Abstract
Protein trafficking requires coat complexes that couple recognition of sorting motifs in transmembrane cargos with biogenesis of transport carriers. The mechanisms of cargo transport through the endosomal network are poorly understood. Here, we identify a sorting motif for endosomal recycling of cargos including the cation-independent mannose-6-phosphate receptor and semaphorin 4C by the membrane tubulating BAR domain-containing sorting nexins SNX5 and SNX6. Crystal structures establish that this motif folds into a β-hairpin that binds a site in the SNX5/SNX6 phox homology domains. Over sixty cargos share this motif and require SNX5/SNX6 for their recycling. These include cargos involved in neuronal migration and a Drosophila snx6 mutant displays defects in axonal guidance. These studies identify a sorting motif and provide molecular insight into an evolutionary conserved coat complex, the ‘Endosomal SNX-BAR sorting complex for promoting exit 1’ (ESCPE-1), which couples sorting motif recognition to BAR domain-mediated biogenesis of cargo-enriched tubulo-vesicular transport carriers.
Introduction
Thousands of transmembrane cargo proteins routinely enter the endosomal network where they transit between two fates: retention within the network for degradation in the lysosome or export from the network for recycling and reuse at the cell surface, the trans-Golgi network (TGN) and the autophagosome1,2. While mechanistic details of degradative sorting are well established, those events that define the export pathways remain poorly described2. Central to understanding endosomal recycling is the identification of sorting motifs within the cytosolic tails of transmembrane proteins that provide the signature for export, and those membrane re-modelling complexes that recognising these signatures and drive packaging into transport carriers3.
While sorting motifs for endocytic and Golgi-directed transport are well known, those that control export from the endosomal network are poorly understood2–4. The characterisation of AP-1 and the CCC (CCDC22, CCDC93, COMMD) complex5–7, Retromer and Retriever and their associated sorting nexin (SNX) cargo adaptors8–16, has identified some export motifs for the sorting of hundreds of integral proteins. However, additional sorting motifs are likely to be required and, moreover, for the majority of identified sorting motifs how their recognition is coordinated with membrane re-modelling to form cargo-enriched transport carriers remains largely unknown17,18.
Here we define the role of a SNX-BAR (SNX containing a carboxy-terminal Bin, Amphiphysin, Rvs (BAR) domain) membrane re-modelling complex in sequence-dependent sorting of integral proteins19–24. Through structural and functional analysis we identify a sorting motif that is required for endosomal recycling through direct interaction with these SNX-BARs. Our study provides insight into an endosomal coat complex that couples sequence-dependent cargo recognition with BAR domain-mediated biogenesis of tubulo-vesicular transport carriers.
Results
Structure of SNX5 bound to the CI-MPR
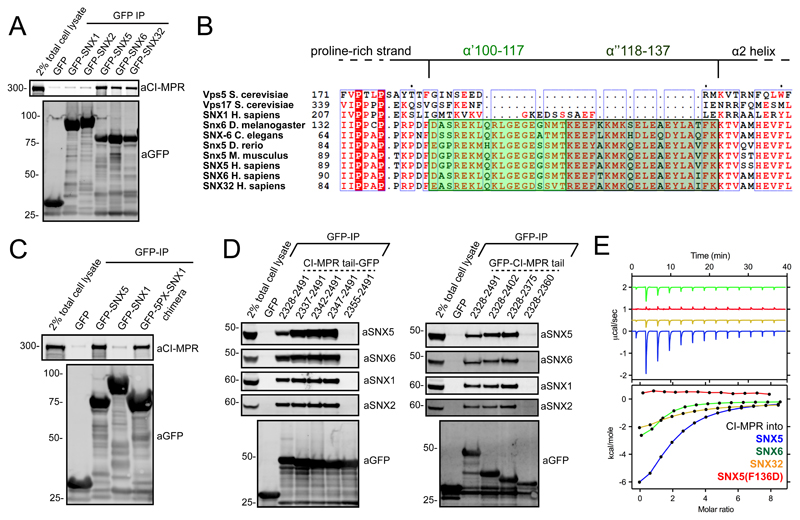
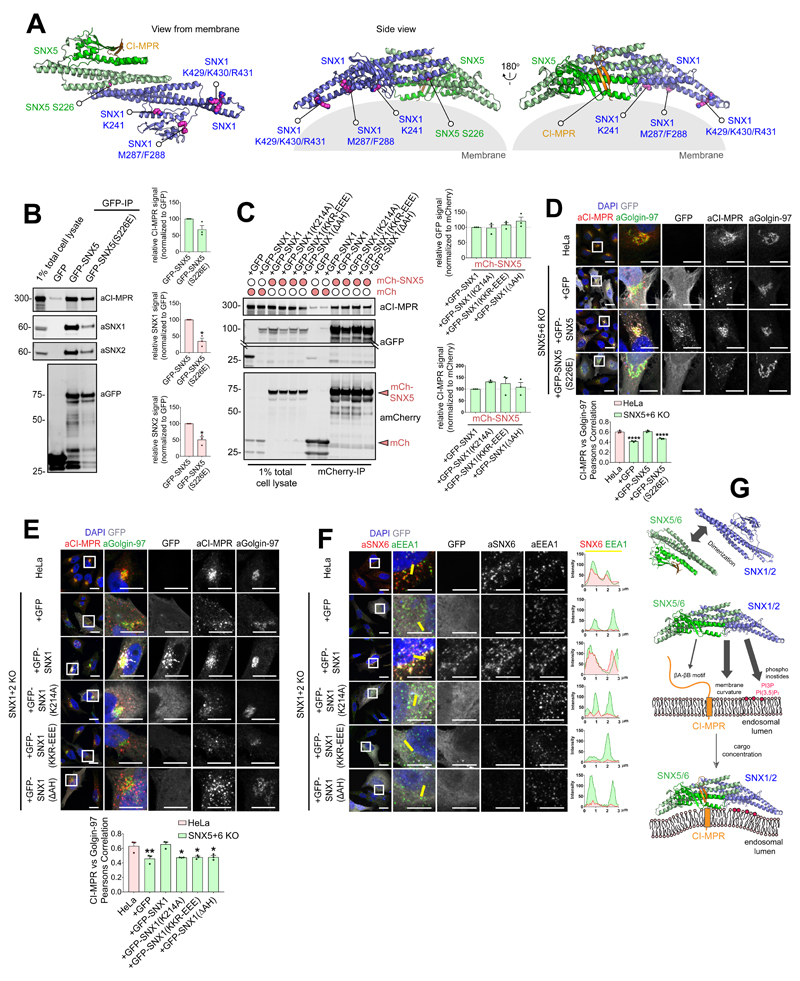
Recently, we observed a role for sorting nexin-5 (SNX5), SNX6 and SNX32, a sub-set of SNX-BAR proteins, in the endosome sorting of the CI-MPR to the TGN23,24. To provide molecular insight into these observations (Figure 1A), we mapped the region of SNX5 necessary for CI-MPR binding. As noted by others25, protein alignment revealed a 38 amino acid insert within the PX domains of these SNX-BAR proteins that is absent in the PX domain of SNX1 or SNX2, functionally related SNX-BAR proteins that form a heterodimeric complex with SNX520 but fail to associate with the CI-MPR (Figures 1A and 1B). Indeed, CI-MPR binding could be transferred from SNX5 to SNX1 by engineering a SNX1:SNX5 switch chimera (Figure 1C).
Figure 1. Mapping the interaction between SNX5 and CI-MPR.
(A) Co-immunoprecipitation of GFP-tagged SNX-BARs transiently transfected in HEK293T cells. (B) Alignment of yeast Vps17 and Vps5, together with SNX1, SNX5, SNX6, SNX32, and the SNX5 homologues from different species. The α’ and α” helices that compose the unique helix-turn-helix extension are highlighted in green. (C) Co-immunoprecipitation of GFP-tagged SNX1, SNX5 and a SNX1 chimera generated by replacement of the SNX1 PX domain with that of SNX5. (D) Co-immunoprecipitation of GFP-tagged CI-MPR tail deletion mutants transiently transfected in HEK293T cells. (E) CI-MPR peptide was injected into either SNX5, SNX5(F136D), SNX6 or SNX32 PX domains and binding measured by ITC. Top panels show the raw data and bottom panels represent the integrated and normalized data fit with a 1:1 binding model. Binding of CI-MPR with SNX5 or SNX6 was measured over three independent experiments, binding of CI-MPR with SNX32 was measured once. ITC binding parameters, including Standard deviation (SD) where calculated, are provided in Table S1. In each case 1A, 1C and 1D are representative blots of three independent GFP traps and the unprocessed original scans of immunoblots are shown in Supplementary Figure 3.
A GFP-nanotrap screen of deletion mutants across the CI-MPR cytosolic tail (covering residues 2328-2491) established that residues 2347-2375 were essential for SNX5 and SNX6 association (Figure 1D). A peptide corresponding to these residues bound directly (Kd ~ 25 μM) to the isolated SNX5 PX domain (Figure 1E) and the corresponding domains of SNX6 and SNX32 (Kd’s of 24 μM and 12 μM respectively) (Figure 1E; Table S1).
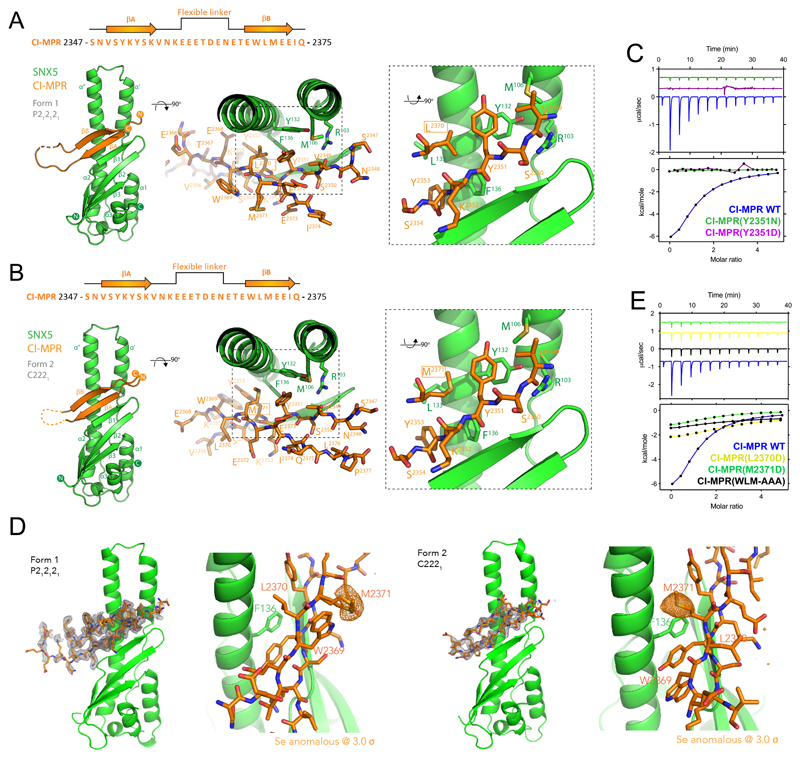
Next, we designed a fusion protein of the SNX5 PX domain coupled through a flexible linker to the CI-MPR sequence, crystallized the protein in two different crystal forms and determined their structures by X-ray crystallography (Figure 2A and 2B; Table S2). The CI-MPR peptide interacted with the SNX5 PX domain via a β-sheet augmentation, forming two anti-parallel β-strands (βA and βB) connected by a long flexible linker. Although the sequences and specific-side-chain interactions are different, the bound CI-MPR structures resemble that of the IncE protein from Chlamydia trachomatis26–28 (Figure S1A and S1B).
