Significance
Anthocyanins are flavonoid plant pigments that may be important for coping with environmental stresses, such as high light, drought, and nutrient deprivation. It has been thought that anthocyanin biosynthesis arose when land plants evolved approximately 450 million y ago, and extant basal plants groups, such as liverworts, have been reported to have “primitive” anthocyanins bound to their cell walls. However, here we report that the red pigments of the liverwort Marchantia are not anthocyanins, but rather a unique class of phenylpropanoids, for which we propose the name “auronidins.” Auronidins have similar colors as anthocyanins but distinct biosynthesis and color properties, and they may contribute to the remarkable ability of liverwort to survive in extreme environments.
Keywords: anthocyanin, CRISPR, flavonoid, liverwort, Marchantia
Abstract
Anthocyanins are key pigments of plants, providing color to flowers, fruit, and foliage and helping to counter the harmful effects of environmental stresses. It is generally assumed that anthocyanin biosynthesis arose during the evolutionary transition of plants from aquatic to land environments. Liverworts, which may be the closest living relatives to the first land plants, have been reported to produce red cell wall-bound riccionidin pigments in response to stresses such as UV-B light, drought, and nutrient deprivation, and these have been proposed to correspond to the first anthocyanidins present in early land plant ancestors. Taking advantage of the liverwort model species Marchantia polymorpha, we show that the red pigments of Marchantia are formed by a phenylpropanoid biosynthetic branch distinct from that leading to anthocyanins. They constitute a previously unreported flavonoid class, for which we propose the name “auronidin,” with similar colors as anthocyanin but different chemistry, including strong fluorescence. Auronidins might contribute to the remarkable ability of liverworts to survive in extreme environments on land, and their discovery calls into question the possible pigment status of the first land plants.
The flavonoid pathway is thought to be unique to land plants. It is hypothesized to have arisen when plants were first colonizing the land, as an adaptation to cope with the additional abiotic stresses faced from a terrestrial lifestyle (1–4). Both seed plants and basal land plant groups produce a variety of flavonoid types, reflecting adaptations of the biosynthetic pathway during evolution to facilitate colonization of the wide range of environments that plants now occupy (2, 4, 5). However, although more than 8,000 individual flavonoid structures have been characterized (6), the great majority belong to a small number of major flavonoid classes. All land plants studied to date, including the liverworts, considered the probable basal plant group, produce nearly colorless, vacuolar-located flavone or flavonol glycosides, which absorb UV light and are important for ameliorating the damaging effects of UV-B radiation (1, 7–12). Seed plants also produce a variety of vacuolar-located anthocyanidin glycosides (anthocyanins) (13) that provide the majority of water-soluble plant colors, ranging from orange to blue.
Anthocyanins have various functions in plant–environment interactions; they aid pollination and seed dispersal through coloration of flowers and fruit and are also thought to help plants cope with different abiotic stresses (3, 14, 15). The flavonoid pigments of basal land plant groups have not been extensively characterized (16), but liverworts, in response to abiotic stresses, produce red nonglycosylated cell wall-bound riccionidin pigments that are proposed to be early evolved anthocyanidin forms (13, 17–19). The flavonoid biosynthetic pathway to flavones and flavonols, which is part of the larger phenylpropanoid pathway, appears to be conserved across land plants (17, 20). Anthocyanins share the same initial biosynthetic steps as flavones/flavonols but require additional enzymatic steps that are well defined in seed plants but not in nonseed plants. It is assumed that the branches of the flavonoid biosynthetic pathway that lead to flavones/flavonols and anthocyanins arose during the evolutionary transition of plants from aquatic to land environments (1, 3, 7–9).
