Significance
X-ray crystallographic analyses of mitochondrial cytochrome c oxidase (CcO) have been based on its dimeric form. Recent cryo-electron microscopy structures revealed that CcO exists in its monomeric form in the respiratory supercomplex. This study, using amphipol-stabilized CcO, shows that the activity of monomer is higher than that of the dimer. The crystal structure of monomer determined here shows that the local structure of one of the proton transfer pathways differs from that in the dimer. The crystal structure also shows that cardiolipins are located at the interface region in the supercomplex. Taken together, these results suggest that CcO in the monomeric state, dimeric state, and supercomplex state depending on cardiolipins are involved in regulation of respiratory electron transport.
Keywords: cytochrome c oxidase, X-ray crystallography, phospholipids, monomer, dimer
Abstract
Cytochrome c oxidase (CcO), a membrane enzyme in the respiratory chain, catalyzes oxygen reduction by coupling electron and proton transfer through the enzyme with a proton pump across the membrane. In all crystals reported to date, bovine CcO exists as a dimer with the same intermonomer contacts, whereas CcOs and related enzymes from prokaryotes exist as monomers. Recent structural analyses of the mitochondrial respiratory supercomplex revealed that CcO monomer associates with complex I and complex III, indicating that the monomeric state is functionally important. In this study, we prepared monomeric and dimeric bovine CcO, stabilized using amphipol, and showed that the monomer had high activity. In addition, using a newly synthesized detergent, we determined the oxidized and reduced structures of monomer with resolutions of 1.85 and 1.95 Å, respectively. Structural comparison of the monomer and dimer revealed that a hydrogen bond network of water molecules is formed at the entry surface of the proton transfer pathway, termed the K-pathway, in monomeric CcO, whereas this network is altered in dimeric CcO. Based on these results, we propose that the monomer is the activated form, whereas the dimer can be regarded as a physiological standby form in the mitochondrial membrane. We also determined phospholipid structures based on electron density together with the anomalous scattering effect of phosphorus atoms. Two cardiolipins are found at the interface region of the supercomplex. We discuss formation of the monomeric CcO, dimeric CcO, and supercomplex, as well as their role in regulation of CcO activity.
Cytochrome c oxidase (CcO), which resides in the inner membrane of mitochondria, is the terminal enzyme of the respiratory electron transport chain; it performs proton pumping through coupling with reduction of dioxygen to water (1, 2). The enzyme is a supramolecular complex composed of 3 subunits encoded by mitochondrial genes (subunits I, II, and III), and 10 subunits derived from nuclear genes. Heme a, heme a3, and CuB are the active centers of the transmembrane region of subunit I, whereas CuA is localized in the hydrophilic domain on the mitochondrial intermembrane side of subunit II. Electrons received from cytochrome c bonded to the hydrophilic surface of subunit II are transferred via CuA and heme a, to the heme a3–CuB site. Dioxygen, the substrate, is transported from the transmembrane surface of subunit III, via a hydrophobic cavity, to the heme a3–CuB site. Protons used in the oxygen reduction are transported from the mitochondrial matrix side to the heme a3–CuB site via 2 hydrogen bond networks (K- and D-pathways) in the transmembrane region of subunit I. In CcO derived from the bovine heart (bovine CcO), it has been proposed that pumped protons are also transported via the H-pathway. Molecular phylogenetic and structural analyses show that CcO has an evolutionary relationship with the heme–copper oxygen reductases (HCORs) and nitric oxide reductases (NORs) in the respiratory electron transport chain of prokaryotes; this group of enzymes is collectively referred to as the HCOR superfamily (2, 3). Based on phylogenetic and structural analysis of the HCOR superfamily, mitochondrial CcO is classified in the A-type subfamily. Crystal structures of A-, B-, and C-type HCORs and NORs have been reported previously (4–12).
In this study, we took the structural analysis of bovine CcO a step further. Among all crystals reported thus far, CcO has existed as a dimer with the same intermonomer contacts (13). Detailed structural analysis of the dimer has revealed that subunits derived from nuclear genes are involved in intermonomer contact (SI Appendix, Fig. S1). In addition, clear electron density corresponding to cholate, as well as a long, thin electron density corresponding to the hydrocarbon chain of detergent, has been observed on the transmembrane surface of CcO, which serves as the association surface of the dimer, implying that these molecules are involved in dimer formation (14). In addition, mass spectrometry analysis has shown that the dimer, as well as the monomer, is present in samples of purified bovine CcO, suggesting that lipid molecules are involved in the intermolecular associations of CcO (15). However, enzymes of the HCOR superfamily derived from prokaryotes exist as monomers in all crystal structures reported to date (5–12). These enzymes consist only of subunits corresponding to subunits I and II, and in some cases III, of mitochondrial CcO. These structural observations and allosteric ATP inhibition suggest that mitochondrial CcO is subjected to some functional control via dimerization involving subunits encoded by nuclear genes (16, 17).
On the other hand, in the mitochondrial membrane, CcO also associates with complex I (CI) and cytochrome bc1 (complex III; CIII), which are members of the respiratory electron transport chain, and forms a supercomplex, i.e., an even higher-order structure. In recent years, multiple studies have described these structures (18–24). The supercomplex can be isolated when the mitochondrial membrane is solubilized with digitonin. In the supercomplex, CcO monomer associates with complex I and complex III. If the membrane is solubilized with DDM, Triton X-100, or similar detergents, most CcO is isolated as independent monomers. Purified CcO consists of a mixture of monomers and dimers, but monomerization proceeds due to a rise in pH or a drop in protein concentration or phospholipid content (25). As indicated above, it is possible that the monomeric state of mitochondrial CcO is functionally important; previously, however, it was not possible to strictly compare the activities of monomeric and dimeric enzymes. Moreover, the high-resolution structure of the monomer and the structural differences between the free monomer and the monomer within dimeric CcO remain to be elucidated.
