Significance
Multidrug resistance (MDR) constitutes a major obstacle in the treatment of infectious diseases, as well as human cancers, even in developed countries. This study deciphers the structural mechanism that dictates the promiscuous multidrug binding to the variable transcriptional regulation of QacR, the major multidrug-binding transcriptional repressor in pathogenic bacteria Staphylococcus aureus. The transcriptional level of the downstream transporter gene, qacA, is dynamically regulated by QacR via the relative populations of inducive versus repressive conformations in the molecule. The conformational equilibrium also defines the basal activity of the MDR system, as well as the elevated transcription levels with the constitutively active mutants. Our structural definition provides an important advance in understanding and potentially targeting the MDR system.
Keywords: multidrug resistance, transcription factor, structure, dynamics, NMR
Abstract
QacR, a multidrug-binding transcriptional repressor in pathogenic bacteria Staphylococcus aureus, modulates the transcriptional level of the multidrug transporter gene, qacA, in response to engaging a set of diverse ligands. However, the structural basis that defines the variable induction level remains unknown. Here, we reveal that the conformational equilibrium between the repressive and inducive conformations in QacR defines the induction level of the transporter gene. In addition, the unligated QacR is already partly populated in the inducive conformation, allowing the basal expression of the transporter. We also showed that, in the known constitutively active QacR mutants, the equilibrium is shifted more toward the inducive conformation, even in the unligated state. These results highlight the unexpected structural mechanism, connecting the promiscuous multidrug binding to the variable transcriptional regulation of QacR, which provide clues to dysfunctioning of the multidrug resistance systems.
Multidrug resistance (MDR) systems are ubiquitous in life, as the efflux of toxic compounds is essential for survival. MDR activities are often achieved by increasing the expression level of multidrug transporters (1–3). Robust expression of multidrug transporters constitutes a major roadblock in the treatment of infectious diseases, as well as human cancers, as it reduces the curative effects of antibiotics, antifungal agents, disinfectants, and chemotherapeutic compounds (4–7). The expression of a multidrug transporter is often regulated by a multidrug-binding transcriptional repressor or activator, which has the ability to bind a range of toxic compounds (8–10). Thus, MDR systems, consisting of a multidrug transcriptional regulator and transporter pair, can efficiently efflux toxic compounds by elevating the expression level of the required multidrug transporter.
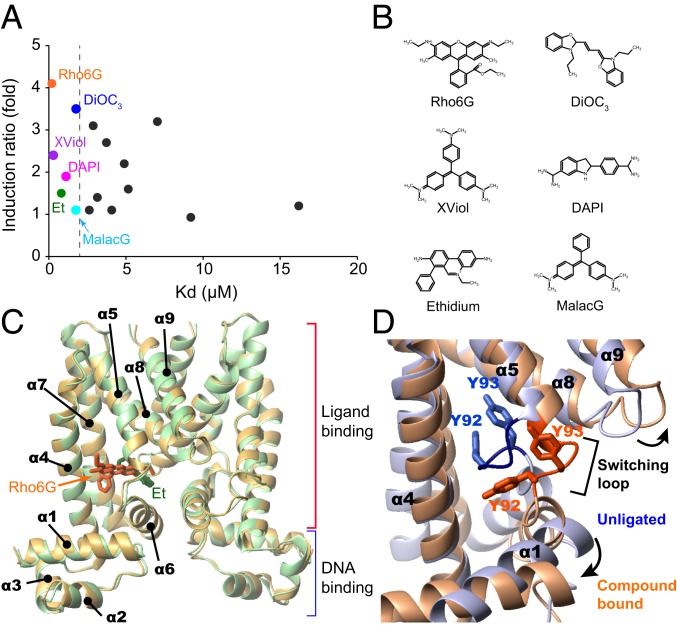
Quaternary ammonium compound transcriptional repressor, QacR, is a multidrug-binding transcriptional repressor in pathogenic bacteria Staphylococcus aureus that represses the expression of the multidrug transporter gene qacA (11) (SI Appendix, Fig. S1A). Although only quaternary ammonium compounds were originally considered to bind to QacR, more-diverse lipophilic monovalent and bivalent cationic toxic compounds that are different in size and shape are also currently known to bind to QacR (12) (SI Appendix, Table. S1). QacR binds to the 28-base pair (bp) inverted repeat 1 (IR1) operator that overlaps with the qacA promoter, for the repression of the gene expression (13, 14) (SI Appendix, Fig. S1A). Nevertheless, there is basal expression of the qacA gene, which presumably provides a minimal degree of resistance to the toxic compounds (SI Appendix, Fig. S1B) (11). The full induction level of the qacA promoter was estimated by deleting the IR1 operator, with a corresponding induction ratio (the ratio between the induced and basal expression levels of the qacA gene) of 4.7-fold (SI Appendix, Fig. S1B) (11). Binding of the agonistic compound to QacR releases the repression of the qacA gene to various extents (12). The induction ratio varies significantly, ranging from near noninduction up to 4.1-fold (12) (SI Appendix, Table S1 and Fig. S1B), with the maximum compound induction ratio being observed for rhodamine 6G (Rho6G), close to the full induction level of the promoter (SI Appendix, Fig. S1B). Note that we refer to the loss of the repressive ability of QacR as inducive, since the loss of the repressive ability of QacR upon the binding to ligands results in the induction of the transporter gene expression. Notably, the induction level does not correlate with the reported affinity of the compounds, suggesting that the compound binding regulates the transcriptional activity of QacR in an allosteric manner (Fig. 1 A and B).
Fig. 1.
Affinities and induction activities of compounds and X-ray structures of QacR−compound complexes. (A) Induction ratios and affinities of compounds to QacR. The induction ratios and KD values are taken from Grkovic et al. (12). Each dot represents one compound and the compounds used in this study are color coded and labeled. (B) Chemical structures of compounds used in this study. (C) The structures of QacR in complex with high (Rho6G) and low (Et) inductive compounds. (D) Expansion of the MDB pocket via the “coil-to-helix” transition. Structures of unligated (light blue) and Rho6G-bound (orange) QacR are overlaid. Rho6G is omitted from the structure for clarity.
QacR belongs to the TetR family and functions as a homodimer. Each subunit of QacR is composed of 9 α-helices, in which the N-terminal 3 helices (α1 to α3) comprise the helix−turn−helix DNA binding domain (DBD) and the last 6 helices (α4 to α9) form the ligand binding domain (LBD), including the dimerization interface and the multidrug binding (MDB) pocket (8, 12, 14–16) (Fig. 1C). The conformational difference between the unligated and compound-bound QacR reveals that the protein undergoes a coil-to-helix transition in the region termed the “switching loop” (residues 89 to 93) (16) (Fig. 1D). The coil-to-helix transition is associated with the expulsion of the Tyr-92 and Tyr-93 sidechains from the interior of the QacR MDB pocket and the expansion of the MDB pocket from ∼400 Å3 to 1,100 Å3 (Fig. 1D and SI Appendix, Fig. S2A) (14, 16). The formation of additional hydrogen bonds and the dissolution of a Ramachandran outlier at Tyr-92 by the coil-to-helix transition (SI Appendix, Fig. S2B) might compensate for the energetically unfavorable Tyr expulsion. The expansion of the MDB pocket leads to the reorganization of the DBDs, which extends the distance between the 2 DNA-binding helices, rendering them too far apart to fit into the DNA major grooves (SI Appendix, Fig. S2A). Therefore, QacR with the expanded MDB pocket reduces the binding to the IR1 operator and induces the expression of the downstream qacA (16).
