Introduction:
In an era where pharmacogenetic testing is becoming more common, the processes of return of results and update of results need to be laid out and established in order for patients to truly benefit from testing. This becomes particularly important in a preemptive setting because testing is being performed independently of medication use. We describe herein the process that St. Jude Children’s Research Hospital has undertaken for the return and update of pharmacogenetic test results.
St. Jude Children’s Research Hospital (St. Jude) has been offering preemptive pharmacogenetic testing to all patients who consent for enrollment on the PG4KDS protocol (www.stjude.org/pg4kds) since 2011. [1] PG4KDS uses a rational, stepwise process to integrate select pharmacogenetic test results into the electronic health record (EHR) to guide safe prescribing of relevant medications. Gene-drug pair implementations are prioritized based on medication use at St. Jude, availability of CPIC guidelines, primary literature review, and with approval of an institutional Pharmacogenetics Oversight Committee. The primary reason for PG4KDS being a clinical trial is to obtain informed consent for withholding results of many of the genes interrogated by the PharmacoScan™ array (ThermoFisher Scientific, Waltham, MA), and for returning incidental findings (two findings of Klinefelter’s in the first 5,100 patients). Genotype results are viewable in a dedicated pharmacogenetics section of the EHR, and are coupled with an interpretative consultation note entered by a clinical pharmacist. A medication profile assessment is performed to check if patients are receiving a medication affected by high-risk results at the time they are posted in the EHR. Phenotype specific problem list entries trigger customized clinical decision support (CDS) alerts, which provide specific recommendations to tailor pharmacotherapy when a medication affected by host pharmacogenetics (high-risk medication) is ordered. Extensive patient and clinician education accompanies each new implementation.
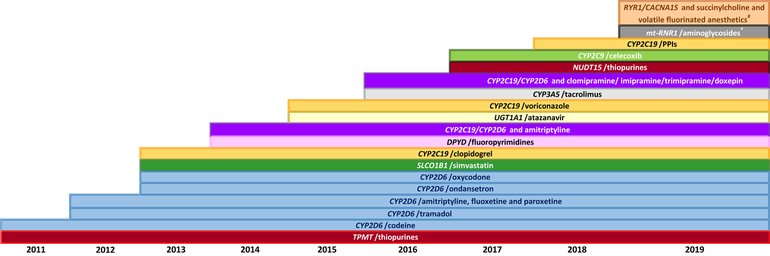
As of June 2019, 11 genes and 35 drugs have been implemented in the EHR of nearly 5,100 patients (Figure 1). A fundamental tenet of preemptive pharmacogenetics is that results are useful over a patient’s lifetime and must therefore be easily retrievable to be used whenever high-risk medications are prescribed. Fully implementing preemptive pharmacogenetics requires that patients have access to their results and for clinicians to have the most updated information when using these results. With 8 years of experience in implementing preemptive pharmacogenetics across our institution, we have learned valuable lessons on the requirements to truly implement a preemptive model.
Figure 1:
Gene/Drug Pair Implementation Timeline. Pharmacogenes and relevant medications have been systematically implemented in the patients’ health record over time. As additional genes and drugs are implemented the list of gene/drug pairs will increase over time.
*Aminoglycosides include: amikacin, gentamicin, plazomicin, streptomycin, and tobramycin.
#Volatile anesthetic agents include: enflurane, methoxyflurane, desflurane, halothane, isoflurane, and sevoflurane.
Return of Results:
To realize the ongoing value of preemptive pharmacogenetics, results must be easily retrievable by both clinicians and patients for years after they become available. Given the complexity of the return of result process, many institutions are electing not to return results to patients. For example, only 3 out of 12 IGNITE network sites reported returning CYP2C19 genotype test results to their patients, whereas healthcare professionals at the testing institutions all had access to the results.[2] The PG4KDS patient consent process initially asked participants if they wanted to be informed of their pharmacogenetic results. Because most patients (96%) requested to be notified of their results, our practice changed in April 2019 to notify all patients when their results are posted in the EHR; if patients do not want to be notified, they are advised not to enroll on the protocol.