In our previous structural analysis of bovine CcO, we solubilized the enzyme from the mitochondrial membrane using cholate, performed ammonium sulfate fractionation in the presence of cholate, and then purified the enzyme by replacing the detergent with n-decyl-β-d-maltoside (DM). Cholate and its analogs, deoxycholate and 3-[(3-cholamidopropyl)dimethylammonio]propanesulfonate (CHAPS) (SI Appendix, Fig. S2), induce dimerization of monomeric CcO solubilized with DDM or phospholipid (25, 26). Therefore, the fact that all CcO structures previously determined by our group are dimers may depend on use of cholate in CcO purification. By contrast, in this study, we first prepared monomers and dimers stabilized using amphipol, and then showed that the monomer exhibited high activity. In addition, we synthesized a new detergent, 3-oxatridecyl-α-d-mannoside (3OM) and used it for purification and crystallization. In this manner, we determined the oxidized and reduced structures of bovine CcO in the monomeric state with resolutions of 1.85 and 1.95 Å, respectively. These results indicate that monomeric CcO is the activated form in the mitochondrial membrane.
Results and Discussion
Monomeric Bovine CcO.
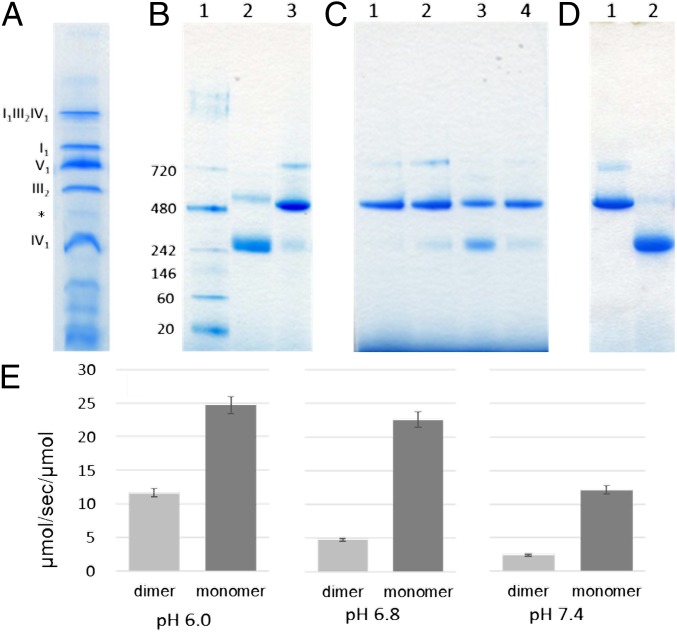
When bovine mitochondrial membrane was solubilized with digitonin, CcO was present as an independent monomer and as a monomer in the supercomplex (Fig. 1A). CcO is also present in the extremely thin band indicated by *, which based on its size likely represents a dimer (19, 24). Because CcO is present as a monomer in the supercomplex, it was previously thought that in the mitochondrial membrane, almost all CcO functions as a monomer. On the other hand, in previously determined crystal structures, CcO forms a dimer. For this purpose, an enzyme is solubilized from the mitochondrial membrane using cholate, and then stabilized and crystallized using DM as a detergent. In a preparation obtained by dissolving a microcrystal preparation crystallized at pH 6.8 at a protein concentration of at least 15 mg/mL and pH 6.8, the dimer almost always formed (Fig. 1C, lane 1). However, when the protein concentration was reduced to 1/10 at pH 6.8, a monomer was clearly evident (Fig. 1C, lane 2). To further decrease the protein concentration, the dimeric enzyme was subjected to gel filtration chromatography using Sephacryl S-300 resin (GE Healthcare Life Sciences) with 100-mM Na-Pi buffer (pH 6.8) containing 0.2% (wt/vol) DM as an elution buffer. The peak molecular mass was 210 kDa, almost the same mass as the monomer (SI Appendix, Fig. S3). These findings indicate that dilution of the sample caused dissociation of the dimer to the monomer. When the pH was raised to 7.4 or 8.5, the protein also shifted from the dimer to the monomer (Fig. 1C, lanes 3 and 4). These observations imply that in solution, CcO is in an equilibrium state between the dimer and monomer.
Fig. 1.
Comparison of the ratio of monomeric vs. dimeric CcO by BN-PAGE (A–D), and cytochrome c oxidation activity of monomeric and dimeric CcOs stabilized with amphipol (E). (A) Supernatant obtained by solubilization of bovine mitochondria in digitonin at 10 times the amount of protein (30 μg per lane). Composition of the respiratory chain complex is shown to the side of each band. The asterisk indicates the position of dimeric CcO. (B and C) 8 μg of protein preparations for each condition was applied to each lane. (B) Lane 1: marker proteins; lane 2: preparation stabilized with 3OM (1.5 mg protein/mL in 40 mM sodium phosphate [Na-Pi] buffer [pH 6.4] containing 0.2% 3OM); lane 3: preparation stabilized with DM (1.5 mg protein/mL in 40 mM Na-Pi buffer [pH 6.8] containing 0.2% DM). (C) Preparations stabilized with DM under the indicated conditions. Lane 1: 15 mg protein/mL in 40 mM Na-Pi buffer (pH 6.8) containing 0.2% DM. Lane 2: 1.5 mg protein/mL in 40 mM Na-Pi buffer (pH 6.8) containing 0.2% DM. When 8 μg of the sample was applied to the lane, the sample could be diluted up to 10-fold (from 15 to 1.5 mg protein/mL). Lane 3: 1.5 mg protein/mL in 40 mM Na-Pi buffer (pH 8.5) containing 0.2% DM. Lane 4: 1.5 mg protein/mL in 40 mM Na-Pi buffer (pH 7.4) containing 0.2% DM. (D) Preparations of dimer (lane 1) and monomer (lane 2) were stabilized with amphipol, 10 μg each. (E) Activity measured at the indicated pH in the presence of 10 μM reduced cytochrome c.