Figure 2. Crystal structure of the SNX5:CI-MPR complex.
(A and B) The SNX5–CI-MPR Form 1 crystal structure (spacegroup P212121) (A) and the SNX5–CI-MPR Form 2 crystal structure (spacegroup C2221) (B) are shown in ribbon diagram (left panels), with a close-up of the bound CI-MPR peptide shown in stick representation (right panels). (C) Binding of SNX5 to CI-MPR peptide and CI-MPR βA-strand mutants Y2351N and Y2351D. Top panels show the raw data and bottom panels represent the integrated and normalized data fit with a 1:1 binding model. (D) The SNX5–CI-MPR Form 1 (spacegroup P212121) and Form 2 (spacegroup C2221) crystal structures are shown with corresponding CI-MPR peptide fo-fc omit maps in the left panels (grey, 3σ level), while the right panels show the anomalous difference maps for the Se atoms in the SeMet labeled protein (orange, 3σ level). This identifies the Met2371 side-chain unambiguously in each structure. (E) Binding of SNX5 to CI-MPR βB-strand mutants L2370D, M2371D and WLM-AAA by ITC. Top panels show the raw data and bottom panels represent the integrated and normalized data fit with a 1:1 binding model. Binding of SNX5 with CI-MPR Wt was measured over three independent experiments, binding of SNX5 with CI-MPR βA/βB-strand mutants was measured once. ITC binding parameters, including Standard deviation (SD) where calculated, are provided in Table S1
In both structures, the first β-strand consists of residues 2349VSYKYSK2355 (Figure 2A and 2B). The key side-chains of SNX5 that mediate binding include Arg103, Met106, Tyr132 and Phe136. In the βA-strand Val2349, Tyr2351 and Tyr2353 side-chains are in close contact with SNX5, with Tyr2351 forming a stacking interaction with SNX5 Phe136 and Val2349 forming a hydrogen bond with SNX5 Arg103. This mirrors a similar stacking interaction of Phe116 from IncE (Figure S1A). Arg103 from SNX5 forms a hydrogen bond with the backbone carbonyl of Val2349, causing it to adopt a substantially different orientation to its pose in the IncE-bound SNX5 structure. Confirming the importance of the 2349VSYKYSK2355 residues, Y2351N or Y2351D mutations completely disrupted the interaction (Figure 2C).
Interestingly, the second βB-strand included a WLM sequence previously identified as required for CI-MPR endosome-to-TGN sorting29. In the two SNX5–CI-MPR crystal forms there was a major difference in the register of this second βB-strand (Figure 2A and 2B), which we confirmed by anomalous difference maps of the SeMet-labelled proteins to unambiguously place the CI-MPR Met2371 side-chain (Figure 2D). In Form 1 the βB strand includes residues 2369WLMEEI2374, while in Form 2 it consists of 2368EWLMEE2373. In Form 1 therefore, Leu2370 engages a hydrophobic pocket composed of CI-MPR side chains Tyr2351, Tyr2353 and SNX5 side chains Tyr132, Leu133, and Phe136; in Form 2 the flip in the βB strand orientation results in CI-MPR Met2371 occupying this pocket (Figure 2A and 2B). Which of these peptide configurations is preferred? We theorized that mutation of either Leu2370 or Met2371 to an aspartate side-chain would block interaction with SNX5 depending on which residue was most favoured. In ITC experiments, neither mutations had a major effect on the binding affinity, although there was a reduced enthalpy of interaction, while mutation of the entire WLM sequence resulted in a larger reduction in binding affinity (Figure 2E; Table S1). This suggests that the two configurations of the βB strand are both equally able to sustain binding; what is important is not the sequence per se, but that a correctly positioned hydrophobic side chain is the main requirement (either Leu2370 or Met2371 in CI-MPR). Notably in the SNX5–IncE complex this site is occupied by a valine side-chain (Figure S1A). The model therefore is that SNX5 (and SNX6) can be engaged by a ‘promiscuous’ β-hairpin structure, where essential features include aromatic residues in the third and fifth positions of the βA strand, a loop region, and a hydrophobic side-chain in the third position of the βB strand.
The CI-MPR sorting motif is required for retrograde transport
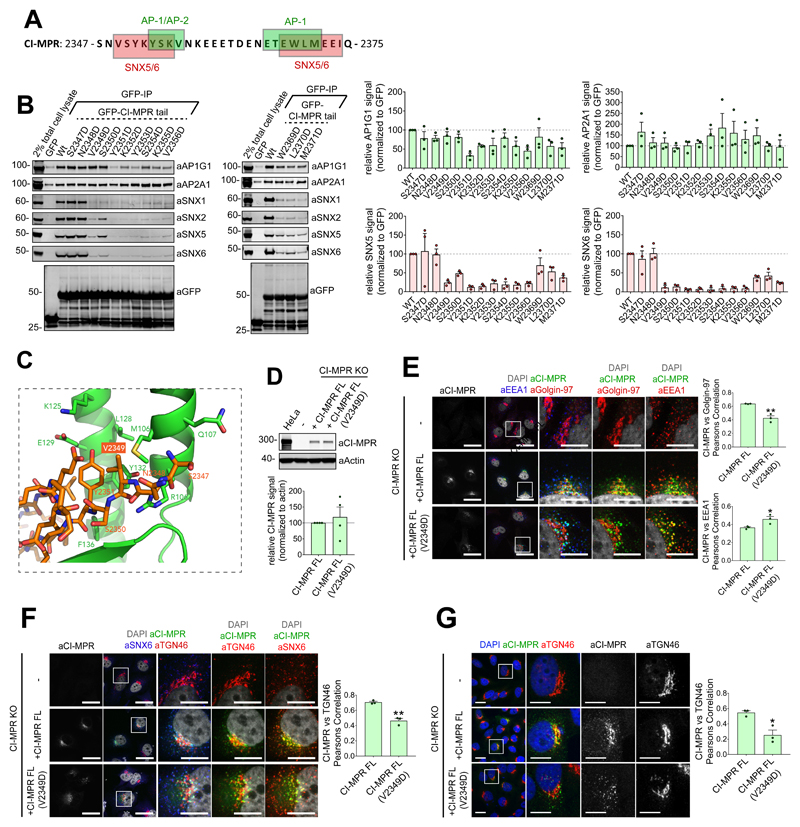
Validation of the 2349VSYKYSK2355 and 2369WLM2371 sequences as sorting motifs is complicated by their overlap with sequences recognised by other cargo adaptors, most notably AP-1 and AP-2, which mediate TGN-to-endosome transport and CI-MPR endocytosis respectively30 (Figure 3A). To identify a mutant that negated SNX5/SNX6 binding while retaining AP-1/AP-2 association we performed aspartate mutagenesis across the 2349VSYKYSK2355 and 2369WLM2371 sequences (Figure 3B). Quantitative western analysis revealed that the V2349D mutant conveying loss of SNX5/SNX6 binding while having no discernible effect on AP-2/AP-1 binding (Figure 3B). Confirming that the 2349VSYKYSK2355 motif constitutes the major contributor to the interaction, mutations of the βB-strand caused a minor loss of SNX5/SNX6 binding (Figure 3B).
Figure 3. The interaction between SNX5 and the CI-MPR β-hairpin structure is required for CI-MPR retrograde trafficking.
(A) Schematic of interactions reported to overlap with CI-MPR β-hairpin structure. (B) Co-immunoprecipitation of GFP-tagged CI-MPR tail constructs transiently transfected in HEK293T cells; summary of relative binding to indicated proteins. Band intensities measured from n=3 independent experiments using Odyssey software. Band intensities normalized to GFP expression presented as average fraction of GFP-CI-MPR Wt control (C) Bound CI-MPR peptide in stick representation highlighting Val2349 position. (D) Re-expression of full-length CI-MPR and CI-MPR(V2349D) mutant in HeLa CI-MPR KO clonal line. CI-MPR levels analyzed by quantitative fluorescence-based western blotting. Band intensities measured using Odyssey software and normalized to β-Actin before calculating percentage of protein compared with full-length CI-MPR control. Bars represent mean of n=4 independent experiments. (E and F) HeLa CI-MPR KO clonal line transiently transfected with full-length CI-MPR or CI-MPR(V2349D) mutant. Steady state colocalization of CI-MPR with EEA1, Golgin-97 and TGN46, analyzed using Pearsons correlation. Cell numbers analysed for CI-MPR vs Golgin-97 colocalisation (E): CI-MPR KO+CI-MPR: 64 cells; CI-MPR KO+CI-MPR(V2349D): 56 cells across n=3 independent experiments. Pearsons coefficient values compared with +CI-MPR using two-tailed Unpaired t-test (**P<0.01; P=0.0036). Cell numbers analysed for CI-MPR vs EEA1 Pearsons correlation: CI-MPR KO+CI-MPR: 69 cells; CI-MPR KO+CI-MPR(V2349D): 49 cells across n=3 independent experiments. Pearsons coefficient values compared with +CI-MPR using two-tailed Unpaired t-test (**P<0.05; P=0.0385). Cell numbers analysed for CI-MPR vs TGN46 colocalisation (F): CI-MPR KO+CI-MPR: 70 cells; CI-MPR KO+CI-MPR(V2349D): 66 cells across n=3 independent experiments. Pearsons coefficient values compared with +CI-MPR using two-tailed Unpaired t-test (**P<0.01; P=0.0019). (G) HeLa CI-MPR KO clonal line transfected with full length CI-MPR or CI-MPR(V2349) mutant and CI-MPR colocalization with TGN46, analyzed after 40 min chase of surface CI-MPR. Cell numbers analysed: CI-MPR KO+CI-MPR: 45 cells; CI-MPR KO+CI-MPR(V2349D): 53 cells across n=3 independent experiments. Pearson’s coefficient values were compared with +CI-MPR using Unpaired t test (*P<0.05; P=0.0133). (E-G) Scale bars 20 μm (micrographs) and 10 μm (insets). (B, D-F), Bars, mean; error bars, SEM, circles represent individual data points. Statistics source data: Supplementary Table 4. Unprocessed original scans of immunoblots: Supplementary Figure 3.
To establish the 2349VSYKYSK2355 motif as the dominant element of the sorting signal we generated a HeLa cell line knocked out for CI-MPR. Next we incorporated a Val2349Asp mutation (Figure 3C) into an expression construct encoding for full-length CI-MPR (Figure 3D). In contrast to expression of a full-length wild-type CI-MPR, which displayed a steady-state enrichment at the TGN46-labelled and Golgin-97-labelled TGN, full-length CI-MPR(V2349D) failed to enrich in the TGN and displayed a more prominent endosomal distribution (Figure 3E and 3F). This is consistent with the V2349D mutant retaining the ability to undergo endocytosis and transport from the TGN while having a defect in retrograde transport. Confirming this, antibody uptake experiments established that wild-type CI-MPR and the V2349D mutant underwent endocytosis, but only the V2349D mutant failed to undergo endosomal export as defined by enrichment in the TGN (Figure 3G). Thus 2349VSYKYSK2355 sorting motif, alongside the 2369WLM2371 motif29, is required for the SNX5/SNX6-dependent endosome-to-TGN transport of the CI-MPR.