Regulation of flavonoid production has been extensively characterized in angiosperms, with R2R3MYB and bHLH family transcription factors found to be the key regulators (21–23). It was recently reported that an R2R3MYB transcription factor, MpMYB14, activates flavonoid biosynthesis in the model liverwort Marchantia polymorpha (17, 24). Transgenic Marchantia plants overexpressing MYB14 under the 35SCaMV gene promoter display strong activation of the flavonoid pathway (17, 24), yielding greatly increased amounts of flavone O-glycosides and a red pigment (2) assumed to be cell wall-bound riccionidin (18). Using the Marchantia model system (25), we show here that the red pigments are formed by a flavonoid biosynthetic branch distinct from that leading to anthocyanins and representing a previously unreported pigment class that raises questions about our current understanding of flavonoid pathway evolution.
Results
In addition to the water-insoluble pigment bound to the cell wall, the 35S:MYB14 Marchantia plants also produce a water-soluble reddish pigment (1) in considerable amounts, previously only barely detectable in wild-type (WT) plants (17). We took advantage of these transgenic plants to obtain sufficient quantities of the 2 pigments for isolation and full structural analysis. After preparation of pure compounds, their structures were determined to be 2,3,6,8-tetrahydroxybenzofuro[3,2-b]chromen-5-ium-6-O-(2-O-α-rhamnopyranosyl-β-glucopyranoside) (termed auronidin 4-neohesperidoside) (1) and its aglycone (2), using various 1D and 2D NMR experiments (Fig. 1 and SI Appendix, Table S1). The high-resolution electrospray ionization mass spectrum of 1 showed a molecular ion at m/z 593.15099 corresponding to the empirical formula C27H29O15+ (calc. 593.15064), in agreement with the predicted structure based on NMR. During the elucidation, we recognized that the 2 pigments represented a pigment class distinct from anthocyanidins but probably also derived from the flavonoid pathway. Therefore, we propose the term “auronidin” for this reddish class to reflect its structural resemblance to aurones and anthocyanidins and common color properties with anthocyanidins. Thus, these pigments are auronidin 4-O-neohesperidoside (1) and its aglycone, auronidin (2). Accordingly, riccionidin A is not an anthocyanidin, although it has been previously characterized as such (17–19, 24).
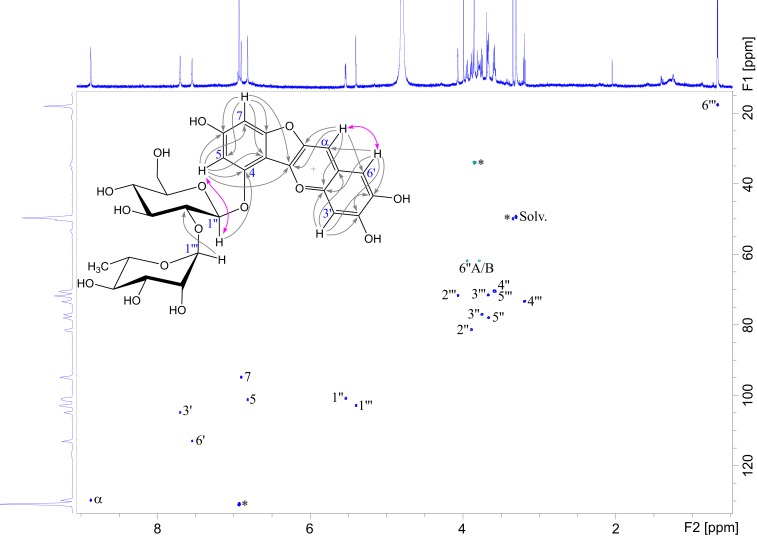
Fig. 1.
Selected NMR information from various 1D and 2D NMR spectra used for structural elucidation of auronidin 4-neohesperidoside (1). The main annotated spectrum (1H-13C heteronuclear single quantum coherence [HSQC]) shows 1JCH cross-peaks representing the carbon atoms, which are directly connected to hydrogen atoms. The gray arrows in the included structure show selected long-range 1H-to-13C bonding correlations observed as cross-peaks in the heteronuclear multiple bond correlation spectrum, while the 2-sided pink arrows represent 1H-to-1H through-space neighborships observed as cross-peaks in the rotating frame Overhauser spectroscopy spectrum. The 1H NMR spectrum is shown on top and the 13C projection of the HSQC spectrum on the left. Pigment 1 is dissolved in CD3OD–CF3CO2D (95:5, vol/vol), and the NMR spectra are recorded at 25 °C. SI Appendix, Table S1 provides the chemical shift values for the proton and carbon atoms.
