study highlights how adverse environmental factors impact diabetes risk through generations.
- 74.Kirchner H, Sinha I, Gao H, Ruby MA, Schonke M, Lindvall JM et al. Altered DNA methylation of glycolytic and lipogenic genes in liver from obese and type 2 diabetic patients. Mol Metab. 2016;5(3):171–83. doi: 10.1016/j.molmet.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jacobsen SC, Gillberg L, Bork-Jensen J, Ribel-Madsen R, Lara E, Calvanese V et al. Young men with low birthweight exhibit decreased plasticity of genome-wide muscle DNA methylation by high-fat overfeeding. Diabetologia. 2014;57(6):1154–8. doi: 10.1007/s00125-014-3198-8. [DOI] [PubMed] [Google Scholar]
- 76.Gillberg L, Perfilyev A, Brons C, Thomasen M, Grunnet LG, Volkov P et al. Adipose tissue transcriptomics and epigenomics in low birthweight men and controls: role of high-fat overfeeding. Diabetologia. 2016;59(4):799–812. doi: 10.1007/s00125-015-3852-9. [DOI] [PubMed] [Google Scholar]
- 77. ••.Roh HC, Tsai LTY, Shao M, Tenen D, Shen Y, Kumari M et al. Warming Induces Significant Reprogramming of Beige, but Not Brown, Adipocyte Cellular Identity. Cell Metab. 2018;27(5):1121–37 e5. doi: 10.1016/j.cmet.2018.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study identified the epigenetic basis of adipose cellular plasticity in response to changes in ambient temperature.
- 78. ••.Sun W, Dong H, Becker AS, Dapito DH, Modica S, Grandl G et al. Cold-induced epigenetic programming of the sperm enhances brown adipose tissue activity in the offspring. Nat Med. 2018;24(9):1372–83. doi: 10.1038/s41591-018-0102-y. [DOI] [PubMed] [Google Scholar]; This study illustrates how seasonal variations in temperature can reprogram the sperm epigenome to confer adaptive advantage to the offspring.
- 79.Barres R, Yan J, Egan B, Treebak JT, Rasmussen M, Fritz T et al. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab. 2012;15(3):405–11. doi: 10.1016/j.cmet.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 80. •.Fabre O, Ingerslev LR, Garde C, Donkin I, Simar D, Barres R. Exercise training alters the genomic response to acute exercise in human adipose tissue. Epigenomics. 2018;10(8):1033–50. doi: 10.2217/epi-2018-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrates that the human sWAT epigenome is sensitive to acute exercise regimen.
- 81.Sassone-Corsi P The Epigenetic and Metabolic Language of the Circadian Clock In: Sassone-Corsi P, Christen Y, editors. A Time for Metabolism and Hormones. Cham (CH) 2016. p. 1–11. [Google Scholar]
- 82.Benegiamo G, Brown SA, Panda S. RNA Dynamics in the Control of Circadian Rhythm. Adv Exp Med Biol. 2016;907:107–22. doi: 10.1007/978-3-319-29073-7_5. [DOI] [PubMed] [Google Scholar]
- 83.Colwell CS, Matveyenko AV. Timing is everything: implications for metabolic consequences of sleep restriction. Diabetes. 2014;63(6):1826–8. doi: 10.2337/db14-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chaix A, Lin T, Le HD, Chang MW, Panda S. Time-Restricted Feeding Prevents Obesity and Metabolic Syndrome in Mice Lacking a Circadian Clock. Cell Metab. 2018. doi: 10.1016/j.cmet.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zimmet P, Shi Z, El-Osta A, Ji L. Epidemic T2DM, early development and epigenetics: implications of the Chinese Famine. Nat Rev Endocrinol. 2018;14(12):738–46. doi: 10.1038/s41574-018-0106-1. [DOI] [PubMed] [Google Scholar]
- 86.Kato M, Natarajan R. Diabetic nephropathy--emerging epigenetic mechanisms. Nat Rev Nephrol. 2014;10(9):517–30. doi: 10.1038/nrneph.2014.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ko YA, Mohtat D, Suzuki M, Park AS, Izquierdo MC, Han SY et al. Cytosine methylation changes in enhancer regions of core pro-fibrotic genes characterize kidney fibrosis development. Genome Biol. 2013;14(10):R108. doi: 10.1186/gb-2013-14-10-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. ••.Chu AY, Tin A, Schlosser P, Ko YA, Qiu C, Yao C et al. Epigenome-wide association studies identify DNA methylation associated with kidney function. Nat Commun. 2017;8(1):1286. doi: 10.1038/s41467-017-01297-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study identified key DNA methylation changes in peripheral blood cells that are associated with a decline of renal function in chronic kidney disease, and are also recapitulated in the kidney cortex biopsies from patients with kidney disease.