SNX5 recognition of the CI-MPR sorting motif is required for endosomal export
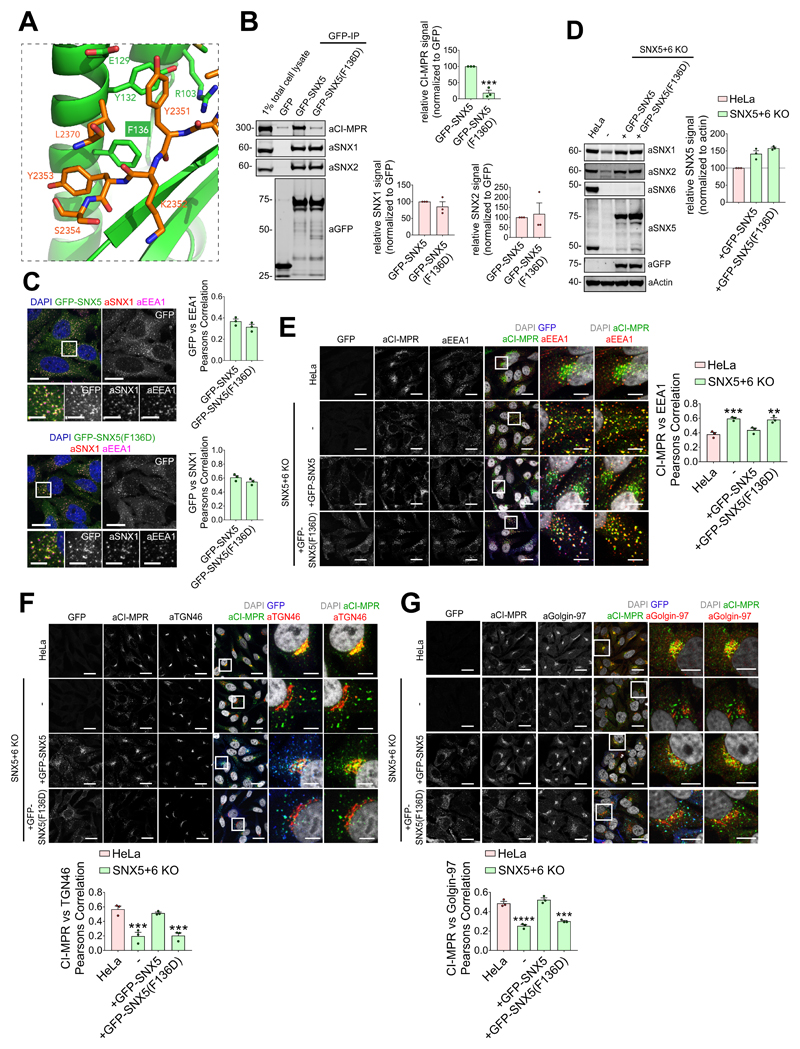
Next we examined the interaction of GFP-tagged SNX5 and an aspartate mutant of the key Phe136 residue (F136D) (Figure 4A) with full-length CI-MPR. In co-immunoprecipitation experiments, while SNX5 displayed CI-MPR binding, the GFP-SNX5(F136D) mutant failed to interact (Figure 4B). Importantly, the GFP-SNX5(F136D) mutant retained endosomal association and formed functional BAR domain-mediated heterodimers with SNX1 and SNX220,21 (Figure 4B and 4C). In a knockout cell line, loss of SNX5 and SNX6 expression results in the steady-state localisation of the CI-MPR shifting from the TGN to peripherally dispersed endosomes23,24. In this background, transduction of wild-type GFP-SNX5 rescued CI-MPR sorting, while GFP-SNX5(F136D) expression failed to do so (Figure 4D-G). Overall, in identifying a sorting motif and revealing its mode of recognition by the SNX5/SNX6 components of the SNX-BAR membrane tubulating complex, these data establish the mechanistic basis of tubular-based endosome-to-TGN export of the CI-MPR.
Figure 4. Interaction between CI-MPR and the hydrophobic groove of SNX5 PX domain is required for CI-MPR retrograde trafficking.
(A) CI-MPR peptide bound to SNX5 hydrophobic groove highlighting Phe136 position. (B) Co-immunoprecipitation of GFP-tagged SNX5 and SNX5(F136D) transiently transfected in HEK293T cells, summary of relative binding. Quantification of band intensities measured from n=3 independent experiments using Odyssey software. Band intensities normalized to GFP expression, presented as average fraction of GFP-SNX5 control. GFP-SNX5(F136D) mutant compared with GFP-SNX5 using two-tailed Unpaired t-test (***P<0.001; P values for SNX1: P=0.3655, SNX2: P=0.7646; CI-MPR: P=0.0006). (C) F136D mutation in SNX5 PX domain does not affect SNX5 endosomal localization. HeLa cells lentivirally transduced with GFP-SNX5 or GFP-SNX5(F136D) viral particles. Transduced cells fixed and stained for EEA1 and SNX1. Cell numbers analysed for colocalization between GFP and EEA1: GFP-SNX5: 50 cells; GFP-SNX5(F136D): 57 cells across n=3 independent experiments. Pearson’s coefficient values compared using two-tailed Unpaired t-test (P=0.1862). Cell numbers analysed for colocalization between GFP and SNX1: GFP-SNX5: 51 cells; GFP-SNX5(F136D): 59 cells across n=3 independent experiments. Pearson’s coefficient values compared using Unpaired t-test (P=0.1771). Scale bars 20 μm (non-zoom images) and 10 μm (insets). (D) GFP-SNX5 and GFP-SNX5(F136D) reintroduced in SNX5+SNX6 KO cells at levels comparable to endogenous. HeLa SNX5+SNX6 KO clonal line lentivirally transduced with GFP-SNX5 or GFP-SNX5(F136D) viral particles and SNX5-levels analyzed by quantitative fluorescence-based western blotting. Band intensities measured using Odyssey software and normalized to β-Actin before calculating percentage of protein compared with parental HeLa control. Bars represent mean of n=3 independent experiments. (E, F and G) Distribution of endogenous CI-MPR in HeLa cells, HeLa SNX5+SNX6 KO clonal line and HeLa SNX5+SNX6 KO clonal line transduced with GFP-SNX5 or GFP-SNX5(F136D). Colocalization analysis between CI-MPR and EEA1, CI-MPR and TGN46, and CI-MPR and Golgin-97. Cell numbers analysed across n=3 independent experiments for CI-MPR and EEA1 colocalisation (E): HeLa: 129 cells, SNX5+SNX6 KO: 144 cells, KO+GFP-SNX5: 159 cells; KO+GFP-SNX5(F136D): 122 cells. Pearson’s coefficient values compared with HeLa control using one-way ANOVA and Dunnett’s test (***P<0.001, **P<0.01; SNX5+SNX6 KO: P=0.0007, KO+GFP-SNX5: P=0.2886, KO+GFP-SNX5(F136D): P=0.0010). Cell numbers analysed across n=3 independent experiments for CI-MPR and TGN46 colocalisation (F): HeLa: 126 cells, SNX5+SNX6 KO: 111 cells, KO + GFP-SNX5: 121 cells; KO+GFP-SNX5(F136D): 110 cells. Pearson’s coefficient values compared with HeLa control using one-way ANOVA and Dunnett’s test (***P<0.001; SNX5+SNX6 KO: P=0.0005, KO+GFP-SNX5: P=0.6897, KO+GFP-SNX5(F136D): P=0.0006). Cell numbers analysed across n=3 independent experiments for CI-MPR and Golgin-97 colocalisation (G): HeLa: 105 cells, SNX5+SNX6 KO: 127 cells, KO+GFP-SNX5: 122 cells; KO+GFP-SNX5(F136D): 131 cells. Pearson’s coefficient values compared with HeLa control using one-way ANOVA and Dunnett’s test ****P<0.0001, ***P<0.001; SNX5+SNX6 KO: P=<0.0001, KO+GFP-SNX5: P=0.3482, KO+GFP-SNX5(F136D): P=0.0002). Scale bars 20 μm (micrographs) and 5 μm (insets). (B-D,G) Bars, mean; error bars, SEM; circles represent individual data points. Unprocessed original scans of immunoblots: Supplementary Figure 3. Statistics source data: Supplementary Table 4.
CI-MPR retrograde trafficking requires multiple features of SNX5-containing heterodimers
Through their BAR domains SNX5 (SNX6 and SNX32) form functional heterodimers with SNX1 (and SNX2)20,22. The resulting SNX1:SNX5 heterodimers are associated with the cytosolic leaflet of endosomes through multiple features of these organelles, including specific phosphoinositides and the sensing/induction of membrane curvature. To explore the relationship between membrane association and CI-MPR sorting we performed rescue experiments with previously validated mutants that perturb aspects of the membrane binding activity of the SNX1:SNX5 heterodimer: SNX5(S226E) displays a reduced ability to form heterodimers with SNX1 (and SNX2)31; SNX1(K214A) prevents association with phosphoinositides19 (the SNX5 PX domain does not bind phosphoinositides32), the BAR domain targeted SNX1(K429E:K430E:R431E) triple mutant (SNX1(KKR-EEE)) is unable to sense or induce membrane curvature19; and the SNX1(M287E:F288E) double mutant (SNX1(ΔAH)22) has a defective amphipathic helix required for membrane insertion (Figure 5A).
Figure 5. CI-MPR retrograde trafficking requires functional SNX5 heterodimers and co-incidence detection of multiple membrane features.
(A) Model of SNX5:SNX1 heterodimer. (B) Co-immunoprecipitation of GFP-tagged SNX5 and SNX5(S226E) transiently transfected in HEK293T cells and summary of relative binding. Quantification of band intensities measured from n=3 independent experiments using Odyssey software. Band intensities normalized to GFP expression, presented as average fraction of GFP-SNX5 control. GFP-SNX5(S226E) mutant compared with GFP-SNX5 using two-tailed Unpaired t-test (*P<0.05; P values for SNX1: P=0.0193, SNX2: P=0.0324; CI-MPR: P=0.1160). (C) Co-immunoprecipitation of mCherry and mCherry-SNX5 transiently transfected in HEK293T cells alongside GFP, GFP-SNX1, GFP-SNX1(K214A), GFP-SNX1(KKR-EEE) and GFP-SNX1(ΔAH); summary of relative binding. Quantification of GFP and CI-MPR band intensities measured from n=3 independent experiments using Odyssey software. Band intensities normalized to mCherry expression, presented as average fraction of the mCherry-SNX5+GFP-SNX1 control. Interactions of mCherry-SNX5+GFP-SNX1 mutants were compared with mCherry-SNX5+wild type GFP-SNX1 using one-way ANOVA and Dunnett’s test (GFP P values for SNX5+GFP-SNX1(K214A): 0.9969, SNX5+GFP-SNX1(KKR-EEE): P=0.8473, SNX5+GFP-SNX1(ΔAH): P=0.2983; CI-MPR P values for SNX5 + GFP-SNX1(K214A): 0.4109, SNX5+GFP-SNX1(KKR-EEE): P=0.6042, SNX5+GFP-SNX1(ΔAH): P=0.9648. (D) Distribution of endogenous CI-MPR in HeLa cells, HeLa SNX5+SNX6 KO clonal line and HeLa SNX5+SNX6 KO clonal line transfected with GFP, GFP-SNX5 or GFP-SNX5(S226E). Colocalization analysis between CI-MPR and the TGN marker Golgin-97. Cell numbers analysed: HeLa: 55 cells, SNX5+SNX6 KO+GFP: 62 cells, KO+GFP-SNX5: 63 cells; KO+GFP-SNX5(S226E): 68 cells across n=3 independent experiments. Pearson’s coefficient values compared with HeLa control using one-way ANOVA and Dunnett’s test, ****P<0.0001 (P values for SNX5+SNX6 KO+GFP: P=<0.0001, KO+GFP-SNX5: P=0.9173, KO+GFP-SNX5(S226E): P=<0.0001). (E) Distribution of endogenous CI-MPR in HeLa cells, HeLa SNX1+SNX2 KO clonal line and HeLa SNX1+SNX2 KO clonal line transfected with GFP, GFP-SNX1, GFP-SNX1(K214A), GFP-SNX1(KKR-EEE) and GFP-SNX1(ΔAH). Colocalization analysis between CI-MPR and Golgin-97. Cell numbers analysed: HeLa: 60 cells, SNX1+SNX2 KO +GFP: 53 cells, KO+GFP-SNX1: 58 cells; KO+GFP-SNX1(K214A): 72 cells, KO+GFP-SNX1(KKR-EEE): 77 cells, KO+GFP-SNX1(ΔAH): 73 cells across n=3 independent experiments. Pearson’s coefficient values compared with HeLa control using one-way ANOVA and Dunnett’s test, *P<0.05,*P<0.01 (P values for SNX1+SNX2 KO+GFP: P=0.0055, KO+GFP-SNX1: P=0.9752; KO+GFP-SNX1(K214A): P=0.0106, KO+GFP-SNX1(KKR-EEE): P=0.0126, KO+GFP-SNX1(ΔAH): P=0.0132). (F) Distribution of endogenous SNX6 in HeLa cells, HeLa SNX1+SNX2 KO clonal line and HeLa SNX1+SNX2 KO clonal line transfected with GFP, GFP-SNX1, GFP-SNX1(K214A), GFP-SNX1(KKR-EEE) and GFP-SNX1(ΔAH). SNX6 colocalization with EEA1. Figure representative of three independent experiments with similar results. (G) Model for how SNX5/6:SNX1/2 heterodimers sense multiple features of endosomal membranes, including presence of specific phosphoinositides, local membrane curvature, and cytosolic tails of cargoes. By co-incident sensing these feature SNX5/6:SNX1/2 heterodimers assemble into functional membrane-associated complexes that couple cargo recognition with membrane remodeling for formation of cargo-enrich transport carriers. (B,C) Bars, mean; error bars, SEM. Circles represent individual data points. Statistics source data: Supplementary Table 4. Unprocessed original scans of immunoblots: Supplementary Figure 3.