The auronidin 4-glycoside (1) and its aglycone (2) have the basic structure of an aurone plus a pyrylium ring (D-ring) similar to the C-ring of the flavylium cationic form of anthocyanidins (Fig. 2A). The conjugation within these rings enables them to facilitate orange through red to magenta colors not far from the color nuances of many anthocyanins (Fig. 2B), in contrast to the more yellow colors (λmax 370 to 400 nm) reported for most aurones. However, we observed that the color palette expressed by auronidin 4-glycoside when exposed to different pH environments and solvents was based on various proportions of the auronidin and its anionic form in a simple acid-base equilibrium (Fig. 3 A and B). This is in contrast to what is observed for anthocyanins, which display a similar but more extended color palette based on various proportions of individual structural forms (chalcones, hemiketals, flavylium, and quinoidal forms) with different colors. The auronidins can be added, alongside anthocyanins, as a flavonoid class that provides coloration to plants suitable for screening in the visible part of the light spectrum but based on distinct chemical expression. In addition, the auronidins 1 and 2 showed strong fluorescence (Fig. 3 C and D) at wavelengths that may provide opportunities for new chemistry-based practical applications, for example, as chemosensors in dye-sensitized solar cells (26) or in the base compound of a line of pharmaceutical derivatives (27).
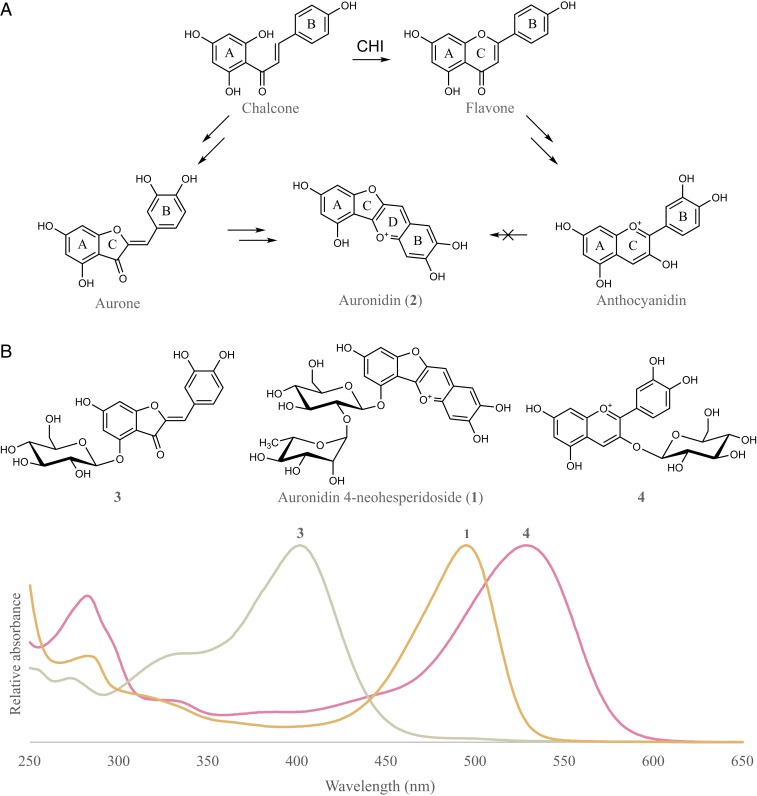
Fig. 2.
Proposed biosynthetic relationships among aurones, auronidins, and anthocyanidins and comparison of the UV/visible-light spectra of corresponding glycosides. (A) Marchantia plants with a knockout mutation of the single chi gene showed no detectable flavones but were unaltered in auronidin production compared with WT plants. Plants with a knockout mutation of the candidate ppo aurone biosynthetic gene showed reduced auronidin content but were unaltered in flavone production. (B) Structures and corresponding UV/visible-light spectra recorded in acidified methanol of an aurone, aureusidin 4-glucoside (3) (λmax = 402 nm); an auronidin, auronidin 4-neohesperidoside (1) (λmax = 495 nm); and an anthocyanin, cyanidin 3-glucoside (4) (λmax = 529 nm).
