Significance
Cells respond to the mechanical properties of their environment, altering a range of cellular functions important to normal tissue function, disease progression, and repair. This mechanosensing ability has been studied predominantly with planar substrates which, while experimentally advantageous, do not capture the complexity of cellular interfaces. This report demonstrates that T cells respond to the mechanical resistance of microfabricated elastomer structures, identifying an aspect of mechanosensing that has implications from understanding cell–cell interactions to design of biomaterials.
Keywords: mechanobiology, T cell, microstructure
Abstract
Cells have the remarkable ability to sense the mechanical stiffness of their surroundings. This has been studied extensively in the context of cells interacting with planar surfaces, a conceptually elegant model that also has application in biomaterial design. However, physiological interfaces are spatially complex, exhibiting topographical features that are described over multiple scales. This report explores mechanosensing of microstructured elastomer surfaces by CD4+ T cells, key mediators of the adaptive immune response. We show that T cells form complex interactions with elastomer micropillar arrays, extending processes into spaces between structures and forming local areas of contraction and expansion dictated by the layout of microtubules within this interface. Conversely, cytoskeletal reorganization and intracellular signaling are sensitive to the pillar dimensions and flexibility. Unexpectedly, these measures show different responses to substrate rigidity, suggesting competing processes in overall T cell mechanosensing. The results of this study demonstrate that T cells sense the local rigidity of their environment, leading to strategies for biomaterial design.
T cells are key agents of the adaptive immune response, providing robust protection against pathogens, but in different contexts contributing to a range of diseases. Contemporary approaches in biomaterials design have provided enhanced control over this branch of immunity, leading to new tools for medicine including vaccination and cellular immunotherapy. Intriguingly, tuning the mechanical rigidity of a substrate used to activate T cells can modulate their subsequent functions including cytokine secretion, proliferation, and population expansion (1–5). These studies have been carried out predominantly in the context of bulk materials that present to cells planar interfaces that are geometrically and mechanically simplistic. However, interactions between T cells and antigen-presenting cells (APCs) are topographically complex, involving cellular protrusions, extensions, and other features (6, 7); the effects of these spatially complex, out-of-plane interactions are not well understood. This report examines how features defined at the scale of micrometers affect T cell function and mechanosensing of deformable materials. To provide a complex yet controllable topography, we focus on arrays of microscale elastomer pillars (8–10), a system that has been central to measurement of cell traction forces (11). In that application, cells laterally deflect the tips of individual pillars (Fig. 1A) and the resultant displacements are used to estimate the force applied to the end of each structure, providing a dynamic map of cellular forces. A standard geometry of 6-μm-tall pillars, each measuring 1 μm in diameter, arranged in hexagonal arrays with 2-μm center-to-center spacing was chosen for this study. These were cast in a polydimethylsiloxane (PDMS, Sylgard 184) elastomer, yielding pillars that are sufficiently flexible to allow deflection by T cells (a spring constant of 0.77 nN/μm) while avoiding extensive collision between adjacent structures (8). The arrays were coated with antibodies that bind to CD3 (epsilon subunit) and CD28, providing activating and costimulatory signaling, respectively, to T cells; engagement of these two receptor systems is associated with cell activation in vivo and initiates expansion in vitro.
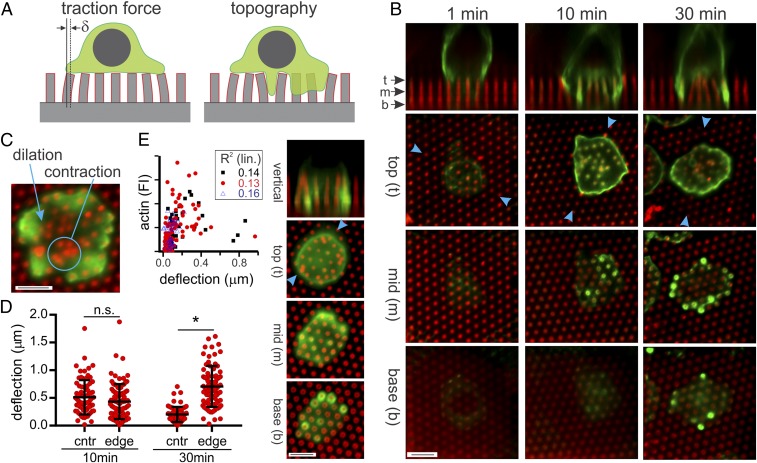
Fig. 1.