More than 20 structures of QacR−compound complexes have been reported to date, including those for QacR mutants (8, 14–17). The structures clearly showed that the MDB pocket of QacR has at least 2 distinct subpockets that can accommodate structurally dissimilar compounds in different positions (16) (SI Appendix, Fig. S2C). However, the overall structures of the available QacR−compound complexes are almost identical, with <1 Å overall pairwise RMSD values for all Cα atoms, while the QacR-driven induction of qacA varies significantly among the compounds (Fig. 1 A and C) (8, 16, 17). There are only a few noticeable differences in the sidechain conformers in the MDB pocket of the compound-bound QacR structures. Therefore, while the X-ray structures provide the structural details of the inducive and repressive conformations that are implemented in the QacR molecule, the structural basis that defines the various levels in the transcriptional induction upon binding to agonistic compounds remains unknown.
Here, we carried out the structural and dynamical analyses of QacR in complex with 6 agonistic compounds with different induction activities (Fig. 1A), along with its cognate DNA in solution, by NMR spectroscopy. NMR analyses reveals that there is a preexisting conformational equilibrium between the repressive and inducive conformations in QacR, and the populations of the inducive conformations in the equilibrium define the induction levels of QacR. Compound binding to QacR shifts the equilibrium toward the inducive conformation, and the extent to which this equilibrium is shifted matches the induction ratios observed with the respective ligand. In contrast, interaction with the operator DNA shifts the equilibrium of QacR toward the repressive conformation, which is suitable for the DNA binding. It should be noted that unligated QacR is already partially populated in the inducive conformation, allowing the basal expression of the transporter. These results provide the molecular mechanism, by which QacR functions as a variable sensor for multiple drugs with different sizes and shapes in the MDR system.
Results
Selection of Compounds for the NMR Analyses of QacR.
Six compounds, which reported to show low-micromolar dissociation constant (KD) values to QacR were selected to ensure that a sufficient population of QacR subunits is occupied by compounds under NMR experimental conditions (Fig. 1 A and B). The selected compounds are Rho6G, 3′,3′-dipropyloxacarbocyanine (DiOC3), crystal violet (XViol), 4',6-diamidino-2-phenylindole (DAPI), ethidium (Et), and malachite green (MalacG), in the order of the induction activity (the highest to the lowest; Fig. 1 A and B and SI Appendix, Table S1) (12). These compounds have different sizes and shapes, with calculated molecular volumes between 227 Å3 and 375 Å3 and molecular weights ranging from 277 to 444. Rho6G, XViol, Et, MalacG, and DiOC3 are monovalent cation compounds, and DAPI is a bivalent compound. The binding affinity of these compounds to QacR was determined by isothermal titration calorimetry (ITC) experiments (SI Appendix, Fig. S3). The experiments verified that the KD values of the compounds are in the low micromolar range, while the KD value for DAPI was not able to be determined (SI Appendix, Fig. S3 legend). A detergent-like compound, cetylpyridinium, reportedly binds to QacR with 0.47 μM affinity (12). However, the compound is expected to form a micelle under the experimental conditions and thus was not used in the present study.
While the selected compounds share high affinities with the reported and measured KD values in the low micromolar range to QacR, the induction ratios upon binding to these compounds differ substantially (Fig. 1A and SI Appendix, Table S1) (12). Rho6G is the strongest inducer and has an induction ratio of 4.1, which is almost equal to the full induction level of the promoter (4.7-fold; SI Appendix, Fig. S1B). In contrast, the weakest MalacG shows almost no induction (induction ratio of 1.1). Thus, the panel of ligands selected allows dissection of the molecular basis for the variable induction levels and the associated conformations of the compound-bound QacR in solution.
Conformational Change of QacR upon Rho6G Binding.
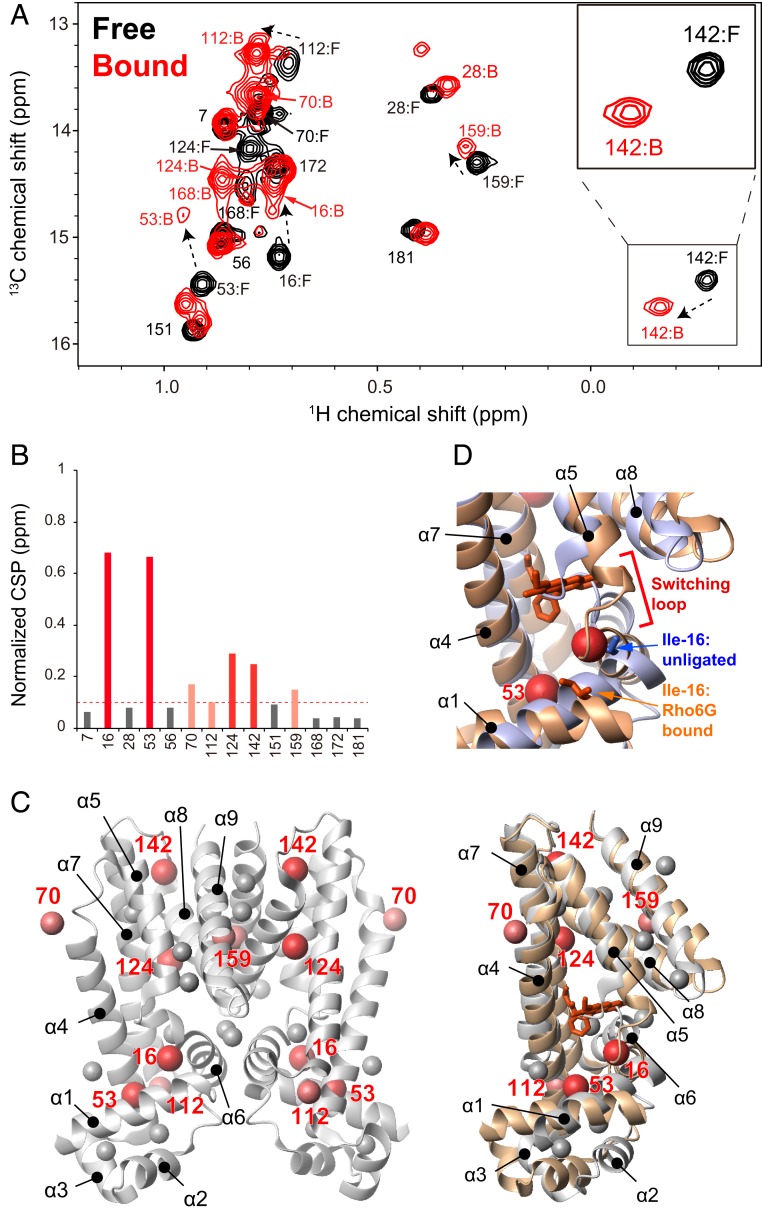
To investigate the structural changes of QacR upon binding to the compounds, we used the methyl resonances of Ile as reporters of binding and conformational state of QacR. Selective Ile δ1 methyl labeling under perdeuterated condition and the 1H-13C heteronuclear multiple quantum coherence (HMQC) spectra have been effectively applied to high-molecular-weight proteins (18). The assignments were established by the disappearance of corresponding NMR signals by introducing specific Ile to Leu mutations (SI Appendix, Fig. S4). An overlay of the 1H-13C HMQC spectra of QacR (100 μM, subunit concentration) in the absence (black) and presence of 120 μM Rho6G (red) is shown in Fig. 2A. All 17 Ile δ1 signals were visible in the unligated QacR spectrum; however, the signals from Ile-99 and Ile-100 in the α6 helix, and Ile-130 in the α7 helix, which constitute the MDB pocket, exhibited weaker intensities compared to other resonances (SI Appendix, Fig. S5). These indicate the presence of a substantial conformational exchange at these sites in the millisecond to microsecond time scale that causes line broadening of the signals from the MDB pocket.
Fig. 2.