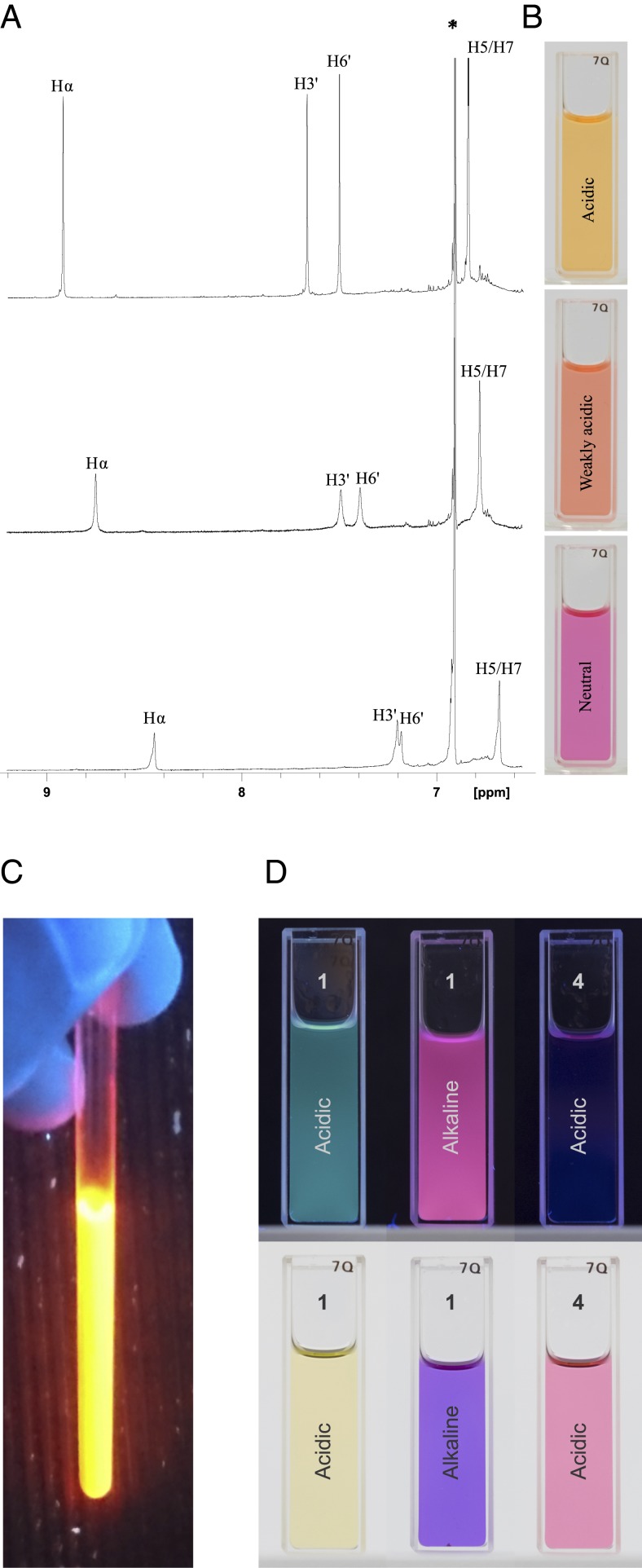
Fig. 3.