T cells embed into micropillar arrays. (A) Schematic comparison of conventional interaction assumed for traction force microscopy and that observed for preactivated CD4+ T cells. (B) Time course of cell interaction with pillar arrays (red) coated with anti-CD3/anti-CD28. Cells (green) were visualized using anti-CD45, and blue arrowheads indicate the position at which the vertical slices were extracted. (C) The cell–substrate interface is complex, often showing concurrent dilation and contraction of local pillar clusters. (D) Time-dependent resolution of force generation within the cell–substrate interface. Each data point represents an individual pillar. Data are mean ± SD, *P < 0.0005 (n > 50 pillars for each condition, collected from 5 cells at each timepoint over 2 independent experiments), while n.s. indicates no statistically significant difference detected (α = 0.05). (E) Actin localization (green, phalloidin stain) does not correlate with regions of strong pillar deflection. The graph compares actin surrounding each pillar with tip deflection for 3 representative cells, collected from independent experiments. R2 values for each cell are reported for a linear regression. (Scale bar, 5 µm for all images.)
Results
T Cells Form Extensive, Mechanically Complex Interactions with Microstructured Surfaces.
During the first minutes of contact, preactivated CD4+ T cells form a compact interface with the tops of the pillars (Fig. 1B). These cells begin to infiltrate the arrays over the next minutes and extend protrusions between the pillars (Fig. 1 A and B), a configuration not associated with traction force microscopy. A timepoint of 10 min (Fig. 1B) will be used to capture this dynamic phase of cell–substrate interaction. Cells embed extensively within the pillar arrays by 30 min, surrounding and reaching the bottom of individual pillars (6 μm below the pillar tips); this interaction is not limited to cell membrane, as cell nuclei also exhibit extensive deformation and embedding (SI Appendix, Fig. S1 A and B). The elastomer pillar arrays thus capture topographical aspects of the natural T cell–APC interaction that are missing with conventional, planar substrates. Notably, naive cells exhibited a similar infiltration into the arrays, but this behavior was preceded by an initial phase in which cells remained on the pillar tops (8). This initial phase in naive cells lasts typically between 30 and 90 min and shows substantial variability between cells; to provide a more consistent model of cell interaction with the topographically complex arrays, this report will focus on preactivated, CD4+ T cells. Like other cell types, preactivated T cells generate centripetal forces along the cell edge (Fig. 1B). However, these cells also show complex manipulations of the substrate across the cell–substrate interface suggesting local contraction or dilation of small groups of pillars (Fig. 1C). These manipulations are strongest during the initial periods of cell invasion (10 min, Fig. 1D), and decrease to a more quiescent state as the cells fully embed within the arrays (30 min). These interactions are more complex than the single point load model used in traction force microscopy, as infiltrating cells can apply forces at multiple points along the length of the pillar. Consequently, pillar deflections, while indicative of applied force, will not be used to calculate traction forces in this report.
Cell-Level Mechanical Interactions Are Directed by Microtubules.
To understand the mechanisms guiding localized contraction and dilation of pillars under the cell body, this study focuses on two complementary components of the cell cytoskeleton. The first is actin, which is central to generation of cellular forces and extension of processes. Cell infiltration and embedding was associated with extensive envelopment of individual pillars by polymerized actin (Fig. 1E). However, there was minimal correlation between the amount of actin associated with a pillar and its deflection (Fig. 1E). In addition, cells on surfaces containing vertical walls that did not present anti-CD3/anti-CD28 still elaborated actin-containing processes into microscale pits (SI Appendix, Fig. S2), suggesting that envelopment and generation of force by the cells does not require adhesion to the substrate. Actin is thus central to extension of processes between pillars, but does not coordinate the overall cellular-level mechanics of T cell interaction with the pillars. As such, a role of microtubules in directing cell interaction with the pillar arrays was examined in this study.