Co-immunoprecipitations confirmed that SNX5(S226E) displayed a reduced interaction with SNX1 (and SNX2), but importantly had little, if any, effect on CI-MPR association (Figure 5B). For the SNX1 mutants, all retained SNX5 heterodimer formation and CI-MPR association (Figure 5C). When expressed in the SNX5+SNX6 knockout HeLa cell line, SNX5(S226E) displayed a cytosolic localisation and failed to rescue the CI-MPR mis-sorting phenotype (Figure 5D). Extending this, in rescue experiments performed in a double SNX1 and SNX2 knock-out HeLa cell line23,24, the individual SNX1 mutants all localised to the cytosol and all failed to rescue the defect in CI-MPR retrograde transport (Figure 5E). Consistent with these data, all SNX1 mutants failed to drive the enrichment of SNX6 to endosomes (an antibody to detect endogenous SNX5 is presently lacking) (Figure 5F). The SNX5 (SNX6)-dependent CI-MPR retrograde transport therefore requires formation of heterodimers with SNX1 (or SNX2) and association of the resulting SNX1:SNX5 complex to the endosomal membrane through sensing of this organelle. With the SNX5(F136D) mutant retaining heterodimer formation and maintaining a localisation to endosomes (Figure 4C), cargo engagement does not appear to be the major driver for endosomal association of the SNX1:SNX5 complex. We speculate that cargo recognition may aid cargo clustering and the SNX1:SNX5 complex as a pre-requisite for the biogenesis of tubular profiles and transport carriers (Figure 5G).
SEMA4C and IGF1R bind to SNX5 through a similar mechanism
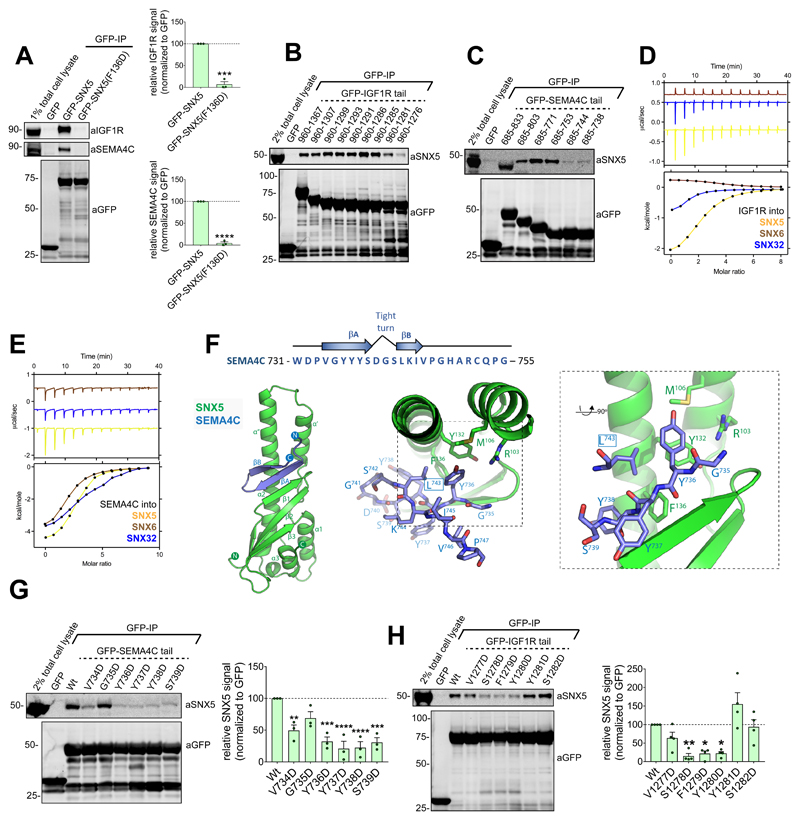
Do other cargo proteins utilise the same molecular mechanism for SNX5/SNX6-mediated endosomal sorting? Besides CI-MPR, two cell surface transmembrane proteins that interact with SNX5 are the IGF1R (a receptor critical in malignant transformation) and SEMA4C (a plexin B2 receptor involved in axon guidance)24. Co-immunoprecipitations of GFP-SNX5 and GFP-SNX5(F136D) established that SNX5 binds to these proteins through the conserved CI-MPR binding site within the PX domain (Figure 6A). Truncation mutagenesis identified residues 731-755 of SEMA4C and residues 1275-1302 of IGF1R for SNX5 binding (Figure 6B and 6C). Using the corresponding peptides, binding of SEMA4C and IGF1R to the PX domain of SNX5 was direct and with micromolar affinities (Kd’s of 26 and 15 μM respectively) comparable to those observed for CI-MPR (Figure 6D and 6E; Table S1).
Figure 6. Mechanism of SEMA4C and IGF1R cargo binding to SNX5.
(A) Co-immunoprecipitation of GFP-tagged SNX5 and SNX5(F136D) transiently transfected in HEK293T cells; summary of relative binding. Quantification of band intensities measured from n=3 independent experiments using Odyssey software. Band intensities normalized to GFP expression, presented as average fraction of GFP-SNX5 control. GFP-SNX5(F136D) mutant compared with GFP-SNX5 using two-tailed Unpaired t-test, ***P<0.001, ****P<0.0001 (P values for IGF1R: P=0.0001, SEMA4C: P=<0.0001). (B) Co-immunoprecipitation of GFP-tagged IGF1R tail truncation mutants transiently transfected in HEK293T cells. Representative blot of three independent GFP traps. (C) Co-immunoprecipitation of GFP-tagged SEMA4C tail truncation mutants transiently transfected in HEK293T cells. Representative blot of three independent GFP traps. (D and E) Binding of SNX5, SNX6 and SNX32 PX domain to IGF1R peptide (D) and SEMA4C peptide (E) by ITC. Top panels show raw data; bottom panels represent integrated and normalized data fit with a 1:1 binding model. Binding of IGF1R and SEMA4C with SNX5 measured over three independent experiments, binding of IGF1R and SEMA4C with SNX6 or SNX32 measured once. ITC binding parameters, including Standard deviation (SD) where calculated, provided in Table S1. (F) SNX5–SEMA4C crystal structure shown in ribbon diagram (left panel), with bound SEMA4C peptide shown in stick representation (right panel). (G) Co-immunoprecipitation of GFP-tagged SEMA4C tail constructs transiently transfected in HEK293T cells. Band intensities measured from n=3 independent experiments using Odyssey software. Band intensities normalized to GFP expression, presented as average fraction of GFP-SEMA4C wild-type control. GFP-SEMA4C mutants compared with GFP-SEMA4C wild-type control using one-way ANOVA and Dunnett’s test; **P<0.01, ***P<0.001, ****P<0.0001 (P values for V734: P=0.0045,G735D: P=0.0929, Y736D: P=0.0003, Y737D: P=<0.0001, Y738D: P=<0.0001, S739D=0.0003). (H) Co-immunoprecipitation of GFP-tagged IGF1R tail constructs transiently transfected in HEK293T cells. Quantification of band intensities measured from n=4 independent experiments using Odyssey software. Band intensities normalized to GFP expression, presented as average fraction of GFP-IGF1R wild-type control. GFP-IGF1R mutants compared with GFP-IGF1R wild-type control using one-way ANOVA and Dunnett’s test; *P<0.05, **P<0.01 (P values for V1277D: P=0.4031, S1278D: P=0.0049, F1279D: P=<0.0101, Y1280D: P=0.0107, Y1281D: P=0.0941, S1282D: P=0.9996). (A, G, H) Bars, mean; error bars, SEM; circles represent individual data points. Unprocessed original scans of immunoblots: Supplementary Figure 3. Statistics source data: Supplementary Table 4.
While we were unable to obtain diffracting crystals for a SNX5-IGF1R complex, crystals of the SNX5 PX domain with the SEMA4C sequence were grown and the structure determined at 2.45 Å resolution (Figure 6F; Table S2). SEMA4C interacts with SNX5 via a similar anti-parallel βA and βB-sheet augmentation, with the 734VGYYYS739 peptide making the first β-strand, followed by a tight turn (residues 740DGS742), then the Leu743 side chain engaging the hydrophobic pocket where Leu2370 or Met2371 are bound in the CI-MPR structures (Figure 6F). Immunoprecipitations of GFP-tagged SEMA4C cytosolic tail mutants, probing for endogenous SNX5 association, confirmed that the 731VGYYYS736 sequence was required for the SNX5-SEMA4C association (Figure 6G).
In examining the sequence of the IGF1R peptide (residues 1275-1302), we noted a 1277VSFYYS1282 sequence very similar to the βA strand of SEMA4C and CI-MPR (731VGYYYS736 and 2349VSYKYS2355, respectively). Immunoprecipitation using wild-type GFP-tagged IGF1R tail and mutants targeting the 1277VSFYYS1282 sequence confirmed that this is required for the SNX5 interaction (Figure 6H).
