Abstract
Chinese indigenous chickens have experienced strong selective pressure in genes or genomic regions controlling critical agricultural traits. To exploit the genetic features that may be useful in agriculture and are caused by artificial selection, we performed whole-genome sequencing of six Pengxian Yellow Chickens and downloaded the sequence data of five Red Jungle fowls from the NCBI. Through selective sweep analysis, we detected several regions with strong selection signals, containing 497 protein-coding genes. These genes were involved in developmental processes, metabolic processes, the response to external stimuli and other biological processes including digestion (ABCG5, ABCG8 and ADRB1), muscle development and growth (SMPD3, NELL1, and BICC1) and reduced immune function (CD86 and MTA3). Interestingly, we identified several genes with extremely strong selection signals associated with the loss of visual capability of domestic chickens relative to their wild ancestors. Amongst them, we propose that CTNND2 is involved in the evolutionary changes of domestic chickens toward reduced visual ability through the diopter system. VAT1 was also likely to contribute to these processes through its regulation of mitochondrial fusion. In summary, these data illustrate the patterns of genetic changes in Pengxian yellow chickens during domestication and provide valuable genetic resources that facilitate the utilization of chickens in agricultural production.
Keywords: Pengxian Yellow Chicken, Whole genome, Resequencing, Selection, SNP
Introduction
At least 3500 years ago, domestic fowls were ubiquitous in China and poultry breeds were raised by locals for exhibitions and food, including eggs and meat production (Wu 2001). The Pengxian Yellow Chicken (PYC) is a quality meat-type breed with yellow feather, best known for the palatability of their meat. They are mainly raised in the foothills of the northwest region of the Chengdu plain, particularly in and around Peng county, Sichuan province, China, which are particularly popular for food in certain areas, thanks in part to their promotion by local authorities (Wang et al. 2016b).
Although few reports regarding PYC have been published, recent studies indicate that PYC genetically resembles domesticated chickens to a greater degree than Red Jungle fowl (RJF; Gallus gallus) and thus may have undergone artificial selective breeding, leaving detectable signatures within the PYC genome (Li et al. 2017). The genetic features caused by selective breeding strongly correlate with some important economical traits that are of particular value to the poultry industry, including eggs and meat yields. We, therefore, considered that, compared to their wild ancestor RJF, PYC, like other commercial animals, should exhibit characteristics that are ideal for production purposes, including egg production and growth performance (Table 1). These differences in phenotypes and chemical composition are associated with genomic differences, suggesting the validity of employing whole-genome resequencing for analysis (Huang et al. 2009b).
Table 1.
Egg production and growth performances between Red Jungle fowl and Pengxian Yellow Chickens
| Traits | AFE (days) | BWFE (g) | FEW (g) | EP1Y | EP43w | 98 d wt (g) | 120 d wt (g) | 140 d wt (g) |
|---|---|---|---|---|---|---|---|---|
| RJF | 298.2 ± 38.0 | 887.2 ± 42.4 | 33.4 ± 2.6 | 28.1 ± 16.1 | – | 538.8 | 614 | 764.2 |
| PYC | 163.2 ± 3.2 | 2292 ± 36.8 | 40.2 ± 0.9 | – | 98.7 ± 2.6 | 1352.4 | 1635 | 2465.5 |
AFE age at first egg, BWFE body weight at first egg, FEW first egg weight, EP1Y total egg production from year 1, EP43 W total egg production from 43 weeks, 98 d wt body weight at 98 days, 120 d wt body weight at 120 days, 140 d wt body weight at 140 days
In recent years, genomic variants and selective sweeps have been identified through resequencing chickens. For example, Rubin et al. re-sequenced the genomes of commercial chickens (broiler and layer) and RJF, and reported more than 7,000,000 single nucleotide polymorphisms (SNPs), almost 1300 deletions, and a number of putative selective sweeps, several of which were associated with growth, reproduction, appetite and metabolic regulation (Rubin et al. 2010). Wang et al. assembled a de novo genome of a Tibetan chicken and re-sequenced the whole genomes of 32 additional chickens. Further analyses revealed that several candidate genes that regulate calcium (Ca2+) signaling are involved in the adaptation to a hypoxia environment, suggesting a potential genetic mechanism underlying high altitude adaptation in Tibetan chickens (Wang et al. 2015). Subsequently, a genome-wide analysis of domestic chickens and Red Jungle fowl genomes showed significant enrichment for positively selected genes involved in the development of vision, revealing positive selection, as opposed to relaxation of purifying selection, contributing to the evolution of vision in domestic chickens (Wang et al. 2016a).
To identify the selection signatures of PYCs resulting from domestication, we performed whole-genome resequencing of 6 PYCs (~ 104.20 Gb in total; ~ 16.80 × coverage per chicken). Using obtained data together with five publicly available genomes of RJF downloaded from the NCBI, we performed a comprehensive analysis of genetic variants and sought to identify genomic regions under selection in PYC. This could provide genetic information for the use of breeding and shed light on the evolutionary history of the chickens.
Materials and methods
Materials
A total of six 150-day male pureblood PYCs were randomly selected from the same generation population in Pengxian yellow chicken conservation farms (Chengdu, Sichuan, China). Genomic DNA was extracted from blood samples and sequenced on an Illumina Hiseq 2500 platform to an average of 16.8-fold coverage depth for each individual. For downstream analysis, we downloaded previously published genomic data of five wild red jungle fowls available from the NCBI (GenBank accession number PRJNA241474).