NMR analysis of the interaction between QacR and Rho6G. (A) Ile δ1 resonances of 100 μM QacR (subunit concentration) in unligated (black) and excess amount of Rho6G (120 μM, red). Inset is the enlarged view of the Ile-142 signal in unligated and the Rho6G-bound states. (B) Normalized CSPs for each residue upon binding to a 2:2 stoichiometric amount of Rho6G. (C) (Left) Mapping of substantially perturbed (normalized CSP > 0.1 ppm) residues on the ribbon representation of QacR in the unligated condition. (Right) Mappings to the overlaid structures of the unligated (white) and Rho6G-bound (brown) subunits are also shown. Rho6G is shown in a stick representation. (D) Enlarged view of the switching loop, along with the positions of Ile-16 in the unligated and Rho6G-bound X-ray structures.
Titration of Rho6G to QacR showed chemical shift perturbations (CSPs) in the slow exchange time regime (SI Appendix, Fig. S6). The titration indicated a 2:2 stoichiometry (each subunit of a QacR dimer binds to one Rho6G molecule; Fig. 2A and SI Appendix, Fig. S6), which is consistent with the result from the ITC experiment. The residues that experienced >0.1 parts per million (ppm) normalized CSPs upon binding to Rho6G were Ile-16, Ile-53, Ile-70, Ile-112, Ile-124, Ile-142, and Ile-159 (Fig. 2B). The distribution of the residues that experienced substantial CSPs was located in the region with the structural differences between unligated and compound-bound crystal structures (Fig. 2C) (8, 14, 16). Ile-70, Ile-112, and Ile-142 are in the hinge region between α4/α5, α6/α7, and α7/α8, respectively, and Ile-159 is located at the dimeric interface. In both the hinge regions and dimer interface, the relative orientations of the helices (α5, α6, α8, and α9) against the 2 backbone helices (α4 and α7) differ substantially between the unligated and compound-bound structures (Fig. 2 C, Right). Ile-124 resides in between the α7 and α8 helices, with the α8 helix being one of the helices that are displaced upon the expansion of the MDB pocket. The CSPs are also distributed to the residues that are distal from the compound-binding site, including those in the interface between the LBD and DBD (Ile-16 and Ile-53). The CSP in Ile-16, located near the “switching loop,” would be of importance (Fig. 2D), as the structural transition at the site is critical for the transcriptional regulation. Given that the maximum compound induction ratio was observed upon binding to Rho6G, these results suggest that the CSPs induced by Rho6G correspond to conformational changes from the repressive to the fully inducive conformation, which are associated with the coil-to-helix transition of the “switching loop” near Ile-16 and the expansion of the MDB pocket (Fig. 2 and SI Appendix, Fig. S2).
Conformational Equilibrium in the QacR−Compound Complexes Defines the Variable Induction Levels by Compounds.
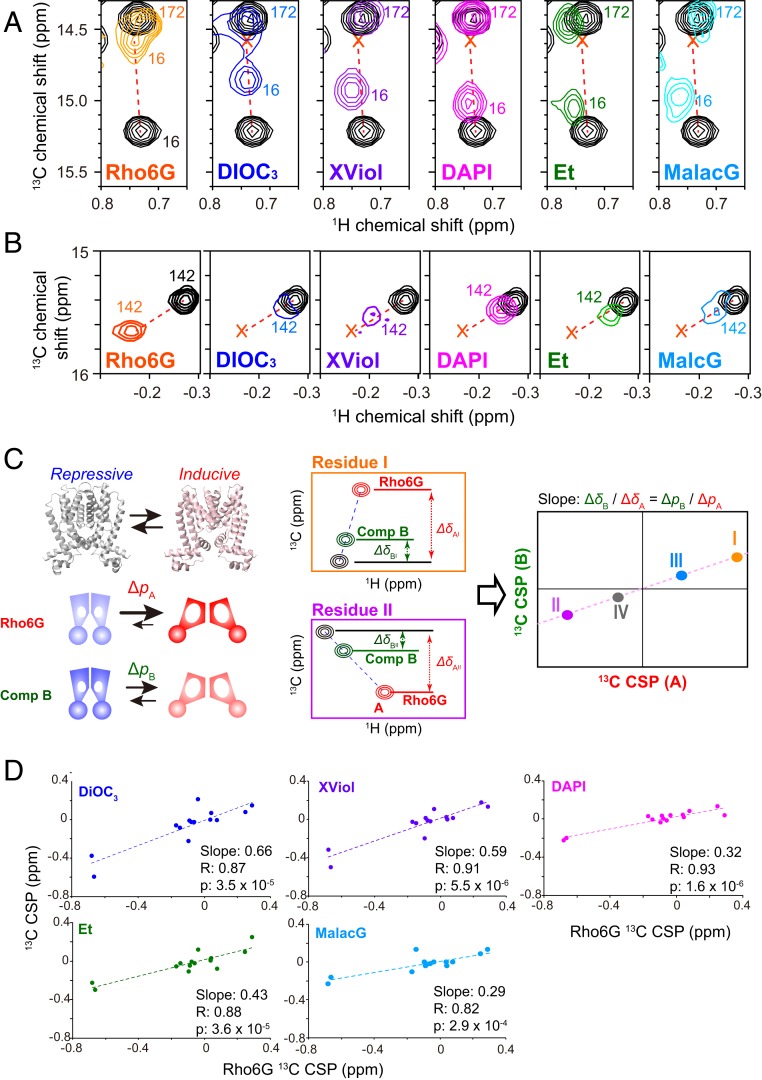
To analyze the conformation of QacR upon binding to the compounds that results in various induction levels, the HMQC spectra of QacR (100 μM subunit concentration) in the absence and presence of an excess amount of compounds (DiOC3, XViol, DAPI, Et, or MalacG; 120 μM, >80% occupancy) were recorded (SI Appendix, Fig. S6). The mapping of the normalized CSPs on the structure of QacR indicates that the strong CSPs induced by the ligands were distributed on a similar position on QacR to those induced by Rho6G (SI Appendix, Fig. S7). In addition, the overlay of the compound-bound spectra with unligated spectrum indicates that several resonances exhibit similar patterns in their chemical shift changes, in which the compound-bound signals are distributed in a line connecting Rho6G-bound and unligated state signals (Fig. 3 A and B). Fig. 3 A and B shows the examples for Ile-16 near the “switching loop” and Ile-142 at the hinge of the α7 and α8 helices (SI Appendix, Fig. S9), respectively. As both of the methyl groups are distant from the compound binding sites, the observed CSPs are not caused by the direct binding interface of the compounds but rather reflect an allosteric conformational change induced by the binding (19).
Fig. 3.
Comparison of the compound-induced chemical shift change to QacR. (A and B) The Ile δ1 resonances of QacR in the unligated state (black) were overlaid with those with the 2:2 stoichiometric amount of the indicated compounds for (A) Ile-16 in the switch region and (B) Ile-142 at the hinge of the compound-binding α7 and α8 helices. The peak position of the Rho6G-bound state is indicated by “x.” Unligated and Rho6G-bound peak positions are connected by the broken line. (C) (Left) The anticipated conformational equilibrium between the repressive and inductive conformations. ΔpA and ΔpB are the amplitude of the population shift induced by Rho6G and another compound (compound B), respectively. (Middle) The amplitude of the allosteric 13C CSPs induced by Rho6G and compound B for residue I and II (ΔδAx and ΔδBx, respectively). Assuming the conformational equilibrium between the 2 conformations, which is fast in NMR time scale, the ratio of compound B CSPs relative to that of Rho6G for each signal (ΔδBi/ΔδAi) would be equal to ΔpB/ΔpA. (Right) Therefore, if the CSPs induced by Rho6G and those induced by the compound B are plotted on the horizontal and the vertical axis, respectively, the CSPs should correlate with each other in a linear fashion, and the slope obtained from the global linear fit should be ΔpB/ΔpA. (D) Correlation between Ile δ1 13C CSPs upon binding of Rho6G, relative to those upon binding of the indicated compounds. Horizontal and vertical axes are for Rho6G and the indicated compound 13C CSPs, respectively. Only CSPs originating from the methyl moiety that are allosteric (>5 Å) from the compound are plotted.