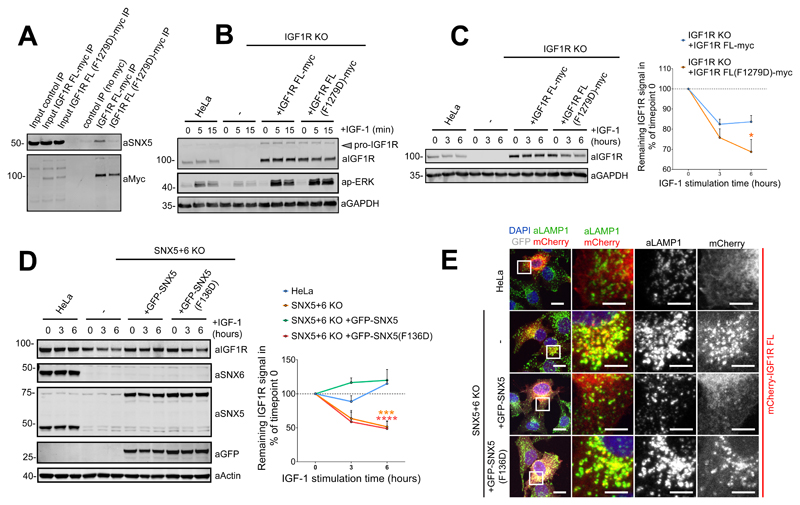
In contrast to the role of SNX5/SNX6 in endosome-to-TGN transport of the CI-MPR, the role of these SNX-BARs in sorting of the IGF1R is distinct24. Following ligand activation at the cell surface, the IGF1R undergoes internalisation and enters the endosomal network for its SNX5/SNX6-dependent recycling to the cell surface. Thus, in SNX5 and SNX6 double knockout cells the internalised IGF1R fails to engage the recycling itinerary and instead enters the lysosomal degradative pathway24. To extend this analysis into its functional role during endosome-to-plasma membrane recycling we introduced the F1279D mutant into full-length IGF1R and confirmed that this displayed a clear loss in endogenous SNX5 binding (Figure 7A). In an IGF1R knockout HeLa cell line we introduced wild-type IGF1R or IGF1R(F1279D). Importantly, the mutation did not affect receptor function as defined by ERK/MAPK activation upon IGF-1 stimulation (Figure 7B). Consistent with the 1277VSFYYS1282 sequence being a sorting motif, long term IGF-1 stimulation of IGF1R(F1279D) resulted in mis-sorting into the lysosomal degradative pathway (Figure 7C). In addition, IGF1R was degraded in SNX5 and SNX6 double knockout cells, and re-expression of SNX5, but not the SNX5(F136D) mutant, rescued IGF1R degradative fate and promoted cell surface recycling (Figure 7D and 7E). These data establish that SNX5 (as well as SNX6/SNX32) engage the cytosolic tail of cargoes through recognition of a ‘promiscuous’ β-hairpin structure.
Figure 7. Interaction between the IGF1R cytosolic tail and the SNX5 PX domain hydrophobic groove is required for the endosome-to-plasma membrane recycling of the receptor.
(A) Co-immunoprecipitation of myc-tagged full length IGF1R and IGF1R(F1279D) in HEK293T cells. Representative blot of three independent myc-IPs. (B) HeLa cells, HeLa IGF1R KO line un-transduced, or transduced with full length IGF1R or IGF1R(F1279D), serum starved and treated with 10 nM IGF-1 for indicated periods. Level of IGF1R and p-ERK analyzed by immunoblotting. Representative blot of three independent experiments. (C) HeLa cells, IGF1R KO line, un-transfected or transfected with full length IGF1R or IGF1R(F1279D) serum starved and treated with 10 nM IGF-1 for indicated periods and level of IGF1R analyzed by quantitative fluorescence-based western blotting. Quantification of IGF1R band intensities measured from n=3 independent experiments using Odyssey software. Band intensities normalized to GAPDH expression, presented as average fraction of the IGF1R signal relative to time point 0. IGF1R(F1279D) levels compared with the levels of IGF1R using two-way ANOVA and Sidak test. *P<0.05 (P values for 3h: P=0.4742, P values for 6h: P=0.0219). (D) HeLa SNX5+SNX6 KO clonal line un-transduced or transduced with GFP-SNX5 or GFP-SNX5(F136D) serum starved and treated with 10 nM IGF-1 for indicated periods and level of endogenous IGF1R analyzed by quantitative fluorescence-based western blotting. Quantification of endogenous IGF1R band intensities measured from n=3 independent experiments using Odyssey software. Band intensities normalized to β-actin expression and expressed as relative fraction of the IGF1R signal to timepoint 0. Levels of IGF1R in different cell lines were compared with levels of IGF1R in HeLa control using two-way ANOVA and Dunnett’s test. ***P<0.001, ****P<0.0001 (P values for 3h KO: P=0.1418, 3h KO+GFP-SNX5: P=0.0919, 3h KO+GFP-SNX5(F136D) P=0.0663, 6h KO: P=0.0001, 6h KO+GFP-SNX5: P=0.9622, 6h KO+GFP-SNX5(F136D) P=<0.0001). (E) Distribution of mCherry-IGF1R in HeLa SNX5+SNX6 KO clonal line un-transduced or transduced with GFP-SNX5 or GFP-SNX5(F136D). Cells were serum starved and treated with 10 nM IGF-1 for 2 hours, prior to fixation and immunostaining. (C,D) Circle data points, mean; error bars, SEM. Representative image of three independent experiments. Unprocessed original scans of immunoblots: Supplementary Figure 3. Statistics source data: Supplementary Table 4.
Numerous cargoes contain a ФxΩxФ(x)nФ motif necessary for SNX5 mediated endosomal sorting
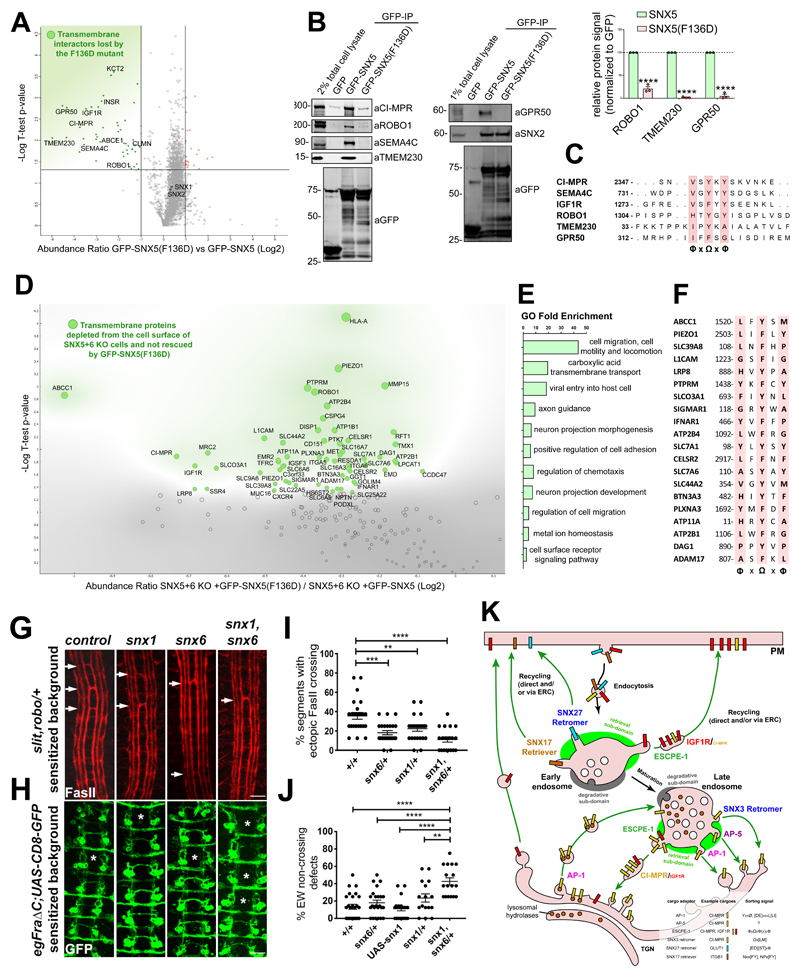
Next, we considered whether other cargoes utilise the same mechanism in binding to SNX5/SNX6 to mediate their endosomal sorting. To analyse this we designed a comparative proteomic screen that involved transient transfection of either GFP, GFP-SNX5 or GFP-SNX5(F136D) in HEK293T cells followed by GFP-nanotrap immunoprecipitation. Interacting proteins were identified by LC-MS/MS and enrichment quantified through isobaric tandem mass tagging (TMT) (Supp. Figure 2A). Compared with the wild-type SNX5 interactome, this identified 48 proteins significantly lost in the SNX5(F136D) interactome, several of which were transmembrane proteins (Figure 8A, Supp. Figure 2A and 2B). Of the transmembrane proteins, we selected for further validation: ROBO1 - a receptor in axon guidance and neuronal precursor cell migration33; TMEM230 - a protein linked to Parkinson disease34; and GPR50 - a regulator of melatonin and TGFβ signalling35. Immunoprecipitation of GFP-tagged SNX5 and the F136D mutant validated the Phe136-dependent association to SNX5 (Figure 7B). Moreover, alignment of their cytosolic tails with CI-MPR, SEMA4C and IGF1R revealed the presence of 1309HTYGY1313, 40IPYKA44 and 317IFFSG321 sequences in ROBO1, TMEM230 and GPR50 respectively, which conform with a general ФxΩxФ consensus motif for the βA strand (Ф = hydrophobic and Ω = aromatic side-chains) (Figure 8C).
Figure 8. Identification of a ФxΩxФ consensus motif for SNX5-mediated cargo recruitment.
(A) Analysis of comparative interactome of wild-type SNX5 vs SNX5(F136D) mutant across n=3 independent experiments using One-sample t-test and Benjamini-Hochberg FDR. (B) Co-immunoprecipitation of GFP-tagged SNX5 and SNX5(F136D) transiently transfected in HEK293T cells and summary of relative binding. Band intensities measured from n=3 independent experiments using Odyssey software. Band intensities normalized to GFP expression and presented as the relative fraction of the GFP-SNX5 control. Bars, mean; error bars, SEM; circles represent individual data points. Unprocessed scans of immunoblots: Supplementary Figure 3. (C) Alignment of the cytosolic tail of SNX5 cargoes identified through comparative interactome of wild-type SNX5 vs SNX5(F136D) mutant. (D) Comparative proteomic analysis of transmembrane proteins that rely on the ability to engage the SNX5 hydrophobic groove for their plasma-membrane localisation. Analysis was performed across n=3 independent experiments using One-sample t-test and Benjamini-Hochberg FDR. (E) List of most represented GO of the SNX5 cargoes, established by comparative surfaceosome analysis. (F) Alignment of the cytosolic tail of the SNX5 cargoes that require the SNX5 hydrophobic groove for their endosomal retrieval and recycling as established by comparative surfaceosome analysis. (G) Drosophila Snx1 and Snx6 promote axon growth across the midline. Representative images of stage 17 Drosophila embryos stained with anti-fasciclin II (FasII) antibodies to reveal a subset of ipsilaterally projecting interneurons in the CNS. All embryos are heterozygous for mutations in slit and robo. Arrows indicate segments in which axon bundles are abnormally crossing the midline. In embryos that are also heterozygous for either snx1, snx6 or both snx1 and snx6 the percentage of segments showing ectopic midline crossing is reduced compared to control embryos. (H) Representative images of stage 16 embryos stained with anti-GFP to reveal the Eagle subset of commissural interneurons, comprised of Eg axons, which cross the midline in the anterior commissure and Eg axons, which cross in the posterior commissure. Asterisks (*) indicate segments where the EW axons have failed to cross the midline. All embryos selectively express the Fra∆C transgene, which results in the failure of a portion of EW axons to cross the midline. In embryos heterozygous for mutations in both snx1 and snx6 there is a significant reduction in the percentage of EW axons that cross the midline. (I) Quantification of ectopic midline crossing in the indicated genotypes; n=number embryos; wild-type control (n=26), snx6/+ (n=26), snx1/+ (n=24), snx1 (n=23), snx6/+ (n=23). 11 segments were scored in each embryo. Statistical significance was determined using one-way ANOVA (followed by Tukey’s test), **P<0.01, ***P<0.001, ****P<0.0001. (J) Quantification of failed midline crossing in the indicated genotypes; n=number embryos; wild-type control (n=26), snx6/+ (n=24), snx1/+ (n=16), snx1 (n=17), snx6/+ (n=17) and UASSnx1 (n=22). The eight abdominal segments were scored in each embryo. Statistical significance was determined using one-way ANOVA (followed by Tukey’s test), **P<0.01, ***P<0.001, ****P<0.0001. In (I) and (J) each embryo (in which segments were quantified) was considered an independent trial. Scale bar in (G) and (H) 10 microns. (K) A model of the role of the ESCPE-1 complex, which consists of heterodimeric combinations of either SNX5 or SNX6 dimerised to either SNX1 or SNX2, in retrieving and recycling transmembrane cargo protein on the cytosolic facing surface of endosomes. The cartoon also represents the other machineries involved in the retrograde transport of cargoes to the TGN or in their recycling to the plasma membrane. Known sorting motifs within the cytosolic domain of the prototypical cargoes are listed in the figure legend. (H, J) Bars, mean; error bars, SEM; circles represent individual data points. Statistics source data: Supplementary Table 4. Proteomic datasets: Supplementary Table 3.