Therefore, when using a preparation stabilized with DM, it is not possible to compare the activities of monomeric and dimeric CcO. We generated preparations consisting exclusively of the monomeric or dimeric form using amphipol (Fig. 1D), an amphiphilic polymer as previously reported (19), and compared their activities (Fig. 1E). At all pH values tested (6.0, 6.8, and 7.4), monomeric CcO exhibited high activity. To confirm the stability of the monomeric and dimeric forms, the samples diluted for the activity measurements were concentrated again and subjected to blue native polyacrylamide gel electrophoresis (BN-PAGE). Almost all CcO in the monomer sample maintained the monomeric form, and more than 80% of CcO in the dimer sample maintained the dimeric form. Using monomers and dimers stabilized with amphipol, it became possible to rigorously compare the 2 forms. As shown in Fig. 1E and SI Appendix, Fig. S4, the activities were all lower than 100 μmol/s/μmol, the typical activity measured in the presence of DM at pH 6.0 using the enzyme stabilized with DM. This is thought to be because in the presence of amphipol, an enzyme is dissolved with its charged groups oriented toward the solution, hindering CcO functions such as the electrostatic interaction between cytochrome c and CcO, electron transfer from cytochrome c to CcO, and uptake of ligand oxygen.
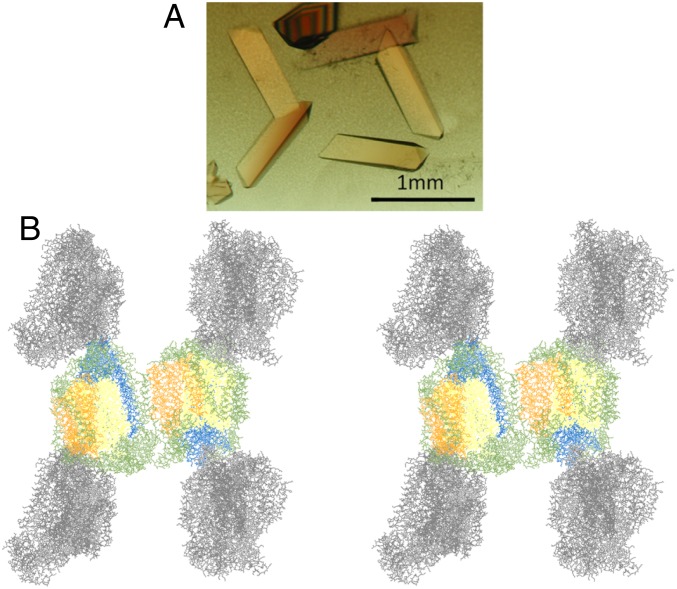
Under these conditions, the monomer exhibited higher activity. Because almost all CcO in the mitochondrial membrane is present as monomeric CcO, this implies that monomer represents the activated form. To understand the difference in activity between the monomer and dimer in terms of the molecular structure, it is of interest to compare the crystal structures of the 2 forms regarding which progress has been made thus far. Previously, we performed crystallization using various detergents, but the enzyme formed dimers in all crystals (13). Hence, we synthesized an even wider variety of detergents, and then sought to achieve crystallization by preparing CcO using the new detergents (SI Appendix, Fig. S2). We obtained a prismatic crystal when 3OM was used as a detergent (Fig. 2A). This detergent differs greatly from detergents previously used, in that an α-bond is formed between the acyl group and sugar. Crystallization was obtained when 3OM was bound to mannose, but not when the detergent was bound with maltose. At present, these newly synthesized detergents are commercially available.
Fig. 2.
Crystals grown with 3OM (A) and crystal packing of monomeric CcO (B). Subunits I, II, III, and other subunits in the 2 CcOs are shown in yellow, blue, orange, and green, respectively. Other CcOs are shown in gray.
The purified sample generated using 3OM was almost all monomeric at pH 6.4, the conditions under which crystallization was carried out (Fig. 1B, lane 2). A band was present at the position thought to represent dimeric CcO, but it had a somewhat higher molecular weight than the corresponding band when DM was used. As explained below, this may be because the dimerization configuration is different, resulting in a difference in the amount of detergent bound to the enzyme. The absorption spectra of purified CcO dissolved in 3OM were the same as those in DM (SI Appendix, Fig. S5 A and B). The activity of CcO with 3OM at pH 6.0 was about 20% higher, not significantly higher, than that of the enzyme with DM at pH 6.0. In the activity measurement, concentration of CcO is very low (about 0.5 nM). Because CcO with DM is a monomer at low enzyme concentration (SI Appendix, Fig. S3), observed activities with both 3OM and DM should be mainly due to monomeric CcO.
Structure of Monomeric Bovine CcO.
X-ray diffraction data with a resolution of 1.8 Å was obtained from CcO in the resting oxidized state, crystallized using 3OM (SI Appendix, Table S1, PDB ID code 6JY3) (27). Structural analysis revealed association of 2 monomers at the transmembrane surface; however, in contrast to the previously reported dimeric CcO structure, the orientation of the transmembrane helix was reversed (Fig. 2B). Because this association does not reflect the physiological state, we concluded that the crystal consisted of monomeric CcO.