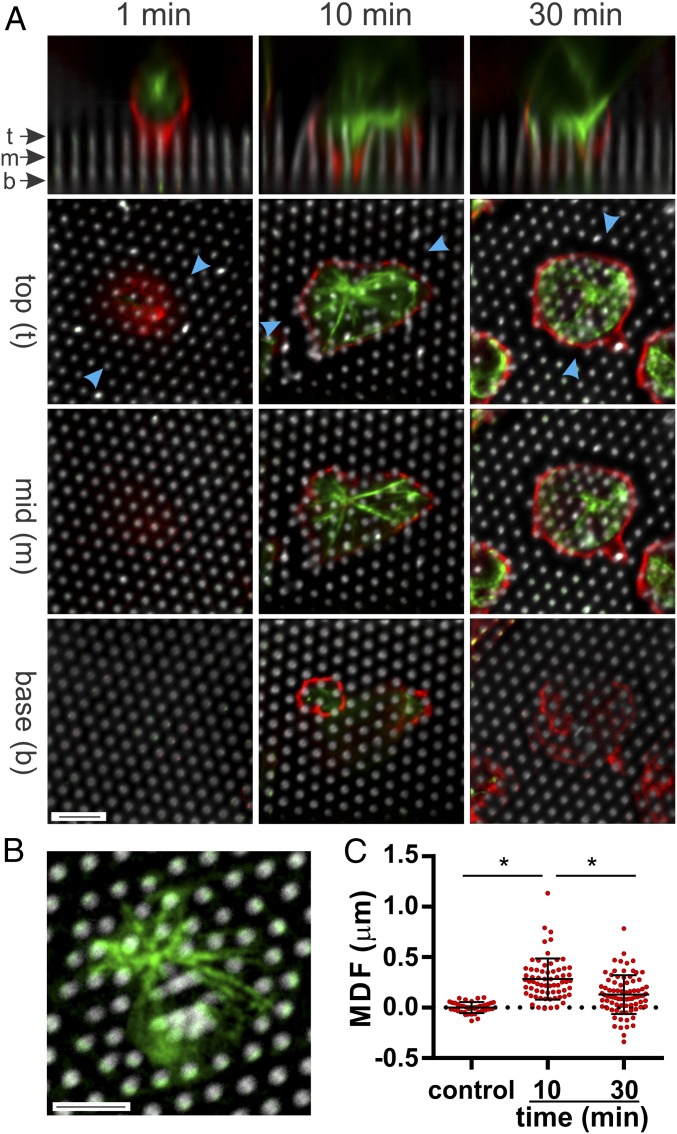
Similar to actin and the cell outer membrane, microtubules infiltrate regions between and surrounding pillars showing significant embedding at both intermediate (10 min) and late (30 min) timepoints (Fig. 2A). However, microtubules do not penetrate fully into the arrays, stopping at a point intermediate between the pillar tip and the depth of the cellular processes (Fig. 2A). Strikingly, the position of microtubules and the centrosome/microtubule-organizing center (MTOC) in the plane of the cell–array interface reflects the mechanical state of this structure. The MTOC is associated with regions of local pillar dilation (Fig. 2 A and B), quantified here by a MTOC-displacement factor (MDF), the average tip deflection away from the MTOC of the three closest pillars. As reported in Fig. 2C, MDF for cells at 10 and 30 min after seeding is increased compared to controls (calculated from a reference, nondeflected region of the array), indicating deflection of pillars away from the MTOC. The microtubules radiating from the MTOC also provide organization of the cell–substrate interface, encircling groups of pillars (Fig. 2B) and are thus associated with the regions of contraction shown in Fig. 1C.
Fig. 2.
Microtubules direct lateral organization of the cell–array interface. (A) Microtubules (green, β-tubulin immunostaining) extend into spaces between pillars, coming to rest at a depth between the pillar top and ends of the cell processes. Cell membranes (red) were visualized by staining for CD45. Pillars are shown in gray. (B) Microtubule layout matches the lateral organization of force generation. The MTOC is associated with sites of local pillar dilation, while extended microtubules surround regions of contraction. (C) Time-dependent dilation of pillars away from the MTOC. Local dilation/contraction is quantified as the average tip deflection for the 3 pillars closest to the MTOC. Each data point represents an individual cell, collected over 3 independent experiments. Data are mean ± SD *P < 0.0001 between conditions spanned by bar (n > 40 cells per condition). (Scale bar, 5 µm for all images.)
T Cells Are Sensitive to the Local Stiffness of Microstructured Surfaces.
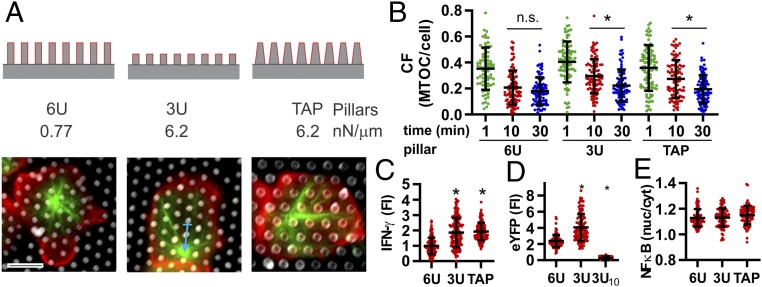
The observation that microtubules organize the interaction of cells with the micropillar arrays raised the intriguing possibility that the local rigidity of these structures could modulate T cell cytoskeletal organization and subsequent cellular function. This was tested by reducing the pillar height from 6 to 3 μm (the 6U and 3U structures in Fig. 3A), thereby increasing the associated spring constant 8-fold from 0.77 to 6.2 nN/μm. The stiffer pillars delayed transport of the MTOC toward the center of the cell–array interface (Fig. 3A), a key step in immune synapse formation and T cell activation (12–15). This was quantified by a centralization factor, CF, the distance from the cell centroid to the MTOC divided by the distance from centroid to cell edge (Fig. 3B). The process of MTOC transport on the 6U pillars is seen as a decrease in CF over the first 10 min of interaction with no further change detected over the next 20 min. In contrast, cells on the 3U arrays show a higher CF at 10 min compared to 30 min indicating a delay in MTOC transport. To test whether MTOC centralization is responding to pillar height [mirroring an ability of other types of cells to sense depth of microscale pits (16)] rather than pillar flexibility, a third array geometry was created consisting of 6-μm-tall pillars that tapered from 1.8 μm in diameter at the base to 1 μm at the tip (TAP in Fig. 3). This tapering produced pillars with height equal to the 6U arrays and spring constant similar to the 3U system. This approach also allowed the use of a single elastomer formulation, avoiding unknown factors associated with changing pillar composition. Centralization on TAP arrays followed that of the 3U system, being delayed at the 10-min timepoint, indicating that cells respond to the cellular-level stiffness of the substrate rather than pillar height alone.