1H NMR spectra, colors, and fluorescence of auronidin 4-neohesperidoside from Marchantia in solvents with different acidities, demonstrating lack of some typical anthocyanin properties. (A and B) Auronidin 4-neohesperidoside (1) dissolved in methanol containing various amounts of TFA. The aromatic regions of the corresponding 1H NMR spectra (A) show that the color changes from orange to pink-purple (B) are in accordance with increased amounts of the anionic form of 1 in an acid-base equilibrium when the acid content is reduced. The lack of additional peaks in the NMR spectra prove the absence of various anthocyanin equilibrium forms expressing different colors. (C) Auronidin 4-neohesperidoside dissolved in deuterated dimethyl sulfoxide containing 5% TFA in 365-nm UV light. (D) Comparison of auronidin 4-neohesperidoside and the anthocyanin cyanidin 3-glucoside dissolved in methanol containing 0.5% TFA (acidic) or 0.5% 0.2 M NaOH (alkaline). The samples were illuminated by a 365-nm UV lamp (Top) or ambient light (Bottom). The anthocyanin in alkaline is not shown, as it degraded in that solvent.
We observed that the hydroxyl substitution pattern of riccionidin A (2) reported as an anthocyanidin (18) did not fit with the standard pattern of anthocyanidins, where the A-ring is derived from the acetate/malonate pathway and the B- and C-rings from the shikimate/phenylalanine pathway. However, this pattern of hydroxyls would be obtained if ring closure occurred between the B- and C-rings of an aurone with 4,6,3′,4′-hydroxylation (4,6,3′,4′-tetrahydroxyaurone, termed aureusidin). In this case, the oxygenation pattern of the A-ring is in accordance with condensation of 3 acetate units, and the B-, C- and D-rings fit with the oxygenation pattern of their C9 skeleton coming from the shikimate/phenylalanine pathway (Fig. 2A). Marchantia has been shown to produce aureusidin 6-O-glucuronide during the reproductive phase (28) and thus possesses the biosynthetic route to the proposed substrate for auronidin formation. Thus, a biosynthetic route to 1 and 2 via aurones was theorized (Fig. 2A and SI Appendix, Fig. S1). In this case, the biosynthetic pathway to auronidins and anthocyanins is common through the early steps of the phenylpropanoid pathway (phenylalanine ammonia lyase [PAL], cinnamate 4-hydroxylase, and 4-coumaroyl CoA ligase) and the first steps of the flavonoid specific branch that result in the formation of chalcones (chalcone synthase [CHS] and chalcone isomerase-like [CHIL]). However, the pathways to each compound from chalcones then diverge. The enzyme chalcone isomerase (CHI) that conducts the second committed step of flavonoid biosynthesis is required for flavone and anthocyanin biosynthesis but not for the biosynthesis of aurones (SI Appendix, Fig. S1). Aurones are formed from chalcones by variant polyphenol oxidase (PPO) enzymes, with different types of PPOs, termed aureusidin synthase (AUS) and aurone synthase (AS), forming aurones in Antirrhinum majus and Coreopsis grandiflora, respectively (29–34). The AUS and AS conduct hydroxylation at the B-ring of chalcones, followed by oxidative cyclization into aurones, either before (CgAS) or after (AmAUS) glycosylation. The majority of PPOs characterized in angiosperms are localized to the plastid and exert an oxidative action on phenolic substrates that can lead to, for example, the browning that occurs during cell rupture (35). The CgAS has the characteristic N-terminal chloroplast transit peptide and thylakoid transfer domain and so might direct aurone biosynthesis in plastids (33, 34). However, the AmAUS lacks the plastid localization signal and is localized to the vacuole (32). As auronidins are a newly identified phenylpropanoid group, there is no information on the possible subsequent biosynthetic steps from aurones to the cell wall-bound or soluble glycosylated auronidin products.