A limitation of the interactome-based analysis is the reliance on sufficient interacting protein being retained through the immunoprecipitation methodology for identification and quantification by LC-MS/MS, key determinants of which are the abundancy of the interacting protein and its SNX5 binding affinity. To expand the cargoes and identify low abundance and/or low affinity cargos, we designed a functionally-orientated proteomic analysis to quantify how the altered cell surface proteome in SNX5+SNX6 knockout cells was rescued by wild-type SNX5 or SNX5(F136D). SNX5+SNX6 knockout cells, or knockout cells transduced with wild-type SNX5 or SNX5(F136D) were subjected to surface biotinylation and TMT-labelling to quantify the proteomic data (Supp Figure 2C). Of the transmembrane proteins that displayed decreased levels in the SNX5+SNX6 knockout cells compared to the cells rescued with SNX5, 61 were significantly depleted from the plasma membrane of cells expressing SNX5(F136D) compared to the wild-type (Figure 8D and Supp Figure 2D). This is consistent with these proteins requiring the SNX5 hydrophobic groove for their endosomal escape and recycling from the lysosomal degradative fate. A Panther gene ontology analysis revealed enrichment for proteins involved in ‘axonal guidance’, ‘neuronal morphogenesis’ (e.g. L1CAM, ROBO1, SEMA4C, PLXNA3, CELSR2) and ‘cell migration’ (e.g. ITGA6, ADAM17) (Figure 7E), and importantly, sequence analysis revealed that 20 proteins contain a putative ФxΩxФ motif in their cytosolic tails (Figure 7F).
In vivo studies establish a role for Drosophila snx-6 in axonal guidance
Finally, we considered the evolutionary conserved helix-turn-helix extension in the PX domain (Figure 1B) and the aforementioned gene ontology analysis through examining axon guidance in the Drosophila embryonic central nervous system (CNS). We initially tested whether loss-of-function mutations in either snx1 or snx6 result in defects in axon guidance at the midline by examining late stage embryos with markers to label all axons (HRP) or subsets of ipsilaterally projecting interneurons (Fasciclin II-FasII) (Drosophila encode a single SNX5/SNX6 ortholog, snx6). Neither snx1 nor snx6 zygotic mutants resulted in dramatic defects in this coarse-level analysis; however, Snx1 and Snx6 are maternally deposited (https://flybase.org/) and this could potentially prevent the detection of phenotypes36. To more carefully analyze roles for Snx1 and Snx6 proteins, we turned to sensitized genetic backgrounds. First, we analyzed reducing snx1 and snx6 genes in embryos heterozygous for mutations in slit, the major midline repellant, and its neuronal receptor robo37 (Figure 8G and 8I). In this background, a small percentage of FasII positive axons abnormally cross the midline and mutations in other genes involved in midline guidance can modify this phenotype to reveal roles in promoting or inhibiting midline crossing38. Removing one copy of either snx1 or snx6 significantly attenuated midline crossing defects in this background, with simultaneous reduction of both snx1 and snx6 leading to a greater reduction in ectopic crossing (Figure 8G and 8I). Next, we made use of a second genetic background where normal midline crossing of commissural axons is partially disrupted. In this case, expression of a truncated Frazzled receptor (Fra∆C) in a subset of commissural axons resulted in the failure of a small percentage of axons to cross the midline. Like in the slit, robo background, mutations in other genes involved in midline guidance can modify this phenotype to reveal roles in either promoting or inhibiting midline crossing39. For embryos in this background that are heterozygous for either snx1 or snx6 midline crossing defects were not significantly altered, while simultaneous reduction of both snx1 and snx6 led to a striking enhancement in the percentage of axons that fail to cross the midline (Figure 8H and 8J). Our preliminary genetic analysis is suggestive of a role of these snx genes in promoting the normal axonal growth across the midline.
Discussion
Through formation of heterodimeric assemblies with SNX1 or SNX2, the BAR domain-containing SNX5 and SNX6 drive the biogenesis of tubular profiles and tubulo-vesicular transport carriers19,22. Here, we establish that a conserved hydrophobic groove in the PX domains of SNX5 and SNX6 (and presumably the related neuronal SNX32) associates with a ФxΩxФ(x)nФ motif present in the cytosolic tails of numerous integral proteins (Ф, hydrophobic; Ω, aromatic; x, any amino acid; n, variable linker region). Our study therefore establishes that SNX1/SNX2:SNX5/SNX6 heterodimers co-ordinate sequence-dependent cargo recognition with the biogenesis of tubulo-vesicular transport carriers for cargo export from the endosomal network.
The degradative capacity of lysosomes relies on the delivery of over 60 acid hydrolases from the biosynthetic pathway by means of membrane transport40. Their lysosomal delivery requires association with the CI-MPR at the TGN30. Transport of the CI-MPR:hydrolase complex from the TGN is mediated by clathrin-coated transport vesicles30. Upon fusion with the endosome, exposure of the CI-MPR:hydrolase complex to the acidic environment leads to hydrolase dissociation, which move into the lysosome and become enzymatically active upon endosome-lysosome fusion41. For continued hydrolase delivery the unbound CI-MPR is sorted from the endosome into tubulo-vesicular transport carriers for transport back to the TGN42,43. The details of this step in CI-MPR sorting have remained elusive44. A number of molecules have been implicated, including TIP47 and the Rab9 GTPase45, PACS-146, AP-147, AP-548 and the Retromer complex44, but for many their precise role remains controversial. Our structural identification and validation of the sorting motif in the CI-MPR and its recognition by the SNX5 and SNX6 components of the SNX1/SNX2:SNX5/SNX6 membrane tubulating complex has provided a detailed mechanistic framework on which to elucidate the early sorting steps of this important pathway.
How this insight interfaces with existing models of CI-MPR sorting, most notably with regard to Retromer, remains controversial44. Identified in yeast as a coat complex involved in endosome-to-Golgi transport8, the yeast Retromer is a stable pentamer of two sub-complexes: a Vps26, Vps35 and Vps29 heterotrimer, and a heterodimer of SNX-BAR proteins Vps5 and Vps17. In higher metazoans, the VPS26:VPS35:VPS29 heterotrimer9,10 does not form a stable pentameric assembly with the corresponding SNX1/SNX2:SNX5/SNX6 heterodimer (SNX1 and SNX2 being Vps5 orthologs, with SNX5 and SNX6 being Vps17 orthologs)19,20. Moreover, the human VPS26:VPS35:VPS29 heterotrimer can function independently of the SNX1/SNX2:SNX5/SNX6 heterodimer in cargo sorting23,24, a conclusion recently extended in vivo through the endosomal sorting of planar cell polarity proteins in the Drosophila wing49. Whilst it cannot be excluded that under certain specific condition(s) a metazoan yeast-like pentameric Retromer complex may assemble, the divergence in the association and functional interaction between the two complexes would argue that such a pentameric complex may form only a minor functional population. In metazoans, the divergence away from the yeast pentameric complex has led to a refined nomenclature where the VPS26:VPS35:VPS29 heterotrimer is referred to as ‘Retromer’50.
The evolution of Retromer and the SNX1/SNX2:SNX5/SNX6 complex into separate functional complexes is paralleled by Retromer acquiring a number of interactions with proteins not found in yeast, including sorting nexin-27 (SNX27)13,14,51, the actin polymerising WASH complex52–54, and ANKRD5055,56. Equally, Retromer’s functional role has expanded to include the sorting of hundreds of cargoes to the cell surface2. Evolution in the diversity can now be extended to the SNX1/SNX2:SNX5/SNX6 heterodimer. The extended helix-loop-helix cargo-binding groove in SNX5 and SNX6 is absent from yeast Vps17 (Figure 1B). This mode of sequence-dependent cargo recognition is therefore not a feature shared with the yeast pentameric Retromer. However, the conservation of the hydrophobic groove in C. elegans, Drosophila melanogaster and Danio rerio (Figure 1B), indicates that an equivalent mode of sequence-dependent endosomal sorting is likely to exist across these organisms, a conclusion supported by our preliminary analysis of axonal guidance in Drosophila.
A number of studies have highlighted the mammalian Retromer in endosome-to-TGN transport of the CI-MPR44. Here, cargo selection is proposed to occur through recognition of the 2369WLM2371 motif at an interface between VPS26 and the non-BAR domain containing sorting nexin, SNX315. Although structural evidence remains to be established, both Retromer and SNX3 can mediate CI-MPR transport in carriers that depend on the Golgin GCC88 for tethering at the TGN57. In contrast SNX1 and SNX2 (and by extension SNX5 and SNX6) mediate CI-MPR trafficking in carriers dependent on the Golgin-245 tether, leading to the proposal of two independent pathways for CI-MPR sorting to the TGN: a SNX3-Retromer pathway57 and a separate SNX1/SNX2:SNX5/SNX6-dependent pathway23,24. In addition, AP-1 and AP-5 adaptors mediate endosome-to-TGN CI-MPR transport43,58,59 (Figure 8K). Disentangling the sequential versus concurrent relationship of these pathways will provide invaluable insight into the global organisation of endosome-to-TGN transport pathways.