Fig. 3.
T cells respond to local mechanics imposed by the pillar arrays. (A) Pillar geometry was altered to test the effect of 3-dimensional structure and spring constant on T cell activation. Images illustrate microtubule structure (green, β-tubulin) and cell morphology (red, CD45) for cells fixed 10 min after seeding onto surfaces. (Scale bar, 5 µm.) (B) Transport of the MTOC to the center of the cell interface was delayed on surfaces of higher spring constant (3U and TAP). CF was calculated as the distance from the cell centroid to MTOC (blue arrow), divided by the distance from centroid to the cell edge. Each data point represents an individual cell, collected over 3 independent experiments. Data are mean ± SD *P < 0.05 between conditions spanned by bar (n > 90 cells per condition). These and additional comparisons are discussed in the main text. (C) IFN-γ secretion increased with increasing spring constant. Cytokines measured 4 h after seeding. Data are mean ± SD; each data point is an individual cell. *P < 0.001 compared to 6U surface (n > 100 cells per condition). (D) IFN-γ transcription, as measured by eYFP expression in GREAT mouse cells, also increased with increasing spring constant. Fluorescence intensity of eYFP was measured 6 h after seeding. Measurement of eYFP 10 min after seeding onto 3U arrays (3U10) was used to control for background at the start of the experiment. Data are mean ± SD; each data point is an individual cell. *P < 0.001 compared to 6U surface (n > 65 cells per condition). (E) NF-κB translocation to the cell nucleus, measured 10 min after seeding onto arrays, was independent of spring constant. Data are mean ± SD; each data point represents an individual cell, α = 0.05 (n > 100 cells per condition).
The effect of pillar stiffness on downstream signaling and T cell activation was examined by measuring secretion of IFN-γ over 4 h, using a surface capture assay (17, 18). In contrast to MTOC localization, IFN-γ secretion increased with rising pillar spring constant (Fig. 3C). IFN-γ production was also compared using the IFN-gamma reporter with endogenous polyA transcript (GREAT) mouse model, in which eYFP expression is under control of IFN-γ promoter/enhancer (19, 20). Similar to that seen for the surface capture assay, eYFP in cells from the GREAT mouse was higher on the stiffer 3U arrays than 6U (Fig. 3D). Notably, the eYFP signal does not involve secretion from the cell, indicating that regulation of IFN-γ occurs at the point of transcription or translation, rather than polarization/secretion. In further support of this idea, polarization of vesicles in T cells was independent of pillar height (SI Appendix, Fig. S3). Surprisingly, nuclear localization of NK-κB was similar across the 6U, 3U, and TAP series (Fig. 3E). The contrasting cellular responses from MTOC centralization to cytokine secretion are unexpected as these steps are often considered to be sequential stages in cell activation. These results thus reveal a complex effect of substrate mechanics that is not captured on traditional, planar surfaces.