The linearity in the CSPs with Rho6G and other compounds suggest a model in which QacR−compound complexes are in exchange between the repressive and inducive conformations (Fig. 3 C, Left). Since there is no significant line broadening of the bound signals, the exchange should be fast in NMR time scale, and the observed CSPs would linearly correlate with the shift in population of the 2 exchanging conformations (Fig. 3 C, Left and Middle). In this respect, it is notable that the amplitude of CSPs upon binding to each compound is in good agreement with their induction activities (the panels in Fig. 3 A and B are shown in the order of the induction activity of the compounds).
If QacR−compound complexes share conformational equilibrium between the repressive and inducive conformations, a linear correlation between the CSPs induced by Rho6G and those by other compounds would be expected for other residues that are distant from the MDB pocket as well (see Fig. 3C and the legend for detailed discussion). Fig. 3D shows the correlation between the 13C CSPs induced by Rho6G and those by other compounds. Significant correlations exist between the 13C CSPs induced by Rho6G and all of the other compounds, supporting a model that a conformational equilibrium is shared among the QacR−compound complexes, whereas the amplitude of the shift in the conformational equilibrium varies among compounds.
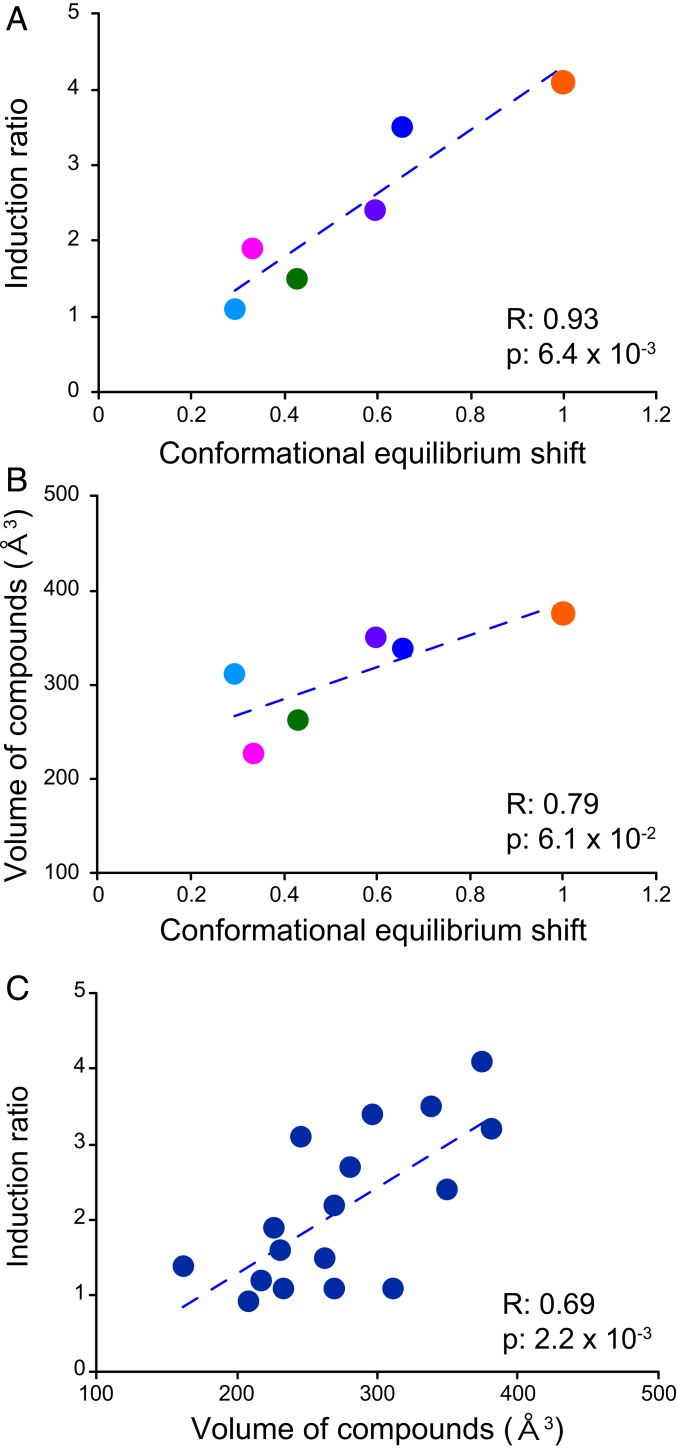
To further explore the functional relevance of the correlated 13C CSPs, we compared the amplitude of the allosteric CSPs for each compound relative to the full inducer Rho6G (i.e., the slopes of the correlation plots in Fig. 3D). Notably, the amplitudes of the compound-induced CSPs showed significant correlations with their reported induction ratios (Fig. 4A) (12). This finding clearly indicates that a shift in equilibrium toward an inducive conformation, which is reflected in the amplitudes of the allosteric 13C CSPs, defines the induction activity of QacR in each compound-bound state.
Fig. 4.
Identification and characterization of the functional conformational equilibrium that defines QacR transcriptional regulation. (A) Correlation between the compound induction ratio and the conformational equilibrium shift, deduced from the slope of the 13C CSPs shown in Fig. 3. (B) Correlation between the compound volume and the conformational equilibrium shift. (C) Correlation between the compound volume and the compound induction ratio. The compound induction ratios were obtained from Grkovic et al. (12). The compound volumes were calculated using the Chem Sketch software (Advanced Chemistry Development). Each dot represents one compound and the compounds used in this study are color coded: Rho6G, orange; DiOC3, dark blue; XViol, purple; DAPI, pink; Et, green; MalacG, light blue. The broken lines in the figure panels are the approximation straight lines for the data.
Notably, the amplitude of the shift in the conformational equilibrium also correlated with the calculated compound volume (Fig. 4B). This indicates that the volume of the bound compound defines the populations of the repressive and inducive conformations in the equilibrium. Larger compounds tend to push the equilibrium more toward the inducive conformation with an expanded MDB in the bound state, thus showing larger induction ratios. In contrast, QacR is still able to adopt the repressive conformation when it binds to small compounds, and thereby only partially induce the transcription. If this notion is valid, a correlation should exist between the compound volume and their induction ratio, for other compounds that were not used in this NMR study (12). Consistent with this, a significant correlation is apparent between the compound volumes and the reported induction ratios in the literature (12) (Fig. 4C).
Mutually Exclusive Compound/DNA Binding by Opposite Shifts in the Conformational Equilibrium.