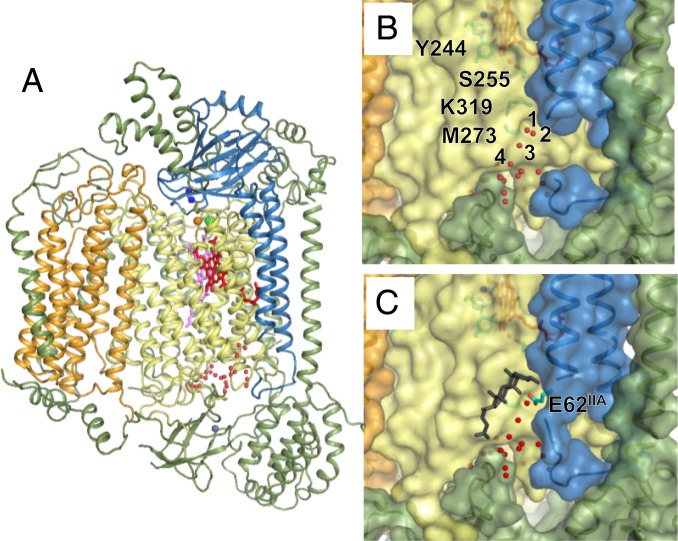
When we made a comparison between the overall protein structure of monomeric CcO determined in this study and the monomeric structures present in previously reported dimeric CcO, we noted a generally good match in the internal structures of subunits I, II, and III, which carry out the functions of the enzyme (SI Appendix, Fig. S6). However, structural changes were evident in several regions. The most conspicuous of these were evident at the transmembrane surface, which serves as the association interface. Involved in the dimerization were helix II (helix IIIIA) of subunit II; helix VII (helix VIIIA) and helix VIII (helix VIIIIA) of subunit I in one monomer (monomer A); and helix I (helix IVIaB) in subunit VIa of the other monomer (monomer B) (SI Appendix, Fig. S7 A and B). In the structure of dimeric CcO, there is a clear electron density corresponding to cholate (CH2) between these helices (SI Appendix, Fig. S7B). CH2 is fixed through hydrogen bonding with Glu62, Thr63 of helix IIIIA, and Arg14 and Arg17 of helix IVIaB. In addition, the aromatic ring of CH2 interacts with the aromatic ring of Trp275 in helix VIIIA and Phe18 of helix IVIaB (SI Appendix, Fig. S2A). Together, these observations imply that these interactions contribute to stabilization of the dimer. However, in the structure of monomeric CcO (Fig. 3A), CH2 was not bound to either subunit II or subunit VIa, and the electron density derived from 3OM was not observed at this CH2-binding site (SI Appendix, Fig. S7C). The electron density for another cholate (CH1), located outside the association interface, was clearly observed in both dimeric and monomeric CcOs, as described in detail below, implying that the binding affinity of CH2 is decreased by monomerization of CcO. Furthermore, in monomeric CcO, the electron density of the segment containing Glu62 of helix IIIIA was markedly reduced (SI Appendix, Fig. S7C). When refinement calculation was performed, this segment exhibited an extremely high temperature factor (SI Appendix, Fig. S8A). This means that, in monomeric CcO, the helix IIIIA segment structure is very unstable. Moreover, a water molecule (Wat2) was positioned in monomeric CcO at a location where there is a Glu62IIA carboxyl group in dimeric CcO (Fig. 3 B and C).
Fig. 3.
Whole structure of monomeric bovine CcO (A), and structure of the proton transfer K-pathway in monomeric (B) and dimeric (C) CcOs. Subunits I, II, III, and other subunits in the 2 CcOs are shown in yellow, blue, orange, and green, respectively. Hemes a and a3 are shown as pink and red sticks, respectively. Copper, magnesium, sodium, and zinc ions are shown as blue, orange, green, and gray spheres, respectively. Water molecules are shown as small red spheres. (B) Heme a3–CuB site and amino acid residues in the K-pathway are shown as stick models. Water molecules are shown as small red spheres. (C) Cholate molecule is shown as a black stick model.
In other regions, differences were observed in the structures of monomeric CcO and dimeric CcO at the transmembrane helix of subunits VIa and VIc, as well as the domain where subunit VIb protrudes into the intermembrane region. In these subunits, a structural shift of 3 to 4 Å was observed between monomeric CcO and dimeric CcO (SI Appendix, Fig. S9). Subunits VIa and VIc act as contact surfaces between monomeric CcO in the 3OM crystal, and this may cause the structure shift. In dimeric CcO, the domain where subunit VIb protrudes acts as the contact surface between monomers, and thus contributes to stabilization of the dimer structure. Despite the structural shift, the conformation of this domain was maintained even in monomeric CcO. These observations suggest that this domain plays a role in determining CcO orientation at the time of dimerization.
Next, to characterize the relationship among the structural changes accompanying dimerization, and to determine why the monomer has higher activity than the dimer, we performed a detailed comparison between monomeric and dimeric CcO, focusing on the structures of sites crucial for enzyme function (i.e., cytochrome c–binding site, heme a3–CuB site, dioxygen transfer pathway, and the K-, D-, and H-pathways for proton transfer). No dimerization-associated structural changes were evident at sites other than one near the entry point to the K-pathway, described in detail below. In previous research, we conducted structural analysis of dimeric CcO in the presence of substrate or inhibitor, and determined several structures in which cytochrome c was bound to the hydrophilic surface of subunit II (28); an inhibitor such as carbon monoxide or cyanide ions was bound to the heme a3–CuB site (29); the inhibitor N,N′-dicyclohexylcarbodiimide (DCCD) was bound to the hydrophobic surface in the dioxygen transfer pathway (14); or an inhibitory cadmium or zinc ion was bound to the entrance of the D-pathway for proton transfer (30). These experimental results imply that even when CcO is dimerized, the substrate (electron donor, dioxygen, or proton) can access the cytochrome c–binding site, heme a3–CuB site, dioxygen transfer pathway, and D-pathways (i.e., these sites are functioning). Moreover, there is no conflict with the fact that, in this study, no dimerization-associated structural change was evident at these sites.
On the other hand, the structure at the entry point of the K-pathway differed between monomeric and dimeric CcOs. Wat2 in the structure of monomeric CcO determined in this study is located at the entry point of the K-pathway, and is within hydrogen bonding distance of other water molecules (Wat1 and Wat3 in Fig. 3B), implying that a hydrogen bond network is formed from Wat4, Wat3, Wat2, and Wat1 to M273 in the K-pathway. In dimeric CcO, on the other hand, the carboxyl group of Glu62IIA is hydrogen bonded with CH2 and Wat1 (Fig. 3C); thus, there is a break in the hydrogen bond network connected with the K-pathway seen in monomeric CcO. Consequently, in monomeric CcO, it is possible to carry out proton transport via a hydrogen bond network of water molecules, whereas in dimeric CcO, the CH2 bond can inhibit proton uptake (31); i.e., structure of the hydrogen bond network in the K-pathway changes due to the CH2 bond. As shown in SI Appendix, Fig. S2A, CH2 interacts with amino acid residues of both monomers in dimeric CcO. Dissociation of dimer to monomers decreases the number of these interactions, and likely decreases the affinity of CH2 for the entry point of the K-pathway.
