Summary
Although olfactory mucosa possesses long-lived horizontal basal stem cells (HBCs) and remarkable regenerative capacity, the function of human olfactory neuroepithelium is significantly impaired in chronic inflammatory rhinosinusitis. Here we show that while inflammation initially damages olfactory neurons and activates HBC-mediated regeneration, continued inflammation locks HBCs in an undifferentiated state. Global gene expression in mouse HBCs reveals broad upregulation of NF-κB-regulated cytokines and chemokines including CCL19, CCL20, and CXCL10, accompanied by enhancement of “stemness”-related transcription factors. Loss-of-function studies identify an NF-κB-dependent role of HBCs in amplifying inflammatory signaling, contributing to macrophage and T-cell local proliferation. Chronically-activated HBCs signal macrophages to maintain immune defense and prevent Treg development. In diseased human olfactory tissue, activated HBCs in a P63+ undifferentiated state similarly contribute to inflammation through chemokine production. These observations establish a mechanism of chronic rhinosinusitis-associated olfactory loss, caused by a functional switch of neuroepithelial stem cells from regeneration to immune defense.
Graphical Abstract

In Brief
Chen et al. identify the immune function of long-lived olfactory stem cells to regulate inflammatory cell recruitment and local proliferation by releasing cytokines and chemokines. Chronically activated stem cells shut down regenerative function and signal macrophage to maintain epithelial immune defense.
INTRODUCTION
Adult stem cells residing in the basal layer of epithelial tissue interact with their niche components and signal to maintain epithelial integrity. The interplay of the local immune system and stem cells in regulating endogenous epithelial regeneration is beginning to be elucidated in several systems(Karin and Clevers, 2016; Naik et al., 2018; Wells and Watt, 2018). In aged skin, impaired epithelial regeneration is associated with defects in immune-basal cell crosstalk, and inflammation-experienced stem cells retain an epigenetic memory that accelerates subsequent wound healing (Keyes et al., 2016; Naik et al., 2017). Repair of the olfactory epithelium after injury necessarily involves a self-limited inflammatory response to initiate regenerative signals through NF-κB activation in basal stem cell (Chen et al., 2017). These observations suggest an essential role of the local immune system in facilitating epithelial stem cell regeneration (Hsu et al., 2014; Lane et al., 2014). However, persistent inflammation can be deleterious and contributes to a variety of chronic epithelial diseases. How the local immune system interacts with the long-lived stem cells to influence the progression of inflammation and tissue regeneration is largely unknown.
The sense of smell is mediated in olfactory epithelium by primary sensory neurons (OSNs) that detect odorants and directly transmit activity to the brain(Buck and Axel, 1991; Krautwurst et al., 1998). The anatomic location of OSNs in the nasal cavity renders them directly vulnerable to external environmental insults. Neural stem cells/progenitors, including horizontal and globose basal cells (HBCs and GBCs), which reside in the basal layer of olfactory epithelium, possess robust regenerative capacity to replenish OSNs lost throughout life (Holbrook et al., 2011; Leung et al., 2007; Schwob et al., 2017). Recent evidence based on genetic strategies has demonstrated that stem cell intrinsic signals, including transcription factors (Fletcher et al., 2011; Schnittke et al., 2015), Wnt (Chen et al., 2014; Fletcher et al., 2017), Notch (Herrick et al., 2017) and epigenetic components (Lin et al., 2017), contribute to mouse olfactory epithelium regeneration after chemical lesioning. Despite the remarkable regenerative capacity of long-lived olfactory stem cells, human olfactory deficits are common, especially in the setting of chronic inflammation, and the molecular basis remains elusive.
Chronic rhinosinusitis (CRS) is a heterogeneous disease associated with persistent inflammation of the sinonasal mucosa. Affecting approximately 12.5% of the US population, CRS is the most common cause of olfactory dysfunction (Hamilos, 2011). The pathogenesis of CRS is multifactorial, but dysregulation of host innate and adaptive immune responses appears to be a common feature (Ramanathan and Lane, 2007; Stevens et al., 2015; Van Crombruggen et al., 2011). Inflammatory mediators modulate sinonasal epithelial cell innate immune activity, and epithelial cell-derived cytokines (Nagarkar et al., 2013; Shaw et al., 2013) have been implicated as driving or sustaining sinonasal inflammation. Human chronic olfactory inflammation is typically associated with CRS and characterized by local immune cell infiltration and inflammatory mediator production, loss of sensory neurons, and decreased olfactory function (Kern, 2000; Victores et al., 2018; Yee et al., 2010). We observed in a mouse genetic model that chronic olfactory mucosal inflammation results not only in olfactory neuronal depletion, but also impaired neurogenesis (Lane et al., 2010). This finding indicates that interaction between olfactory stem cells and immune system might critically underlie ongoing olfactory impairment in CRS.
Here, through detailed analysis of morphology, proliferation dynamics, immunologic profile, and transcriptomics in genetically-modified mice, we identify previously unrecognized roles of stem cells in orchestrating immune cell infiltration and local proliferation, and we elucidate mechanisms through which HBCs switch off their regenerative function in response to prolonged inflammation
RESULTS
Horizontal basal cell dynamics during inflammation
Human chronic olfactory inflammation typically occurs in the context of CRS, a heterogeneous condition of unknown etiology that does not occur naturally in animals. To mimic the key features of human olfactory inflammation, including an immune cell infiltrate, local inflammatory mediator production, loss of olfactory sensory neurons, and decreased olfactory function, we created an inducible olfactory inflammation (IOI) mouse model (Lane et al., 2010). In this model, temporally-controlled, olfactory-specific expression of TNF initiates a robust inflammatory response in the lamina propria, transforming over the course of weeks from an acute phase to into an unresolving and uniformly intense leukocytic (CD45+) infiltrate (Figures 1A, S1A–S1C).
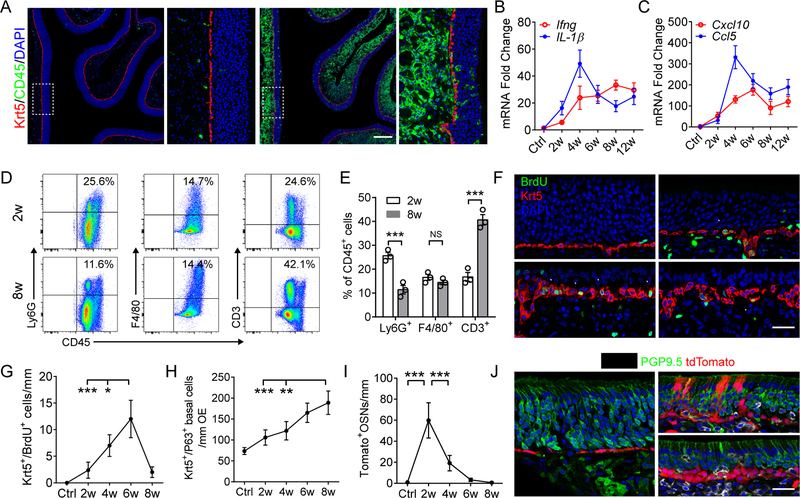
Figure 1. Continuous inflammation impedes olfactory neurogenesis.
(A) Representative images of no Dox control and olfactory inflammation after 3 w (weeks) of Dox treatment.
(B and C) Quantitative PCR (qPCR) analysis of mRNA expression of cytokines (B) and chemokines (C) in whole olfactory mucosa after indicated time periods of Dox induction. (D and E) Percentage of inflammatory cell subpopulation in olfactory mucosa (E) and representative FCS plots (D). A total of 8 × 104 CD45+ cells were recorded in each OE sample. n = 3 mice in (B–E).
(F and G) Quantification of dividing Krt5+ stem cells (arrowheads) in olfactory epithelium (G), and representative images (F) of BrdU incorporation assay.
(H) Quantification of P63+/Krt5+ olfactory stem cells. n = 5 mice per group in (F–H).
(I and J) Lineage tracing of Krt5+ basal cells in inflammatory microenvironment, n = 3 mice.
Data are represented as mean ± SEM, *P < 0.05, **P < 0.01, ***P < 0.001, P values were calculated by unpaired two tailed Student’s t-test (E) or by one-way ANOVA (G–I); NS, not significant. Scale bars, 200 μm (A) and 25 μm (F and J). See also Figure S1.
To understand the impact of chronic local inflammation on the olfactory neuroepithelium, we first performed a detailed characterization of the immune cell profile and downstream mediators that develop in the IOI model in response to continuous TNF stimulation. As in chronic inflammatory diseases, where multiple immune mediators are upregulated, mRNA expression of pro-inflammatory cytokines IL-1β and IFNγ, as well as chemokines Cxcl10, Ccl5, Cxcl2, and Ccl2, were dramatically increased, peaking at 4 weeks of Dox treatment and then maintained at a chronically elevated level (Figures 1B, 1C, S1D, and S1E). The early peak corresponds to the inflammatory reaction to mediators and debris from dying/dead cells, which, after being cleared, is followed by development of a stable chronic phase. Expression of the Th17 cytokine IL-22 mRNA in the chronic inflammatory stage was low (Figure S1F) and IL-17a was undetectable. Immunostaining for eosinophil major basic protein showed rare to no eosinophil infiltration in the IOI mouse olfactory mucosa.
Flow cytometry and immunostaining revealed a significant increase in CD3+ T cells with a reduction of Ly6G+ granulocytes (neutrophils) in the olfactory mucosa over an 8-week dox exposure period (Figures 1D, 1E, S1G, and S1H), indicative of a shift to a T cell-dominated chronic inflammatory microenvironment. A prominent effect of the inflammatory state was neuronal apoptosis and eventually near complete loss of PGP9.5+ OSNs. Survival of CK18+ sustentacular cells was unaffected, allowing an unresolving source of TNF transgene expression (Figures S1I, S1J).
We then assessed olfactory stem cell dynamics as the inflammation progressed. Adult quiescent HBCs normally appear as a flat single layer residing above the basal lamina (Leung et al., 2007). By 2 weeks of continuous doxycycline (Dox)-regulated genetic initiation of inflammation, the morphology of Krt5+ HBCs became rounded or triangular, and some incorporate BrdU (Figure 1F). The number of Krt5+/BrdU+ dividing cells increased remarkably by 6 weeks, then regressed at a later time point (Figures 1F and 1G). The increased Krt5+ cell population (Figure 1H) formed stacked layers, co-expressing the canonical HBC marker ∆Np63 (Figure 4E), confirming an olfactory stem cell identity (Fletcher et al., 2011; Packard et al., 2011).
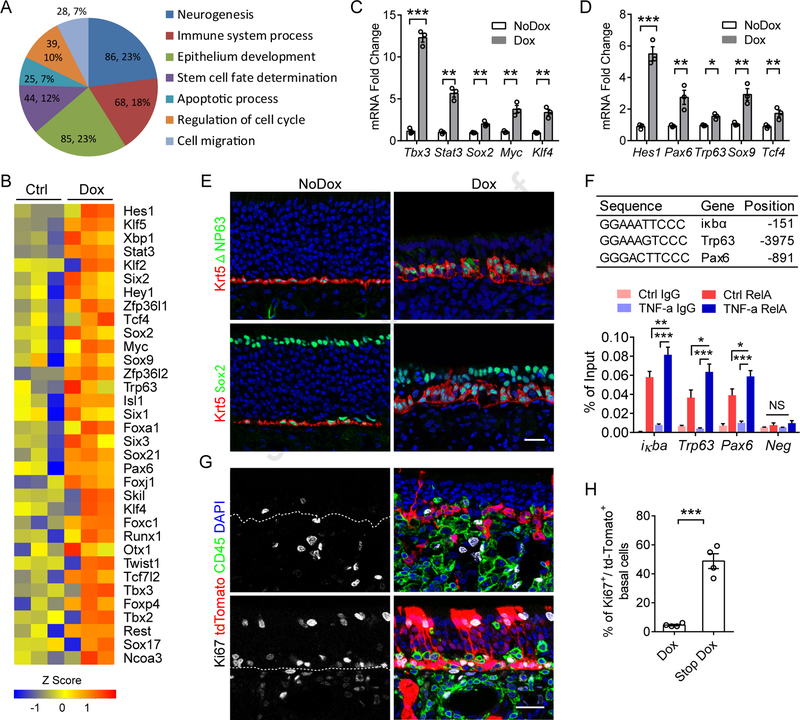
Figure 4. Transcriptional regulation switches off the HBC regenerative phenotype.