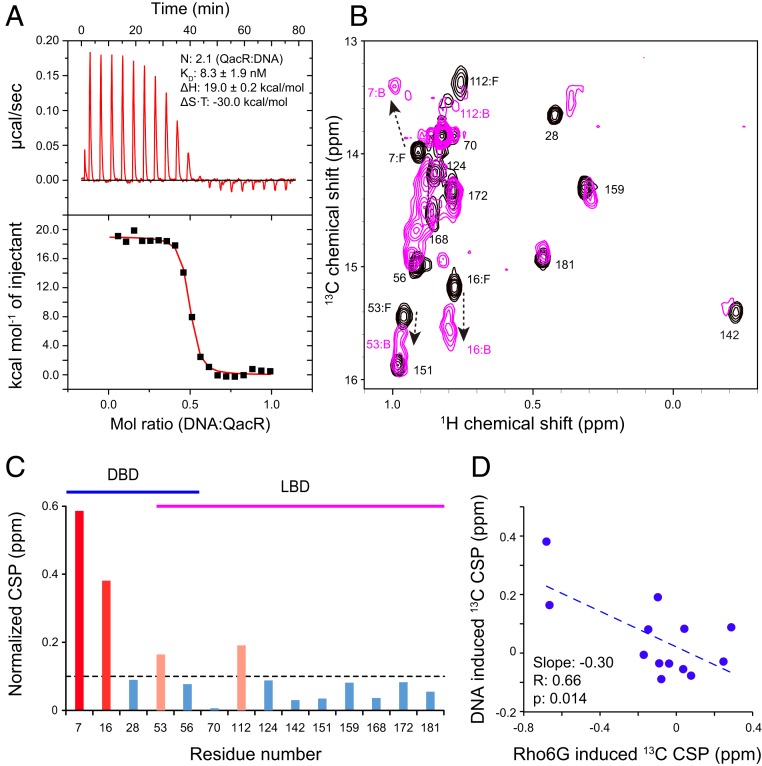
The proposed model anticipates that QacR would show mutually exclusive compound- and DNA-binding properties to exert transcriptional repression activity (8, 14, 16). To gain structural insights into the mutually exclusive interactions, we studied the conformation of QacR in complex with the IR1 operator DNA by NMR. A pair of QacR dimers reportedly binds to a single IR1 operator sequence in the double-strand DNA (14). Interaction between QacR and the IR1 oligo was assayed by ITC, which showed an endothermic interaction with an apparent KD of 8.3 ± 1.9 nM and a stoichiometry of 2.1, in which 2 QacR dimers bind to a single DNA duplex (Fig. 5A). The data are consistent with the previously proposed interaction (13, 14). Although the apparent KD is tighter compared to the value reported in literature (52 nM) (14), it is presumably due to the lower NaCl concentration used in our experimental condition (100 mM rather than 300 mM in the literature). This is consistent with the significant contribution of the electrostatic interactions in the QacR−DNA interaction (14).
Fig. 5.
NMR analysis of the interaction between QacR and the IR1 DNA. (A) ITC experiment of QacR and the IR1 DNA interaction. The IR1 DNA was titrated to the protein. N is the stoichiometry of the QacR dimer to the IR1 DNA duplex (i.e., N = 2 corresponds to the 2 QacR dimer:1 DNA duplex interaction). (B) The Ile δ1 resonances of QacR in the unligated state (black) were overlaid with those with the 2:1 (QacR:DNA) stoichiometric amount of the IR1 DNA. (C) Plot of the magnitudes of the normalized chemical shift change for each residue. Residues showed normalized CSP >0.1 ppm (black broken line) and >0.2 ppm are shown in pink and red, respectively. Residues that belong to DBD and LBD are indicated by blue and magenta lines, respectively. (D) Correlation between 13C CSPs upon binding to Rho6G and those induced by the IR1 DNA. The broken line is the approximation line for the data.
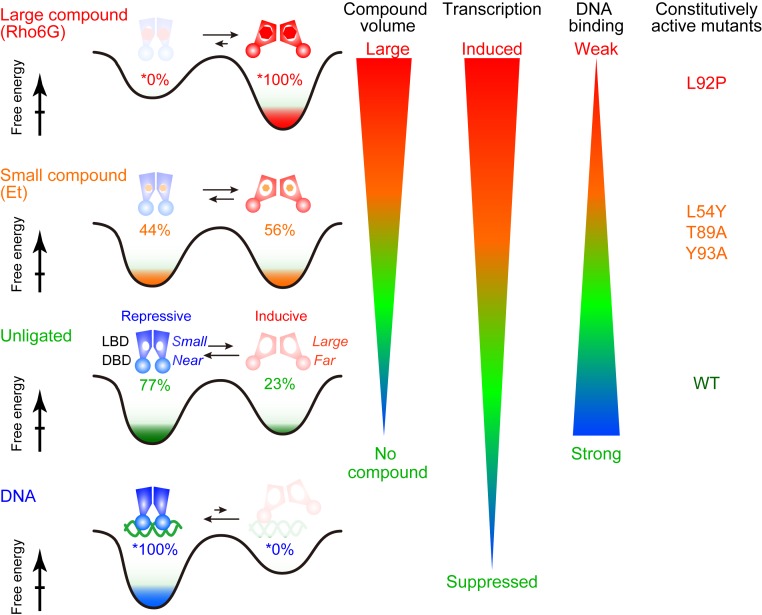
The titration of IR1 to QacR showed CSPs in the slow exchanging regime saturating at the 2:1 QacR dimer:IR1 duplex ratio (Fig. 5B). The CSPs were not only localized to the DNA binding domain but were found in the LBD and the domain interfaces, indicating allosteric conformational changes (Fig. 5C). Notably, 13C CSPs induced by the IR1 DNA binding were anticorrelated with those by Rho6G (Fig. 5D), indicating that the compounds and DNA induce the opposing shifts to the conformational equilibrium, which is fully consistent with the proposed model of the mutually exclusive interaction by the conformational equilibrium (Fig. 6) (14, 16). The results also indicate that the unligated QacR is not fully populated in the repressive conformation. Assuming that the DNA-bound conformation is fully populated in the suppressive conformation (slope = −0.3) and Rho6G is in the fully inducive conformation (slope = 1), ∼23% (= 0.3/1.3) of the unligated QacR has adopted the inducive conformation (Fig. 6). This value quantitatively matches the reported basal expression level of the qacA gene (SI Appendix, Fig. S1; 24%).
Fig. 6.
Schematic representations of the conformational equilibrium in QacR for variable transcriptional regulation. The populations of the conformation in each condition were calculated from the slope of the CSP relative to Rho6G, assuming that the Rho6G- and the IR1 DNA-bound states are fully populated at either end of the conformation in the equilibrium.
Constitutively Active QacR Mutants Exhibit a Conformational Equilibrium Biased toward the Inducive Conformation.
It has been shown that some of the QacR mutants, which have substitution in the MDB pocket, have shown unexpected impairment in repression capabilities (20). The basal levels of transcriptional activity were increased up to ∼4-fold, as compared to those of the wild type (WT) (SI Appendix, Fig. S10A). Especially, L54Y, T89A, Y92P, and Y93A exhibited more than 2-fold increases in the basal transcriptional activity, as compared to that of WT QacR. These substitutions are distributed in the LBD, and are all far from the DNA binding sites (SI Appendix, Fig. S10B). Thus, the substitutions should allosterically impair the repression activity of QacR.