Sequence quality and filtering
To exclude bias from low-quality paired reads that arise from the process of base-calling or adapter contamination, reads with any of the following features were filtered: (1) ≥ 10% of unidentified nucleotides; (2) > 10 nt aligned to the adapter; (3) > 50% of bases with Phred quality < 33; and (4) putative PCR duplicates generated during library construction. The remaining filtered data were used for subsequent analysis.
Read mapping
We mapped high-quality data per individual to the reference chicken genome using Burrows–Wheeler Aligner (BWA) software (version 0.7.8) with the parameters “mem -t 10 -k 32”. SAM tools (version 0.1.19) were used to convert sequence alignments (SAM) and binary versions of the SAM (BAM) formats, sorting, and indexing alignment. The alignments were further improved by several steps: (1) filtering the alignment reads with mismatches ≥ 5 and mapping quality = 0; (2) alignment results were corrected using Picard (version 1.96; http://broadinstitute.github.io/picard/) with two core commands. The “Add or Replace Read Groups” command was used to replace all read groups in the INPUT file, and new read groups were assigned to all reads in the OUTPUT BAM. The “Fix Mate Information” command was used to ensure that all mate pair information was synchronized between each read and its mate pair; (3) PCR duplications were removed when multiple read pairs had identical external coordinates. Only pairs with the highest map quality were retained.
Single-nucleotide polymorphism (SNP) calling and variant annotation
After mapping, we performed SNP calling for each group (6 PYC and 5 RJF, respectively) using a Bayesian approach as implemented in the package SAM tools. Individual SNPs were detected by the command “mpileup” with the parameters “-C 50 -D -S -m 2 -F 0.002 -d 1000” (“-C 50” as recommended parameters, “-D” and “-S” as default parameters, “-m 2”, “-F 0.002” and “-d 1000” as required parameters). To eliminate the SNP calling errors caused by incorrect mapping and InDels, only high-quality SNPs were used, namely those with a coverage depth ≥ 4 and ≤ 200, RMS mapping quality ≥ 20, distance between adjacent SNPs ≥ 5 bp, with no InDels within a 3 bp window and missing ratios within each group ≥ 50%. SNPs matching these criteria were maintained for subsequent analysis. ANNOVAR (version May 20, 2013) was used to annotate synonymous and non-synonymous SNPs.
The distribution of the SNPs within the genomic region was summarized, and the functional enrichment analysis in genes identified for each candidate region under selection was also performed using the Enrichr web server.
Genome-wide selective sweeping
The sequence diversity statistic (θπ) of each population and the population differentiation statistic (Fst) between PYC and RJF were quantified using a sliding window using a 40-kb window with a 10-kb step across the whole genome. We reasoned that 40-kb was the most appropriate window size and windows ≤ 20 SNPs were filtered. In each window, we calculated the aθπ values for each population and representative Fst values of the two populations. After calculation of the Fst values and log2 (θπ(RJF)/θπ(PYC)) of each window, we adopted an outlier method with significantly low log2 (θπ(RJF)/θπ(PYC); 5% right tail) and significantly high Fst values (5% left tail) based on genome-wide empirical distributions to identify regions with strong selective sweep signals (Akey et al. 2002).
An outlier method based on genome-wide empirical distributions of the statistics was employed to detect genomic divergence between the two populations. Windows with top 5% Fst and log2 (θπ(RJF)/θπ(PYC)) were identified as candidate regions that could account for biological differences between the two populations. We annotated genes in these regions to the reference chicken genome and submitted all data to Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases for enrichment analyses on the Enrichr web server. We searched for genes significantly overrepresented in GO biological processes (GO-BP), molecular functions (GO-MF) and KEGG pathways. Known genes in the whole genome were appointed as backgrounds for significance testing. We simultaneously considered p values, adjusted p values of false-discovery rates (FDR) corrected for binomial distribution probability approaches (Chen et al. 2013) as the criteria for significance tests of gene enrichment, with significant levels of p ≤ 0.05 and adjusted p ≤ 0.2.
Results and discussion
Sequencing, SNP calling, and annotations
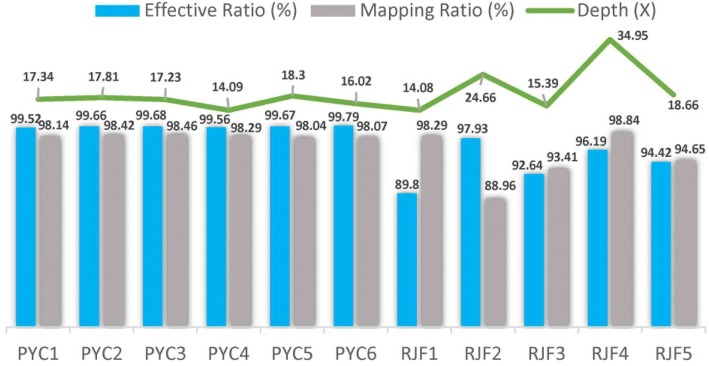
A total of 6 PYCs genomes were sequenced, which generated 104.20 Gb of paired-end DNA sequences, and data were deposited in the SRA database (accession number SRP067615). Within these data, 103.83 Gb (99.65%) of high-quality paired-end reads remained after filtration and were used for mapping to assemble the reference chicken genome (Galgal 4.78). Approximately, 98.24% of reads in each individual were mapped to the reference with an average depth of 16.80-fold using the Burrow–Wheeler Aligner (BWA) tool. To accurately detect genomic footprints, the publicly available genomic data of 5 RJFs were downloaded from the NCBI database. These downloaded datasets had an average depth of 21.55-fold which mapped to ~ 94.83% of the reads of the assembled reference genome (Fig. 1).