For the activity measurement, amphipol-stabilized monomeric and dimeric CcOs were prepared from crystalline samples of dimeric CcO containing CH2. Because monomerization of CcO reduces the affinity of CH2, as described above, the structural alteration at the entry point of the K-pathway caused by release of CH2 is likely responsible for the differences in the activities of monomeric and dimeric CcO. In other words, the monomeric form determined in this study is likely to represent the activated structure of CcO. It has long been known that cholate inhibits the activity of bovine CcO (32, 33). The results of our structural analyses support this idea. On the other hand, the lack of activity in the presence of higher concentrations of cholate may be due to some additional effect. In addition, it has been suggested that ATP, which has a structure similar to cholate, can also associated with the CH2-binding site (17, 34, 35), suggesting that under physiological conditions, solutes such as ATP bind to the protein and regulate its activity.
In structural terms, the K-pathway of monomeric bovine CcO matches that of the Rhodobacter sphaeroides and Paracoccus denitrificans enzymes (RsCcO and PdCcO, respectively), both of which are A-type members of the HCOR superfamily. In the glutamate residue variant at the entry point of the RsCcO K-pathway, enzymatic activity increases in the presence of bile salts such as cholate or amphipathic carboxylates and decreases in the presence of cholesterol (36–38). In addition, in the structures of RsCcO and PdCcO, helix II of subunit II containing the K-pathway entry point has a higher temperature factor than other parts (SI Appendix, Fig. S8). In addition, in the structure of RsCcO, deoxycholate is bound to the K-pathway entry point, and there is drop in temperature factor in the helix part (37). On the basis of these observations, we speculate that the flexibility of this segment structure is important for taking up protons from the K-pathway, a feature shared with A-type HCORs.
Structural analysis of the oxidized and reduced forms of dimeric CcO revealed changes dependent on the redox state in 4 regions inside subunits I and II: 1) the heme a3–CuB site in subunit I, 2) the Val380–Met383 segment in helix X between hemes a and a3, 3) the Gly49–Asn55 segment in the loop of helices I–II in subunit I, and 4) the water cluster around Mg2+ site between subunits I and II (4, 39, 40). These structural changes may be involved in expressing the function of this enzyme. To explore this idea, we generated a crystal in which monomeric CcO was reduced (SI Appendix, Fig. S5 C–E), and determined its structure at a resolution of 1.95 Å (SI Appendix, Table S1, PDB ID code 6JY4) (41). The structural changes observed for monomeric CcO were essentially the same as those observed previously for dimeric CcO (SI Appendix, Fig. S10). This fact indicates that these structural changes are not dependent on crystal packing. In addition, the difference in activity between monomeric and dimeric CcO does not depend on the difference in redox-dependent structural changes between monomeric and dimeric CcO, supporting the idea that there is a difference in proton uptake function at the entrance of the K-pathway.
Lipid Structures in Monomeric Bovine CcO.
In the electron density map of monomeric CcO determined in this study, we observed a long, thin electron density that could indicate lipids or 3OM. To determine the structure of the phospholipid, we carried out X-ray diffraction measurement at a wavelength of 1.75 Å. This enabled detection of the anomalous scattering effect of phosphorus atoms, and allowed us to obtain an anomalous difference Fourier map with 2.76-Å resolution (SI Appendix, Table S1). On this map, we observed 9 peaks corresponding to phosphorus atoms in the phospholipid, in addition to peaks for sulfur atoms contained in many Met and Cys residues (SI Appendix, Fig. S11). Combining this result with the FO–FC difference electron density map, we determined the structures of the 3 cardiolipins (CLs) in monomeric CcO (SI Appendix, Fig. S11 A–C). At the position of CL1, previously modeled in dimeric CcO, the electron density of cardiolipin was not present in monomeric CcO. At the position of CL2 modeled in dimeric CcO, the electron density of cardiolipin was also observed in monomeric CcO. Two other electron densities for cardiolipins (CL3 and CL4) were detected in monomeric CcO. In a previous analysis of dimeric CcO, the lipids bound at the CL3 and CL4 positions were regarded as triglycerides. In previous analyses, identification of phosphorus atoms using anomalous difference Fourier map was not performed (14); therefore, for dimeric CcO, the identification of phospholipids will need to be reexamined.
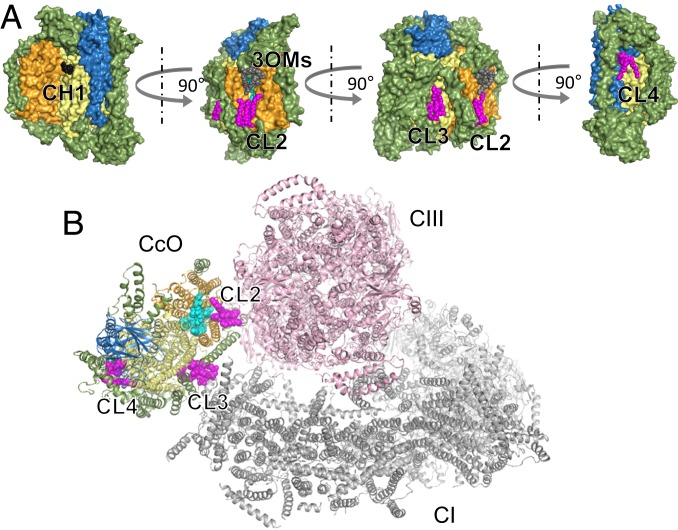
CL2 bound to the matrix side of the transmembrane surface of subunit III (Fig. 4A), and of the 2 phosphate groups PA1 and PB2, PA1 formed hydrogen bonds with NH2 and Nε of Arg63 in subunit III. CL3 bound to the matrix side of the transmembrane surfaces of subunit I and VIIc (Fig. 4A), and PB2 formed hydrogen bonds with the OH of Ser14 and the NH2 of Arg20 in subunit VIIc. CL4 bound to the intermembrane side of the transmembrane surfaces of subunit I, II, and VIc (Fig. 4A), and PA1 formed hydrogen bonds with the NH2 of Arg43 of subunit I. As indicated above, hydrogen bonds of phosphate groups play important role in the binding stability of the 3 cardiolipins. In addition, of the 4 fatty acid tails of CL2, CL3, and CL4, 1 or 2 were strongly bound to the transmembrane helix via hydrophobic interactions. The electron density of the remaining fatty acid tails was comparatively low, and it is likely that these structures are unstable.