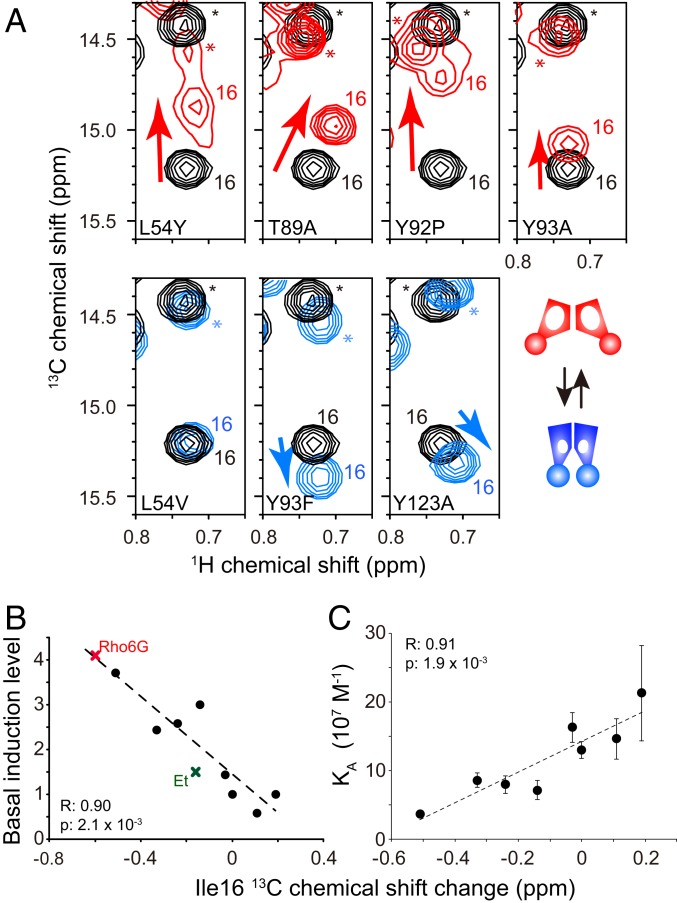
Fig. 7A shows the expanded regions of the 1H-13C HMQC spectra of the constitutively active mutants (L54Y, T89A, Y92P, and Y93A; Fig. 7 A, Top), along with 3 less affected mutations (L54V, Y93F, and Y123A; Fig. 7 A, Bottom), in the unligated state. We analyzed the conformation of the constitutively active mutants that showed more than 2-fold increases in the basal transcriptional activity, as compared to those of WT QacR, along with those of the less affected mutants with marginal increase, equality, or decrease in the basal transcriptional activity as compared to WT (SI Appendix, Fig. S10A). The Ile-16 peak positions of the constitutively active mutants were shifted away from those of WT toward the inducive conformation (Fig. 7A, red). The chemical shift changes induced by the less affected mutations were much smaller (Fig. 7A, blue). In addition, the chemical shift changes significantly correlated with the increase in the basal transcriptional activity (Fig. 7B) (20). It should also be noted that the Ile-16 CSPs induced by the constitutively active mutants were similar to those induced by compounds (Fig. 7A), and the correlations between Ile-16 13C CSPs and basal induction levels matched those for the compound-induced induction levels, indicating that the compound-bound WT QacR and the unligated constitutively active mutants share the same functional conformational equilibrium (Fig. 7B).
Fig. 7.
NMR analysis of constitutively active QacR mutants. (A) Ile-16 δ1 resonances of QacR WT (black) were overlaid with that with those of the constitutively active mutants (red) and the less affected mutants (cyan). Asterisks highlight signals originating from Ile-172. (B) Correlation between the Ile-16 13C chemical shift changes of the mutants relative to WT QacR and basal induction levels. Those for the Rho6G- and Et-bound states are also indicated. (C) Correlation between the 13C chemical shift changes of mutants relative to the WT QacR and the binding affinity to the IR1 DNA. Basal transcription levels of QacR mutants reported in Peters et al. (20) were used.
Furthermore, the ITC data indicate that the constitutively active mutants display an IR1 DNA affinity reduced by ∼4-fold. This affinity reduction correlates with the population of the inducive conformation in the equilibrium, reflected in the 13C chemical shift difference of the constitutively active mutant relative to WT (Fig. 7C). These findings clearly indicate the functional relevance of the conformational equilibrium to the transcriptional regulation of QacR, in which the population of the inducive conformation in each state defines the transcriptional activity (Fig. 6).
Discussion
Our results directly support the notion that an equilibrium between repressive and inducive conformations regulates the variable transcriptional activity of QacR upon binding to the agonistic compounds in a conformational selection manner (Fig. 6). Whereas X-ray structures have defined the high-resolution coordinates of the unligated, DNA-bound, and compound-bound states (8, 14–17), the population of each conformation in the equilibrium can be quantified by NMR analyses to define the variable induction levels of QacR. Although other solution spectroscopic approaches are conceivable, NMR is very powerful, as it provides multiple atom-specific probes in the complex for simultaneous monitoring to define the structural features of the functional conformational equilibrium. Although the conformational equilibrium encompasses the whole molecule for QacR, the analysis of the CSP correlation utilized here is also able to define the distribution of the functional conformational equilibrium in a molecule. In the case that the conformational equilibrium involves only a part of a molecule, the correlation between CSPs would be observed only for those residues originating from the positions. Indeed, a similar approach was taken to define the distribution of the conformational equilibrium in protein kinase A (21).
Assuming that the DNA-bound and the Rho6G-bound QacR are fully populated in the repressive and inducive conformations, respectively, 23% of QacR in unligated state adopt the inducive conformation to allow a basal level of qacA expression (Fig. 6 and SI Appendix, Fig. S1). This would be functionally important, since the basal expression of the multidrug transporters is reported to contribute toward maintaining a basic level of resistance to toxic compounds (22, 23). Free energy difference (∆∆G) between the conformations can be described as ∆∆G = |−RT × ln(pr/pi)|, where pr and pi are the populations of the repressive and inducive conformations, respectively. In unligated QacR, the ∆∆G between repressive and inducive conformations is small (∼0.7 kcal/mol), which may facilitate the shift of the conformational equilibrium upon the compound binding.
Our study reveals that the populations of the inducive conformations in the equilibrium significantly correlates with the volume of the compounds (Figs. 4C and 6). Larger compounds force QacR to populate more in the inducive conformation for stronger induction. In contrast, smaller compounds are able to bind to the repressive conformation, and the shift of the conformational equilibrium to the inducive conformation is smaller. Thus, the QacR in complex with small compound is still able to bind to DNA to repress the transcription. The MDB pocket in the repressive conformation already has a volume of ∼400 Å3, which is sufficiently large to accommodate small compounds and provide an initial interaction site for large compounds (compound volume are between 160 Å3 and 380 Å3; SI Appendix, Table S1) (8, 17).
The conformational equilibrium of QacR between repressive and inducive conformations is fast in the NMR time scale. From the chemical shift difference between repressive and inducible conformations (∼40 Hz for Ile-16), the expected exchange rate should be >100 s−1. The fast conformational equilibrium enables a rapid transcriptional regulation of QacR, as a sensory module in the MDR systems. It has also been shown that more than one compound can simultaneously bind to QacR (17). Thus, if the volume-dependent transcriptional regulation is in effect for the total volume of multiple compounds in the MDR pocket, QacR might be able to provide a larger induction upon the simultaneous exposure of 2 toxic compounds. Our study also imprecates the strategy to make the QacR unfunctional via an inhibitor; the size of the molecule would matter, and small but strong binder should be selected to stabilize the conformation of the protein to the repressive conformation.
The NMR analyses reveal that the constitutively active mutants populate more in the inducible conformation even in the unligated state (Figs. 6 and 7). The 13C chemical shift of active mutants were quantitatively correlated with the increase in the basal transcriptional activity. Furthermore, the population of the inducive conformation was inversely correlated to the affinity reduction of QacR to the IR1 operator (Fig. 7C). These findings clearly indicate the functional relevance of the conformational equilibrium to the transcriptional regulation of QacR. The reduction of the affinity was not as significant as with other transcriptional repressors. For example, the lac repressor reduces the affinity to the operator by ∼1,000-fold upon binding to its specific inducers (24). However, the affinity reduction by a few-fold was also observed for another multidrug transcriptional repressor, LmrR, in which the affinity to its operator was reduced by ∼3-fold upon binding to the compound (25). Thus, several fold of reduction in the affinity would be sufficient for regulating the MDR systems.
LmrR also couples the multidrug recognition to transcriptional regulation via a preexisting conformational equilibrium (26). As transcriptional regulations by preexisting conformational equilibria have been suggested to act in bacterial repressors as well as eukaryotic nuclear receptors (27, 28), the conformational selection mechanisms would be shared widely among the ligand-inducible types of transcriptional regulators. The conformational equilibrium also allows various levels of responses in different biological systems such as G protein-coupled receptors (GPCRs), ion channels, kinases, and molecular chaperons (29–38), which might be a fundamental mechanism shared by biological systems that require rapid responses to exert their functions.
The NMR analyses provided the way to potentially dysfunctioning of the MDR systems. It is now suggested that a compound that is small but tightly binds to QacR might be able to stop the MDR system or at least keep it less active, by biasing the equilibrium to the repressive conformation. Therefore, not only revealing the conformational difference between the repressive and inducive conformations but defining the population of each conformation in the function-related equilibrium would be of importance to target such a dynamic biological system.