Although a biosynthetic pathway to auronidins via aurones seemed probable, based on the shared structural characteristics with aurones and the characterized function of MpMYB14 in activating flavonoid biosynthesis, alternative biosynthetic routes could not be excluded. Thus, RNA-seq differential gene expression analysis was conducted to identify candidate genes for auronidin biosynthesis. Marchantia produces auronidins when placed under nutrient deprivation stress, and this response is lost in the myb14 mutant (17). RNA-seq comparisons of the gene expression response of WT and myb14 plants under nutrient deprivation identified 260 differential genes with a cutoff of an adjusted P value <0.001 and a log2 fold increase >1.0 (Fig. 4A and SI Appendix, Table S2). Of these, 121 were up-regulated in WT but not myb14 plants and thus included candidates for auronidin biosynthetic genes. Of the 20 up-regulated transcripts with lowest adjusted P values, 7 encoded known phenylpropanoid biosynthetic enzymes, including those corresponding to PAL (Mapoly0005s0086, Mapoly0005s0088, and Mapoly0014s0211), CHS (Mapoly0021s0159), and CHIL (Mapoly0175s0004). However, CHI (Mapoly0167s0012) was not 1 of these 260 genes, being up-regulated by only 0.5 log2 fold with an adjusted P value >0.001 and ranked at 1,627 out of 11,937 gene models on adjusted P value. In addition to the 7 transcript models for characterized phenylpropanoid biosynthetic enzymes, 4 others corresponded to candidates for additional auronidin biosynthetic or localization steps. These included a transcript for a PPO (Mapoly0021s0041), which had the lowest adjusted P value of any gene model (2.2 × 10−308) and one of the highest fold differences (8.5 log2 fold, a baseMean change from 42 to 15,622 reads). To further resolve candidate auronidin biosynthetic genes, comparisons were made to genes up-regulated in 35S:MYB14 transgenics compared with unstressed WT plants, as the 35S:MYB14 Marchantia transgenics have greatly increased auronidin content (SI Appendix, Fig. S3). Applying the same adjusted P value <0.001 and log2 fold increase >1.0 cutoff gave 160 transcripts as up-regulated in 35S:MYB14 transgenics (Fig. 4B). Twenty-two transcripts were common across the 2 pairwise comparisons, and 6 of these corresponded to candidate phenylpropanoid biosynthetic enzymes, including the PPO. The pattern of genes up-regulated by MYB14 thus supported a phenylpropanoid biosynthetic route to auronidins but not via chalcones, as CHI was not activated. The proposed route via aurones was supported by the strong and coordinated up-regulation of genes for PAL, CHS, CHIL, and a candidate AUS PPO.
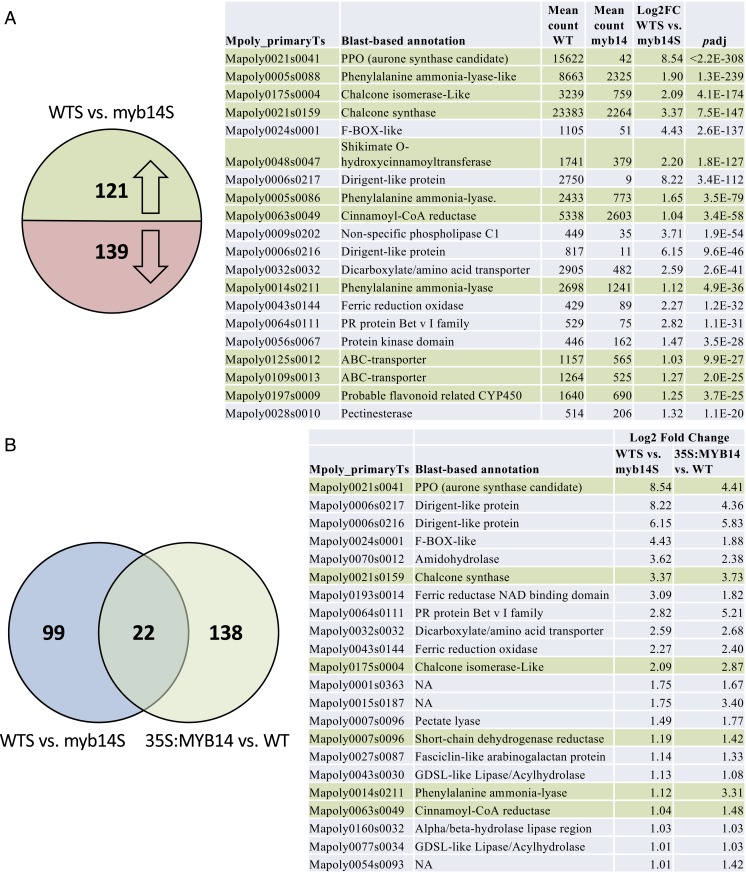
Fig. 4.