Fig. 4.
Lipid structures on monomeric CcO (A) and the contact surface for formation of the respiratory supercomplex (B). The surfaces of subunits I, II, III, and other subunits in CcO are shown in yellow, blue, orange, and green, respectively. CLs are shown as magenta sphere models. PGs and PE, shown as cyan sphere models, are in the cleft between the transmembrane helix bundles of subunit III. CcO is colored as in Fig. 1. (B) Complexes I and III from PDB ID code 5J4Z (56) are shown as gray and pink models, respectively.
Phospholipids identified other than the CLs were 1 phosphatidylethanolamine (PE1) and 2 phosphatidylglycerols (PG1, PG2) (SI Appendix, Fig. S11 D–F). All of these phospholipids were bound in a hydrophobic cavity sandwiched in the transmembrane helices of subunit III (PE1 on the intermembrane side, and PG1 and PG2 on the matrix side) (Fig. 4). All provided a clear anomalous difference Fourier peak of phosphorus atoms, and their phosphate groups formed hydrogen bonds with amino acid residues. Except for a small region at the end of the fatty acid tail, the electron density of the entire lipid was also clear. In addition, even in a previous structural analysis of dimeric CcO, phospholipids of the same types were identified at the positions of these phospholipids (14). Aside from the 6 phospholipids described above, a number of long, thin electron densities were observed, likely representing the hydrophobic tails of lipids or 3OM. Of these 3, the electron density at the head of maltose could be clearly observed and was therefore assigned as 3OM (SI Appendix, Fig. S11 G–I); 3OM was bound to the transmembrane surface of subunit III and positioned in a layer on the opposite side of the lipid bilayer from CL2 (Fig. 4A). This region is thought to be the entry point of the channel that transports substrate dioxygen from subunit III to the heme a3–CuB site. Therefore, some lipid is bound to what was originally the position of 3OM, forming (along with CL2) a hydrophobic environment at the surface of subunit III; this arrangement may promote efficient uptake of dioxygen. A long, thin electron density was also observed on the transmembrane surface that forms the cavity between the 2 monomers of dimeric CcO, but it was difficult to determine whether this was lipid or 3OM. These electron densities were comparatively low, implying that the lipids and 3OM do not have high affinity for these positions. Moreover, clear electron density corresponding to cholate (CH1) was observed in this region (SI Appendix, Fig. S11J and Fig. 4A). CH1 was hydrogen bonded to Thr301I, Trp99III, and His103III, and to Asp298I and Thr301I via a water molecule. CH1 was bonded in the same way with dimeric CcO, and was about 4 Å from Leu127III in the monomer on the other side. The bonding site of CH1 is separated from the functional sites of CcO; therefore, it is unlikely that CH1 binding is involved in enzyme activity.
Cardiolipin is involved in the stability of subunits associated with the structure of bovine CcO, as well as in enzyme activity (42–45). In addition, biochemical experiments and computer simulations have revealed that cardiolipin is involved in the formation of the mitochondrial respiratory supercomplex (46, 47). In recent years, active progress has been made in structural analysis of supercomplexes using cryo-electron microscopy, leading to elucidation of the structure of CcO in the supercomplex (Fig. 4B). Within the structure of the supercomplex, space has been observed between CcO and CI or CIII, suggesting that phospholipids are present in this space and participate in crosslinking between complexes (20, 21). When we superposed the structure of monomeric CcO determined in this study onto the supercomplex structure, we found that CL3 is located between CcO and complex I, and CL2 is located in the space between CcO and complex III (Fig. 4B). Therefore, within the fatty acid tails of CL3 and CL2, the flexible portions corresponding to regions of low electron density form supercomplex through interactions with complex I and complex III, respectively, or with lipids bound to them. In addition, it has been proposed that Higd1a, a membrane protein involved in regulation of CcO activity, binds transmembrane surface near CL4 (48). Therefore, the fatty acid tails of CL4 may be involved in the interaction with Higd1a. In a CIII–CIV supercomplex derived from a Mycobacterium, recently determined by cryo-electron microscopy, CLs were identified between CIII and CIV, and shown to interact with specific residues (49, 50). It is therefore conceivable that CL carries out supercomplex formation in a species-universal fashion.
At the mitochondrial inner membrane in the bovine heart, the enzymes of the respiratory chain (CI, CIII, CcO [complex IV: CIV], and ATP synthase [complex V: CV]) are present in the ratio CI: CIII: CIV: CV = 1: 3: 6–7: 3–5, and it has been suggested that they are organized as a higher order structure conforming to the shape of cristae (51–53). CcO is present as a monomer in the supercomplex, but in this structure, the region corresponding to the association surface for dimerization faces outward; thus, formation of dimeric CcO is possible through association of monomeric CcO. Therefore, it has been proposed that dimerization may contribute to lining up the supercomplex with regularities in the membrane. The structure of the megacomplex CI2CIII2CIV2, an even higher-order structure, has recently been elucidated; in this megacomplex, 2 CcO monomers associate with CI and CIII in the monomeric state, but due to steric hindrance, it is impossible to form dimeric CcO via the other CcO molecule (23). The CcO in this megacomplex is unable to form a dimer, indicating that dimerization is not an effective means to fit the supercomplex compactly within the mitochondrial membrane, allowing it to function efficiently at the membrane. As shown in Fig. 1A, dimeric CcO is present in the mitochondrial membrane, although not as abundantly as monomeric CcO and supercomplex. In addition, recent quantitative analysis of the mitochondrial membrane has revealed the presence of the dimer (54). Based on these results, we postulate that CcO is present in the mitochondrial membrane in 3 states: independent monomer, monomer within the supercomplex, and dimer.