Fig. 1.
Resequencing coverage and mean depth. Bars charts represent both effective and mapping ratios, respectively. Line charts represent the sequencing depths of each sample
The SNP calling for the populations was performed using SAM tools and identified 12.97 Mb of SNPs after variant filtration processes. Of these, PYC and RJF held 6.25 Mb and 6.72 Mb of SNPs, respectively. For the SNPs of PYC, 5.79 Mb was previously annotated in dbSNP and 0.46 Mb was novel, whilst 6.08 Mb of the SNPs of RJF overlapped with dbSNP and 0.64 Mb was novel (Fig. 2). The density of SNPs was determined as ~ 1 per 180 bp, which was sufficient to cover the candidate genomic region for downstream analysis. The qualities of the SNPs were further assessed by calculating the transition-to-transversion ratio (Ti/Tv) for each SNP, regarded as an indicator of potential sequencing errors. The results for this ratio (PYC: 2.465, RJF: 2.447) match previously observed values of ~ 2.45 (Aslam et al. 2012), suggesting that the identified SNPs detected in this study were relatively accurate.
Fig. 2.
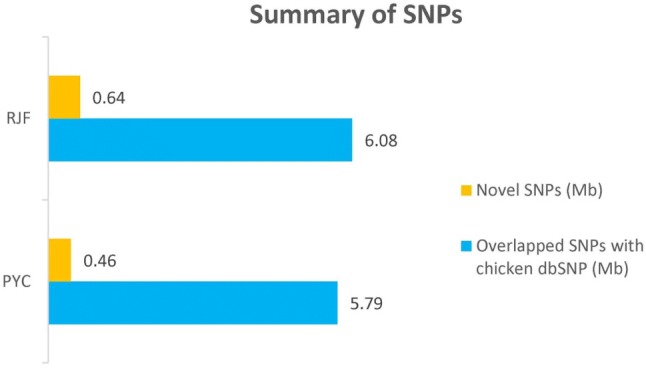
Comparison of the identified SNPs in PYC populations and RJF with the public database of chicken variants. The SNPs identified from the genomes of each population were merged into non-redundant sets
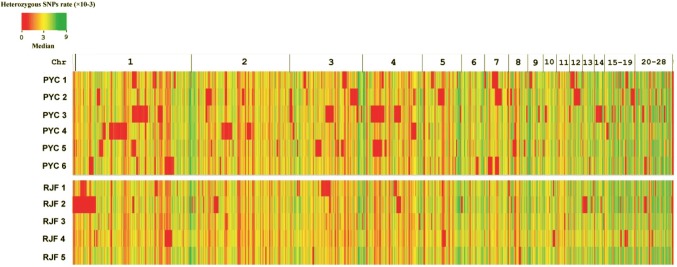
The rates of heterozygosity of the SNPs of each individual were calculated in each non-overlapping 500-kb window across the whole genome to permit estimates of the levels of heterozygosity of the two breeds (Fig. 3). We anticipated that the wild RJF had higher rates of heterozygosity of SNPs than PYC since the indigenous breed of chickens in the latter group that we sequenced had undergone a degree of artificial selection. Indeed, the average ratios of homozygous to heterozygous SNPs were 1:1.28 (PYC) and 1:1.56 (RJF). This shows that the rates of heterozygosity of PYC were slightly lower than those of RJF, as predicted. We also estimated the genetic diversity of 6 PYCs and 5 RJFs (Table 2). As expected, wild RJFs (pairwise nucleotide diversity, θπ = 2.58 × 10−3; average nucleotide polymorphisms, θω = 2.38 × 10−3) had a markedly higher level of genetic diversity than PYCs (θπ = 1.9 × 10−3, θω = 1.66 × 10−3), indicating that PYC experienced stronger selection pressure or more intense inbreeding.
Fig. 3.
Distribution of heterozygous SNPs. Heat maps of the heterozygous SNP rates within each non-overlapping 500 Kb window across the genome
Table 2.
SNP annotation and genetic diversity of PYC and RJF populations
| Type | No. | Total SNP (Mb) | Upstream (kb) | Exonic | Intronic (Mb) | Splicing (bp) | Intergenic (Mb) | Upstream/downstream | Downstream (kb) | θω (× 10−3) | θπ (× 10−3) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Non-synonymous (kb) | Synonymous (kb) | Non syn/syn ratio (ω) | Stop gain (bp) | Stop loss (bp) | |||||||||||
| PYC | 6 | 6.25 | 118.71 | 26.55 | 73.62 | 0.36 | 191 | 30 | 2.37 | 226 | 3.58 | 4.79 | 87.28 | 1.66 | 1.9 |
| RJF | 5 | 6.72 | 124.31 | 26.79 | 78.2 | 0.34 | 192 | 27 | 2.56 | 258 | 3.84 | 4.93 | 92.61 | 2.38 | 2.58 |
‘Upstream’ refers to a variant that overlaps with the 1 kb region upstream of the gene start site
‘Stop gain’ indicates that a non-synonymous SNP that leads to the creation of a stop codon at the variant site
‘Stop loss’ indicates that a non-synonymous SNP that leads to the elimination of a stop codon at the variant site
‘Splicing’ indicates a variant within 2 bp of a splice junction
‘Downstream’ indicates a variant that overlaps with the 1 kb region downstream of the gene end site
‘Upstream/downstream’ indicates that a variant located in the downstream and upstream regions (possibly for two different genes)
(θω): Average nucleotide polymorphism
(θω): Nucleotide diversity
The distribution of SNPs is summarized in Table 2. In PYC, we detected 100.39 kb of coding SNPs, leading to 26.55 kb of non-synonymous nucleotide substitutions. We also found that the majority of SNPs were located in intergenic and intronic regions (57% and 38%, respectively), whilst SNPs in genomic regions (exons, splice sites, and untranslated regions; 5% in total) were rare. These distributions revealed intergenic and intronic regions in the genome that play a role in gene regulation and were responsible for the phenotypic diversity of the chicken breeds.