Identification of transcripts associated with MpMYB14-regulated auronidin production. (A) Differential transcript abundance for WT vs. myb14 mutant Marchantia lines during nutrient deprivation stress, as determined using RNA-seq DESeq2, with an adjusted P value <0.001 and log2 fold increase >1.0 cutoff. The number of transcripts up- or down-regulated are shown on the left, and details of the 20 up-regulated transcripts having the lowest adjusted P value are shown in the table on the right. (B) Comparison of the 121 up-regulated transcripts from the WT-stress (WTS) vs. myb14-stress (myb14S) analysis to those up-regulated between a 35S:MYB14 transgenic and WT, with an adjusted P value <0.001 and log2 fold increase >1.0 cutoff. Details of the 22 transcripts up-regulated in both comparisons are shown on the right. In both A and B, candidate transcripts related to phenylpropanoid biosynthesis are highlighted in green.
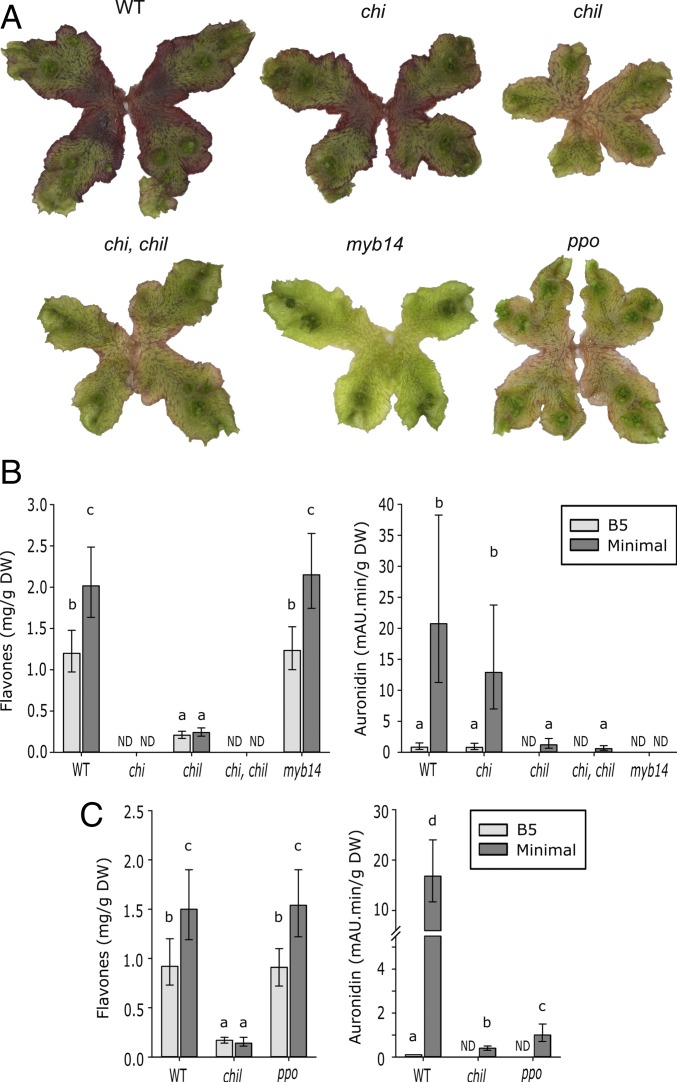
To confirm that CHI was not required for auronidin biosynthesis, we analyzed Marchantia plants that have CRISPR/Cas9-based mutations in the CHI gene (9). Previous studies have identified CHI and CHIL as single-copy genes in M. polymorpha (9, 25). We confirmed this through BLAST analysis of the genome sequence, transcript assemblies of Bowman et al. (25), and our own de novo transcript assemblies based on RNA from the 35S:MYB14 transgenics. In phylogenetic analysis, the MpCHI and MpCHIL sequences fall into the well- characterized separate CHI and CHIL clades (SI Appendix, Fig. S2), with only 25.4% identity between the deduced amino acid sequences. CHI and CHIL sequences from Marchantia paleacea, termed MpalCHI1 and MpalCHI2, have been characterized for their encoded enzyme activities, with MpalCHI1 able to form flavanones from chalcones with a similar efficiency to CHI enzymes from angiosperms but MpalCHI2 lacking any flavanone biosynthetic activity (36). MpCHI and MpCHIL have 90.7% and 92.9% amino acid identity to MpalCHI1 and MpalCHI2, respectively. Wild-type Marchantia plants under nutrient deprivation stress produced both flavone O-glycosides and the red pigment auronidin (2) (Fig. 5). Plants with knockout mutations of the Mpchi gene produced no detectable flavones, confirming that the single MpCHI gene is required for production of flavanone-derived compounds such as flavones and anthocyanins (SI Appendix, Fig. S1). However, the chi mutants were unaltered in auronidin production (Fig. 5). As an additional visualization of the lack of requirement of CHI for auronidin biosynthesis, the chi line was crossed with a 35S:MYB14 transgenic line. The (haploid) F1 lines containing the chi mutated allele and 35S:MYB14 transgene were visually indistinguishable from the 35S:MYB14 parental line, showing substantial overproduction of auronidin and deep purple pigmentation (SI Appendix, Fig. S3).