The NDUFA4 protein bonds with CcO as a 14th subunit (55). In the structure containing NDUFA4, CcO dimerization is impossible due to the occurrence of steric hindrance. Therefore, it is thought that in the physiological environment, monomeric CcO is stabilized when NDFUA4 bonding occurs. In addition, the dimeric CcO structure may be stabilized by binding of physiological ligands structurally similar to cholate, implying that the activity of dimeric CcO is dependent on such ligands (e.g., ATP and cholesterol) in the environment. Because the activity of the dimer in the physiological environment is also expected to be lower than that of the monomer, the monomer and dimer can be regarded as the active form of CcO and the standby form for activation by monomerization, respectively. Moreover, the equilibrium relationship among the 3 states containing CcO in the supercomplex may be involved in regulation of CcO activity.
Materials and Methods
CcO was purified from bovine heart mitochondria as described in SI Appendix, Materials and Methods. Crystallization of CcO dissolved in 3OM was performed by the batch method at 277 K. CcO was mixed with a precipitant, polyethylene glycol 4000. Cytochrome c oxidation activity was measured at 20 °C by monitoring the decrease in absorbance at 550 nm as described in SI Appendix, Materials and Methods. Preparation of crystals for X-ray diffraction experiments under cryogenic conditions is described in SI Appendix, Materials and Methods. X-ray diffraction measurements were performed at BL44XU of SPring-8. Processing of X-ray diffraction data and structural analysis are described in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
Diffraction data were collected at the Osaka University beamline BL44XU at SPring-8 (Harima, Japan) (Proposal No. 2010B6500 and 2011A6500). This work was supported by Japan Society for the Promotion of Science KAKENHI Grants 117048028, 22570122, and 17H03646 (to K.S.-I); and 22370060, 15K07029, and 18K06162 (to K.M.). We acknowledge support from Tomoko Maeda for the sample preparation and Hidenori Fujisawa for data collection.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Atomic coordinates and structure factors have been deposited in the Protein Data Bank, under PDB ID codes 6JY3 and 6JY4).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1907183116/-/DCSupplemental.
References
- 1.Yoshikawa S., Shimada A., Reaction mechanism of cytochrome c oxidase. Chem. Rev. 115, 1936–1989 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Wikström M., Krab K., Sharma V., Oxygen activation and energy conservation by cytochrome c oxidase. Chem. Rev. 118, 2469–2490 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sousa F. L., et al. , The superfamily of heme-copper oxygen reductases: Types and evolutionary considerations. Biochim. Biophys. Acta 1817, 629–637 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Yano N., et al. , The Mg2+-containing water cluster of mammalian cytochrome c oxidase collects four pumping proton equivalents in each catalytic cycle. J. Biol. Chem. 291, 23882–23894 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koepke J., et al. , High resolution crystal structure of Paracoccus denitrificans cytochrome c oxidase: New insights into the active site and the proton transfer pathways. Biochim. Biophys. Acta 1787, 635–645 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Svensson-Ek M., et al. , The X-ray crystal structures of wild-type and EQ(I-286) mutant cytochrome c oxidases from Rhodobacter sphaeroides. J. Mol. Biol. 321, 329–339 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Lyons J. A., et al. , Structural insights into electron transfer in caa3-type cytochrome oxidase. Nature 487, 514–518 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abramson J., et al. , The structure of the ubiquinol oxidase from Escherichia coli and its ubiquinone binding site. Nat. Struct. Biol. 7, 910–917 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Tiefenbrunn T., et al. , High resolution structure of the ba3 cytochrome c oxidase from Thermus thermophilus in a lipidic environment. PLoS One 6, e22348 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buschmann S., et al. , The structure of cbb3 cytochrome oxidase provides insights into proton pumping. Science 329, 327–330 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Hino T., et al. , Structural basis of biological N2O generation by bacterial nitric oxide reductase. Science 330, 1666–1670 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Matsumoto Y., et al. , Crystal structure of quinol-dependent nitric oxide reductase from Geobacillus stearothermophilus. Nat. Struct. Mol. Biol. 19, 238–245 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Lee S. J., et al. , Intermonomer interactions in dimer of bovine heart cytochrome c oxidase. Acta Crystallogr. D Biol. Crystallogr. 57, 941–947 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Shinzawa-Itoh K., et al. , Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. EMBO J. 26, 1713–1725 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liko I., et al. , Dimer interface of bovine cytochrome c oxidase is influenced by local posttranslational modifications and lipid binding. Proc. Natl. Acad. Sci. U.S.A. 113, 8230–8235 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arnold S., Kadenbach B., Cell respiration is controlled by ATP, an allosteric inhibitor of cytochrome-c oxidase. Eur. J. Biochem. 249, 350–354 (1997). [DOI] [PubMed] [Google Scholar]
- 17.Kadenbach B., Hüttemann M., The subunit composition and function of mammalian cytochrome c oxidase. Mitochondrion 24, 64–76 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Schägger H., Pfeiffer K., Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 19, 1777–1783 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shinzawa-Itoh K., et al. , Purification of active respiratory supercomplex from bovine heart mitochondria enables functional studies. J. Biol. Chem. 291, 4178–4184 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Letts J. A., Fiedorczuk K., Sazanov L. A., The architecture of respiratory supercomplexes. Nature 537, 644–648 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Wu M., Gu J., Guo R., Huang Y., Yang M., Structure of mammalian respiratory supercomplex I1III2IV1. Cell 167, 1598–1609. (2016). [DOI] [PubMed] [Google Scholar]
- 22.Letts J. A., Sazanov L. A., Clarifying the supercomplex: The higher-order organization of the mitochondrial electron transport chain. Nat. Struct. Mol. Biol. 24, 800–808 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Guo R., Zong S., Wu M., Gu J., Yang M., Architecture of human mitochondrial respiratory megacomplex I2III2IV2. Cell 170, 1247–1257. (2017). [DOI] [PubMed] [Google Scholar]
- 24.Shimada S., et al. , A unique respiratory adaptation in Drosophila independent of supercomplex formation. Biochim. Biophys. Acta Bioenerg. 1859, 154–163 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Musatov A., Ortega-Lopez J., Robinson N. C., Detergent-solubilized bovine cytochrome c oxidase: Dimerization depends on the amphiphilic environment. Biochemistry 39, 12996–13004 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Musatov A., Robinson N. C., Cholate-induced dimerization of detergent- or phospholipid-solubilized bovine cytochrome C oxidase. Biochemistry 41, 4371–4376 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Shinzawa-Itoh K., Muramoto K., Monomeric form of bovine heart cytochrome c oxidase in the fully oxidized state. Protein Data Bank. https://pdbj.org/mine/summary/6jy3. Deposited 26 April 2019.