- 89. •.Qiu C, Hanson RL, Fufaa G, Kobes S, Gluck C, Huang J et al. Cytosine methylation predicts renal function decline in American Indians. Kidney Int. 2018;93(6):1417–31. doi: 10.1016/j.kint.2018.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study identified DNA methylation changes associated with impaired kidney function in the context of diabetic nephropathy in a cohort of Pima Indians with history of diabetes.
- 90.Pirola L, Balcerczyk A, Tothill RW, Haviv I, Kaspi A, Lunke S et al. Genome-wide analysis distinguishes hyperglycemia regulated epigenetic signatures of primary vascular cells. Genome Res. 2011;21(10):1601–15. doi: 10.1101/gr.116095.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kim ES, Isoda F, Kurland I, Mobbs CV. Glucose-induced metabolic memory in Schwann cells: prevention by PPAR agonists. Endocrinology. 2013;154(9):3054–66. doi: 10.1210/en.2013-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kowluru RA, Mishra M. Epigenetic regulation of redox signaling in diabetic retinopathy: Role of Nrf2. Free Radic Biol Med. 2017;103:155–64. doi: 10.1016/j.freeradbiomed.2016.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Miao F, Wu X, Zhang L, Yuan YC, Riggs AD, Natarajan R. Genome-wide analysis of histone lysine methylation variations caused by diabetic conditions in human monocytes. J Biol Chem. 2007;282(18):13854–63. doi: 10.1074/jbc.M609446200. [DOI] [PubMed] [Google Scholar]
- 94.Villeneuve LM, Reddy MA, Lanting LL, Wang M, Meng L, Natarajan R. Epigenetic histone H3 lysine 9 methylation in metabolic memory and inflammatory phenotype of vascular smooth muscle cells in diabetes. Proc Natl Acad Sci U S A. 2008;105(26):9047–52. doi: 10.1073/pnas.0803623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Leung A, Amaram V, Natarajan R. Linking diabetic vascular complications with LncRNAs. Vascul Pharmacol. 2018. doi: 10.1016/j.vph.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Leung A, Natarajan R. Long Noncoding RNAs in Diabetes and Diabetic Complications. Antioxid Redox Signal. 2018;29(11):1064–73. doi: 10.1089/ars.2017.7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kato M, Natarajan R. MicroRNAs in diabetic nephropathy: functions, biomarkers, and therapeutic targets. Ann N Y Acad Sci. 2015;1353:72–88. doi: 10.1111/nyas.12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kato M, Putta S, Wang M, Yuan H, Lanting L, Nair I et al. TGF-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat Cell Biol. 2009;11(7):881–9. doi: 10.1038/ncb1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. ••.Kato M, Wang M, Chen Z, Bhatt K, Oh HJ, Lanting L et al. An endoplasmic reticulum stress-regulated lncRNA hosting a microRNA megacluster induces early features of diabetic nephropathy. Nat Commun. 2016;7:12864. doi: 10.1038/ncomms12864. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study identified a novel, ER-stress senstive, non-coding RNA regulatory module in mesangial cells under diabetic conditions that induces key features of diabetic nephropathy, highlighting that cellular-stress can drive epigenetic dysregulation to drive the pathogenesis of T2D complications. It also illustrates the use of modified antisense oligonucleotides to target lncRNAs in vitro and in vivo in mice.
- 100. •.Das S, Reddy MA, Senapati P, Stapleton K, Lanting L, Wang M et al. Diabetes Mellitus-Induced Long Noncoding RNA Dnm3os Regulates Macrophage Functions and Inflammation via Nuclear Mechanisms. Arterioscler Thromb Vasc Biol. 2018;38(8):1806–20. doi: 10.1161/ATVBAHA.117.310663. [DOI] [PMC free article] [PubMed] [Google Scholar]; The data presented in this study showcase the importance of altered lncRNA regulation in promoting inflammation and macrophage dysfunction in diabetes complications.