Materials and Methods
All chemicals were purchased from Wako or Sigma, unless otherwise stated. All stable isotope-labeled materials were acquired from Cambridge Isotope Laboratories or Isotec.
DNA Oligo Sequences.
Oligonucleotide sequences used in this study were as follows:
IR1 sequence in the qacR operator (36 bp):
5′- AATCCTTATAGACCGATCGCACGGTCTATAAGGATT -3′
5′- AATCCTTATAGACCGTGCGATCGGTCTATAAGGATT-3′.
Preparation of QacR.
The expression contract of QacR was prepared in a similar way to that described in previous literature (26). The resultant expression vector encodes QacR with a C-terminal His6-tag. In short, the codon-optimized synthetic DNA fragment (5′- CAT ATG GCC ATG GGC ATG AAC TTG AAA GAC AAA ATT CTC GGT GTT GCG AAA GAA CTT TTC ATC AAG AAT GGC TAT AAT GCG ACA ACT ACT GGT GAA ATT GTG AAG CTG TCT GAA TCG TCC AAA GGC AAC TTG TAC TAT CAC TTC AAA ACC AAA GAG AAT CTG TTT CTG GAG ATT CTC AAC ATC GAA GAG AGC AAA TGG CAA GAA CAA TGG AAG AAA GAA CAG ATC AAA TGC AAA ACG AAT CGT GAG AAA TTT TAT CTG TAT AAT GAA CTG AGT CTT ACC ACC GAA TAC TAC TAT CCG TTA CAG AAT GCC ATC ATT GAG TTT TAT ACG GAA TAC TAC AAA ACG AAT AGC ATT AAC GAG AAG ATG AAC AAG CTG GAA AAC AAA TAC ATT GAT GCG TAT CAT GTC ATT TTC AAA GAA GGG AAC CTG AAT GGT GAA TGG TGT ATT AAC GAT GTT AAC GCC GTA TCG AAA ATT GCT GCA AAT GCA GTG AAT GGA ATT GTG ACC TTT ACC CAT GAA CAG AAC ATC AAC GAA CGC ATC AAA CTG ATG AAC AAA TTC TCA CAG ATC TTT CTG AAC GGC TTA AGC AAA CTC GAG TGA GTC GAC-3′) encoding S. aureus QacR that is suitable for the expression in Escherichia coli was ligated into the pET28b vector (Novagen). Mutants of QacR were produced using the QuikChange strategy (Agilent Technology), following the procedure provided by the manufacturer.
The expression of QacR was carried out in a similar way to that described in previous literature for LmrR (26). E. coli strain BL21(DE3) was utilized for the QacR expression. For inducing QacR expression, 0.6 mM Isopropyl-β-d-thiogalactopyranoside was added at an optical density at 600-nm wavelength (OD600) of 0.8. The protein expression was induced at 32 °C for 16 h. For the expression of deuterated QacR, 3 g/L [2H7]-d-glucose and D2O was used. For selective 13CH3 labeling of the Ile (Ile-δ1) methyl groups, 100 mg/L [methyl-13C, 3,3-2H2]-α-ketobutyric acid was added to the media.
The purification of QacR was carried out in a similar way to that described in previous literature for LmrR (26). The QacR-expressing cells were lysed by sonication in the lysis buffer, consisting of 50 mM Tris⋅HCl (pH 8.0), 300 mM NaCl, 10 mM imidazole, and 5 mM β-mercaptoethanol. A 3-mL COSMOGEL His-Accept column, equilibrated with the lysis buffer, was used for purification. QacR was eluted with 20 mL of 50 mM Tris⋅HCl (pH 7.5), 300 mM NaCl, 300 mM imidazole, and 5 mM β-mercaptoethanol. The eluate was concentrated and was further purified by size exclusion chromatography (SEC) using a HiLoad Superdex 75 prep grade column (GE Healthcare). A 50 mM sodium phosphate buffer (NaPi; pH 6.8) containing 300 mM NaCl and 2 mM DTT was used as the SEC buffer. The QacR-containing fraction was buffer-exchanged into the NMR buffer containing 10 mM NaPi (pH 6.8), 100 mM NaCl, and 2 mM DTT.
NMR Experiments.
All titration experiments were performed at the same field strength using the Bruker Avance 600-MHz spectrometer. In contrast, the experiments for assignments were performed using both the 600-MHz spectrometer and the Bruker Avance III 800-MHz spectrometers. The both spectrometers were equipped with cryogenic triple-resonance probes. All spectra were recorded in the previously mentioned NMR buffer. The typical subunit concentration of QacR was 0.1 mM. For compound titration experiments, 5% of deuterated dimethyl sulfoxide (DMSO) (D6) was added to the buffer, in order to increase the solubility of the compound. All NMR experiments were performed at 298 K unless otherwise noted. TOPSPIN (Bruker Biospin) was used to process NMR spectra, and Sparky was used for spectral analysis. The assignments of Ile δ1 resonances were established by introducing Ile to Leu mutations (SI Appendix, Figs. S4 and S11). The mutations were introduced to all 17 Ile residues in QacR (I7L, I16L, I28L, I53L, I56L, I70L, I99L, I100L, I112L, I124L, I130L, I142L, I151L, I159L, I168L, I172L, and I181L), with the assignments of some residues being further confirmed by introducing double mutations (I70L/I99L and I70L/I100L) (SI Appendix, Fig. S4). In addition, resonances that shifted in slow-exchange fashion upon binding to Rho6G (Ile-16, Ile-53, Ile-70, Ile-112, Ile-124, Ile-142, and Ile159) and DNA (Ile-7, Ile-16, Ile-53, and Ile-112) were assigned individually by recording the corresponding mutant spectra in the bound states (SI Appendix, Fig. S11). We did not establish the assignment for Ile-99, Ile-100, and Ile-130 in bound states, as these residues are not discussed in the manuscript and are intrinsically too weak in intensity to assign them unambiguously. As the compounds showed similar chemical shift changes, the assignments of compounds other than Rho6G were established by referencing the corresponding assignments of the Rho6G-bound state.
The normalized CSP values (Δδnorm) were calculated as follows:
Δδnorm was used to compare the amplitude of CSPs in each condition. In contrast, 13C CSPs were used to compare the correlation between different conditions, as the 13C chemical shift is known to more directly reflect the local conformation (20).
ITC Measurements.
Calorimetric titrations were performed using a VP-ITC microcalorimeter (MicroCal) at 25 °C. A mutant QacR (C72A/C141S) was used to prevent the cross-linking and oxidation during the experiment that were observed in the WT protein (13). The double-mutant QacR is reportedly fully active, and its compounds and DNA binding properties are essentially identical to those of the WT protein (13). For the QacR−compound interaction, the ITC experiment was performed in 10 mM NaPi buffer (pH 6.8) containing 100 mM NaCl and 5% DMSO, which is the same as the NMR buffer except for the addition of DTT. For the QacR−DNA interaction, 10 mM Tris⋅HCl buffer (pH 8.0) with 100 mM NaCl was used, as the interaction was not exothermic or endothermic in the phosphate buffer. Protein samples were extensively dialyzed against ITC buffers before the experiments. For the QacR−compound interaction, the sample cell was filled with 10 µM compound, and 200 µM QacR in the syringe was titrated. For the QacR−DNA interaction, the sample cell was filled with 5 µM QacR, and 50 µM oligo DNA in the syringe was titrated. After a preliminary 3-µL injection, 24 to 27 subsequent 10-µL injections were performed. The data were fitted using the one-site binding model embedded in Origin 7.0 (MicroCal).