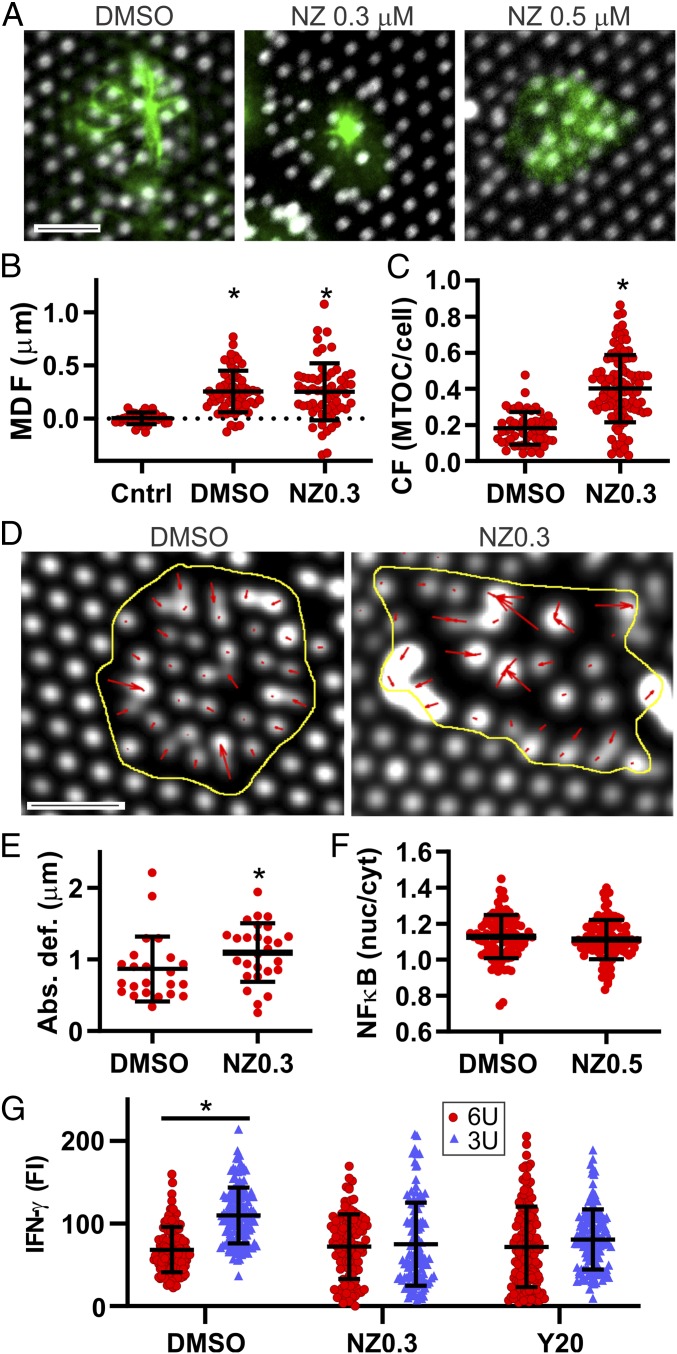
To better understand the role of the MTOC and microtubules in T cell mechanosensing, tubulin polymerization was inhibited with nocodazole (NZ). At a concentration of 0.3 µM, NZ disrupted cell-level microtubule structure but left the MTOC morphologically intact (Fig. 4A). Higher concentrations (0.5 µM) disrupted MTOC (Fig. 4A). Inhibition at 0.3 μM had no effect on pillar deflection around the MTOC (MDC, Fig. 4B) interaction, indicating that this organelle guides local infiltration into the array independent from the larger microtubule organization. However, NZ inhibition results in a larger CF at the 10-min timepoint (Fig. 4C), indicating that microtubules extending throughout the cell–substrate interface assist centralization of the MTOC. NZ inhibition disrupted the centripetal organization of forces, producing local areas of contraction (Fig. 4D), and also increased the absolute force generated by each cell (Fig. 4E), suggesting that microtubules serve to coordinate force generation across the cell–array interface, locally inhibiting actin-based contractility. Inhibition of MTs with NZ at even 0.5 μM had no effect on NF-κB translocation. Looking to longer-term function, NZ inhibition (at 0.3 µM) abrogated regulation of IFN-γ secretion as a function of pillar geometry (Fig. 4G) indicating a role of microtubules in mechanosensing. Notably, inhibition of actomyosin contractility with Y-27632 also eliminated regulation of IFN-γ secretion indicating that both cytoskeletal networks are needed for mechanosensing.
Fig. 4.
Microtubule architecture coordinates cellwide organization of cellular function. (A) Inhibition of microtubule (green, β-tubulin) polymerization by NZ, 10 min after seeding. (Scale bar, 5 µm.) (B) Inhibition with 0.3 µM NZ did not affect MTOC dilation of neighboring pillars. *P < 0.0001 compared to Cntrl (n > 500 cells per condition). (C) NZ inhibition decreased MTOC centralization, 10 min after seeding. *P < 0.0001 compared to dimethyl sulfoxide (DMSO) control (n > 500 cells per condition). (D) NZ inhibition disrupted cellular-scale organization of forces in T cells. This image illustrates pillar deflections (red arrows) 15 min after cell seeding. [Scale bar (black bar), 5 µm.] (E) NZ inhibition increased absolute pillar deflection imparted by each cell. Each data point represents displacement averaged over all pillars under an individual cell. *P < 0.05 compared to DMSO control (n = 25 cells per condition). (F) NZ inhibition has no effect on NF-κB translocation (α = 0.05, n > 100 cells per condition). (G) Inhibition of cytoskeletal function with NZ or Y-27632 (20 μM, Y20) abrogated pillar mechanosensing. *P < 0.05 compared to DMSO control (n > 100 cells per condition).
Local Structure of Deformable Materials Influences T Cell Response.