(A) GO analysis of the differentially expressed transcription factors in HBCs between control and 6w Dox treated group (up or downregulation > 1-fold, p < 0.05). Each slice of the pie chart represents the number of genes and percentage that belong to the indicated GO terms.
(B) Upregulated transcription factors in HBCs of 6w Dox treated group that are involved in maintaining stem cell capacity.
(C and D) qPCR validation of upregulated transcription factors in sorted HBCs. n = 3 independent samples.
(E) Immunostaining of Krt5, Sox2 and ΔNP63 in sections from Control or 6 w Dox treated mice. Some ΔNP63− cells located between Krt5+/ΔNP63+ stem cells are immune cells (Figures 4G and S1G).
(F) Chip-qPCR analysis of RelA binding sites using primary cultured HBCs. Sequence and location are indicated in above table. A region lacking a binding site was examined as negative control (Neg). n = 3 biological replicates.
(G and H) Immunostaining of Ki67 and CD45 in Krt5cre-IOI-Rosa26-stopflox/flox-tdTomato mice (G), and quantification of Ki67+/tdTomato+ proliferating basal cells (H). n = 4 mice.
Data are represented as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001. P values were calculated by unpaired two tailed Student’s t-test (C and D) or by one-way ANOVA (F); NS, not significant. Experiments were replicated at least twice except (A and B). Scale bars, 25 μm (E and G). See also Figure S4.
To track the fate of HBCs and their progeny under conditions of chronic inflammation, we performed lineage-tracing experiments by crossing IOI mouse with a Krt5cre/Rosa26-stopflox/flox-td-Tomato strain. In control mice, td-Tomato labeled HBCs rarely proliferate. Notably, after 2 weeks of Dox treatment, we observed abundant td-Tomato+ cells distributed in the neuroepithelium, including differentiated PGP9.5+ OSNs, suggesting ongoing neurogenesis at early stages of inflammation. However, the newborn olfactory-epithelial lineage cells were largely eliminated as inflammation continued (Figures 1I and 1J). Taken together, these data demonstrate a contributing role of HBCs in maintaining olfactory epithelium homeostasis during early stages of inflammation. Prolonged inflammation that is detrimental to newly generated olfactory neuron development and simultaneously inhibitive to activated HBC differentiation results in a state of neuronal depletion.
NF-kB activation in HBCs induces a broad spectrum of cytokines and chemokines
To explore the core transcriptional signature of HBCs affected by chronic inflammation, we sorted HBCs from Krt17-EGFP (Control) and Krt17-EGFP-IOI mice (Figures S2A–D) and performed RNA-seq to analyze the global expression profile. RT-PCR validated that the sorted HBC RNA was not contaminated by inflammatory cells, endothelial cells or fibroblasts (Figure S2E). Gene annotation enrichment analysis highlighted the TNF/NF-κB signaling pathway among the top hits of broad transcriptional changes in these HBCs (Figure 2A). Other significantly upregulated genes encode molecules involved in cell adhesion, PI3k-Akt signaling, Hippo signaling, MAPK signaling, and the Wnt signaling pathway.
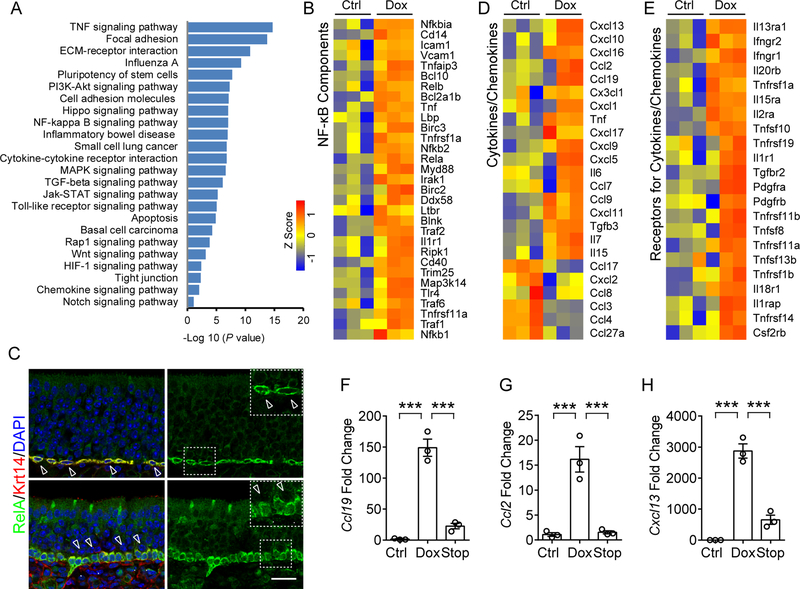
Figure 2. NF-κB activation in HBC targeted broad spectrum cytokines and chemokines.
(A) Pathway enrichment analysis of genes that were significantly upregulated (>2-fold, p < 0.05) in sorted HBCs after 6 w Dox treatment, relative to untreated control. Enriched pathways were presented according to –log10 (p value).
(B) Heat map of NF-κB pathway-related genes that are significantly upregulated in HBCs after Dox treatment.
(C) Immunostaining analysis of RelA localization in HBCs. RelA is highly expressed in Krt14+ HBCs and largely absent in nuclei (upper panel, hollow arrow heads) in the static state. Upon Dox-initiated inflammation, RelA translocated to the nucleus in Krt14+ HBCs (Arrow heads in lower panel, compared to hollow arrow heads). IOI mouse was treated with Dox for 12 days.
(D) Heat map of cytokines or chemokines genes that are differentially expressed in HBCs.
(E) Heat map of upregulated receptor genes of cytokines or chemokines in HBCs. The color scale shows the Z score of each gene.
(F–H) qPCR analysis of mRNA expression of Ccl19 (F), Ccl2 (G), and Cxcl13 (H) in HBCs sorted from control, Dox treated and 3 days post-Dox treatment (Stop) mice.
Data are represented as mean ± SEM, n = 3 independent samples. ***P < 0.001, P values were calculated by one-way ANOVA. Scale bar, 50 μm. See also Figure S2.
We are particularly interested in NF-κB signaling since RelA is markedly highly expressed in HBCs in the static state (Figure 2C). The upregulated expression of NF-κB pathway components/targets (KEGG pathway database) supports the involvement of NF-κB signaling in HBC dysfunction (Figure 2B) in chronic inflammation. Specifically, these genes include the four NF-κB family members (Nfκb1, Nfκb2, Rela, and Relb), IκB family member (Nfκbiα), and other molecules for NF-κB signaling transduction (Tnfrsf1a, Il1r1, Cd14, Irak1, Myd88, Traf) (Li and Verma, 2002; Taniguchi and Karin, 2018). In addition, the translocation of RelA into the nuclei of HBCs upon Dox treatment in vivo or TNF stimulation in vitro suggests NF-κB activation (Figures 2C and S2F).
We observed extensive upregulation of inflammatory cytokines/chemokines by HBCs in Dox-induced inflammation compared with normal controls (Figure 2D). These genes represent chemoattractant molecules controlling neutrophil trafficking (Cxcl1, Cxcl5), T cell recruitment and differentiation (Cxcl9, Cxcl10, Cxcl11, Cxcl13, Cxcl16, Ccl2, Ccl9, and Ccl19), monocytes/ macrophage migration (Ccl2, Ccl7 and Cx3cl1), and B cell positioning (Cxcl13) (Griffith et al., 2014). Interestingly, majority of these cytokines/chemokines (TNF, IL-6, Cxcl1, Cxcl5, Cxcl9, Cxcl10, Cxcl11, Cxcl13, Ccl19, Ccl9, Ccl2, and Ccl7) are known NF-κB targets (Richmond, 2002; Taniguchi and Karin, 2018; Tong et al., 2016). We performed qPCR using sorted HBCs (Figures 2F–H and S2G) as well as immunostaining in olfactory tissue (Figures 3I–3N) further validated the activated expression of chemokines in HBCs. The increased expression of chemokines in HBCs was also observed in primary HBC cultures after TNF stimulation (Figures S2H). A number of chemokines normally expressed by isolated HBCs (Ccl27a, Ccl4, Ccl3, Ccl8, Cxcl2, and Cxcl17) were downregulated in Dox treated mice (Figure 2D), suggesting complexity in neuro-immune signaling.
Figure 3. HBCs amplify inflammatory signals in olfactory mucosa via NF-κB.
(A) Representative images of CD45 and BrdU staining in control (NoDox) or 6 days of Dox treatment (Dox and RelAΔ/Δ) groups. RelAΔ/Δ, IOI mice with selective RelA deletion in HBCs.
(B) Quantification of CD45+ immune cells in (A).
(C and D) Quantification of CD45+/BrdU+ (C) or CD45+/Ki67+ (D) proliferating immune cells at indicated time points of Dox treatment.
(E and F) Representative images of co-staining for Ki67 and the macrophage marker F4/80 (E), and quantification of F4/80+/Ki67+ proliferating macrophages (F) in olfactory mucosa.
(G and H) Co-staining CD3 and Ki67 (H), and quantification of CD3+/Ki67+ proliferating T lymphocytes (G).
(I–N) Immunostaining of CXCL10, CCL19, and CCL20 with Krt5 in Dox treated IOI (I–K) or IOI with RelA deletion (L–N) in HBCs. Below (I–N) show magnified views of boxed area.
Data are represented as mean ± SEM, n = 4 mice. *P < 0.05, *P < 0.01, **P < 0.001; P values were calculated by unpaired two tailed Student’s t-test. All experiments were replicated at least twice. Scale bars, 25 μm. See also Figure S3 and S4.
In addition, a subset of genes which encode products of cytokine receptors were upregulated in dysfunctional HBCs relative to normal control (Figure 2E and Table S4), including interferon gamma receptors (Ifngr1, Ifngr2), interleukin receptors (Il13ra1, Il20rb, Il15ra, Il2a, Il1r1, and Il18r1), and TNF receptor/ligand super family members. Taken together, these data reveal a previously unappreciated function of HBCs in potentially participating in crosstalk with immune cells in the setting of chronic inflammation.
HBCs relay inflammatory signaling via NF-κB
To evaluate the function of NF-κB activation in HBCs during the progression of inflammation, we crossed IOI mice to a Krt5cre-RelAflox/flox strain to generate selective loss of RelA, an essential part of the NF-κB transcription factor complex (Figure S3A). In the early stage of inflammation (6 days Dox treatment), we observed that deletion of RelA in HBCs (RelAΔ/Δ) significantly reduced the infiltration of CD45+ inflammatory cells in the olfactory mucosa when compared with IOI mice (Figures 3A and 3B). The level of Dox-induced TNF expression was comparable between IOI and Krt5cre-RelAflox/flox-IOI mice (Figures S3B and S3C). Normally, the number of CD45+ cells in the olfactory mucosa is low and proliferation is rare as assessed by co-staining with proliferation markers Ki67 or BrdU. Interestingly, Dox treatment-induced inflammation activated a considerable fraction of proliferating CD45+ immune cells (Figures 3A, 3C, and 3D). Selective loss of RelA in HBCs remarkably decreased the percentage of Ki67+ or BrdU+ proliferating immune cells on day 4 and day 6 when compared with IOI control (Figures 3A, 3C, and 3D). The proportions of proliferating immune cells were comparable at 12 days of Dox treatment, indicating that non-NF-κB-dependent HBC-derived signaling molecules (Figure 2A) likely participate in the highly dynamic infiltration process. The number of actively proliferating CD45+ cells was decreased at 8w compared to 1 week in Dox-treated IOI mice (Figures S3D and S3E), suggesting an intrinsic inhibitory response once immune cell infiltration reaches a high density.
We investigated the subpopulations of actively proliferating inflammatory cells by co-staining Ki67 with F4/80, CD3, or Ly6G after 6 day Dox treatment. We observed that a large portion of F4/80+ macrophages (61.8%) were Ki67+ (Figures 3E and 3F), in accordance with reported in situ proliferation of pleural tissue macrophages in response to parasitic infection (Jenkins et al., 2011). The percentage of CD3+Ki67+ cells (31.5%) was remarkably elevated in Dox treated mice, suggesting that T cell local proliferation also contributes to the inflammatory progression (Figures 3G and 3H). As compared to the IOI control, the number of proliferating macrophages and T cells in RelA deleted mice was reduced 40.8% and 36%, respectively (Figures 3E–3H). The number of neutrophils was significantly decreased in RelA-deleted mice, with very little Ki67 co-expression (Figures S3F and S3G). Notably, RelA-deleted mice displayed attenuated olfactory neuroepithlial pathology, with a significant reduction in the number of Caspase-3+ cells in the early stage of inflammation (Figures S3H). In addition, local proliferation of immune cells was also a feature of LPS-induced olfactory inflammation, and a pronounced reduction of immune cell infiltration and proliferation was readily detected in mice lacking RelA in the HBC population (Figures S4A–S4C). These data suggest that NF-κB signaling in HBCs contributes to inflammatory cell infiltration specifically by promoting local proliferation of macrophages and T lymphocytes, and by recruitment of neutrophils.