For the SNX1/SNX2:SNX5/SNX6 coat complex, cargos that are exported from the endosomal network through engaging SNX5/SNX6 can be sorted to (at least) two distinct destinations: for IGF1R (and numerous other cargos) a steady-state cell surface enrichment and a TGN-enrichment for the CI-MPR23,24 (Figure 8K). What principles govern how recognition of similar motifs by the same sorting complex ultimately leads to cargo sorting to the plasma membrane versus the TGN? At present we have no direct experimental data to address this fundamental question. That said, von Zastrow and colleagues have documented that endosomal exit of two cargo proteins, β2-adrenergic receptor and Wntless, occurs through a shared tubular profile even though ultimately their steady-state enrichments are defined as the cell surface and TGN respectively60. While the mechanistic basis of these sorting events are distinct, sorting of β2-adrenergic receptor and Wntless is Retromer-dependent23,24, conceptually the central problem is the same. Similar to the mechanism suggested for the β2-adrenergic receptor and Wntless60, we believe that the relative enrichment of the two cargoes is an important element in their sorting itineraries. Upon activation, IGF1R enters the endosomal network from the cell surface whilst the majority of the CI-MPR undergoes continual rounds of TGN-to-endosome and endosome-to-TGN transport from maturing endosomes. The relative enrichment of the two cargoes therefore reflects the maturation status of the endosome: endocytosed IGF1R (and other cell surface cargoes) will be enriched in newly formed endosomes while CI-MPR will be enriched within the ‘late’ endosomes. SNX1/SNX2:SNX5/SNX6 promoted tubular exit across the endosomal maturation pathway will therefore display distinct cargo enrichments: early exit will favour IGF1R over CI-MPR, while later exit favours CI-MPR over IGF1R. If early exit promotes entry into cell surface recycling pathways (either directly or via the endocytic recycling compartment (ERC)) and late exit promotes entry into retrograde transport pathways then such a sorting ‘refinery’ would achieve differential sorting. Such sorting may not immediately result in the correct steady-state localisation for all cargoes, for example some IGF1R may reach the TGN, but the inter-connected nature of membrane trafficking pathways provides a route for cell surface delivery through re-secretion (Figure 8K). Indeed there is evidence for cell surface cargoes, such as β1-integrin, being recycled through the TGN61. The presence of such ‘sub-refineries’, including the TGN and the ERC, would provide further platforms for differential sorting.
Finally, the interaction between Chlamydia trachomatis effector IncE and SNX5/SNX6 serves to manipulate host mechanisms to promote pathogen survival and replication in the host’s intracellular environment62. The secreted IncE is associated with the cytosolic face of the C. trachomatis inclusion body from where it recruits SNX5 and SNX6 and overrides the normal endosome association of these SNX-BARs62. This leads to a defect in CI-MPR endosome-to-TGN transport26,27,28,62. Our structural description of CI-MPR binding to SNX5/SNX6 and its resemblance to that of IncE, establishes a molecular explanation for this phenotype. With the importance of CI-MPR recycling for hydrolase delivery and lysosomal degradative health, C. trachomatis may induce perturbation in this pathway in order to reduce the restrictive role of this organelle. Our analysis has identified a number of signalling receptors that require SNX5/SNX6 for their endosomal sorting, which are likely to provide insight into the significance of IncE perturbed SNX5/SNX6 trafficking. One way C. trachomatis secures survival of the host cell is by modulating the MEK-ERK and PI 3-kinase survival pathway downstream of EPHA2 receptor activation. This receptor is internalised with, and retained on the maturing inclusion from where it triggers long-lasting anti-apoptotic signaling63. It is tempting to speculate that the secreted IncE, by preventing the SNX5/SNX6 mediated transport of active signalling receptors away from the inclusion, may ensure retention of pro-survival signalling receptors during replication.
Overall, we have identified a mechanism through which endosome-associated SNX1/SNX2:SNX5/SNX6 coat complex coordinates sequence-dependent cargo recognition with membrane re-modelling to generate cargo-enriched tubulo-vesicular transport carriers. We have provided evidence that this mechanism is required for the export of numerous cargoes from the endosomal network for onward transport to the TGN and the cell surface. To highlight this function and to reflect that the mammalian VPS26:VPS35:VPS29 Retromer and the SNX1/SNX2:SNX5/SNX6 coat complex have functionally diversified, we propose to refer to the SNX1/SNX2:SNX5/SNX6 coat complex as the ‘Endosomal SNX-BAR sorting complex for promoting exit-1’ (ESCPE-1) (Figure 8K). We speculate that additional ESCPE complexes may function in sequence-mediated tubulo-vesicular based endosomal sorting.
Supplementary Material
Acknowledgements
Work was supported by the MRC (MR/L007363/1 and MR/P018807/1), the Wellcome Trust (104568/Z/14/2) and the Lister Institute of Preventive Medicine to P.J.C, and the Wellcome Trust PhD Studentship for the Dynamic Cell Biology programme (083474) to B.S. B.M.C. is supported by an NHMRC Senior Research Fellowship (APP1136021) and the National Health and Medical Research Council (NHMRC) (APP1099114), the Australian Research Council (ARC) (DP160101743) and Bright Focus Foundation (A2018627S), while G.J.B is supported by the National Institutes of Health grant (R35 NS097340). We thank the University of Queensland Remote Operation Crystallisation and X-ray (UQ ROCX) facility and the staff for their support with crystallisation experiments; the staff of the Australian Synchrotron for assistance with X-ray diffraction data collection; and the University of Bristol Wolfson Bioimaging Facility.
Footnotes
Author Contributions
Initial conceptualization, B.S., F.S., B.M.C. and P.J.C.; Evolution of conceptualization, B.S., B.P., S.W., F.S., G.J.B., B.M.C. and P.J.C.; Formal analysis, K.J.H.; Investigation, B.S., B.P., K.C., S.W., M.G., F.S. and K.J.H. Writing – Original Draft, B.S., B.M.C. and P.J.C.; Writing – Review and Editing, all authors; Funding Acquisition, F.S., G.J.B., B.M.C. and P.J.C.; Supervision, F.S., G.J.B., B.M.C. and P.J.C.
Competing Interests Statement
The authors declare no competing financial interests.
References
- 1.Goldenring JR. Recycling endosomes. Curr Opin Cell Biol. 2015;35:117–122. doi: 10.1016/j.ceb.2015.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cullen PJ, Steinberg F. To degrade or not to degrade: mechanisms and significance of endocytic recycling. Nat Rev Mol Cell Biol. 2018;19:679–696. doi: 10.1038/s41580-018-0053-7. [DOI] [PubMed] [Google Scholar]
- 3.Behnia R, Munro S. Organelle identity and the signposts for membrane traffic. Nature. 2005;438:597–604. doi: 10.1038/nature04397. [DOI] [PubMed] [Google Scholar]
- 4.McNally KE, Cullen PJ. Endosomal retrieval of cargo: Retromer is not alone. Trends Cell Biol. 2018;28:807–822. doi: 10.1016/j.tcb.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 5.Phillips-Krawczak CA, Singla A, Starokadomskyy P, Deng Z, Osborne DG, Li H, Dick CJ, Gomez TS, Koenecke M, Zhang JS, et al. COMMD1 is linked to the WASH complex and regulates endosomal trafficking of the copper transporter ATP7A. Mol Biol Cell. 2015;26:91–103. doi: 10.1091/mbc.E14-06-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartuzi P, Billadeau DD, Favier R, Rong S, Dekker D, Fedoseienko A, Fieten H, Wijers M, Levels JH, Huijkman N, et al. CCC- and WASH-mediated endosomal sorting of the LDLR is required for normal clearance of circulating LDL. Nat Commun. 2016;7 doi: 10.1038/ncomms10961. 10961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyer C, Zizioli D, Lausmann S, Eskelinen EL, Hamann J, Saftig P, von Figura K, Schu P. μ1A-adaptin-deficient mice: lethality, loss of AP-1 binding and rerouting of mannose 6-phosphate receptors. EMBO J. 2000;19:2193–2203. doi: 10.1093/emboj/19.10.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seaman MNJ, McCaffery JM, Emr SD. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J Cell Biol. 1998;142:665–681. doi: 10.1083/jcb.142.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seaman MN. Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J Cell Biol. 2004;165:111–122. doi: 10.1083/jcb.200312034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arighi CN, Hartnell LM, Aguilar RC, Haft CR, Bonifacino JS. Role of the mammalian retromer in sorting of the cation-independent mannose 6-phosphate receptor. J Cell Biol. 2004;165:123–133. doi: 10.1083/jcb.200312055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strochlic TI, Setty TG, Sitaram A, Burd CG. Grd19/Sns3p functions as a cargo-specific adapter for retromer-dependent endocytic recycling. J Cell Biol. 2007;177:115–125. doi: 10.1083/jcb.200609161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harterink M, Port F, Lorenowicz MJ, McGough IJ, Silhankova M, Betist MC, van Weering JRT, van Heesbeen RGHP, Middelkoop TC, Basler K, et al. A SNX3-dependent retromer pathway mediates retrograde transport of the Wnt sorting receptor Wntless and is required for Wnt secretion. Nat Cell Biol. 2011;13:914–923. doi: 10.1038/ncb2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Temkin P, Lauffer B, Jäger S, Cimermancic P, Krogan NJ, von Zastrow M. SNX27 mediates retromer tubule entry and endosome-to-plasma membrane trafficking of signalling receptors. Nat Cell Biol. 2011;13:715–721. doi: 10.1038/ncb2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steinberg F, Gallon M, Winfield M, Thomas EC, Bell AJ, Hessom KJ, Tavare JM, Cullen PJ. A global analysis of SNX27-retromer assembly and cargo specificity reveals a function in glucose and metal ion transport. Nat Cell Biol. 2013;15:461–471. doi: 10.1038/ncb2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lucas M, Gershlick DC, Vidaurrazaga A, Rojas AL, Bonifacino JS, Hierro A. Structural mechanism for cargo recognition by the retromer complex. Cell. 2016;167:1623–1635. doi: 10.1016/j.cell.2016.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McNally KE, Faulkner R, Steinberg F, Gallon M, Ghai R, Pim D, Langton P, Pearson N, Danson CM, Nägele H, et al. Retriever is a multiprotein complex for retromer-independent endosomal cargo recycling. Nat Cell Biol. 2017;19:1214–1225. doi: 10.1038/ncb3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purushothaman LK, Ungermann C. Cargo induces retromer-mediated membrane remodelling on membranes. Mol Biol Cell. 2018;29:2709–2719. doi: 10.1091/mbc.E18-06-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kovtun O, Leneva N, Bykov YS, Ariotti N, Teasdale RD, Schaffer M, Engel BD, Owen DJ, Briggs JAG, Collins BM. Structure of the membrane-assembled retromer coat determined by cryo-electron tomography. Nature. 2018;561:561–564. doi: 10.1038/s41586-018-0526-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carlton J, Bujny M, Peter BJ, Oorschot VM, Rutherford A, Mellor H, Klumperman J, McMahon HT, Cullen PJ. Sorting nexin-1 mediates tubular endosome-to-TGN transport through coincidence sensing of high-curvature membranes and 3-phosphoinositides. Curr Biol. 2004;14:1791–1800. doi: 10.1016/j.cub.2004.09.077. [DOI] [PubMed] [Google Scholar]