The development of systems that promote desirable biological responses from living systems involves interplay of knowledge between cellular physiology and material design. Inspired by advances in other cellular systems, leveraging of T cell mechanosensing into new materials has focused predominantly on flat surfaces such as hydrogels, elastomers, and supported lipid bilayers which present interfaces that are conceptually straightforward and convenient for materials processing. The current study demonstrates that topographical features not captured in conventional planar formats also modulate cellular mechanosensing, offering both strategies for biomaterial design and insight into how cell–cell interface topography controls T cell–APC communication. Distinct from earlier studies demonstrating that T cells can sense rigid topographical features (10, 21, 22), a key conclusion of this report is that cells respond to mechanical resistance imparted by both the substrate material and geometry. Increasing the spring constant of pillars delayed MTOC centralization (Fig. 3 B and C), suggesting that a stiffer 3D microenvironment hinders lateral motion of microtubules and the MTOC, which are typically under the control of dynein motors. Given the role of MTOC centralization in immune synapse function, this would suggest that a stiffer microenvironment leads to reduced activation. This would be opposite that observed for mouse T cells on planar gel surfaces (2) as well as other types of cells in studies that used decreased micropillar height to mimic increasing bulk modulus (23, 24). Intriguingly, we found that IFN-γ secretion follows the predictions of those earlier studies, increasing with greater pillar stiffness (Fig. 3C). Together, our results on MTOC function and cytokine secretion suggest competing roles of the microtubule and actin cytoskeleton systems in T cell activation, which are differentially modulated by the topographically complex pillar array structure. These may converge at the point of IFN-γ transcription and translation, supported by our results with the GREAT mouse (Fig. 3D) and the large nuclear deformations seen on these systems (SI Appendix, Fig. S1B).
Finally, studies in other cell systems demonstrate cell sensing of the depth of microscale pits in a material through podosomes (16), providing a precedent for the effects of microscale topography shown here with T cells. This does suggest that there may be a minimum pillar dimension below which the contrasting effects of microscale rigidity are not observed; below some height, the MTOC might start to rise above and lose interaction with the pillars. Such a cutoff would be less than 3 µm, since the 3U and TAP geometries had similar effects. This cutoff, as well as the mechanisms that localize the microtubule structure between the pillar tops and full depth of descent of the cells into the pillars, remain unresolved.
Methods
Substrate Fabrication.
Arrays of microscale pillars and trenches were created in PDMS elastomer (Sylgard 184, Dow Corning) using established techniques (8, 9, 11, 25, 26). Briefly, positive masters containing arrays of pillars were created using standard micro- and nanofabrication techniques (8, 26). Negative molds, containing microscale pits, were cast in PDMS from these masters, silanized overnight with (tridecafluoro-1,1,2,2,-tetrahydrooctyl)-1-trichlorosilane (United Chemical Technologies), and then used to cast pillar arrays onto glass coverslips (thickness 0, Thermo Fisher Scientific). A subset of experiments focused on cell response to micropit arrays, which were created as previously described (16). Micropits were designed to be 2 µm in diameter and 9 µm in depth, with 4-µm center-to-center spacing.
Unless otherwise specified, pillar and micropit arrays were coated with Alexa 568-labeled streptavidin (Thermo) at a concentration of 20 µg/mL, rinsed, then incubated with biotinylated antibodies against CD3 and CD28 (clones 145–2C11 and 37.51, eBioscience) at a concentration of 20 µg/mL each. Each step was performed for 1 h at room temperature.
Cell Preparation and Assays of Cell Response.
Mouse CD4+ T cells were isolated from the spleens of C57BL/6 mice, age 6–10 wk. After filtering through a 40-μm mesh, naive CD4+ cells were enriched via negative selection using Dynalbeads Untouched kit (Invitrogen). To prepare preactivated cells, naive CD4+ T cells were seeded on cell culture-treated plastic plates previously coated with 10 μg/mL anti-CD3 (clone 145–2C11, eBioscience). Cells were maintained for 48 h under standard cell culture conditions (37 °C, 5% CO2/95% air) in complete culture media (consisting of RPMI 1640 supplemented with 10% FBS, 10 mM Hepes, 2 mM l-glutamine, 50 µM β-mercaptoethanol [Sigma], 50 U/mL penicillin, 50 µg/mL streptomysin, and 50 U/mL rhIL-2 [Peprotech], all reagents from Thermo unless otherwise noted), which was further supplemented with 2 μg/mL anti-CD28. Cells were then collected and recovered overnight in complete culture media. supplemented with 10% FBS (Thermo), 10 mM Hepes Gibco), 2 mM l-glutamine (Gibco), which was further supplemented with 50 U/mL rhIL-2 (Peprotech) and 2 μg/mL anti-CD28 (eBioscience) for 48 h under standard cell culture conditions (37 °C, 5% CO2/95% air). Cells were then collected and recovered overnight in complete culture media.