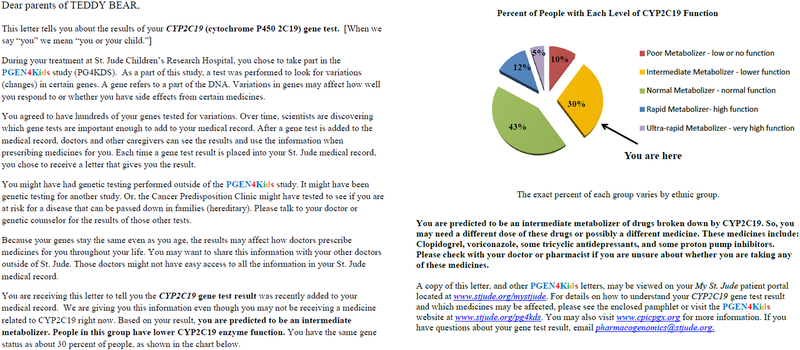
Beyond accessing the pharmacogenetics results and their associated interpretive consult note from a dedicated flowsheet view in the EHR, St. Jude clinicians are actively notified (through an automated email) when high-risk pharmacogenetic test results are entered in the EHR of their patients. Phenotype specific letters are provided to patients for each pharmacogenetic test result (Figure 2). Each letter contains five main sections: 1) A phenotype assignment for the gene test result; 2) a brief explanation defining the phenotype; 3) a pie chart representing the phenotype frequencies for that gene and which piece of the pie the patient belongs to; 4) a list of some of the medications affected by variations in the gene; and 5) a “for more information” section providing contact information to the St. Jude Pharmacogenomics Service and links to PG4KDS educational materials, and the Clinical Pharmacogenetics Implementation Consortium (CPIC-www.cpicpgx.org) website. Patients are encouraged to contact a member of our team with questions or to discuss the results in person. Since July 2015, the letters are accessible to patients and available for printing on the St. Jude online portal (Cerner Health, North Kansas City, MO, USA). In addition, for results generated before April 2019, patients also received a copy of the letter via USPS mail to their home address. As of June 2019, nearly 30,500 gene and phenotype specific result letters have been posted in the EHR and mailed to patients in 29 different countries. St. Jude has also created patient and provider educational material for each implemented gene that are easily accessible for online review by clinicians as they are making pharmacotherapy decisions to treat their patients (www.stjude.org/PG4KDS/implemented-genes).
Figure 2:
Example of a pharmacogenetic result letter that is returned to patients (in this case a CYP2C19 normal metabolizer phenotype). Patients receive one gene/phenotype specific letter for each implemented gene.
Due to the fragmented nature of the United States healthcare system, we are forced to rely on our patients to share their pharmacogenetic test results with their non-St. Jude providers. This step becomes crucial when patients grow older and they are no longer routinely receiving care at our hospital. There are many limitations to relying on patients. First, patients must remember to share the pharmacogenetic test results with each new care provider and pharmacy. Second, the clinicians who are the recipients of the letters must have some basic understanding of how to interpret and act on the pharmacogenetic test results. Because > 90% of patients who are genotyped for actionable pharmacogenes will have at least one high-risk phenotype, it is essential for clinicians to understand the implications of a high-risk result.[1, 3] Most EHR systems require verification of the authenticity of laboratory results before utilizing them. Given the number of disconnected providers and pharmacies that any one patient sees, the importance of delivering interruptive CDS to guide prescribing, and the lack of a common EHR system to facilitate use of CDS, there are many routes by which pharmacogenetic information will be unavailable for proper use throughout a patient’s lifetime.