Loss of RelA reduces immune cell infiltration indicating the potential role of HBC-specific molecules in amplifying inflammatory signaling. We further assessed the localization of NF-κB-regulated chemokines by co-staining with the HBC marker Krt5. Six days of Dox treatment dramatically activated HBC expression of Cxcl10, Ccl19 and Ccl20 (Figures 3I–3K), which are chemokines functioning to enhance immune cell recruitment and proliferation (Dufour et al., 2002; Luther et al., 2002; Marsland et al., 2005). Loss of RelA in HBCs completely eliminated the expression of these chemoattractants (Figures 3L–3M). None of these chemokines can be detected in normal control mice (Figure S4D). Interestingly, HBCs are the primary cellular source of Ccl19 and Ccl20. We found that high levels of Cxcl10 were also expressed by infiltrating inflammatory cells in the lamina propria, but rarely by other neuroepithelial cells. In addition, the production of Ccl20 and Cxcl10 by HBCs was observed in LPS induced olfactory inflammation (Figures S4E–S4G). Together, these data highlight the function of HBCs, a neural stem cell population, as a primary contributor to inflammatory cell infiltration and local proliferation in neuroepithelium via release NF-κB targeted chemokines.
Transcriptional regulation switches off the HBC regenerative phenotype
Given the essential role of transcription factors in regulating neurogenesis and regeneration (Fletcher et al., 2017; Schwob et al., 2017), we further analyzed the RNA-seq data sets and identified 315 differentially expressed transcription factors which were significantly up- (219 genes) or down-regulated (98 genes) in sorted HBCs after 6w induced inflammation. Gene ontology (GO) analysis revealed that these genes were enriched in distinct but overlapped subsets mainly involved in regulation of neurogenesis, epithelium development, stem cell fate determination, and immune system process (Figure 4A).
Interestingly, 45 up-regulated transcription factors encode products associated with negative regulation of stem cell differentiation (Figure 4B and Table S5). These “stemness”-related transcription factors closely match the listed genes which were selectively expressed by quiescent HBCs (Fletcher et al., 2017). The upregulated genes Tcf7l2, Rest, and Otx1 have been shown to maintain quiescent neural stem cells (Shin et al., 2015) or control embryonic neurogenesis (Gao et al., 2014) in the brain. In agreement with the RNA-seq data, qPCR results validated the up-regulation of transcription factors (Stat3, Sox2, Myc, Klf4, and Tbx3) which are linked to pluripotent stem cells (Figure 4C), and transcription factors (Pax6, Trp63, Hes1, Sox9, and Tcf4) that are necessary for maintaining HBCs (Fletcher et al., 2011; Herrick et al., 2017) or epithelial stem cells (Adam et al., 2015; Lien et al., 2014) in a quiescent state (Figure 4D). Co-staining of Krt5 with ΔNP63 (Trp63) or Sox2 revealed that chronic inflammation activated HBCs to form stacked layers but maintained them in ΔNP63+/Sox2+ undifferentiated state (Figure 4E). Notably, among those down-regulated genes, Insm1, Olig1, and Lhx2 encode products promoting neurogenesis (Figure S4H).
To address whether NF-κB directly regulate the “stemness” transcription factors, we performed Chip-qPCR using primary cultured HBCs (Chen et al., 2017). Compared to IgG and negative region controls, highly enriched RelA binding to the NF-κB motif was readily detected at the promoter regions of Trp63 and Pax6. Activation of NF-κB by TNF treatment modestly, but significantly, increased the binding of RelA compared to the unstimulated control (Figure 4F), suggesting baseline NF-κB activation in cultured HBCs. Increased RelA binding to Trp63, related to inflammatory signaling, could also be detected in sorted HBCs from 6w Dox treated IOI mice (Figure S4I). Consistent with the observations in Chip, TNF treatment slightly, but dosage dependently, increased the mRNA expression of Trp63 and Pax6 in HBC cultures (Figures S4J and S4K). These data, together with previous evidence that NF-κB directly targets Myc and Sox9 (Taniguchi and Karin, 2018), raise the possibility that inflammation-induced NF-κB activation regulates transcription factors to maintain or enhance hierarchy of HBC “stemness”, which is necessary for subsequent epithelial repair upon resolution of inflammation. Indeed, when Dox administration is stopped for 4 days, Td-Tomato-labeled HBCs are released to undergo dramatic proliferation and regeneration of OSN lineage and sustentacular cells (Figure 4G, 4H). Together, these data reveal that HBCs turn off their primary regenerative repair function via previous unknown mechanism in which NF-κB contributes prominently by targeting core transcription factors in response to chronic inflammation.
HBC signaling activates macrophage immune defense
The inflammatory response is essential for initiation of protective immunity, including the recruitment classically-activated M1 macrophages that mediate antimicrobial defense (Afonina et al., 2017; Murray and Wynn, 2011). However, dysregulated immune activation can result in chronic inflammatory or autoimmune diseases with consequent tissue damage. Intrinsic anti-inflammatory signals from Foxp3+ regulatory T cells (Treg) play critical role in preventing excessive inflammatory reactions and maintaining immune homeostasis (Sakaguchi et al., 2008). The expression of Tgfb1, which promotes Treg development (Korn et al., 2009), as well as foxp3, were strongly upregulated upon Dox induction. Il6, an acute phase pro-inflammatory cytokine that negatively regulates Foxp3+ Treg differentiation (Bettelli et al., 2006) was maintained at an elevated level after reaching a peak at 4w Dox treatment (Figure 5A). These data suggest the initiation of intrinsic signaling to limit the immune response, and the fluctuating pattern of Tgfb1 expression, in parallel with foxp3, characteristic of opposing pro- and anti-inflammatory processes in the chronic stage.
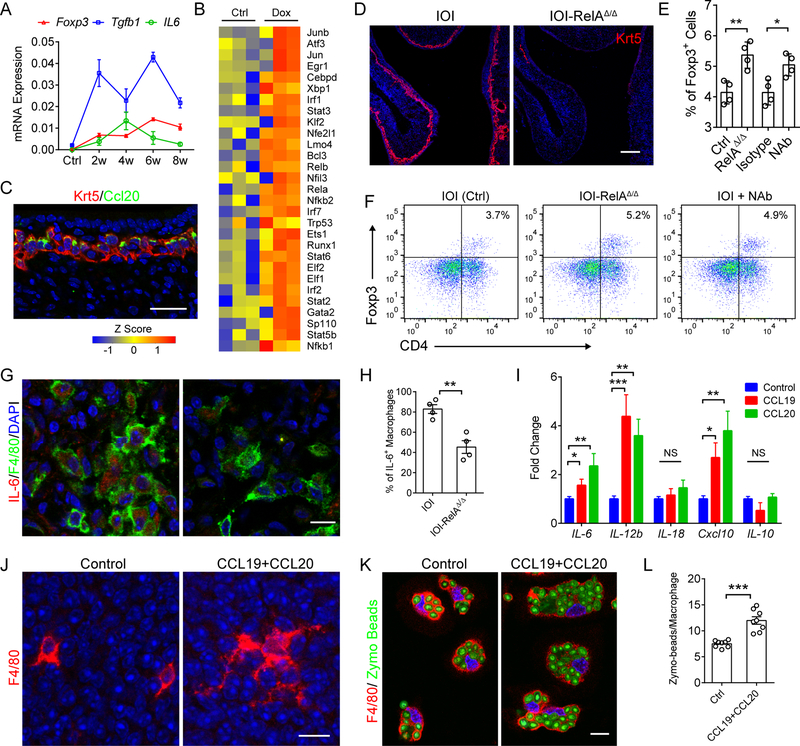
Figure 5. HBC signaling activates immune defense by macrophages.
(A) qPCR analysis of Tgfb1, Foxp3, and Il6 mRNA expression in whole olfactory mucosa after Dox induction. Data at indicated time periods was collected from 3 mice.
(B) Upregulated immune defense-related transcription factors in sorted HBCs after 6 w Dox treatment.
(C) Representative image of Krt5 and CCL20 staining in olfactory epithelium after 8 weeks of Dox treatment.
(D) Immunostaining of Krt5 in RelA ablated IOI mice after 6 w Dox treatment.
(E and F) Quantification of CD4+/Foxp3+ Treg (E) and representative flow cytometric plots (F). IOI mice (Ctrl, Isotype, and NAb) and IOI with HBC RelA deletion (RelAΔ/Δ) were administered Dox for 6 weeks. To block HBC chemokines, isotype IgG or anti-CCL19 and anti-CCL20 neutralizing antibody (NAb) were administered for 5 consecutive days. A total of 1 × 104 CD45+ immune cells were presented.
(G and H) Representative images of F4/80 and IL-6 co-staining (G), and the percentage of IL-6 expressing macrophages (H). n = 4 mice.
(I) qPCR analysis of indicated genes expression in macrophage cultures after 3 h of CCL19 or CCL20 treatments. n = 3 mice.
(J) Morphology of F4/80+ cells in whole mount olfactory mucosa with or without CCL19 and CCL20 stimulation. Tissue was resting in DMEM without serum for 2 hours before chemokines stimulation.
(K and L) Representative images of F4/80+ macrophages ingesting Zymosan beads (K), and quantification of Zymosan beads in each cell (L). n = 8 fields of 40 x images.
Data are represented as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001. P values were calculated by unpaired two tailed Student’s t-test (H and L) or by one-way ANOVA (E and I). Scale bars, 10 μm (G, J, and K), 25 μm (C), 100 μm (D). See also Figure S5.
The halted regeneration but enhanced production of inflammatory mediators in HBCs raised the possibility of a functional switch at the transcriptional level in the chronic inflammatory setting. Interestingly, systemic analysis of RNA-seq data revealed a subset of up-regulated transcription factors in HBCs that encode products involved in regulation of immune system process, including the NF-κB pathway components, STAT family members (Stat2, Stat3, Stat5b, and Stat6), JNK pathway components (Jun and Junb), as well as cytokines/chemokines related transcription factors (Enpp2, Irf1, Irf2, and Irf7) (Figure 5B). Indeed, prolonged inflammation induced persistent expression of chemokines CCL20, CXCL10 and CCL19 in Krt5+ HBCs of Dox-treated animals (Figures 5C, S5A and S5B). Notably, both CCL19 (MIP-3β) and CCL20 (MIP-3ɑ) are chemoattractants that promote macrophage/dendritic cell migration (Le Borgne et al., 2006; Rodda et al., 2018). These data suggested that an immune-related transcriptional network was switched on in HBCs. As dox-induced TNF transgene expression remained robust at later stages of inflammatory progression, the number of total CD45+ immune cells and their proliferation rates are comparable between HBC RelA-deleted and control (Figures S5C–S5E). Strikingly, constitutive deletion of RelA in 6w Dox treated Krt5cre-RelAflox/flox-IOI mice caused extensive loss of HBCs, suggesting the intact NF-κb pathway is critical for maintaining HBC survival in the chronic inflammatory environment (Figure 5D).
Given the essential role of HBCs in orchestrating infiltration and proliferation of macrophages and T cells in the acute phase of inflammation, we therefore asked whether HBC signaling functionally regulates immune defense in the chronic setting. The depletion of HBCs after RelA ablation allowed us to discern the impact of HBC signaling on the recruited chronic inflammatory cell infiltrate. Interestingly, RelA deletion and subsequent loss of HBCs significantly increased the ratio of Foxp3+ Tregs (Figures 5E, 5F, S5F, and S5G) compared to control, while proliferating Foxp3+ Tregs remained unchanged (Figure S5H). This suggests that HBC-derived signals regulate Treg development but not local proliferation. When inflammation was induced with intranasal LPS, a pronounced increase in the Foxp3+ Treg population could be readily detected in HBC RelA-deleted mice (Figures S5I and S5J). In contrast, the proportion of the F4/80+ cell population was reduced in RelA-deleted IOI mice when compared to control (Figure S5K and S5L).