- 20.Wassmer T, Attar N, Bujny MV, Oakley J, Traer CJ, Cullen PJ. A loss-of-function screen reveals SNX5 and SNX6 as potential components of the mammalian retromer. J Cell Sci. 2007;120:45–54. doi: 10.1242/jcs.03302. [DOI] [PubMed] [Google Scholar]
- 21.Wassmer T, Attar N, Harterink M, van Weering JR, Traer CJ, Oakley J, Goud B, Stephens DJ, Verkade P, Korswagen HC, Cullen PJ. The retromer coat complex coordinates endosomal sorting and dynein-mediated transport with carrier recognition by the trans-Golgi network. Dev Cell. 2009;17:110–122. doi: 10.1016/j.devcel.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Weering JR, Sessions RB, Traer CJ, Kloer DP, Bhatia VK, Stamou D, Carlsson SR, Hurley JH, Cullen PJ. Molecular basis for SNX-BAR-mediated assembly of distinct endosomal sorting tubules. EMBO J. 2012;31:4466–4480. doi: 10.1038/emboj.2012.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simonetti B, Danson CM, Heesom KJ, Cullen PJ. Sequence-dependent cargo recognition by SNX-BARs mediates retromer-independent transport of CI-MPR. J Cell Biol. 2017;216:3695–3712. doi: 10.1083/jcb.201703015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kvainickas A, Jimenez-Orgaz A, Nägele H, Hu Z, Dengjel J, Steinberg F. Cargo-selective SNX-BAR proteins mediate retromer trimer independent retrograde transport. J Cell Biol. 2017;216:3677–3693. doi: 10.1083/jcb.201702137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teasdale RD, Loci D, Houghton F, Karlsson L, Gleeson PA. A large family of endosome-localised proteins related to sorting nexin 1. Biochem J. 2001;358:7–16. doi: 10.1042/0264-6021:3580007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elwell CA, Czudnochowski N, von Dollen J, Johnson JR, Nakagawa R, Mirrashidi K, Krogan NJ, Engel JN, Rosenberg OS. Chlamydia interfere with an interaction between the mannose-6-phosphate receptor and sorting nexins to counteract host restriction. Elife. 2017;6:e22709. doi: 10.7554/eLife.22709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paul B, Kim HS, Kerr MC, Huston WM, Teasdale RD, Collins BM. Structural basis for the hijacking of endosomal sorting nexin proteins by Chlamydia trachomatis. Elife. 2017;6:e22311. doi: 10.7554/eLife.22311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun Q, Yong X, Sun X, Yang F, Dai Z, Gong Y, Zhou L, Zhang X, Niu D, Dai L, et al. Structural and functional insights into sorting nexin 5/6 interaction with bacterial effector IncE. Signal Transduct Target Ther. 2017;2:17030. doi: 10.1038/sigtrans.2017.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seaman MN. Identification of a novel conserved sorting motif required for retromer-mediated endosome-to-TGN retrieval. J Cell Sci. 2007;120:2378–2389. doi: 10.1242/jcs.009654. [DOI] [PubMed] [Google Scholar]
- 30.Coutinho MF, Prata MJ, Alves S. Mannose-6-phosphate pathway: a review on its role in lysosomal function and dysfunction. Mol Genet Metab. 2012;105:542–550. doi: 10.1016/j.ymgme.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 31.Itai N, Shimazu T, Kimura T, Ibe I, Yamashita R, Kaburagi Y, Dohi T, Tonozuka T, Takao T, Nishikawa A. The phosphorylation of sorting nexin 5 at serine 226 regulates retrograde transport and macropinocytosis. PLoS One. 2018;13(11) doi: 10.1371/journal.pone.0207205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chandra M, Chin YK, Mas C, Feathers J, Paul B, Datta S, Chen KE, Jia X, Yang Z, Norwood SJ, Mohanty B, et al. Classification of the human phox homology (PX) domains based on their phosphoinositide binding specificities. Nat Commun. 2019 doi: 10.1038/s41467-019-09355-y. 1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blockus H, Chedotal A. Slit-Robo signaling. Development. 2016;143:3037–3044. doi: 10.1242/dev.132829. [DOI] [PubMed] [Google Scholar]
- 34.Deng HX, Shi Y, Yang Y, Ahmeti KB, Miller N, Huang C, Cheng L, Zhai H, Deng Sm, Nuytemans K, et al. Identification of TMEM230 mutations in familial Parkinson’s disease. Nat Genet. 2016;48:733–739. doi: 10.1038/ng.3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wojciech S, Ahmad R, Belaid-Choucair Z, Journe AS, Gallet S, Dam J, Daulat A, Ndiaye-Lobry D, Lahuna O, Karamitri A, et al. The orphan GPR50 receptor promotes constitutive TGFβ receptor signaling and protects against cancer development. Nat Commun. 2018;9 doi: 10.1038/s41467-018-03609-x. 1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fan X, Labrador JP, Hing H, Bashaw GJ. Slit stimulation recruits dock and pak to the Roundabout receptor and increases Rac activity to regulate axon repulsion at the CNS midline. Neuron. 2003;40:113–127. doi: 10.1016/s0896-6273(03)00591-9. [DOI] [PubMed] [Google Scholar]
- 37.Kidd T, Bland KS, Goodman CS. Slit is the midline repellent for the robo receptor in Drosophila. Cell. 1999;96:785–794. doi: 10.1016/s0092-8674(00)80589-9. [DOI] [PubMed] [Google Scholar]
- 38.Coleman HA, Labrador JP, Chance RK, Bashaw GJ. The Adam family metalloprotease Kuzbanian regulates the cleavage of the Roundabout receptor to control axon repulsion at the midline. Development. 2010;137:2417–2426. doi: 10.1242/dev.047993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hernandez-Fleming M, Rohrbach E, Bashaw GJ. Sema-1a reverse signaling promotes midline crossing in response to secreted Semaphorins. Cell Reports. 2017;18:174–184. doi: 10.1016/j.celrep.2016.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klumperman J, Raposo G. The complex ultrastrutcure of the endolysosomal system. Cold Spring Harb Perspect Biol. 2014;6:a016857. doi: 10.1101/cshperspect.a016857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bright NA, Davis LJ, Luzio JP. Endolysosomes are the principal intracellular sites of acid hydrolase activity. Curr Biol. 2016;26:2233–2245. doi: 10.1016/j.cub.2016.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mari M, Bujny MV, Zeuschner D, Geerts WJ, Griffith J, Petersen CM, Cullen PJ, Klumperman J, Geuze HJ. SNX1 defines an early endosomal recycling exit for sortilin and mannose 6-phosphate receptors. Traffic. 2008;9:380–393. doi: 10.1111/j.1600-0854.2007.00686.x. [DOI] [PubMed] [Google Scholar]
- 43.Johannes L, Wunder C. Retrograde transport: two (or more) roads diverged in an endosomal tree? Traffic. 2011;12:956–962. doi: 10.1111/j.1600-0854.2011.01200.x. [DOI] [PubMed] [Google Scholar]
- 44.Seaman MNJ. Retromer and the cation-independent mannose 6-phosphate receptor – Time for a trial separation? Traffic. 2018;19:150–152. doi: 10.1111/tra.12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carroll KS, Hanna J, Simon I, Krise J, Barbero P, Pfeffer SR. Role of Rab9 GTPase in facilitating receptor recruitment by TIP47. Science. 2001;292:1373–1376. doi: 10.1126/science.1056791. [DOI] [PubMed] [Google Scholar]
- 46.Wan L, Molloy SS, Thomas L, Liu G, Xiang Y, Rybak SL, Thomas G. PACS-1 defines a novel gene family of cytosolic sorting proteins required for trans-Golgi network localisation. Cell. 1998;94:205–216. doi: 10.1016/s0092-8674(00)81420-8. [DOI] [PubMed] [Google Scholar]
- 47.Robinson MS, Sahlender DA, Foster SD. Rapid inactivation of proteins by rapamycin-induced rerouting to mitochondria. Dev Cell. 2010;18:324–331. doi: 10.1016/j.devcel.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hirst J, Borner GH, Edgar J, Hein MY, Mann M, Buchholz F, Antrobus R, Robinson MS. Interaction between AP-5 and the hereditary spastic paraplegia proteins SPG11 and SPG15. Mol Biol Cell. 2013;24:2558–2569. doi: 10.1091/mbc.E13-03-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strutt, Langton PF, Pearson N, McMillan KJ, Strutt D, Cullen PJ. Retromer controls planar polarity protein levels and asymmetric localization at intercellular junctions. Current Biology. 2019:484–491. doi: 10.1016/j.cub.2018.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burd C, Cullen PJ. Retromer: a master conductor of endosome sorting. Cold Spring Harb Perspect Biol. 2014;6:a016774.44. doi: 10.1101/cshperspect.a016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gallon M, Clairfeuille T, Steinberg F, Mas C, Ghai R, Sessions RB, Teasdale RD, Collins BM, Cullen PJ. A unique PDZ domain and arrestin-like fold interaction reveals mechanistic details of endocytic recycling by SNX27-retromer. Proc Natl Acad Sci USA. 2014;111:E3604–3613. doi: 10.1073/pnas.1410552111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gomez TS, Billadeau DD. A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev Cell. 2009;17:699–711. doi: 10.1016/j.devcel.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harbour ME, Breusegem SY, Seaman MN. Recruitment of the endosomal WASH complex is mediated bu the extended ‘tail’ of Fam21 binding to the retromer protein Vps35. Biochem J. 2012;442:209–220. doi: 10.1042/BJ20111761. [DOI] [PubMed] [Google Scholar]
- 54.Jia D, Gomez TS, Billadeau DD, Rosen MK. Multiple repeat elements within the FAM21 tail link the WASH actin regulatory complex to the retromer. Mol Biol Cell. 2012;23:2352–2361. doi: 10.1091/mbc.E11-12-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McGough IJ, Steinberg F, Gallon M, Yatsu A, Ohbayashi N, Heesom KJ, Fukuda M, Cullen PJ. Identification of molecular heterogeneity in SNX27-retromer-mediated endosome-to-plasma membrane recycling. J Cell Sci. 2014;127:4940–4953. doi: 10.1242/jcs.156299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kvainickas A, Orgaz AJ, Nägele H, Diedrich B, Heesom KJ, Dengjel J, Cullen PJ, Steinberg F. Retromer- and WASH-dependent sorting of nutrient transporters requires a multivalent interaction network with ANKRD50. J Cell Sci. 2017;130:382–395. doi: 10.1242/jcs.196758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cui Y, Carosi JM, Yang Z, Ariotti N, Kerr MC, Parton RG, Sargeant TJ, Teasdale RD. Retromer has a selective function in cargo sorting via endosome transport carriers. J Cell Biol. 2019;218:615–631. doi: 10.1083/jcb.201806153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hirst J, Borner GH, Antrobus R, Peden AA, Hodson NA, Sahlender DA, Robinson MS. Distinct and overlapping roles for AP-1 and GGAs revealed by the ‘knocksideways’ system. Curr Biol. 2012;22:1711–1716. doi: 10.1016/j.cub.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hirst J, Itzhak DN, Antrobus R, Borner GHH, Robinson MS. Role of the AP-5 adaptor protein complex in late endosome-to-Golgi retrieval. PLoS Biol. 2018;16:e2004411. doi: 10.1371/journal.pbio.2004411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Varandas KC, Irannejad R, von Zastrow M. Retromer endosome exit domains serve multiple traffickingdestinations and regulate local G protein activation by GPCRs. Curr Biol. 2016;26:3129–3142. doi: 10.1016/j.cub.2016.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shafaq-Zadah M, Gomes-Santos CS, Bardin S, Maiuri P, Maurin M, Iranzo J, Gautreau A, Lamaze C, Caswell P, Goud B, Johannes L. Presistent cell migration and adhesion rely on retrograde transport of β1-integrin. Nat Cell Biol. 2016;18:54–64. doi: 10.1038/ncb3287. [DOI] [PubMed] [Google Scholar]
- 62.Mirrashidi KM, Elwell CA, Verschueren E, Johnson JR, Frando A, Von Dollen J, Rosenberg O, Gulbahce N, Jang G, Johnson T, et al. Global mapping of the Inc-human interactome reveals that retromer restricts Chlamydia infection. Cell Host Microbe. 2015;18:109–121. doi: 10.1016/j.chom.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Subbarayal P, Karunakaran K, Winkler AC, Rother M, Gonzalez E, Meyer TF, Rudel T. EphrinA2 receptor (EphA2) is an invasion and intracellular signalling receptor for Chlamydia trachomatis. PLoS Pathog. 2015;11:e1004846. doi: 10.1371/journal.ppat.1004846. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.