Genome-wide selective sweep signals
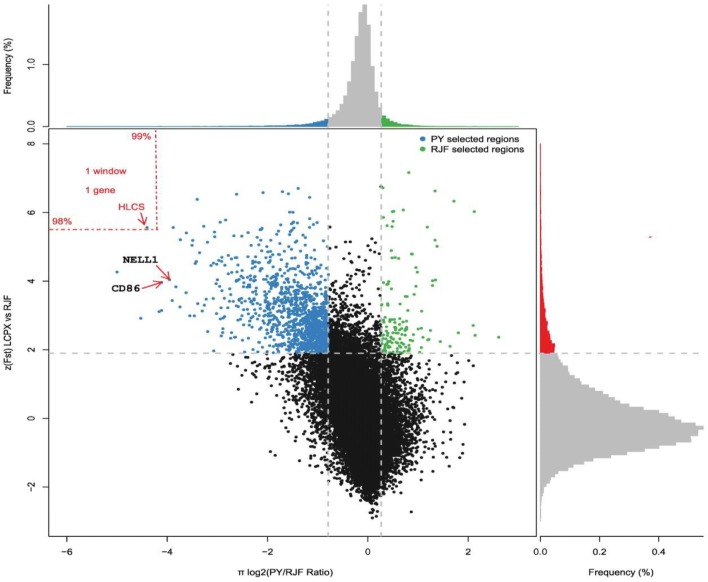
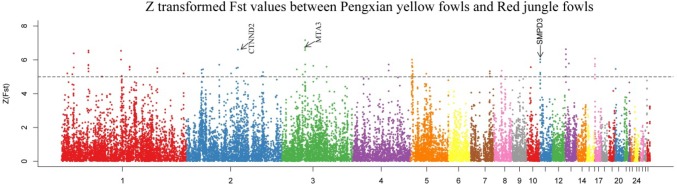
The candidate genes of PYC under selective pressure were identified by scanning the whole genome and exploring the genomic landscape using two parameters: population differentiation (Fst) and the ratio of pairwise nucleotide diversity (θπ). For the 62,433 windows (except W/Z/MT) of 40 kb (25% overlapping) across the whole genome, we filtered out windows containing ≤ 10 SNPs, leaving 46,060 windows for subsequent analyses. Finally, we identified genomic regions with a total size of ~ 36.80 Mb (3.42% of the genome, 1148 windows) that showed significant differences (p < 10−16, Mann–Whitney U test) in the log2 θπ ratios and Fst values compared with the genomic background of those under selection pressure (Fig. 6). Amongst the selected windows, the log2 θπ ratio ranged from − 5.00 to 2.60 and the Fst values fluctuated between 0.28 and 0.68 (zFst ranged from 1.90 to 6.62).
Fig. 6.
Distribution of Log2 (θπ ratio) and zFst values plotted to show candidate selective sweeping regions. Data points shown on the left of the vertical dashed line (corresponding to the 5% left tails of the empirical θπ ratio distribution) and above the horizontal dashed line (the 5% up tail of the empirical zFst distribution) are selected regions for PYC. Data points on the left of the vertical red dashed line (corresponding to the 1% left tails of the empirical θπ ratio distribution) and above the horizontal red dashed line (the 2% up tail of the empirical zFst distribution) represent windows containing only HLCS
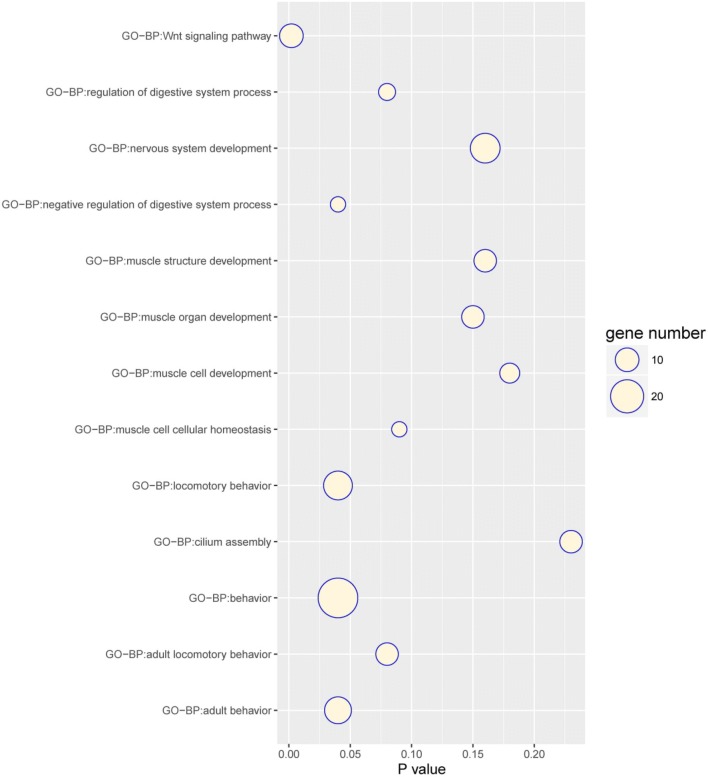
The SNPs detected in the candidate windows were located within 497 genes. We used the gene IDs as queries in the GO and KEGG databases of the Enrichr web server for enrichment analyses (Huang et al. 2009a). Only terms with p values ≤ 0.05 were considered as significant and listed. Subsequently, 497 genes were identified as associated with the digestive system [“negative regulation of digestive system process” (4 genes, p = 0.00004) and “regulation of digestive system process” (5 genes, p = 0.00039)], muscle development-related categories [“muscle organ development” (9 genes, p = 0.001) and “muscle cell development” (7 genes p = 0.003)], nerve development-related categories (“nervous system development,” p = 0.002), and vision-related categories [“Retinal dysplasia” (2 genes, p = 0.03) and “Microcornea” (7 genes, p = 0.001)] (Table 3 and Fig. 4). These categories may account for phenotypic changes in PYC over the course of domestication, including meat yields and body size.