Co-staining revealed that the F4/80+ macrophages are the major source of IL-6 (Murray and Wynn, 2011), and the percentage of F4/80+/IL-6+ cells was significantly reduced in HBC RelA-deleted mice (Figures 5G and 5H). Therefore, the HBCs derived chemokines could deliver essential signal to maintain M1 macrophage activation and IL-6 production, which consequently prevent Treg differentiation. As expected, CCL19 or CCL20 treatment clearly induced the expression of IL-6 and M1 cytokine IL-12b, and Cxcl10 in peritoneal macrophage cultures in vitro (Figure 5I). Furthermore, we utilized neutralizing antibodies to block the activity of CCL19 and CCL20 in Dox-treated IOI mice. Compared to mice treated with isotype control IgG, intranasal administration of anti-CCL19 and anti-CCL20 for 5 consecutive days significantly increased the population of CD4+/Foxp3+ Treg, and reduced CD11b+/F4/80+ macrophages (Figures 5E, 5F, S5K, and S5L).
We then assessed the morphology change in F4/80+ cells in response to CCL19 and CCL20 stimulation in transiently cultured mice olfactory mucosa. On day 1 post intranasal LPS stimulation, tissue retention but resting F4/80+ cells displayed spindle shape with short branches (Figure 5J). Remarkably, CCL19 and CCL20 treatment induced dramatic morphologic change with long processes involved in sensing of cues in microenvironment (McWhorter et al., 2013). We further evaluated the macrophage phagocytic activity using Zymosan bioparticles. Indeed, CCL19 and CCL20 pre-treatment significantly enhanced the ability of F4/80+ macrophages to ingest Zymosan bioparticles (Figures 5K and 5L). Together, these data reveal a previous unrecognized function of HBCs in maintaining immune defense by activating macrophages and preventing Treg generation in the process of chronic inflammation.
Olfactory inflammation in human CRS is associated with neural stem cell dysfunction
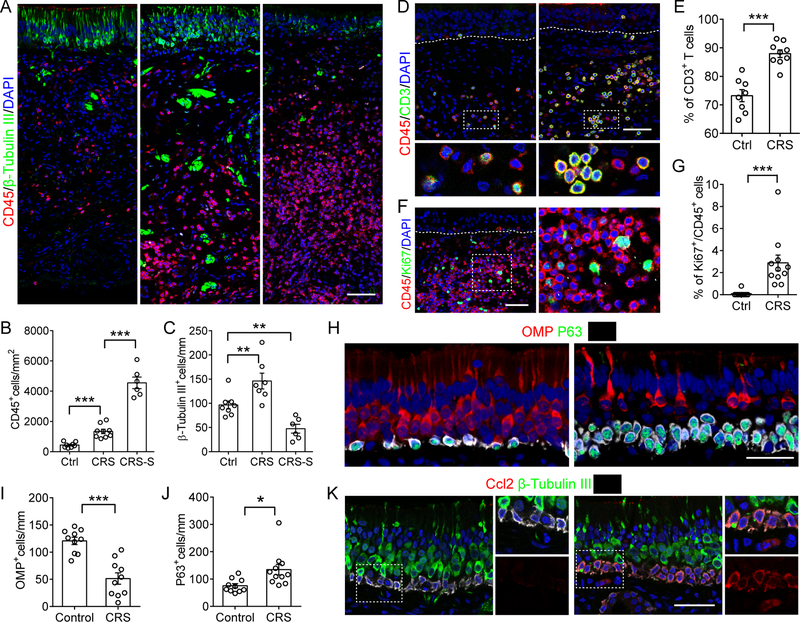
To explore whether the molecular signatures observed in our genetic model could be extended to human disease, we analyzed epithelial tissue removed from olfactory regions of CRS patients and normal controls during endonasal surgical procedures. Immunostaining revealed a robust increase of CD45+ inflammatory cells in the olfactory mucosa of CRS patients. Surprisingly, the number of β-Tubulin III+ immature neurons was slightly increased in CRS patients with moderate inflammation. In a subset of 24% CRS patients with severe inflammation characterized by a dense CD45+ cell infiltrate, a dramatically reduced number of β-Tubulin III+ cells were present in the olfactory epithelium (Figures 6A–C).
Figure 6. Olfactory inflammation in CRS is associated with neural stem cell dysfunction.
(A–C) Quantification of CD45+ cells (B) or β-Tubulin III+ immature OSNs (C) in human olfactory biopsy (A). CRS-S, CRS subjects with severe inflammatory cell infiltration.
(D and E) Immunostaining of CD3 and CD45 (D), and percentage of CD3+ T lymphocytes in total immune cells (E).
(F and G) Immunostaining analysis of immune cell local proliferation. Graph shows the percentage of Ki67+ subpopulation in total CD45+ cells.
(H–J) Triple-staining of Krt5, P63 and OSNs marker OMP(H). OMP+ OSNs (I) and P63+ HBCs (J) in control and CRS were counted.
(K) Representative images of CCL2 expression in olfactory biopsy.
Dots in graph represent independent patients. Data are represented as mean ± SEM. *P < 0.05; **P < 0.01, ***P < 0.001. P values were calculated by unpaired two tailed Student’s t-test (E, G, I, and J) or by one-way ANOVA (B and C). Scale bars, 25 μm. See also Figure S6.
Co-staining of CD45 and CD3 revealed that ~72% of resident immune cells are CD3+ lymphocytes in the normal human olfactory mucosa. The CD3+ cell population was significantly increased to ~90% of inflammatory cells in CRS biopsies, suggesting that T lymphocytes are the predominant immune cell population that is involved in CRS-associated olfactory loss (Figures 6D and 6E). We further determined that 65% of those CD3+ T cells in CRS tissue are CD4+ helper T cells (Figures S6A and S6B). Compared to normal controls, a larger proportion of CD45+ immune cells is actively proliferating in CRS patients (Figures 6F and 6G), consistent with our observations in the genetic model that inflammatory cells undergo local proliferative expansion contributing to the progression of inflammation.
We next studied the response of human olfactory neural stem cells in the microenvironment of chronic inflammation. In agreement with evidence from the IOI model, a significant increase of Krt5+P63+ stem cell number in CRS was found in comparison to normal controls, and this was accompanied by a decline of OMP+ olfactory sensory neurons (Figures 6H–J). Greater than 25% of biopsies from CRS patients displayed stacked Krt5+ cell layers, with persistent expression of P63 indicating restriction to an undifferentiated state. The number of proliferating basal cells in the neuroepithelium was sharply decreased in CRS subjects (Figures S6C and S6D). A higher expression of RelA was observed in Krt5+ basal cells with nuclear translocation in olfactory tissue samples from CRS patients (Figures S6E). Relative to control neuroepithelium, we observed remarkable activation of Ccl2 expression in Krt5+ cells of CRS subjects (Figure 6K), a chemokine that recruits monocytes and T lymphocytes (Carr et al., 1994). Activated expression of CCL20 in HBCs can also be detected in CRS olfactory tissue (Figure S6F). Thus, features of olfactory stem cell switching identified in the mouse models, enhancing immune defense activity while down-regulating regenerative function, are also displayed in human olfactory tissue, suggesting the mechanism of CRS-associated olfactory deficits described here is conserved in human disease.
DISCUSSION
The molecular pathology of olfactory loss in CRS is poorly understood. While airflow obstruction contributes to sensory deficits in some cases, OSN death and olfactory stem cell dysfunction may play major roles (Lane et al., 2010). In the current study, our findings elucidate the immune capacity of long-lived olfactory stem cells to regulate inflammatory cell recruitment and local proliferation by releasing cytokines and chemokines. Simultaneously, activated stem cells in a chronic inflammatory milieu shut down regenerative function by enhancing “stemness”-related transcription factors. Our findings highlight the central role of NF-κB in this stem cell functional switch, a process which directly blocks neurogenesis, leading to olfactory loss in CRS.
Recent studies of immune cell dynamic and function have changed our understanding of inflammation. In peripheral tissues, resident macrophages repopulate locally, independent of circulating monocytes (Hashimoto et al., 2013). The contribution of macrophage local proliferation to inflammatory progression has been established (Jenkins et al., 2011). Emerging evidence suggests stem cell-like properties of antigen-experienced memory T cells (Gattinoni et al., 2017). In skin, tissue-resident memory T cells undergoing local proliferation in response to viral infection has recently been recognized (Park et al., 2018). These observations highlight the importance of immune cell proliferation to inflammatory diseases and indicate potential strategies for therapeutic intervention. However, the type and origin of local proliferative signals are largely unknown. Our observation that the loss of RelA in HBCs reduces the infiltration and proliferation of immune cells reveals the contribution of guidance and proliferation signaling by long-lived basal stem cells.
Chemokines are chemotactic cytokines that control the migratory patterns and positioning of all immune cells (Griffith et al., 2014). In response to stimuli, activated NF-κB targets a broad-spectrum of cytokine and chemokine genes(Richmond, 2002; Taniguchi and Karin, 2018). We validated that in mouse olfactory mucosa, HBCs are the primary source of CCL19 and CCL20, both known chemokines that stimulate dendritic cell (Le Borgne et al., 2006; Schumann et al., 2010) and T cell trafficking (Luther et al., 2002) and enhance T cell proliferation in a co-culture system (Marsland et al., 2005). The HBC-released CXCL10 is also functionally related to T cell migration and proliferation in lymph node cell cultures (Dufour et al., 2002; Griffith et al., 2014). Our observations that the monocyte and T lymphocyte chemoattractant CCL2 (Carr et al., 1994; Luther and Cyster, 2001) was predominantly produced by Krt5+ basal cells in CRS subjects suggests that human and mouse HBCs share similar function in regulating inflammatory progression in olfactory mucosa. Accurate immune cell trafficking to peripheral tissues is highly regulated (Islam and Luster, 2012). Chemokines that govern immune cell trafficking constitute the largest family of cytokines (~50 chemokines in mouse and human) (Griffith et al., 2014). Analysis of the interaction between epithelium-derived chemokines and immune cells is relevant to multiple inflammatory diseases.
Mucosal epithelial surfaces such as the gut were once considered to be mainly physical barriers, until their essential roles in active immune defense and maintenance of immune homeostasis were recognized (Artis, 2008). Intestinal epithelial cells regulate immune cell function through release of NF-κB dependent cytokines which directly influence the expression of pro-inflammatory cytokines by dendritic cells/macrophages (Taniguchi and Karin, 2018; Zaph et al., 2007). These properties were in part demonstrated through genetic manipulation of the NF-κB signaling globally in all intestinal epithelial cells. This approach, however, was not sufficient to parse out the contribution specifically of epithelial stem cells in regulating the immune response. Elucidation of the interplay between stem cells and local immune cells, as well as their related signaling molecules, allows greater insight into the pathologic mechanisms underlying impaired regeneration in chronic inflammatory diseases. Emerging evidence has shown that signals derived from local immune cells regulate skin stem cell regeneration (Ali et al., 2017; Keyes et al., 2016). More recently, a role of Lgr5+ intestine stem cells as non-conventional antigen presenting cells has been shown in a co-culture model with CD4+ T helper cells (Biton et al., 2018). In the present investigation, our RelA loss-of-function study specifically dissects out the critical role of Krt5+ olfactory stem cells in recruitment of immune cell infiltration and proliferation. Chronically-activated Krt5+ stem cells signal macrophages continuously to retain them in the tissue and maintain their activated state, while preventing Foxp3+ Treg development. These observations provide direct evidence that basal stem cells functionally regulate immune defense in the setting of chronic olfactory inflammation.
The stem cell potency of HBCs is maintained by a set of core transcription factors (Schwob et al., 2017) consisting of Trp63, Sox2, Pax6, and Hes1. Conditional knockout of Trp63 in quiescent HBCs activates and initiates the differentiation process (Fletcher et al., 2017; Fletcher et al., 2011; Schnittke et al., 2015). The expression of Sox2 and Pax6 in HBCs is related to maintenance of stemness and is essential for neurogenesis upon lesioning (Gadye et al., 2017; Guo et al., 2010; Lin et al., 2017). A role of Notch signaling and its target Hes1 in maintaining HBC dormancy has also been reported recently (Herrick et al., 2017). Notably, the expression of these transcription factors in HBCs of our mouse model or CRS subjects is either elevated or maintained. The abundance of up-regulated transcription factors in our inflammatory model closely matches the list of genes selectively expressed by quiescent HBCs (Fletcher et al., 2017), suggesting roles in the negative regulation of HBC differentiation. Given that newly differentiated OSNs do not survive in the inflammatory environment, HBC maintenance in a stem cell state offers a protective strategy necessary for the efficient repair upon inflammatory resolution. Our observations that activated NF-κB maintains “stemness” in response to chronic inflammation through targeting HBC core transcription factors is reminiscent of previous findings that NF-κB regulates tumor-initiation, a process in which NF-κB signaling is necessary for stem cell gene expression and expansion in intestine (Myant et al., 2013; Schwitalla et al., 2013) or breast cancer (Zhang et al., 2013).