Updating of Results:
Because the implications and potential actionability of pharmacogenetic results will often apply to multiple medications and last for a patient’s lifetime, it is essential to have a system to update results or their interpretation as scientific evidence and consensus groups (such as CPIC) publish new and updated findings. In recent years the debate of how and whether to re-contact patients with updated genomics results has prompted major genomic societies (such as the American College of Medical Genetics and Genomics-ACMG- and the European Society of Human Genetics -ESHG) to weigh in with recommendations. [4, 5] While the recommendations do not specifically address pharmacogenes, they recommend that the clinical team to inform patients of the possibility of future result updates, that reasonable efforts be made to re-contact patients with the updated results. They also state that patient re-contact should be a shared responsibility between healthcare providers and the clinical laboratories that generated performed the genotyping.[4] Because consensus does not exist for updating pharmacogenetic results, site specific decisions must be made to determine when result updates are needed. At St. Jude the threshold for update has been set to when new information emerges that prompts a change in phenotype assignment.
The result update process at St. Jude is led by the Pharmacogenomics Service. It includes updating the genotype (when applicable and feasible), phenotype, interpretive consult note, and the pharmacogenetic problem list entry in the EHR. Clinicians and patients are notified of the updated result if a phenotype change occurs. An updated phenotype-specific result letter is placed in the patient’s EHR and is accessible in the online portal. Results are updated for all living patients who remain enrolled on the PG4KDS study irrespective of when the original result was generated. As of June 2019, we have updated 1,335 (3.3%) of the 39,826 results posted in the EHR to date. The majority of the results updated involved a change in CYP2C19 phenotype assignment (from ultra-rapid metabolizer to a rapid metabolizer phenotype) for patients with a *1/*17 diplotype; in this case, the recommendations for starting doses of voriconazole changed substantially and impacted patients. Similar to the original results, the responsibility of informing the non-St. Jude healthcare providers lies with the patients. Patients are directed to go to the PG4KDS website (www.stjude.org/PG4KDS) to review an updated list of actionable medications as they relate to each pharmacogene.
In summary, most patients wanted to be informed of their pharmacogenetic test results. As access to the online portal grew, we shifted to using it as our primary method for returning results and eliminated the laborious process of mailing thousands of letters to patients; patients have the option to ask to speak to a clinician if they prefer not to use the portal. We expect that some patients will take their pharmacogenetic result letters to future care settings, but our approach remains hindered by our fractured health care system. As the United States continues to embrace interoperability of medical records, pharmacogenetic information must be positioned to be one of the elements transferred across medical record systems. Increasing standardization of pharmacogenetic terms with representation in terminologies such as SNOMED-CT may facilitate transfer of pharmacogenetic data across EHRs. While debate remains among clinical laboratories and healthcare providers about the responsibility to update results, our experience illustrates that using preemptive pharmacogenetic data over time demands selected updates be made to continue to reap the safety benefits of pharmacogenetic testing.
Funding:
The authors are supported by National Cancer Institute grants CA 36401, CA 21765; National Institute of Health (grants U01 GM115279; U24 HG010135); and by the American Lebanese Syrian Associated Charities (ALSAC).
Footnotes
Conflict of Interest:
The authors declared no competing interests for this work.
References:
- 1.Hoffman JM, et al. , PG4KDS: a model for the clinical implementation of pre-emptive pharmacogenetics. Am J Med Genet C Semin Med Genet, 2014. 166C(1): p. 45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Empey PE, et al. , Multisite Investigation of Strategies for the Implementation of CYP2C19 Genotype-Guided Antiplatelet Therapy. Clin Pharmacol Ther, 2018. 104(4): p. 664–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Driest SL, et al. , Clinically actionable genotypes among 10,000 patients with preemptive pharmacogenomic testing. Clin Pharmacol Ther, 2014. 95(4): p. 423–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.David KL, et al. , Patient re-contact after revision of genomic test results: points to consider-a statement of the American College of Medical Genetics and Genomics (ACMG). Genet Med, 2019. 21(4): p. 769–771. [DOI] [PubMed] [Google Scholar]
- 5.Carrieri D, et al. , Recontacting patients in clinical genetics services: recommendations of the European Society of Human Genetics. Eur J Hum Genet, 2019. 27(2): p. 169–182. [DOI] [PMC free article] [PubMed] [Google Scholar]