Table 3.
Main functional gene categories of the identified selection genes
| Category | Description | p value | Adjusted p value | Gene counts |
|---|---|---|---|---|
| GO-BP: 0060457 | Negative regulation of digestive processes | 0.00004 | 0.04 | 4 |
| GO-BP: 0044058 | Regulation of digestive system processes | 0.0004 | 0.08 | 5 |
| GO-BP: 0007610 | Behavior | 0.00002 | 0.04 | 29 |
| GO-BP: 0007626 | Locomotory behavior | 0.00006 | 0.04 | 15 |
| GO-BP: 0030534 | Adult behavior | 0.00005 | 0.04 | 13 |
| GO-BP: 0046716 | Muscle cell cellular homeostasis | 0.0005 | 0.09 | 4 |
| GO-BP: 0008344 | Adult locomotory behavior | 0.0003 | 0.08 | 9 |
| GO-BP: 0007399 | Nervous system development | 0.002 | 0.16 | 16 |
| GO-BP: 0007517 | Muscle organ development | 0.001 | 0.15 | 9 |
| GO-BP: 0061061 | Muscle structure development | 0.002 | 0.16 | 9 |
| GO-BP: 0055001 | Muscle cell development | 0.003 | 0.18 | 7 |
| KEGG: 04310 | Wnt signaling | 0.003 | 0.002 | 10 |
p values indicate significance of the overlap between various gene sets, calculated using combined Fisher’s exact tests and the z score of deviation from the expected rank
Fig. 4.
Most significant categories in functional enrichment analysis. The y coordinates represent the different categories of enrichment analysis. The x-coordinates show the p values calculated through hypergeometric distribution probability approaches that represent the significance of enrichment analysis. Bubble sizes show the gene numbers for each specific category
Among the digestive system-related categories, the ATP-binding cassette sub-family G member 5 and 8 (ABCG5 and ABCG8) are paralogs and have been shown to play an indispensable role in the regulation of dietary cholesterol absorption and excretion (Yu et al. 2002). Other studies identified the selection of rapid growth rates leading to increased food conservation in chickens (Mitchell and Smith 1991), presumably supporting the detection of ABCG5 and ABCG8 that contributes to the efficiency of cholesterol conversion. Previously, the adrenorecrptor beta 1 (ADRB1) polymorphisms were found to contribute to enhanced body weight and fat mass also observed in this category (Dionne et al. 2002). In particular, HLCS (Holocarboxylase Synthetase) that regulates gluconeogenesis, fatty acid synthesis, and branched chain amino acid catabolism through promoting biotin utilization was embedded in the most significantly selected regions Fst value (2% right tail) and log2 θπ ratio (1% right tail; Fig. 6). Since supplemental biotin is added in the poultry industry (Anderson and Warnick 1970), the selection of these genes is likely to improve both growth and metabolism.
Muscle development and growth significantly differ between PYC and RJF owing to artificial selection. Indeed, we identified genes from four categories of muscle development. In addition to the very important muscle developmental regulator myogenic differentiation 1 (MYOD1) (Tapscott et al. 1988), Calpain 3 (CAPN3) was identified, which has been reported to affect chicken muscle growth and carcass traits such as body and breast muscle weight (Zhang et al. 2009). Other identified genes included Four and a Half LIM Domains 2 (FHL2) which regulates Wnt signaling as a coactivator of β-Catenin, contributing to muscle development (Martin et al. 2012). Additionally, candidate regions for growth-related traits harbored several genes such as sphingomyelin phosphodiesterase 3 (SMPD3), detected in a locus with the highest zFst score in chromosome 11 (Fst = 0.63, zFst = 6.04; Fig. 5), which regulates cell growth, differentiation, and apoptosis, contributing to postnatal growth and development (Stoffel et al. 2005). Moreover, the neural EGFL-like 1 (NELL1) that had the lowest log2θπ ratio (log2 θπ = − 3.96) in chromosome 5 (Fig. 6) and encodes a cytoplasmic protein that contains epidermal growth factor (EGF)-like repeats is a novel growth factor that specifically targets the osteochondral lineage (James et al. 2015). In addition, the BicC family RNA-binding protein 1(BICC1), which had the second highest zFst score of 4.10 (Fst = 0.47) in chromosome 6 (Fig. 5), is a genetic determinant of osteoblast genesis and bone mineral density (Mesner et al. 2014). These two bone development-related genes have been reported in human and rats. The SMPD3, NELL1, and BICC1 are also likely to correlate with poultry growth and be responsible for the enhanced body weights of PYC compared to RJF.