- 28.Shimada S., et al. , Complex structure of cytochrome c-cytochrome c oxidase reveals a novel protein-protein interaction mode. EMBO J. 36, 291–300 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muramoto K., et al. , Bovine cytochrome c oxidase structures enable O2 reduction with minimization of reactive oxygens and provide a proton-pumping gate. Proc. Natl. Acad. Sci. U.S.A. 107, 7740–7745 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muramoto K., et al. , A histidine residue acting as a controlling site for dioxygen reduction and proton pumping by cytochrome c oxidase. Proc. Natl. Acad. Sci. U.S.A. 104, 7881–7886 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu J., Hiser C., Ferguson-Miller S., Role of conformational change and K-path ligands in controlling cytochrome c oxidase activity. Biochem. Soc. Trans. 45, 1087–1095 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Buuren K. J., Van Gelder B. F., Biochemical and biophysical studies on cytochrome c oxidase. XIII. Effect of cholate on the enzymic activity. Biochim. Biophys. Acta 333, 209–217 (1974). [DOI] [PubMed] [Google Scholar]
- 33.Sinjorgo K. M. C., et al. , The effect of detergents on bovine cytochrome c oxidase: A kinetic approach. Biochim. Biophys. Acta 893, 241–250 (1987). [DOI] [PubMed] [Google Scholar]
- 34.Tsukihara T., et al. , The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science 272, 1136–1144 (1996). [DOI] [PubMed] [Google Scholar]
- 35.Kadenbach B., et al. , Regulation of energy transduction and electron transfer in cytochrome c oxidase by adenine nucleotides. J. Bioenerg. Biomembr. 30, 25–33 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Qin L., Mills D. A., Buhrow L., Hiser C., Ferguson-Miller S., A conserved steroid binding site in cytochrome C oxidase. Biochemistry 47, 9931–9933 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hiser C., Buhrow L., Liu J., Kuhn L., Ferguson-Miller S., A conserved amphipathic ligand binding region influences k-path-dependent activity of cytochrome C oxidase. Biochemistry 52, 1385–1396 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hiser C., Liu J., Ferguson-Miller S., The K-path entrance in cytochrome c oxidase is defined by mutation of E101 and controlled by an adjacent ligand binding domain. Biochim. Biophys. Acta Bioenerg. 1859, 725–733 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshikawa S., et al. , Redox-coupled crystal structural changes in bovine heart cytochrome c oxidase. Science 280, 1723–1729 (1998). [DOI] [PubMed] [Google Scholar]
- 40.Tsukihara T., et al. , The low-spin heme of cytochrome c oxidase as the driving element of the proton-pumping process. Proc. Natl. Acad. Sci. U.S.A. 100, 15304–15309 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shinzawa-Itoh K., Muramoto K., Monomeric form of bovine heart cytochrome c oxidase in the fully reduced state. Protein Data Bank. https://pdbj.org/mine/summary/6jy4. Deposited 26 April 2019.
- 42.Sedlák E., Robinson N. C., Destabilization of the quaternary structure of bovine heart cytochrome c oxidase upon removal of tightly bound cardiolipin. Biochemistry 54, 5569–5577 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Musatov A., Robinson N. C., Bound cardiolipin is essential for cytochrome c oxidase proton translocation. Biochimie 105, 159–164 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abramovitch D. A., Marsh D., Powell G. L., Activation of beef-heart cytochrome c oxidase by cardiolipin and analogues of cardiolipin. Biochim. Biophys. Acta 1020, 34–42 (1990). [DOI] [PubMed] [Google Scholar]
- 45.Fry M., Green D. E., Cardiolipin requirement by cytochrome oxidase and the catalytic role of phospholipid. Biochem. Biophys. Res. Commun. 93, 1238–1246 (1980). [DOI] [PubMed] [Google Scholar]
- 46.Mileykovskaya E., Dowhan W., Cardiolipin-dependent formation of mitochondrial respiratory supercomplexes. Chem. Phys. Lipids 179, 42–48 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arnarez C., Marrink S. J., Periole X., Molecular mechanism of cardiolipin-mediated assembly of respiratory chain supercomplexes. Chem. Sci. 7, 4435–4443 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hayashi T., et al. , Higd1a is a positive regulator of cytochrome c oxidase. Proc. Natl. Acad. Sci. U.S.A. 112, 1553–1558 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gong H., et al. , An electron transfer path connects subunits of a mycobacterial respiratory supercomplex. Science 362, eaat8923 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Wiseman B., et al. , Structure of a functional obligate complex III2IV2 respiratory supercomplex from Mycobacterium smegmatis. Nat. Struct. Mol. Biol. 25, 1128–1136 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Hatefi Y., The mitochondrial electron transport and oxidative phosphorylation system. Annu. Rev. Biochem. 54, 1015–1069 (1985). [DOI] [PubMed] [Google Scholar]
- 52.Allen R. D., Schroeder C. C., Fok A. K., An investigation of mitochondrial inner membranes by rapid-freeze deep-etch techniques. J. Cell Biol. 108, 2233–2240 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davies K. M., et al. , Macromolecular organization of ATP synthase and complex I in whole mitochondria. Proc. Natl. Acad. Sci. U.S.A. 108, 14121–14126 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chorev D. S., et al. , Protein assemblies ejected directly from native membranes yield complexes for mass spectrometry. Science 362, 829–834 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zong S., et al. , Structure of the intact 14-subunit human cytochrome c oxidase. Cell Res. 28, 1026–1034 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Letts J. A., Fiedorczuk K., Sazanov L. A., Architecture of tight respirasome. Protein Data Bank. https://www.rcsb.org/structure/5J4Z. Deposited 1 April 2016.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.