In summary, our results provide significant insights into the role of basal stem cells as direct participants in the progression of chronic inflammation and identify a concomitant functional switch away from neuroregeneration. This has important implications for olfactory dysfunction in CRS. The immune activity of basal cells in communicating with infiltrating inflammatory cells may play a more generalized role in mucosal immunity at epithelial barrier surfaces in health and disease. Insights from our mouse model and human tissue have potential to inform novel treatment strategies for CRS-related olfactory deficits.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Andrew P. Lane (alane3@jhmi.edu).
MATERIALS AVAILABILITY STATEMENT
This study did not generate new unique reagents.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animal Models
The Cyp2g1-rtTA stain, which carries the reverse tetracycline transactivator (rtTA) gene under the control of olfactory sustentacular cell-specific Cyp2g1 gene, was created as previously described (Lane et al., 2010). The TRE-TNF transgene strain (Vuillemenot et al., 2004) was crossed with Cyp2g1-rtTA line to generate the IOI (Induced Olfactory Inflammation) mouse, based on the Tet-on genetic system. 6–8 weeks old mice were treated with 0.2g/kg doxycycline food to induce TNF expression specifically by olfactory sustentacular cells. The RelAflox/flox mouse (Luedde et al., 2008) possesses loxp sites flanking exons 2–4 of RelA gene was kindly provided by Dr. Mollie K. Meffert; Krt17-EGFP strain (Bianchi et al., 2005) which carries EGFP reporter directed by HBCs specific Krt17 gene promoter (Figure S2C) was kindly provided by Dr. Pierre Coulombe. Krt5-Cre transgenic line in which Cre recombinase is driven by the HBC specific Krt5 promoter (Leung et al., 2007) was breed with Rosa26-stopflox/flox-tdTomato strain to label HBCs and their progenies. Gender matched animals from same litter were assigned randomly to different experimental groups based on genotypes. The number of animals shown in each figure is indicated in the legends as n = x mice per group. Mice were bred and maintained under specific-pathogen-free conditions in compliance with established ethical guidelines. Animal experimental procedures were approved by the Animal Care and Use Committee at the Johns Hopkins University.
Intranasal LPS Administration
For LPS induced olfactory inflammatory model (Hasegawa-Ishii et al., 2017), 6–8 weeks old control mice and Krt5cre-RelAflox/flox mice were anesthetized by isoflurane,followed by intranasal instillation of 10μl LPS (1mg/ml, Sigma). After each instillation, the animal was restrained on its side to prevent liquid from being aspirated into the lower airway. Macrophage morphologic analysis was performed 1 day after the last LPS treatment. Briefly, mice were perfused with PBS, and then intact septal olfactory mucosa was dissected out and washed in PBS. After incubation in DMEM at 37 °C for 2 hours to allow tissue-retained macrophage to become rested, the tissue explant was treated with chemokines for 1 hour, followed by a 30 minute 4% PFA fixation for subsequent immunostaining.
Neutralizing Antibodies Treatment
For neutralizing experiments (Moon et al., 2018), Dox treated IOI mice were administrated with neutralizing antibodies (NAb) or isotype IgG from day 37 for 5 consecutive days. After anesthetizing by isophorone, animals were intranasally instilled with 500 ng each of anti-CCL19 (R&D Cat# AF880) and anti-CCL20 (R&D Cat# AF760) in 5 μl sterile PBS per nostril. Control mice were treated with equal amount of isotype IgG.
Human Olfactory Tissue
The research protocol involving human specimens was approved by the Johns Hopkins institutional review board, and all subjects provided signed informed consent. Human olfactory epithelial samples were collected from CRS patients and from control subjects undergoing endonasal surgical approaches for non-CRS disease processes. None of the control subjects reported decreased olfactory function. All CRS patients reported fluctuating olfactory loss with disease exacerbations. The specimen was immediately fixed in 4% PFA for 1 hour on ice with gentle rotation, any bone present was dissected out and followed by the similar tissue process described as above. Approximately 130 human samples were cryosectioned, 17 control (8 females and 9 males ranged from 34 to 68 years old) and 32 CRS samples (14 females and 18 males ranged from 33 to 76 years old) with intact morphology of olfactory mucosa were used for detailed immunohistochemistry analysis.
Primary cell culture
Mouse olfactory HBCs were isolated from 3-month-old C57BL/6 mice on day 3 post methimazole lesion as previously described (Chen et al., 2017). The lesion procedure enriches and activates the olfactory stem cells. Briefly, the olfactory neuroepithelium was dissected out and dissociated in 2mg/mL Collagenase IA (Gibco) and 20 U/mL DNase I at 37°C. Dissociation was stopped in DMEM containing 1% BSA, 1% FBS and 0.1 mM EDTA. After washing and resuspending in growth media, cells were filtered through 100 µm, 70 µm cell strainer sequentially then seeded in Laminin-coated plates or chamber slides at 2–3 × 104/cm2. The growth media contains NeuroCult medium plus 10% Proliferation Supplement (Stem Cell Technology), 20 ng/mL EGF (Peprotech), and 10 ng/mL bFGF (Peprotech). Before TNF stimulation experiment, cell media was replaced with NeuroCult containing 2% Proliferation Supplement for overnight starvation. TNF stimulation was performed on day 4 when HBC colonies reached 80% confluence.
Macrophage culture and phagocytosis analysis
Mouse macrophages were collected by peritoneal lavage then seeded in DMEM/F12 media supplemented with 10% FBS at 1×106/cm2. Nonadherent cells were removed after 2 hours by replacement with fresh media. Phagocytosis analysis (Georgouli et al., 2019) was performed on day 6 after overnight serum starvation. After CCL19 (20ng/ml) and CCL20 (20ng/ml) pretreatment for 30 minutes, macrophages were incubated with Zymosan bioparticles Alexa Fluor 488 conjugated (Molecular Probes) for 30 minutes. Extracellular particles were removed by PBS wash and then fixed in 4% PFA for 10 minutes. F4/80+ cells and ingested zymosan beads were quantified by confocal images randomly obtained from 8 fields.
METHOD DETAILS
Immunohistochemistry
The tissue collection and staining process was performed as previously described (Chen et al., 2017). Briefly, animals were anesthetized and transcardially perfused with PBS and 4% paraformaldehyde (PFA). The whole nasal cavity with attached cribriform plate was dissected out and post-fixed in 4% PFA on ice for 1 hour with gentle rotation. After being washed in PBS, tissues were equilibrated sequentially in 15% and 30% sucrose, the tissue was then embedded in Optimum Cutting Temperature (OCT, Tissue-Tek) for sectioning. 12-μm frozen sections were processed from cribriform plate side. For immunostaining, sections were washed in PBS and then blocked in 10% normal serum containing 0.1% Triton X-100, followed by incubation with primary antibodies at 4 °C overnight. After washing in PBS, the tissue sections were incubated with Alexa Fluor conjugated secondary antibodies along with DAPI for nuclear counterstaining.
The following primary antibodies were used: Rabbit anti-Krt5 (1:800, PRB-160P; Covance), Rabbit anti-Ki67 (1:500, Ab16667; Abcam), Rat anti-BrdU (1:400, Ab6326; Abcam), Mouse anti-Krt14 (1:800, MA5–11599; Thermo Fisher), Mouse anti-β-Tubulin III (1:200, MAB1637; Millipore), Rabbit anti-∆NP63 (1:1,000, 619001; BioLegend), Mouse anti-P63 (1:200, sc-8431; Santa Cruz), Rabbit anti-RelA (1:200, Sc-372; Santa Cruz), Rat anti-CD45 (1:200, 14–0451-81; Ebioscience), Rat anti-F4/80(1:500, MCA497GA; Bio-Rad), Rat anti-Ly6G (1:500, 127601; Biolegend), Rat anti-CD3 (1:200, 14–0032-81; Ebioscience), Goat anti-OMP (1:1000, 544–10001; Wako), Goat anti Sox2 (1:200, sc-17320; Santa Cruz), Goat anti-CCL19 (1:50, AF880; R&D), Goat anti-CXCL10 (1:100, AF-466-NA; R&D), Mouse anti Human CCL2 (1:100, MAB2791; R&D), Rabbit anti Human-CD45 (1:400, Ab40763; Abcam), Mouse anti-CD45 (1:500, 304002; Biolegend), Mouse anti-human CD3 (1:300, 300413; Biolegend), Alexa Fluor® 488 anti-human CD3 (1:200, 300454; Biolegend), Mouse anti-Human CD4 (1: 200, 555344, BD Pharmingen), and Alexa Fluor® 488 anti-β-Tubulin III (1:1000, 801203; Biolegend).
BrdU Incorporation
To label dividing cells, animals were injected i.p. with BrdU (Sigma) at 50 μg/g of body weight two times at 12 hours intervals. Olfactory tissue was collected and processed as described above. After washing in PBS, frozen sections were treated with 1 M HCl 10 min on ice, 2 M HCl for 10 at room temperature, followed by 15 min at 37 °C. After neutralizing in 0.1 M borate buffer for 10 min tissue sections were proceeded with the immunostaining procedure. BrdU incorporation was detected with anti-BrdU antibody.
Confocal Imaging and Quantification
Immunostaining images were obtained using a Zeiss LSM 780 confocal microscope. For quantification, consecutive images that cover approximately one half to two-thirds of septum OE were collected using 40x objectives under the tile scan mode. Positive cells in olfactory mucosa were quantified per mm of surface epithelium. For each sample, images were acquired at least from three different levels (Chen et al., 2017). Cells were counted from three to five independent animals.
ELISA
The expression of TNF in the olfactory mucosa was determined by ELISA as previously described (Lane et al., 2010). Nasal lavage fluid was collected by perfusing 500 µL PBS over the bisected nasal cavity. After spinning to remove blood cells, 100 µL supernatant was used for ELISA (eBioscience).
Flow Cytometry
Olfactory neuroepithelium tissue was dissected and dissociated as described below in the FACS methods. After incubating with anti-CD16/CD32 (BD Bioscience, Cat #553142) for 10 min to block Fc-receptor binding, cells were stained with fluorophore-conjugated CD45 (BioLegend 103139), F4/80 (BD 565410), CD3 (BD 560527), and Ly6G (BD 561236). Inflammatory cells were gated on FCS/SSC area and with live PI−, CD45+. Neurophils (CD45+, Ly6G+; CD3−, F4/80−), T lymphocytes (CD45+, CD3+; Ly6G−, F4/80−), and macrophages (CD45+, F4/80+; Ly6G−, CD3−). For intracellular Foxp3 detection, cells were prepared using True-Nuclear™ Transcription Factor Buffer Set (Biolegend, Cat#424401). For multicolor staining, gate setting was established with fluorescence minus one controls. Phenotypic analysis of inflammatory cell subpopulations was performed by flow cytometry using BD LSR II system. Compensation was performed with single antibody staining controls. Data were analyzed with the FlowJo software. Dead cell and doublet cell exclusion criteria were included for all flow cytometry analysis.
Fluorescence-Activated Cell Sorting (FACS)
Animals were anesthetized and transcardially perfused with ice cold PBS. Olfactory mucosa tissue was dissected out under stereo microscope (Figure S2B). Tissue was minced and dissociated in 2mg/mL Collagenase IA (Gibco) and 20 U/mL DNase I at 37°C. Dissociation was stopped in cold PBS containing 5% FBS, 5 U/mL DNase I, and 0.1mM EDTA. After washing in PBS containing 1% FBS, 1% BSA, and 0.1mM EDTA, the cell suspension was passed through 100 µm, 70 µm cell strainer sequentially for flow cytometry or cell sorting. Control HBCs were isolated from Krt17-EGFP reporter mice. Inflammation-stimulated HBCs were isolated from Krt17-EGFP-IOI mice after 6 weeks of Dox treatment. HBCs from Dox treated and 3 days post-6 weeks Dox treatment (Stop) mice were also isolated as recovery control. PI was added to cell suspension to eliminate dead cells. EGFP− cells were collected as negative control for FACS validation (Supplementary Fig. 2d). Cell sorting was performed at Johns Hopkins School of Public Health Flow Cytometry Core Facility.