Fig. 5.
Manhattan plot of the zFst values between PYC and RJF. The Fst values were calculated for each 40-kb autosomal window. Dashed lines denote a threshold of zFst = 5
Additionally, we discovered two genes associated with the immune response. One gene within a locus with the second lowest log2 θπ ratio value in chromosome 1 (log2 θπ = − 4.12; Fig. 6), the CD86 antigen (CD86), is a well-known immune-related gene with known associations to cytotoxic T-lymphocyte antigen 4 (CTLA-4), which is an essential negative regulator of T cell immune responses (Omar et al. 2011). The second gene was the metastasis-associated 1 family member 3 (MTA3) that was detected through the second highest zFst score of 5.43 (Fst = 0.58) in chromosome 3 (Fig. 5) and interacts with the transcriptional repressor B-cell CLL/lymphoma 6 (BCL-6) to negatively regulate B-lymphocyte differentiation (Fujita et al. 2004). These findings support the occurrence of a trade-off between the selection of rapid growth and immunity during the domestication of chickens (Most et al. 2011).
Interestingly, we observed a locus with extremely strong sweeping signals, with no relationship to muscle development, growth, or other economic traits. The locus had the highest zFst score in chromosome 2 (Fst = 0.59, zFst = 5.5, the 26th highest Fst value in the whole genome; Fig. 5) and included a gene of the β-Catenin superfamily implicated in brain and eye development, and that was catenin delta 2 (CTNND2). Domestic animals including dogs, horses, and chickens exhibit diminished vision, most likely due to the reduced selection pressure to natural predators, reducing the requirement for sharp vision (Roth and Lind 2013). Important studies have uncovered the genetic mechanisms behind reduced visual capability, identifying that selection rather than the relaxation of functional constraints leads to the deterioration of vision over the course of evolution in domestic chickens (Wang et al. 2016a). This means that vision-related genes under selective pressure, such as those detected in selective sweeps, may be responsible for the deterioration in vision. Previous studies assessing the optical prowess of domestic chickens have mainly focused on critical flicker fusion frequencies and the sensitivity of retina-like phototransduction and photoreceptors (Lisney et al. 2012; Osorio et al. 1999). However, the development of the dioptric system of domestic chickens was also impacted by human activities. In this regard, studies have verified that the continuous light produced by humans leads to reduced corneal curvature and smaller pupils of fowls, likely influencing their diopter system (Lauber and Mcginnis 1966). According to previous human studies, CTNND2 is strongly associated with myopia (Li et al. 2011); meanwhile, CTNND2 has been documented to play a crucial role in retinal morphogenesis, adhesion, and retinal cell architectural integrity in non-human models (Duparc et al. 2006; Paffenholz et al. 1999). Based on the association of CTNND2 with retinal development, for which strong selection signals were detected, it is reasonable to hypothesize that CTNND2 is associated with the retinogenesis regulation in chickens and, thus, presumably gives rise to eye evolution in domestic chicken. The vesicle amine transport 1 (VAT1) had the lowest log2 θπ ratio in chromosome 27 (log2 θπ ratio = − 3.27, and the 20th lowest log2 θπ ratio in the whole genome) and has been reported to negatively regulate mitochondrial fusion in cooperation with the mitofusin proteins 1 and 2 (MFN1 and MFN2) (Eura et al. 2006). Previous studies found that mitochondrial dynamics (fusion, fission, movement, and mitophagy) is altered in neurodegenerative diseases contributing to dominant optical atrophy in humans (Chen and Chan 2009) and it is highly likely that VAT1 is involved in the diminished vision of domestic chickens through its regulation of mitochondrial fusion that influences the early development of the optic nerve.
Meanwhile, we identified seven categories from enrichment analyses (Table 2) including “Retinal dysplasia” (HP: 0007973), “Cilium assembly” (GO: 0042384, p = 0.002) and “Microcornea” (HP: 0000482) that included genes associated with eye maldevelopment and dysfunction, which could provide support for our conclusions. One example is intraflagellar transport 140 (IFT140), which encodes a subunit of the intraflagellar transport complex A reported to play an important role in opsin transportation, with opsin being critical for the detection of light (Crouse et al. 2014). The intraflagellar transport 57 (IFT57) was also reported to be associated with the regulation of photoreceptors (Krock and Perkins 2008). Additionally, the Abelson Helper Integration Site 1 (AHI1) is relevant to photoreceptor outer segment development and retinal dystrophy in humans (Parisi et al. 2006). Moreover, the presence of RPGRIP1 like (RPGRIP1L) in this category confirmed its involvement in vision, coinciding with the results of Wang et al. (2016a).
Conclusions
These findings are beneficial to provide useful genetic information on PYC that is conducive to the screening and utilization of genes related to important economic traits. Although some of the functions and mechanisms of the genes identified remain unclear and require further studies, these results provide a useful foundation on which future studies in chickens can be built.
Acknowledgements
This work was supported by Sichuan Science and Technology Program (2016NYZ0050), and Technology Planning Project of Chengdu (2015-NY01-00036-NC).