For experiments, cells were seeded onto surfaces at a density of 1.3 × 103 cells per mm2 (from a 5 × 106 cells per mL solution) in complete media. Cells were imaged on an Olympus IX81 inverted microscope equipped with an Andor iXon 888 electron multiplying charge-coupled device camera. Temperature and gas concentrations were maintained with a Tokai stage top incubation system. For live tracking of the cell membrane, cells were stained with Alexa 488-labeled anti-CD45.2 Fab antibody fragments. For inhibition experiments, cells were pretreated with either NZ (Sigma) or Y-27632 (Sigma) in complete culture media for 15 min, and then seeded onto experimental surfaces while maintaining the specified concentration in the media.
Cytokine production/secretion was measured using two complementary approaches. Except for the experiment of Fig. 3D, IFN-γ secretion was measured using a surface capture assay (Miltenyi) as previously described (17, 18, 27). Fig. 3D compares IFN-γ production using the GREAT mouse model (19, 20). CD4+ T cells from these mice were isolated, activated, and then allowed to return to rest in uncoated well for 8 d to allow intracellular levels of eYFP, which was not secreted, to decrease. This background level was measured by quantifying eYFP 10 min after seeding of cells on the micropillar arrays.
Pillar deflections were monitored by live cell microscopy (11, 28, 29) or in fixed samples, using the Alexa 568-labeled streptavidin for visualization. The field of view was sufficiently large to include an adequate number of neighboring pillars that were not displaced by cells, which were used to correct for ambient drift and stage movement. Following acquisition, the Fiji software package (30) was used to correct stacks for ambient drift and track pillar movement.
All experiments were carried out under a protocol approved by Columbia University’s Institutional Animal Care and Use Committee.
Immunostaining.
Immunofluorescence microscopy was carried out using standard techniques. At specified timepoints, cells were fixed with 4% paraformaldehyde for 10 min, then permeabilized with 0.1% Triton X-100 in PBS. Samples were then blocked using 5% BSA for 2 h at room temperature or overnight at 4 °C. Samples were stained with primary antibodies targeting CD45 (Biolegend) and β-tubulin (BD Biosciences), followed by appropriate secondary antibodies conjugated with Alexa fluorphores (Invitrogen). Cells were also stained for actin cytoskeleton using fluorescently labeled phalloidin (Invitrogen).
For imaging of NF-κB translocation, cells were fixed and permeabilized using an FOXP3 fix/perm kit (Biolegend). Cells were blocked with 5% BSA for 2 h at room temperature or overnight at 4 °C, and then stained with an antibody against NF-κB subunit p65 (Cell Signaling Technology), followed by secondary antibody Alexa 647-labeled goat anti-rabbit (Invitrogen), nuclear stain Hoechst 33342 (Thermo), and Alexa 388-labeled CD45.2 (Biolegend). NF-κB translocation was calculated as the average staining intensity within the nucleus normalized to that of the entire cell, taken at a plane cutting through the main cell body (17). Image processing was carried out using Fiji (30) and the Deconvolution Lab plugin (31).
Quantification of MTOC Local Pillar Displacement and Centralization.
An MDF was calculated to quantify the local effect of MTOC position on pillar deflection. This was calculated as the average outward displacement of the nearest 3 pillars to an MTOC, measured at the pillars’ tips; an MDF > 0 indicates pushing away from the MTOC while an MDF < 0 shows local contraction. Control regions were calculated from reference regions that were not under a cell.
MTOC position within the cell–substrate interface was quantified in terms of a CF. This was calculated as the distance from the cell–substrate interface centroid to the MTOC divided by the distance to the cell edge.
Statistics and Data Sharing.
All data were analyzed for statistical significance using two-tailed Kruskal–Wallis methods; when justified by this first test, multiple comparisons were carried out using Dunn’s test. A value of α = 0.05 was chosen to identify statistical significance, but values of P that are smaller than this cutoff are also included in the figures. All data have been deposited in figshare and are publicly available (32).
Supplementary Material
Acknowledgments
This study is supported by the NIH (R01AI087644 and R01AI110593) and the NSF (CMMI 1562905). This research used resources of the Center for Functional Nanomaterials, a US DOE Office of Science Facility at Brookhaven National Laboratory operated under Contract DE-SC0012704. The content of this work is solely the responsibility of the authors and does not necessarily represent the official views of the funding institutions.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: All quantitative and image-based data have been deposited in figshare (https://figshare.com/articles/Images/9776126).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1906986116/-/DCSupplemental.