RNA and Library Preparation
For RNA extraction, reporter-positive cells were sorted directly into Trizol LS (Invitrogen). Reporter-negative cells were collected separately as negative control. Completely lysed samples were treated with chloroform to separate phases. The aqueous phase containing RNA was transferred and purified using a RNeasy Micro Kit (Qiagen). On-Column DNase I digestion was conducted to remove genomic DNA contamination. The quality of RNA from sorted cells was verified by Agilent 2100 Bioanalyzer. Three individual RNA samples for each group with high quality (RIN > 7) were enrolled for library preparation. mRNA amplification was performed using 5ng total RNA with a Clontech Smarter RNA seq v4 kit. RNA-seq libraries were prepared using Illumina Nextera library preparation kit.
RNA sequencing (RNA-seq)
After validating the quality of the libraries using Agilent 2100 Bioanalyzer, high-throughput sequencing was performed using a HiSeq 2500 instrument (Illumina) at the Johns Hopkins Medicine Genetic Resource Core facility. Raw reads for each library were trimmed for quality and aligned to the mouse (mm10) genome using TopHat v2.1.1. The mapped reads were converted to FPKM (fragments per kilobase of exon per million fragments mapped) by running Cuffdiff2 to determine the expression levels. Differentially expressed genes were determined with a t-test from the expression levels of each gene between different groups, and genes with both p-value and q-value less than 0.05 were considered significant and reported.
To identify the signaling pathways involved in HBC functional changes, upregulated genes in inflamed HBCs with log2(fold change) > 1 were selected for gene pathway enrichment analysis using DAVID Bioinformatics Resources (Huang da et al., 2009). Heat maps of genes with significantly changed expression were generated using R program. Differentially expressed transcription factors were filtered based on the list of mouse transcription factors database. Transcription factors functional classification was analyzed by Gene Ontology Consortium (Panther Classification System) using the biological process terms (Ashburner et al., 2000). The gene-filtered expression data are available in Table S4 and S5.
RNA isolation, cDNA synthesis and qPCR
Total RNA was isolated from cultured olfactory stem cells or olfactory neuroepithelium tissue using a RNeasy Mini Kit (Qiagen). Equal amounts of RNA were transcribed into cDNA by an Omniscript Reverse Transcription Kit (Qiagen). On-Column DNase I digestion was conducted to remove genomic DNA contamination. Ten nanograms of cDNA was added to a 20-μL PCR reaction using SYBR Green PCR Master Mix or TaqMan Fast Universal PCR Master Mix (Applied Biosystems) on StepOne Plus System (Applied Biosystems). For SYBR Green PCR, post-amplification melting curve analysis was performed to monitor unspecific products. Fold change in mRNA expression was calculated using the comparative cycle method (2−ΔΔCt). SYBR Green PCR primer sequences or TaqMan probes are listed in Table S1 and S2.
Chromatin immunoprecipitation (ChIP) Assay
Olfactory HBC cultures were treated with or without 20ng/mL TNF for 30min. ChIP was performed using the MAGnify ChiP Kit (Life Technologies) according to the manufacturer’s instructions. In brief, 1 × 106 cells were crosslinked for 10 min by adding 37% formaldehyde (Sigma) into culture media to a final concentration of 1%. Reactions were stopped by adding glycine and washed in cold PBS. Cells were harvested using a cell scraper and lysed in buffer containing protease inhibitors. Cell lysates were sonicated using Bioruptor UCD-200 to generate ~200–500 bp chromatin fragments. For each ChIP, diluted chromatin DNA from 2 × 105 cells was incubated with 2 μg rabbit anti-RelA (Sc-372; Santa Cruz) or anti-IgG coupled Dynabeads at 4°C overnight. ChIPed DNA was washed and reversed crosslinked with input control. Real-time qPCR was performed using SYBR Green PCR Master Mix (Applied Biosystems) to evaluate the presence of specific promoter regions. Primer targets DNA region lacking RelA binding site served as negative control. Ct values were normalized to the input control and the amount of bound DNA regions was present as % of input. Chip-PCR primer sequences are listed in Table S3.
QUANTIFICATION AND STATISTICAL ANALYSES
The animal sample size was determined by preliminary experiments and in consideration of ethical reduction of the number of animals used. All images of immunostaining are representative of at least three separate experiments. Other experiments were independently replicated with similar results at least twice except the RNA-seq experiment. The investigators were not formally blinded to allocation during experiments and outcome assessment. Data are expressed as mean ± SEM. as indicated. Data analyses were carried out using GraphPad Prism. For experiments with two groups, P values were calculated using the unpaired two-tailed Student’s t-test. One-way ANOVA was used to compare three or more groups. Differences were considered significant when P < 0.05.
DATA AND SOFTWARE AVAILABILITY
All software tools can be found online (see Key Resources Table). The accession number for the raw data files reported in this paper is GSE135042.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies for Immunohistochemistry | ||
| Rabbit anti-Krt5 | Covance | Cat#PRB-160P; RRID: AB_291581 |
| Rabbit anti-Ki67 | Abcam | Cat#Ab16667; RRID: AB_302459 |
| Rat anti-BrdU | Abcam | Cat#Ab6326; RRID: AB_305426 |
| Mouse anti-Krt14 | Thermo Fisher | Cat#MA5–11599; RRID: AB_10982092 |
| Mouse anti-β-Tubulin III | Millipore | Cat#MAB1637; RRID: AB_1707889 |
| Rabbit anti-∆NP63 | BioLegend | Cat#619001; RRID: AB_2256361 |
| Mouse anti-P63 | Santa Cruz | Cat#sc-8431; RRID: AB_628091 |
| Rabbit anti-RelA | Santa Cruz | Cat#sc-372; RRID: AB_632037 |
| Rat anti-CD45 | Ebioscience | Cat#14–0451-81; RRID: AB_467250 |
| Rat anti-F4/80 | Bio-Rad | Cat#MCA497GA; RRID: AB_323806 |
| Rat anti-Ly6G | Biolegend | Cat#127601; RRID: AB_1089179 |
| Rat anti-CD3 | Ebioscience | Cat#14–0032-81; RRID: AB_467052 |
| Goat anti-OMP | Wako | Cat#544–10001; RRID: AB_2315007 |
| Goat anti Sox2 | Santa Cruz | Cat#sc-17320; RRID: AB_2286684 |
| Goat anti-CCL19 | R&D Systems | Cat#AF880; RRID: AB_2071545 |
| Rabbit anti-TNF-α | Cell Signaling | Cat#3707; RRID: AB_10691460 |
| Goat anti-CXCL10 | R&D Systems | Cat#AF-466-NA; RRID: AB_2292487 |
| Rat anti-Foxp3 | eBioscience | Cat#14–5773-82; RRID: AB_467576 |
| Chicken anti Krt18 | Abcam | Cat# ab14047; RRID: AB_2133173 |
| Rabbit anti PGP9.5 | Ultraclone | Cat# RA95101; RRID: AB_2313685 |
| Goat anti-CCL20 | R&D Systems | Cat# AF760; RRID: AB_355580 |
| Rabbit anti IL-6 | Novus | Cat# NB600–1131; RRID: AB10001997 |
| Mouse anti Human CCL2 | R&D Systems | Cat#MAB2791; RRID: AB_2071560 |
| Rabbit anti Human-CD45 | Abcam | Cat#Ab40763; RRID: AB_726545 |
| Mouse anti-CD45 | Biolegend | Cat#304002; RRID: AB_314390 |
| Mouse anti-human CD3 | Biolegend | Cat#300413; RRID: AB_314067 |
| Alexa Fluor® 488 anti-human CD3 | Biolegend | Cat#300454; RRID: AB_2564149 |
| Mouse anti-Human CD4 | BD Pharmingen | Cat#555344; RRID: AB_395749 |
| Alexa Fluor® 488 anti-β-Tubulin III | Biolegend | Cat#801203; RRID: AB_2564757 |
| Antibodies for Flow Cytometry | ||
| FITC anti-mouse CD45 | Biolegend | Cat#103107; RRID: AB_312972 |
| PE anti-Ms F4/80 | Biolegend | Cat#123109; RRID: AB_893498 |
| Alexa F 647 anti FOXP3 | Biolegend | Cat#320013; RRID: AB_439749 |
| PE/Cy7 anti-Ms CD4 | Biolegend | Cat#100421; RRID: AB_312706 |
| PerCP/Cy5.5 anti-Ms/Hu CD11b | Biolegend | Cat#101227; RRID: AB_893233 |
| PerCP-Cy™5.5 Rat Anti-Mouse CD3 | BD Pharmingen | Cat#560527; RRID: AB_1727463 |
| Alexa Fluor® 700 Rat Anti-Mouse Ly-6G | BD Pharmingen | Cat#561236; RRID: AB_10611860 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Dox food | Bio Serv | Cat#S3888 |
| BrdU | Sigma | Cat#B9285 |
| Lipopolysaccharides (LPS) | Sigma | Cat#L2880 |
| Zymosan A Bioparticles, Alexa Fluor 488 | Molecular Probes | Cat#Z23373 |
| True-Nuclea Transcription Factor Buffer | Biolegend | Cat#424401 |
| Alexa Fluor 488 Tyramide SuperBoost Kit | Life Technologies | Cat#B40912 |
| TNF-α | Peprotech | Cat#315–01A |
| Murine EGF | Peprotech | Cat#315–09 |
| Murine FGF-basic | Peprotech | Cat#450–33 |
| Murine CCL19 | Peprotech | Cat#250–27B |
| Murine CCL20 | Peprotech | Cat#250–27 |
| Critical Commercial Assays | ||
| SYBR Green PCR Master Mix | Applied Biosystems | Cat#4385612 |
| TaqMan Fast Universal PCR Master Mix | Applied Biosystems | Cat#4352042 |
| Mouse TNF alpha ELISA Kit | eBioscience | Cat#88–7324 |
| MAGnify ChiP Kit | Life Technologies | Cat#49–2024 |
| Deposited Data | ||
| Raw data files for RNA sequencing | NCBI | GEO: GSE135042 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Cyp2g1-rtTA | N/A | (Lane et al., 2010) |
| Mouse: TRE-TNF-α | N/A | (Vuillemenot et al., 2004) |
| Mouse: Krt17-EGFP | Jackson lab | Stock Number: 023965 |
| Mouse: Krt5-Cre | N/A | (Leung et al., 2007) |
| Mouse: Rosa26-stopflox/flox-tdTomato | Jackson lab | Stock Number: 007914 |
| Mouse: RelAflox/flox | N/A | (Luedde et al., 2008) |
| Oligonucleotides | ||
| qRT-PCR primers | This paper | Table S1–3 |
| Software and Algorithms | ||
| Prism 6 | Graphpad | https://www.graphpad.com |
| R | r-project.org | R 3.4.4 |
| Zen lite | Zeiss | ZEN 2012 (blue edition) |
| FlowJo v10 | FlowJo, LLC | flowjo.com |
| DAVID Bioinformatics Resources | (Huang da et al., 2009) | david.ncifcrf.gov |
Supplementary Material
Table S4. RNA-seq data of differentially expressed genes, related to Figure 2
Data including significantly upregulated NF-κB components or targets, differentially expressed cytokines or chemokines, significantly upregulated receptor genes for cytokines or chemokines. HBCs were sorted from 6 weeks Dox treated or No-Dox control mice. Reads are shown as average FPKM of 3 individual samples in each group and filtered for genes which Log2 (Fold Change) > 1, or < - 1.
Table S5. RNA-seq data of differentially expressed transcription factors, related to Figure 4 and Figure 5
Data including transcription factors related to regulation of “stemness” and neurogenesis, and transcription factors involved in immune function. HBCs were sorted from 6 weeks Dox treated or No-Dox control mice. Reads are shown as average FPKM of 3 samples in each group and filtered for genes which Log2 (Fold Change) > 1, or < - 1.
Highlights.