Author contributions
Conceptualization, HY and QZ; methodology, HY and YW; software, HY and DL; formal analysis, HY and DL; resources, QZ; writing—original draft preparation, HY; writing—review and editing, HY and QZ; supervision, QZ; project administration, YW; funding acquisition, QZ and HY.
Compliance with ethical standards
Conflict of interest
All the authors report no conflicts of interest in this work.
Ethics approval
All experimental operations were approved by the Animal Ethics Committee of Sichuan Agricultural University (approval number: 20171410401). Relevant guidelines and regulations were followed for all methods.
References
- Akey JM, Zhang G, Zhang K, Jin L, Shriver MD. Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 2002;12(12):1805–1814. doi: 10.1101/gr.631202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JO, Warnick RE. Studies of the need for supplemental biotin in chick rations. Poult Sci. 1970;49(2):569–578. doi: 10.3382/ps.0490569. [DOI] [PubMed] [Google Scholar]
- Aslam ML, Bastiaansen JW, Elferink MG, Megens HJ, Crooijmans RP, Le AB, Fleischer RC, Tassell CPV, Sonstegard TS, Schroeder SG. Whole genome SNP discovery and analysis of genetic diversity in Turkey (Meleagris gallopavo) BMC Genom. 2012;13(1):391. doi: 10.1186/1471-2164-13-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chan DC. Mitochondrial dynamics–fusion, fission, movement, and mitophagy–in neurodegenerative diseases. Hum Mol Genet. 2009;18(R2):169–176. doi: 10.1093/hmg/ddp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Yan K, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013;14(1):128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouse JA, Lopes VS, Sanagustin JT, Keady BT, Williams DS, Pazour GJ. Distinct functions for IFT140 and IFT20 in opsin transport. Cytoskeleton. 2014;71(5):302–310. doi: 10.1002/cm.21173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionne IJ, Garant MJ, Nolan AA, Pollin TI, Lewis DG, Shuldiner AR, Poehlman ET. Association between obesity and a polymorphism in the β1-adrenoceptor gene (Gly389Arg ADRB1) in Caucasian women. Int J Obes Relat Metab Disord. 2002;26(5):633–639. doi: 10.1038/sj.ijo.0801971. [DOI] [PubMed] [Google Scholar]
- Duparc RH, Boutemmine D, Champagne MP, Tetreault N, Bernier G. Pax6 is required for delta-catenin/neurojugin expression during retinal, cerebellar and cortical development in mice. Dev Biol. 2006;300:647–655. doi: 10.1016/j.ydbio.2006.07.045. [DOI] [PubMed] [Google Scholar]
- Eura Y, Ishihara N, Oka T, Mihara K. Identification of a novel protein that regulates mitochondrial fusion by modulating mitofusin (Mfn) protein function. J Cell Sci. 2006;119(Pt 23):4913–4925. doi: 10.1242/jcs.03253. [DOI] [PubMed] [Google Scholar]
- Fujita N, Jaye DL, Geigerman C, Akyildiz A, Mooney MR, Boss JM, Wade PA. MTA3 and the Mi-2/NuRD complex regulate cell fate during B lymphocyte differentiation: cell. Cell. 2004;119(1):75–86. doi: 10.1016/j.cell.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang X, Feng Q, Qian Q, Zhao Q, Wang L, Wang A, Guan J, Fan D, Weng Q, Huang T. High-throughput genotyping by whole-genome resequencing. Genome Res. 2009;19(6):1068. doi: 10.1101/gr.089516.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James AW, Shen J, Zhang X, Asatrian G, Goyal R, Kwak JH, Jiang L, Bengs B, Culiat CT, Turner AS. NELL-1 in the treatment of osteoporotic bone loss. Nat Commun. 2015;6(Suppl 6):7362. doi: 10.1038/ncomms8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krock BL, Perkins BD. The intraflagellar transport protein IFT57 is required for cilia maintenance and regulates IFT-particle–kinesin-II dissociation in vertebrate photoreceptors. J Cell Sci. 2008;121(11):1907–1915. doi: 10.1242/jcs.029397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber JK, Mcginnis J. Eye lesions in domestic fowl reared under continuous light. Vision Res. 1966;6(12):619–626. doi: 10.1016/0042-6989(66)90073-3. [DOI] [PubMed] [Google Scholar]
- Li YJ, Fan Q, Yu M, Han SY, Sim XL, Ong RTH, Wong TY, Vithana EN. Genome-wide association studies reveal genetic variants in CTNND2 for high myopia in Singapore Chinese. Ophthalmology. 2011;118(2):368–375. doi: 10.1016/j.ophtha.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Che T, Chen B, Tian S, Zhou X, Zhang G, Li M, Gaur U, Li Y, Luo M. Genomic data for 78 chickens from 14 populations. Gigascience. 2017;6:1–5. doi: 10.1093/gigascience/gix026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisney TJ, Ekesten B, Tauson R, Håstad O, Odeen A. Using electroretinograms to assess flicker fusion frequency in domestic hens Gallus gallus domesticus. Vision Res. 2012;62(3):125–133. doi: 10.1016/j.visres.2012.04.002. [DOI] [PubMed] [Google Scholar]