References
- 1.Dang A. P., et al. , Enhanced activation and expansion of T cells using mechanically soft elastomer fibers. Adv. Biosyst. 2, 1700167 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Judokusumo E., Tabdanov E., Kumari S., Dustin M. L., Kam L. C., Mechanosensing in T lymphocyte activation. Biophys. J. 102, L5–L7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lambert L. H., et al. , Improving T cell expansion with a soft touch. Nano Lett. 17, 821–826 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Connor R. S., et al. , Substrate rigidity regulates human T cell activation and proliferation. J. Immunol. 189, 1330–1339 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wahl A., et al. , Biphasic mechanosensitivity of T cell receptor-mediated spreading of lymphocytes. Proc. Natl. Acad. Sci. U.S.A. 116, 5908–5913 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ueda H., Morphew M. K., McIntosh J. R., Davis M. M., CD4+ T-cell synapses involve multiple distinct stages. Proc. Natl. Acad. Sci. U.S.A. 108, 17099–17104 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sage P. T., et al. , Antigen recognition is facilitated by invadosome-like protrusions formed by memory/effector T cells. J. Immunol. 188, 3686–3699 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bashour K. T., et al. , CD28 and CD3 have complementary roles in T-cell traction forces. Proc. Natl. Acad. Sci. U.S.A. 111, 2241–2246 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin W., Black C. T., Kam L. C., Huse M., Probing synaptic biomechanics using micropillar arrays. Methods Mol. Biol. 1584, 333–346 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Tamzalit F., et al. , Interfacial actin protrusions mechanically enhance killing by cytotoxic T cells. Sci. Immunol. 4, eaav5445 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan J. L., et al. , Cells lying on a bed of microneedles: An approach to isolate mechanical force. Proc. Natl. Acad. Sci. U.S.A. 100, 1484–1489 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geiger B., Rosen D., Berke G., Spatial relationships of microtubule-organizing centers and the contact area of cytotoxic T lymphocytes and target cells. J. Cell Biol. 95, 137–143 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kupfer A., Dennert G., Singer S. J., Polarization of the Golgi apparatus and the microtubule-organizing center within cloned natural killer cells bound to their targets. Proc. Natl. Acad. Sci. U.S.A. 80, 7224–7228 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quann E. J., Merino E., Furuta T., Huse M., Localized diacylglycerol drives the polarization of the microtubule-organizing center in T cells. Nat. Immunol. 10, 627–635 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Yi J., et al. , Centrosome repositioning in T cells is biphasic and driven by microtubule end-on capture-shrinkage. J. Cell Biol. 202, 779–792 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaudhuri P. K., Pan C. Q., Low B. C., Lim C. T., Differential depth sensing reduces cancer cell proliferation via Rho-Rac-regulated invadopodia. ACS Nano 11, 7336–7348 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Shen K., Thomas V. K., Dustin M. L., Kam L. C., Micropatterning of costimulatory ligands enhances CD4+ T cell function. Proc. Natl. Acad. Sci. U.S.A. 105, 7791–7796 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sims T. N., et al. , Opposing effects of PKCtheta and WASp on symmetry breaking and relocation of the immunological synapse. Cell 129, 773–785 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Stetson D. B., et al. , Constitutive cytokine mRNAs mark natural killer (NK) and NK T cells poised for rapid effector function. J. Exp. Med. 198, 1069–1076 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reinhardt R. L., et al. , A novel model for IFN-γ-mediated autoinflammatory syndromes. J. Immunol. 194, 2358–2368 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu J., et al. , High-throughput mechanobiology screening platform using micro- and nanotopography. Nano Lett. 16, 2198–2204 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwon K. W., Park H., Doh J., Migration of T cells on surfaces containing complex nanotopography. PLoS One 8, e73960 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu J., et al. , Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat. Methods 7, 733–736 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saez A., Buguin A., Silberzan P., Ladoux B., Is the mechanical activity of epithelial cells controlled by deformations or forces? Biophys. J. 89, L52–L54 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang M. T., Fu J., Wang Y. K., Desai R. A., Chen C. S., Assaying stem cell mechanobiology on microfabricated elastomeric substrates with geometrically modulated rigidity. Nat. Protoc. 6, 187–213 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Basu R., et al. , Cytotoxic T cells use mechanical force to potentiate target cell killing. Cell 165, 100–110 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bashour K. T., et al. , Cross talk between CD3 and CD28 is spatially modulated by protein lateral mobility. Mol. Cell. Biol. 34, 955–964 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lemmon C. A., et al. , Shear force at the cell-matrix interface: Enhanced analysis for microfabricated post array detectors. Mech. Chem. Biosyst. 2, 1–16 (2005). [PMC free article] [PubMed] [Google Scholar]
- 29.Dauriac V., Descroix S., Chen Y., Peltre G., Sénéchal H., Isoelectric focusing in an ordered micropillar array. Electrophoresis 29, 2945–2952 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Schindelin J., et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sage D., et al. , DeconvolutionLab2: An open-source software for deconvolution microscopy. Methods 115, 28–41 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Jin W., et al. , T cells sense rigidity of microscale structures. figshare. https://figshare.com/articles/Images/9776126. Deposited 6 September 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.