NF-kB-mediated signals in HBCs direct immune cell infiltration/proliferation
Chronic inflammation blocks HBC regeneration and enhances stemness
HBC signaling activates macrophages, inhibiting local Treg development
Olfactory inflammation in humans is associated with neural stem cell dysfunction
ACKNOWLEDGEMENTS
We thank Liliana Florea and Corina Antonescu (Johns Hopkins School of Medicine) for assistance in RNA-seq data analysis, Hao Zhang (Bloomberg School of Public Health, JHU) for FACS, and the Center for Sensory Biology imaging core facility. This work was funded by NIH Grant R01 DC009026 (A.P.L.), and NIH grant 5R56DC008295 (R.R.R). M.C. is supported by the National Natural Science Foundation of China (81200684).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures and 5 tables and can be found with this article online at https://doi.org/10.1016/j.stem.2019.08.011.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Adam RC, Yang H, Rockowitz S, Larsen SB, Nikolova M, Oristian DS, Polak L, Kadaja M, Asare A, Zheng D, et al. (2015). Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature 521, 366–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonina IS, Zhong Z, Karin M, and Beyaert R (2017). Limiting inflammation-the negative regulation of NF-kappaB and the NLRP3 inflammasome. Nat Immunol 18, 861–869. [DOI] [PubMed] [Google Scholar]
- Ali N, Zirak B, Rodriguez RS, Pauli ML, Truong HA, Lai K, Ahn R, Corbin K, Lowe MM, Scharschmidt TC, et al. (2017). Regulatory T Cells in Skin Facilitate Epithelial Stem Cell Differentiation. Cell 169, 1119–1129 e1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artis D (2008). Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat Rev Immunol 8, 411–420. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, and Kuchroo VK (2006). Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 441, 235–238. [DOI] [PubMed] [Google Scholar]
- Bianchi N, Depianto D, McGowan K, Gu C, and Coulombe PA (2005). Exploiting the keratin 17 gene promoter to visualize live cells in epithelial appendages of mice. Mol Cell Biol 25, 7249–7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biton M, Haber AL, Rogel N, Burgin G, Beyaz S, Schnell A, Ashenberg O, Su CW, Smillie C, Shekhar K, et al. (2018). T Helper Cell Cytokines Modulate Intestinal Stem Cell Renewal and Differentiation. Cell 175, 1307–1320 e1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck L, and Axel R (1991). A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell 65, 175–187. [DOI] [PubMed] [Google Scholar]
- Carr MW, Roth SJ, Luther E, Rose SS, and Springer TA (1994). Monocyte chemoattractant protein 1 acts as a T-lymphocyte chemoattractant. Proc Natl Acad Sci U S A 91, 3652–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Reed RR, and Lane AP (2017). Acute inflammation regulates neuroregeneration through the NF-kappaB pathway in olfactory epithelium. Proc Natl Acad Sci U S A 114, 8089–8094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Tian S, Yang X, Lane AP, Reed RR, and Liu H (2014). Wnt-responsive Lgr5(+) globose basal cells function as multipotent olfactory epithelium progenitor cells. J Neurosci 34, 8268–8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour JH, Dziejman M, Liu MT, Leung JH, Lane TE, and Luster AD (2002). IFN-gamma-inducible protein 10 (IP-10; CXCL10)-deficient mice reveal a role for IP-10 in effector T cell generation and trafficking. J Immunol 168, 3195–3204. [DOI] [PubMed] [Google Scholar]
- Fletcher RB, Das D, Gadye L, Street KN, Baudhuin A, Wagner A, Cole MB, Flores Q, Choi YG, Yosef N, et al. (2017). Deconstructing Olfactory Stem Cell Trajectories at Single-Cell Resolution. Cell Stem Cell 20, 817–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher RB, Prasol MS, Estrada J, Baudhuin A, Vranizan K, Choi YG, and Ngai J (2011). p63 regulates olfactory stem cell self-renewal and differentiation. Neuron 72, 748–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadye L, Das D, Sanchez MA, Street K, Baudhuin A, Wagner A, Cole MB, Choi YG, Yosef N, Purdom E, et al. (2017). Injury Activates Transient Olfactory Stem Cell States with Diverse Lineage Capacities. Cell Stem Cell 21, 775–790 e779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Postiglione MP, Krieger TG, Hernandez L, Wang C, Han Z, Streicher C, Papusheva E, Insolera R, Chugh K, et al. (2014). Deterministic progenitor behavior and unitary production of neurons in the neocortex. Cell 159, 775–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattinoni L, Speiser DE, Lichterfeld M, and Bonini C (2017). T memory stem cells in health and disease. Nat Med 23, 18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgouli M, Herraiz C, Crosas-Molist E, Fanshawe B, Maiques O, Perdrix A, Pandya P, Rodriguez-Hernandez I, Ilieva KM, Cantelli G, et al. (2019). Regional Activation of Myosin II in Cancer Cells Drives Tumor Progression via a Secretory Cross-Talk with the Immune Microenvironment. Cell 176, 757–774 e723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith JW, Sokol CL, and Luster AD (2014). Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annu Rev Immunol 32, 659–702. [DOI] [PubMed] [Google Scholar]
- Guo Z, Packard A, Krolewski RC, Harris MT, Manglapus GL, and Schwob JE (2010). Expression of pax6 and sox2 in adult olfactory epithelium. J Comp Neurol 518, 4395–4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilos DL (2011). Chronic rhinosinusitis: epidemiology and medical management. J Allergy Clin Immunol 128, 693–707; quiz 708–699. [DOI] [PubMed] [Google Scholar]
- Hasegawa-Ishii S, Shimada A, and Imamura F (2017). Lipopolysaccharide-initiated persistent rhinitis causes gliosis and synaptic loss in the olfactory bulb. Sci Rep 7, 11605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto D, Chow A, Noizat C, Teo P, Beasley MB, Leboeuf M, Becker CD, See P, Price J, Lucas D, et al. (2013). Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity 38, 792–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrick DB, Lin B, Peterson J, Schnittke N, and Schwob JE (2017). Notch1 maintains dormancy of olfactory horizontal basal cells, a reserve neural stem cell. Proc Natl Acad Sci U S A 114, E5589–E5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook EH, Wu E, Curry WT, Lin DT, and Schwob JE (2011). Immunohistochemical characterization of human olfactory tissue. Laryngoscope 121, 1687–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YC, Li L, and Fuchs E (2014). Emerging interactions between skin stem cells and their niches. Nat Med 20, 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, and Lempicki RA (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Islam SA, and Luster AD (2012). T cell homing to epithelial barriers in allergic disease. Nat Med 18, 705–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins SJ, Ruckerl D, Cook PC, Jones LH, Finkelman FD, van Rooijen N, MacDonald AS, and Allen JE (2011). Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science 332, 1284–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M, and Clevers H (2016). Reparative inflammation takes charge of tissue regeneration. Nature 529, 307–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern RC (2000). Chronic sinusitis and anosmia: pathologic changes in the olfactory mucosa. Laryngoscope 110, 1071–1077. [DOI] [PubMed] [Google Scholar]
- Keyes BE, Liu S, Asare A, Naik S, Levorse J, Polak L, Lu CP, Nikolova M, Pasolli HA, and Fuchs E (2016). Impaired Epidermal to Dendritic T Cell Signaling Slows Wound Repair in Aged Skin. Cell 167, 1323–1338 e1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn T, Bettelli E, Oukka M, and Kuchroo VK (2009). IL-17 and Th17 Cells. Annu Rev Immunol 27, 485–517. [DOI] [PubMed] [Google Scholar]
- Krautwurst D, Yau KW, and Reed RR (1998). Identification of ligands for olfactory receptors by functional expression of a receptor library. Cell 95, 917–926. [DOI] [PubMed] [Google Scholar]
- Lane AP, Turner J, May L, and Reed R (2010). A genetic model of chronic rhinosinusitis-associated olfactory inflammation reveals reversible functional impairment and dramatic neuroepithelial reorganization. J Neurosci 30, 2324–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane SW, Williams DA, and Watt FM (2014). Modulating the stem cell niche for tissue regeneration. Nat Biotechnol 32, 795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Borgne M, Etchart N, Goubier A, Lira SA, Sirard JC, van Rooijen N, Caux C, Ait-Yahia S, Vicari A, Kaiserlian D, et al. (2006). Dendritic cells rapidly recruited into epithelial tissues via CCR6/CCL20 are responsible for CD8+ T cell crosspriming in vivo. Immunity 24, 191–201. [DOI] [PubMed] [Google Scholar]
- Leung CT, Coulombe PA, and Reed RR (2007). Contribution of olfactory neural stem cells to tissue maintenance and regeneration. Nat Neurosci 10, 720–726. [DOI] [PubMed] [Google Scholar]
- Li Q, and Verma IM (2002). NF-kappaB regulation in the immune system. Nat Rev Immunol 2, 725–734. [DOI] [PubMed] [Google Scholar]
- Lien WH, Polak L, Lin M, Lay K, Zheng D, and Fuchs E (2014). In vivo transcriptional governance of hair follicle stem cells by canonical Wnt regulators. Nat Cell Biol 16, 179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B, Coleman JH, Peterson JN, Zunitch MJ, Jang W, Herrick DB, and Schwob JE (2017). Injury Induces Endogenous Reprogramming and Dedifferentiation of Neuronal Progenitors to Multipotency. Cell Stem Cell 21, 761–774 e765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luedde T, Heinrichsdorff J, de Lorenzi R, De Vos R, Roskams T, and Pasparakis M (2008). IKK1 and IKK2 cooperate to maintain bile duct integrity in the liver. Proc Natl Acad Sci U S A 105, 9733–9738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luther SA, Bidgol A, Hargreaves DC, Schmidt A, Xu Y, Paniyadi J, Matloubian M, and Cyster JG (2002). Differing activities of homeostatic chemokines CCL19, CCL21, and CXCL12 in lymphocyte and dendritic cell recruitment and lymphoid neogenesis. J Immunol 169, 424–433. [DOI] [PubMed] [Google Scholar]
- Luther SA, and Cyster JG (2001). Chemokines as regulators of T cell differentiation. Nat Immunol 2, 102–107. [DOI] [PubMed] [Google Scholar]
- Marsland BJ, Battig P, Bauer M, Ruedl C, Lassing U, Beerli RR, Dietmeier K, Ivanova L, Pfister T, Vogt L, et al. (2005). CCL19 and CCL21 induce a potent proinflammatory differentiation program in licensed dendritic cells. Immunity 22, 493–505. [DOI] [PubMed] [Google Scholar]