- Martin B, Schneider R, Janetzky S, Waibler Z, Pandur P, Kühl M, Behrens J, Kvd Mark, Starzinski-Powitz A, Wixler V. The LIM-only protein FHL2 interacts with β-catenin and promotes differentiation of mouse myoblasts. J Cell Biol. 2012;159(1):113. doi: 10.1083/jcb.200202075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesner LD, Ray B, Hsu YH, Manichaikul A, Lum E, Bryda EC, Rich SS, Rosen CJ, Criqui MH, Allison M. Bicc1 is a genetic determinant of osteoblastogenesis and bone mineral density. J Clin Invest. 2014;124(6):2736–2749. doi: 10.1172/JCI73072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell MA, Smith MW. The effects of genetic selection for increased growth rate on mucosal and muscle weights in the different regions of the small intestine of the domestic fowl (Gallus domesticus) Comp Biochem Phys A. 1991;99(1–2):251–258. doi: 10.1016/0300-9629(91)90268-H. [DOI] [PubMed] [Google Scholar]
- Most PJVD, Jong BD, Parmentier HK, Verhulst S. Trade-off between growth and immune function: a meta-analysis of selection experiments. Funct Ecol. 2011;25(1):74–80. doi: 10.1111/j.1365-2435.2010.01800.x. [DOI] [Google Scholar]
- Omar S, Qureshi YZ, Nakamura Kyoko, Attridge Kesley, Manzotti Claire, Schmidt Emily M, Baker Jennifer, Jeffery Louisa E, Kaur Satdip, Briggs Zoe, Hou Tie Z, Futter Clare E, Anderson Graham, Walker Lucy SK, Sansom David M. Trans-endocytosis of CD80 and CD86: a molecular basis for the cell extrinsic function of CTLA-4. Science. 2011;332(6029):600–603. doi: 10.1126/science.1202947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio D, Vorobyev M, Jones CD. Colour vision of domestic chicks. J Exp Biol. 1999;202(21):2951–2959. doi: 10.1242/jeb.202.21.2951. [DOI] [PubMed] [Google Scholar]
- Paffenholz R, Kuhn C, Grund C, Stehr S, Franke WW. The arm-repeat protein NPRAP (neurojungin) is a constituent of the plaques of the outer limiting zone in the retina, defining a novel type of adhering junction. Exp Cell Res. 1999;250:452–464. doi: 10.1006/excr.1999.4534. [DOI] [PubMed] [Google Scholar]
- Parisi MA, Doherty D, Eckert ML, Shaw DW, Ozyurek H, Aysun S, Giray O, Al SA, Al SS, Dohayan N, Bakhsh E, Indridason OS, Dobyns WB, Bennett CL, Chance PF, Glass IA. AHI1 mutations cause both retinal dystrophy and renal cystic disease in Joubert syndrome. J Med Genet. 2006;43(4):334–339. doi: 10.1136/jmg.2005.036608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth LSV, Lind O. The impact of domestication on the chicken optical apparatus. PLoS One. 2013;8(6):e65509. doi: 10.1371/journal.pone.0065509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin CJ, Zody MC, Eriksson J, Meadows JRS, Sherwood E, Webster MT, Lin J, Ingman M, Sharpe T, Sojeong K. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature. 2010;464(7288):587. doi: 10.1038/nature08832. [DOI] [PubMed] [Google Scholar]
- Stoffel W, Jenke B, Blöck B, Zumbansen M, Koebke J. Neutral sphingomyelinase 2 (smpd3) in the control of postnatal growth and development. Proc Natl Acad Sci USA. 2005;102(12):4554–4559. doi: 10.1073/pnas.0406380102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapscott SJ, Davis RL, Thayer MJ, Cheng PF, Weintraub H, Lassar AB. MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science. 1988;242(4877):405–411. doi: 10.1126/science.3175662. [DOI] [PubMed] [Google Scholar]
- Wang MS, Li Y, Peng MS, Zhong L, Wang ZJ, Li QY, Tu XL, Dong Y, Zhu CL, Wang L, Zhang YP. Genomic analyses reveal potential independent adaptation to high altitude in Tibetan Chickens. Mol Biol Evol. 2015;32(7):1880–1889. doi: 10.1093/molbev/msv071. [DOI] [PubMed] [Google Scholar]
- Wang MS, Zhang R, Su LY, Li Y, Peng MS, Liu HQ, Zeng L, Irwin DM, Du JL, Wu DD, Yao YG, Zhang YP. Positive selection rather than relaxation of functional constraint drives the evolution of vision during chicken domestication. Cell Res. 2016;26(5):556. doi: 10.1038/cr.2016.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MS, Zhang R, Su LY, Li Y, Peng MS, Liu HQ, Zeng L, Irwin DM, Du JL, Yao YG. Positive selection rather than relaxation of functional constraint drives the evolution of vision during chicken domestication. Cell Res. 2016;26(5):556. doi: 10.1038/cr.2016.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CX. The utilization of inheritance resource and local fowls breeders in the poultry producing. China Poult. 2001;23(23):3–4. [Google Scholar]
- Yu L, Lihawkins J, Hammer RE, Berge KE, Horton JD, Cohen JC, Hobbs HH. Overexpression of ABCG5 and ABCG8 promotes biliary cholesterol secretion and reduces fractional absorption of dietary cholesterol. J Clin Invest. 2002;110(5):671–680. doi: 10.1172/JCI0216001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZR, Liu YP, Yao YG, Jiang XS, Du HR, Zhu Q. Identification and association of the single nucleotide polymorphisms in calpain3 (CAPN3) gene with carcass traits in chickens. BMC Genet. 2009;10(1):10. doi: 10.1186/1471-2156-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]