- 101. ••.Das S, Senapati P, Chen Z, Reddy MA, Ganguly R, Lanting L et al. Regulation of angiotensin II actions by enhancers and super-enhancers in vascular smooth muscle cells. Nat Commun. 2017;8(1):1467. doi: 10.1038/s41467-017-01629-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study exemplifies how the signaling by growth factors associated with diabetes complications epigenetically activates the enhancers and super-enhancers in the target cells to regulate the expression of the associated target genes, including lncRNAs.
- 102. •.Das S, Zhang E, Senapati P, Amaram V, Reddy MA, Stapleton K et al. A Novel Angiotensin II-Induced Long Noncoding RNA Giver Regulates Oxidative Stress, Inflammation, and Proliferation in Vascular Smooth Muscle Cells. Circ Res. 2018;123(12):1298–312. doi: 10.1161/CIRCRESAHA.118.313207. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study identifies a novel, Angiotesin II responsive lncRNA that regulates oxidative stress and inflammation in vascular smooth muscle cells in the context of hypertension. A human ortholog is upregulated in arteries of hypertensive subjects.
- 103.Writing Team for the Diabetes C, Complications Trial/Epidemiology of Diabetes I, Complications Research G. Sustained effect of intensive treatment of type 1 diabetes mellitus on development and progression of diabetic nephropathy: the Epidemiology of Diabetes Interventions and Complications (EDIC) study. JAMA. 2003;290(16):2159–67. doi: 10.1001/jama.290.16.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chalmers J, Cooper ME. UKPDS and the legacy effect. N Engl J Med. 2008;359(15):1618–20. doi: 10.1056/NEJMe0807625. [DOI] [PubMed] [Google Scholar]
- 105.Miao F, Chen Z, Genuth S, Paterson A, Zhang L, Wu X et al. Evaluating the role of epigenetic histone modifications in the metabolic memory of type 1 diabetes. Diabetes. 2014;63(5):1748–62. doi: 10.2337/db13-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. ••.Chen Z, Miao F, Paterson AD, Lachin JM, Zhang L, Schones DE et al. Epigenomic profiling reveals an association between persistence of DNA methylation and metabolic memory in the DCCT/EDIC type 1 diabetes cohort. Proc Natl Acad Sci U S A. 2016;113(21):E3002–11. doi: 10.1073/pnas.1603712113. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study uncovered the epigenetic basis of metabolic memory in the landmark DCCT/EDIC diabetes clinical trial. The results identified novel mechanistic targets that display sustained differential epigenetic patterns (DNA methylation) in the same type 1 diabetic subjects over 16 years in asscoiation with glycemic history and an adverse diabetes complications outcome.
- 107. ••.Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J. Cell-free DNA Comprises an In Vivo Nucleosome Footprint that Informs Its Tissues-Of-Origin. Cell. 2016;164(1–2):57–68. doi: 10.1016/j.cell.2015.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows that circulating cell-free DNA faithfully mirrors the epigenetic footprints of tissue-of-origin, highlighting the potential of cell-free DNA as a tool for monitoring tissue specific changes in disease.
- 108. •.Moss J, Magenheim J, Neiman D, Zemmour H, Loyfer N, Korach A et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat Commun. 2018;9(1):5068. doi: 10.1038/s41467-018-07466-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study developed a detailed human cell-type DNA-methylation atlas to advance the development of cell-free DNA based disease biomarkers.
- 109.Akirav EM, Lebastchi J, Galvan EM, Henegariu O, Akirav M, Ablamunits V et al. Detection of beta cell death in diabetes using differentially methylated circulating DNA. Proc Natl Acad Sci U S A. 2011;108(47):19018–23. doi: 10.1073/pnas.1111008108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Husseiny MI, Kaye A, Zebadua E, Kandeel F, Ferreri K. Tissue-specific methylation of human insulin gene and PCR assay for monitoring beta cell death. PLoS One. 2014;9(4):e94591. doi: 10.1371/journal.pone.0094591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ronn T, Volkov P, Gillberg L, Kokosar M, Perfilyev A, Jacobsen AL et al. Impact of age, BMI and HbA1c levels on the genome-wide DNA methylation and mRNA expression patterns in human adipose tissue and identification of epigenetic biomarkers in blood. Hum Mol Genet. 2015;24(13):3792–813. doi: 10.1093/hmg/ddv124. [DOI] [PubMed] [Google Scholar]
- 112. •.Bacos K, Gillberg L, Volkov P, Olsson AH, Hansen T, Pedersen O et al. Blood-based biomarkers of age-associated epigenetic changes in human islets associate with insulin secretion and diabetes. Nat Commun. 2016;7:11089. doi: 10.1038/ncomms11089. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study illustrates the utility of blood-based biomarkers in predicting metabolic health, using islets as an example.