- McWhorter FY, Wang T, Nguyen P, Chung T, and Liu WF (2013). Modulation of macrophage phenotype by cell shape. Proc Natl Acad Sci U S A 110, 17253–17258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon HG, Kim SJ, Jeong JJ, Han SS, Jarjour NN, Lee H, Abboud-Werner SL, Chung S, Choi HS, Natarajan V, et al. (2018). Airway Epithelial Cell-Derived Colony Stimulating Factor-1 Promotes Allergen Sensitization. Immunity 49, 275-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray PJ, and Wynn TA (2011). Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol 11, 723–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myant KB, Cammareri P, McGhee EJ, Ridgway RA, Huels DJ, Cordero JB, Schwitalla S, Kalna G, Ogg EL, Athineos D, et al. (2013). ROS production and NF-kappaB activation triggered by RAC1 facilitate WNT-driven intestinal stem cell proliferation and colorectal cancer initiation. Cell Stem Cell 12, 761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarkar DR, Poposki JA, Tan BK, Comeau MR, Peters AT, Hulse KE, Suh LA, Norton J, Harris KE, Grammer LC, et al. (2013). Thymic stromal lymphopoietin activity is increased in nasal polyps of patients with chronic rhinosinusitis. J Allergy Clin Immunol 132, 593–600 e512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S, Larsen SB, Cowley CJ, and Fuchs E (2018). Two to Tango: Dialog between Immunity and Stem Cells in Health and Disease. Cell 175, 908–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S, Larsen SB, Gomez NC, Alaverdyan K, Sendoel A, Yuan S, Polak L, Kulukian A, Chai S, and Fuchs E (2017). Inflammatory memory sensitizes skin epithelial stem cells to tissue damage. Nature 550, 475–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packard A, Schnittke N, Romano RA, Sinha S, and Schwob JE (2011). DeltaNp63 regulates stem cell dynamics in the mammalian olfactory epithelium. J Neurosci 31, 8748–8759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SL, Zaid A, Hor JL, Christo SN, Prier JE, Davies B, Alexandre YO, Gregory JL, Russell TA, Gebhardt T, et al. (2018). Local proliferation maintains a stable pool of tissue-resident memory T cells after antiviral recall responses. Nat Immunol 19, 183–191. [DOI] [PubMed] [Google Scholar]
- Ramanathan M Jr., and Lane AP (2007). Innate immunity of the sinonasal cavity and its role in chronic rhinosinusitis. Otolaryngol Head Neck Surg 136, 348–356. [DOI] [PubMed] [Google Scholar]
- Richmond A (2002). Nf-kappa B, chemokine gene transcription and tumour growth. Nat Rev Immunol 2, 664–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodda LB, Lu E, Bennett ML, Sokol CL, Wang X, Luther SA, Barres BA, Luster AD, Ye CJ, and Cyster JG (2018). Single-Cell RNA Sequencing of Lymph Node Stromal Cells Reveals Niche-Associated Heterogeneity. Immunity 48, 1014–1028 e1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi S, Yamaguchi T, Nomura T, and Ono M (2008). Regulatory T cells and immune tolerance. Cell 133, 775–787. [DOI] [PubMed] [Google Scholar]
- Schnittke N, Herrick DB, Lin B, Peterson J, Coleman JH, Packard AI, Jang W, and Schwob JE (2015). Transcription factor p63 controls the reserve status but not the stemness of horizontal basal cells in the olfactory epithelium. Proc Natl Acad Sci U S A 112, E5068–5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann K, Lammermann T, Bruckner M, Legler DF, Polleux J, Spatz JP, Schuler G, Forster R, Lutz MB, Sorokin L, et al. (2010). Immobilized chemokine fields and soluble chemokine gradients cooperatively shape migration patterns of dendritic cells. Immunity 32, 703–713. [DOI] [PubMed] [Google Scholar]
- Schwitalla S, Fingerle AA, Cammareri P, Nebelsiek T, Goktuna SI, Ziegler PK, Canli O, Heijmans J, Huels DJ, Moreaux G, et al. (2013). Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell 152, 25–38. [DOI] [PubMed] [Google Scholar]
- Schwob JE, Jang W, Holbrook EH, Lin B, Herrick DB, Peterson JN, and Hewitt Coleman J (2017). Stem and progenitor cells of the mammalian olfactory epithelium: Taking poietic license. J Comp Neurol 525, 1034–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw JL, Fakhri S, Citardi MJ, Porter PC, Corry DB, Kheradmand F, Liu YJ, and Luong A (2013). IL-33-responsive innate lymphoid cells are an important source of IL-13 in chronic rhinosinusitis with nasal polyps. Am J Respir Crit Care Med 188, 432–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Berg DA, Zhu Y, Shin JY, Song J, Bonaguidi MA, Enikolopov G, Nauen DW, Christian KM, Ming GL, et al. (2015). Single-Cell RNA-Seq with Waterfall Reveals Molecular Cascades underlying Adult Neurogenesis. Cell Stem Cell 17, 360–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens WW, Lee RJ, Schleimer RP, and Cohen NA (2015). Chronic rhinosinusitis pathogenesis. J Allergy Clin Immunol 136, 1442–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi K, and Karin M (2018). NF-kappaB, inflammation, immunity and cancer: coming of age. Nat Rev Immunol 18, 309–324. [DOI] [PubMed] [Google Scholar]
- Tong AJ, Liu X, Thomas BJ, Lissner MM, Baker MR, Senagolage MD, Allred AL, Barish GD, and Smale ST (2016). A Stringent Systems Approach Uncovers Gene-Specific Mechanisms Regulating Inflammation. Cell 165, 165–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Crombruggen K, Zhang N, Gevaert P, Tomassen P, and Bachert C (2011). Pathogenesis of chronic rhinosinusitis: inflammation. J Allergy Clin Immunol 128, 728–732. [DOI] [PubMed] [Google Scholar]
- Victores AJ, Chen M, Smith A, and Lane AP (2018). Olfactory loss in chronic rhinosinusitis is associated with neuronal activation of c-Jun N-terminal kinase. Int Forum Allergy Rhinol 8, 415–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuillemenot BR, Rodriguez JF, and Hoyle GW (2004). Lymphoid tissue and emphysema in the lungs of transgenic mice inducibly expressing tumor necrosis factor-alpha. Am J Respir Cell Mol Biol 30, 438–448. [DOI] [PubMed] [Google Scholar]
- Wells JM, and Watt FM (2018). Diverse mechanisms for endogenous regeneration and repair in mammalian organs. Nature 557, 322–328. [DOI] [PubMed] [Google Scholar]
- Yee KK, Pribitkin EA, Cowart BJ, Vainius AA, Klock CT, Rosen D, Feng P, McLean J, Hahn CG, and Rawson NE (2010). Neuropathology of the olfactory mucosa in chronic rhinosinusitis. Am J Rhinol Allergy 24, 110–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaph C, Troy AE, Taylor BC, Berman-Booty LD, Guild KJ, Du Y, Yost EA, Gruber AD, May MJ, Greten FR, et al. (2007). Epithelial-cell-intrinsic IKK-beta expression regulates intestinal immune homeostasis. Nature 446, 552–556. [DOI] [PubMed] [Google Scholar]
- Zhang W, Tan W, Wu X, Poustovoitov M, Strasner A, Li W, Borcherding N, Ghassemian M, and Karin M (2013). A NIK-IKKalpha module expands ErbB2-induced tumor-initiating cells by stimulating nuclear export of p27/Kip1. Cancer Cell 23, 647–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S4. RNA-seq data of differentially expressed genes, related to Figure 2
Data including significantly upregulated NF-κB components or targets, differentially expressed cytokines or chemokines, significantly upregulated receptor genes for cytokines or chemokines. HBCs were sorted from 6 weeks Dox treated or No-Dox control mice. Reads are shown as average FPKM of 3 individual samples in each group and filtered for genes which Log2 (Fold Change) > 1, or < - 1.
Table S5. RNA-seq data of differentially expressed transcription factors, related to Figure 4 and Figure 5
Data including transcription factors related to regulation of “stemness” and neurogenesis, and transcription factors involved in immune function. HBCs were sorted from 6 weeks Dox treated or No-Dox control mice. Reads are shown as average FPKM of 3 samples in each group and filtered for genes which Log2 (Fold Change) > 1, or < - 1.
Data Availability Statement
All software tools can be found online (see Key Resources Table). The accession number for the raw data files reported in this paper is GSE135042.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies for Immunohistochemistry | ||
| Rabbit anti-Krt5 | Covance | Cat#PRB-160P; RRID: AB_291581 |
| Rabbit anti-Ki67 | Abcam | Cat#Ab16667; RRID: AB_302459 |
| Rat anti-BrdU | Abcam | Cat#Ab6326; RRID: AB_305426 |
| Mouse anti-Krt14 | Thermo Fisher | Cat#MA5–11599; RRID: AB_10982092 |
| Mouse anti-β-Tubulin III | Millipore | Cat#MAB1637; RRID: AB_1707889 |
| Rabbit anti-∆NP63 | BioLegend | Cat#619001; RRID: AB_2256361 |
| Mouse anti-P63 | Santa Cruz | Cat#sc-8431; RRID: AB_628091 |
| Rabbit anti-RelA | Santa Cruz | Cat#sc-372; RRID: AB_632037 |
| Rat anti-CD45 | Ebioscience | Cat#14–0451-81; RRID: AB_467250 |
| Rat anti-F4/80 | Bio-Rad | Cat#MCA497GA; RRID: AB_323806 |
| Rat anti-Ly6G | Biolegend | Cat#127601; RRID: AB_1089179 |
| Rat anti-CD3 | Ebioscience | Cat#14–0032-81; RRID: AB_467052 |
| Goat anti-OMP | Wako | Cat#544–10001; RRID: AB_2315007 |
| Goat anti Sox2 | Santa Cruz | Cat#sc-17320; RRID: AB_2286684 |
| Goat anti-CCL19 | R&D Systems | Cat#AF880; RRID: AB_2071545 |
| Rabbit anti-TNF-α | Cell Signaling | Cat#3707; RRID: AB_10691460 |
| Goat anti-CXCL10 | R&D Systems | Cat#AF-466-NA; RRID: AB_2292487 |
| Rat anti-Foxp3 | eBioscience | Cat#14–5773-82; RRID: AB_467576 |
| Chicken anti Krt18 | Abcam | Cat# ab14047; RRID: AB_2133173 |
| Rabbit anti PGP9.5 | Ultraclone | Cat# RA95101; RRID: AB_2313685 |
| Goat anti-CCL20 | R&D Systems | Cat# AF760; RRID: AB_355580 |
| Rabbit anti IL-6 | Novus | Cat# NB600–1131; RRID: AB10001997 |
| Mouse anti Human CCL2 | R&D Systems | Cat#MAB2791; RRID: AB_2071560 |
| Rabbit anti Human-CD45 | Abcam | Cat#Ab40763; RRID: AB_726545 |
| Mouse anti-CD45 | Biolegend | Cat#304002; RRID: AB_314390 |
| Mouse anti-human CD3 | Biolegend | Cat#300413; RRID: AB_314067 |
| Alexa Fluor® 488 anti-human CD3 | Biolegend | Cat#300454; RRID: AB_2564149 |
| Mouse anti-Human CD4 | BD Pharmingen | Cat#555344; RRID: AB_395749 |
| Alexa Fluor® 488 anti-β-Tubulin III | Biolegend | Cat#801203; RRID: AB_2564757 |
| Antibodies for Flow Cytometry | ||
| FITC anti-mouse CD45 | Biolegend | Cat#103107; RRID: AB_312972 |
| PE anti-Ms F4/80 | Biolegend | Cat#123109; RRID: AB_893498 |
| Alexa F 647 anti FOXP3 | Biolegend | Cat#320013; RRID: AB_439749 |
| PE/Cy7 anti-Ms CD4 | Biolegend | Cat#100421; RRID: AB_312706 |
| PerCP/Cy5.5 anti-Ms/Hu CD11b | Biolegend | Cat#101227; RRID: AB_893233 |
| PerCP-Cy™5.5 Rat Anti-Mouse CD3 | BD Pharmingen | Cat#560527; RRID: AB_1727463 |
| Alexa Fluor® 700 Rat Anti-Mouse Ly-6G | BD Pharmingen | Cat#561236; RRID: AB_10611860 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Dox food | Bio Serv | Cat#S3888 |
| BrdU | Sigma | Cat#B9285 |
| Lipopolysaccharides (LPS) | Sigma | Cat#L2880 |
| Zymosan A Bioparticles, Alexa Fluor 488 | Molecular Probes | Cat#Z23373 |
| True-Nuclea Transcription Factor Buffer | Biolegend | Cat#424401 |
| Alexa Fluor 488 Tyramide SuperBoost Kit | Life Technologies | Cat#B40912 |
| TNF-α | Peprotech | Cat#315–01A |
| Murine EGF | Peprotech | Cat#315–09 |
| Murine FGF-basic | Peprotech | Cat#450–33 |
| Murine CCL19 | Peprotech | Cat#250–27B |
| Murine CCL20 | Peprotech | Cat#250–27 |
| Critical Commercial Assays | ||
| SYBR Green PCR Master Mix | Applied Biosystems | Cat#4385612 |
| TaqMan Fast Universal PCR Master Mix | Applied Biosystems | Cat#4352042 |
| Mouse TNF alpha ELISA Kit | eBioscience | Cat#88–7324 |
| MAGnify ChiP Kit | Life Technologies | Cat#49–2024 |
| Deposited Data | ||
| Raw data files for RNA sequencing | NCBI | GEO: GSE135042 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Cyp2g1-rtTA | N/A | (Lane et al., 2010) |
| Mouse: TRE-TNF-α | N/A | (Vuillemenot et al., 2004) |
| Mouse: Krt17-EGFP | Jackson lab | Stock Number: 023965 |
| Mouse: Krt5-Cre | N/A | (Leung et al., 2007) |
| Mouse: Rosa26-stopflox/flox-tdTomato | Jackson lab | Stock Number: 007914 |
| Mouse: RelAflox/flox | N/A | (Luedde et al., 2008) |
| Oligonucleotides | ||
| qRT-PCR primers | This paper | Table S1–3 |
| Software and Algorithms | ||
| Prism 6 | Graphpad | https://www.graphpad.com |
| R | r-project.org | R 3.4.4 |
| Zen lite | Zeiss | ZEN 2012 (blue edition) |
| FlowJo v10 | FlowJo, LLC | flowjo.com |
| DAVID Bioinformatics Resources | (Huang da et al., 2009) | david.ncifcrf.gov |