Supplementary Material
Acknowledgments
This work was supported, in part, by the Japan Agency for Medical Research and Development under Grant JP18ae0101046 (to I.S.) and the Ministry of Education, Culture, Sports, Science and Technology/Japan Society for the Promotion of Science Grants-in-Aid for Scientific Research (KAKENHI) Grant JP17H06097 (to I.S.). Funding was also provided by Precursory Research for Embryonic Science and Technology (PRESTO) Grant JPMJPR14L5 from the Japan Science and Technology Agency (to K.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1906129116/-/DCSupplemental.
References
- 1.Ma D., Cook D. N., Hearst J. E., Nikaido H., Efflux pumps and drug resistance in gram-negative bacteria. Trends Microbiol. 2, 489–493 (1994). [DOI] [PubMed] [Google Scholar]
- 2.Saier M. H., Jr, et al. , Evolutionary origins of multidrug and drug-specific efflux pumps in bacteria. FASEB J. 12, 265–274 (1998). [DOI] [PubMed] [Google Scholar]
- 3.van Veen H. W., Konings W. N., Structure and function of multidrug transporters. Adv. Exp. Med. Biol. 456, 145–158 (1998). [DOI] [PubMed] [Google Scholar]
- 4.Geick A., Eichelbaum M., Burk O., Nuclear receptor response elements mediate induction of intestinal MDR1 by rifampin. J. Biol. Chem. 276, 14581–14587 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Riordan J. R., et al. , Amplification of P-glycoprotein genes in multidrug-resistant mammalian cell lines. Nature 316, 817–819 (1985). [DOI] [PubMed] [Google Scholar]
- 6.Baguley B. C., Multiple drug resistance mechanisms in cancer. Mol. Biotechnol. 46, 308–316 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Gottesman M. M., Pastan I., Ambudkar S. V., P-glycoprotein and multidrug resistance. Curr. Opin. Genet. Dev. 6, 610–617 (1996). [DOI] [PubMed] [Google Scholar]
- 8.Schumacher M. A., Brennan R. G., Structural mechanisms of multidrug recognition and regulation by bacterial multidrug transcription factors. Mol. Microbiol. 45, 885–893 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Thakur J. K., et al. , A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature 452, 604–609 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Nishikawa J. L., et al. , Inhibiting fungal multidrug resistance by disrupting an activator-Mediator interaction. Nature 530, 485–489 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grkovic S., Brown M. H., Roberts N. J., Paulsen I. T., Skurray R. A., QacR is a repressor protein that regulates expression of the Staphylococcus aureus multidrug efflux pump QacA. J. Biol. Chem. 273, 18665–18673 (1998). [DOI] [PubMed] [Google Scholar]
- 12.Grkovic S., Hardie K. M., Brown M. H., Skurray R. A., Interactions of the QacR multidrug-binding protein with structurally diverse ligands: Implications for the evolution of the binding pocket. Biochemistry 42, 15226–15236 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Grkovic S., Brown M. H., Schumacher M. A., Brennan R. G., Skurray R. A., The staphylococcal QacR multidrug regulator binds a correctly spaced operator as a pair of dimers. J. Bacteriol. 183, 7102–7109 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schumacher M. A., et al. , Structural basis for cooperative DNA binding by two dimers of the multidrug-binding protein QacR. EMBO J. 21, 1210–1218 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brooks B. E., Piro K. M., Brennan R. G., Multidrug-binding transcription factor QacR binds the bivalent aromatic diamidines DB75 and DB359 in multiple positions. J. Am. Chem. Soc. 129, 8389–8395 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Schumacher M. A., et al. , Structural mechanisms of QacR induction and multidrug recognition. Science 294, 2158–2163 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Schumacher M. A., Miller M. C., Brennan R. G., Structural mechanism of the simultaneous binding of two drugs to a multidrug-binding protein. EMBO J. 23, 2923–2930 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tugarinov V., Hwang P. M., Ollerenshaw J. E., Kay L. E., Cross-correlated relaxation enhanced 1H[bond]13C NMR spectroscopy of methyl groups in very high molecular weight proteins and protein complexes. J. Am. Chem. Soc. 125, 10420–10428 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Hansen D. F., Neudecker P., Kay L. E., Determination of isoleucine side-chain conformations in ground and excited states of proteins from chemical shifts. J. Am. Chem. Soc. 132, 7589–7591 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Peters K. M., Sharbeen G., Theis T., Skurray R. A., Brown M. H., Biochemical characterization of the multidrug regulator QacR distinguishes residues that are crucial to multidrug binding and induction of qacA transcription. Biochemistry 48, 9794–9800 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Akimoto M., et al. , Signaling through dynamic linkers as revealed by PKA. Proc. Natl. Acad. Sci. U.S.A. 110, 14231–14236 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agustiandari H., Lubelski J., van den Berg van Saparoea H. B., Kuipers O. P., Driessen A. J., LmrR is a transcriptional repressor of expression of the multidrug ABC transporter LmrCD in Lactococcus lactis. J. Bacteriol. 190, 759–763 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agustiandari H., Peeters E., de Wit J. G., Charlier D., Driessen A. J., LmrR-mediated gene regulation of multidrug resistance in Lactococcus lactis. Microbiology 157, 1519–1530 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Barkley M. D., Riggs A. D., Jobe A., Burgeois S., Interaction of effecting ligands with lac repressor and repressor-operator complex. Biochemistry 14, 1700–1712 (1975). [DOI] [PubMed] [Google Scholar]
- 25.Takeuchi K., Imai M., Shimada I., Dynamic equilibrium on DNA defines transcriptional regulation of a multidrug binding transcriptional repressor, LmrR. Sci. Rep. 7, 267 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takeuchi K., Tokunaga Y., Imai M., Takahashi H., Shimada I., Dynamic multidrug recognition by multidrug transcriptional repressor LmrR. Sci. Rep. 4, 6922 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Changeux J.-P., Edelstein S., Conformational selection or induced fit? 50 years of debate resolved. F1000 Biol. Rep. 3, 19 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liguori A., et al. , Molecular basis of ligand-dependent regulation of NadR, the transcriptional repressor of meningococcal virulence factor NadA. PLoS Pathog. 12, e1005557 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kofuku Y., et al. , Functional dynamics of deuterated β2-adrenergic receptor in lipid bilayers revealed by NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 53, 13376–13379 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Okude J., et al. , Identification of a conformational equilibrium that determines the efficacy and functional selectivity of the μ-opioid receptor. Angew. Chem. Int. Ed. Engl. 54, 15771–15776 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Isogai S., et al. , Backbone NMR reveals allosteric signal transduction networks in the β1-adrenergic receptor. Nature 530, 237–241 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Eddy M. T., et al. , Allosteric coupling of drug binding and intracellular signaling in the A2A adenosine receptor. Cell 172, 68–80.e12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minato Y., et al. , Conductance of P2X4 purinergic receptor is determined by conformational equilibrium in the transmembrane region. Proc. Natl. Acad. Sci. U.S.A. 113, 4741–4746 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toyama Y., et al. , Structural basis for the ethanol action on G-protein−activated inwardly rectifying potassium channel 1 revealed by NMR spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 115, 3858–3863 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sekhar A., et al. , Conserved conformational selection mechanism of Hsp70 chaperone-substrate interactions. eLife 7, e32764 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saleh T., Rossi P., Kalodimos C. G., Atomic view of the energy landscape in the allosteric regulation of Abl kinase. Nat. Struct. Mol. Biol. 24, 893–901 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tokunaga Y., Takeuchi K., Takahashi H., Shimada I., Allosteric enhancement of MAP kinase p38α’s activity and substrate selectivity by docking interactions. Nat. Struct. Mol. Biol. 21, 704–711 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Shimada I., Ueda T., Kofuku Y., Eddy M. T., Wüthrich K., GPCR drug discovery: Integrating solution NMR data with crystal and cryo-EM structures. Nat. Rev. Drug Discov. 18, 59–82 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.