- 113.Parrizas M, Novials A. Circulating microRNAs as biomarkers for metabolic disease. Best Pract Res Clin Endocrinol Metab. 2016;30(5):591–601. doi: 10.1016/j.beem.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 114.Lin X, Qin Y, Jia J, Lin T, Lin X, Chen L et al. MiR-155 Enhances Insulin Sensitivity by Coordinated Regulation of Multiple Genes in Mice. PLoS Genet. 2016;12(10):e1006308. doi: 10.1371/journal.pgen.1006308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. ••.Ying W, Riopel M, Bandyopadhyay G, Dong Y, Birmingham A, Seo JB et al. Adipose Tissue Macrophage-Derived Exosomal miRNAs Can Modulate In Vivo and In Vitro Insulin Sensitivity. Cell. 2017;171(2):372–84 e12. doi: 10.1016/j.cell.2017.08.035. [DOI] [PubMed] [Google Scholar]; This study identifies a novel role for exosomal miRNAs in T2D pathogenesis, and highlights their potential as predictive biomarkers.
- 116.Castano C, Kalko S, Novials A, Parrizas M. Obesity-associated exosomal miRNAs modulate glucose and lipid metabolism in mice. Proc Natl Acad Sci U S A. 2018;115(48):12158–63. doi: 10.1073/pnas.1808855115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fu W, Farache J, Clardy SM, Hattori K, Mander P, Lee K et al. Epigenetic modulation of type-1 diabetes via a dual effect on pancreatic macrophages and beta cells. Elife. 2014;3:e04631. doi: 10.7554/eLife.04631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Deeney JT, Belkina AC, Shirihai OS, Corkey BE, Denis GV. BET Bromodomain Proteins Brd2, Brd3 and Brd4 Selectively Regulate Metabolic Pathways in the Pancreatic beta-Cell. PLoS One. 2016;11(3):e0151329. doi: 10.1371/journal.pone.0151329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Huijbregts L, Kjaer Petersen MB, Berthault C, Hansson M, Aiello V, Rachdi L et al. Bromodomain and Extra Terminal Proteins Inhibitors Promote Pancreatic Endocrine Cell Fate. Diabetes. 2019. doi: 10.2337/db18-0224. [DOI] [PubMed] [Google Scholar]
- 120. ••.Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S et al. Editing DNA Methylation in the Mammalian Genome. Cell. 2016;167(1):233–47 e17. doi: 10.1016/j.cell.2016.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study presents a novel, locus specific epigenetic editing approach that can be leveraged to tailor the epigenetic signatures of disease specific epialleles towards potential therapeutic interventions.
- 121. ••.Ou K, Yu M, Moss NG, Wang YJ, Wang AW, Nguyen SC et al. Targeted demethylation at the CDKN1C/p57 locus induces human beta cell replication. J Clin Invest. 2019;129(1):209–14. doi: 10.1172/JCI99170. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study illustrates the power of using locus specific epigenetic tailoring to mimic specific physiological and epigenetic contexts to induce a therapeutically relevant outcome, in this case, the replication of human beta-cells, that are normally resistant to mitogenic cues.
- 122. •.Liu J, Banerjee A, Herring CA, Attalla J, Hu R, Xu Y et al. Neurog3-Independent Methylation Is the Earliest Detectable Mark Distinguishing Pancreatic Progenitor Identity. Dev Cell. 2019;48(1):49–63 e7. doi: 10.1016/j.devcel.2018.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study utilizes epigenetic tailoring to bias endocrine differentiation to beta cell fate.
- 123. •.Liao HK, Hatanaka F, Araoka T, Reddy P, Wu MZ, Sui Y et al. In Vivo Target Gene Activation via CRISPR/Cas9-Mediated Trans-epigenetic Modulation. Cell. 2017;171(7):1495–507 e15. doi: 10.1016/j.cell.2017.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study presents a novel approach to epigenetic engineering, which involves the targeting of transcriptional regulatory complexes to specific loci to induce epigenetic remodeling.