Fig. 5.
Marchantia genetic mutants showing that auronidins are produced by a different branch of the flavonoid pathway than that producing anthocyanins. WT and mutant Marchantia plants were induced to synthesize red auronidin pigments by nutrient deprivation. Lines contain mutations in flavonoid biosynthetic (chi, chil, and ppo) or regulatory (myb14) genes. (A) Representative pigmentation phenotypes. (B and C) Flavone and auronidin contents. n = 3 biological replicates ± 95% confidence interval. ND, not detected. Means that are significantly different are indicated by different letters (a, b, c, and d).
The chi mutant analysis confirmed that the auronidin biosynthetic branch differ from that of anthocyanins and does not require flavanone production. The differential gene expression suggested a flavonoid biosynthetic origin. This was tested by analysis of Marchantia plants with knockout mutations of the chil gene (9). CHIL function is unresolved, but mutant studies in Arabidopsis and Ipomoea nil show that it is generally required for efficient function of the early steps of the flavonoid pathway (37, 38) and may noncatalytically enhance chalcone formation and downstream conversion (39). The Mpchil lines had reduced content of both flavones and auronidin (Fig. 5), supporting a flavonoid origin for auronidins. In accordance with this, plants with a knockout of both the chi and chil genes, generated by crossing the individual mutant lines, contained almost no detectable flavonoids, with both flavones and auronidins reduced (Fig. 5).
The RNA-seq differential gene analysis identified the PPO gene Mapoly0021s0041 as a candidate biosynthetic step for auronidins, potentially as an AUS/AS. To investigate this further, we analyzed the Mapoly0021s0041 predicted amino acid sequence in the context of the Marchantia PPO gene family. We found 63 candidate Marchantia PPO gene models (SI Appendix, Fig. S4), by far the largest PPO gene family identified for any plant species (35), despite the comparatively small number of gene models in the Marchantia genome sequence. Mapoly0021s0041 contained an N-terminal extension, which may be a targeting peptide, and the conserved PPO catalytically active domain of ∼40 kDa. However, it lacked the C-terminal domain of ∼19 kDa that is thought to shield the active site until cleavage at the site of action. The N-terminal targeting peptide did not have the typical characteristics for plastid localization, with the highest prediction being for secretory. We used CRISPR/Cas9 (40) to generate knockout mutants for Mapoly0021s0041 (ppo plants). The haploid status of the dominant liverwort life stage, the gametophyte, allowed for efficient visual screening of the first transgenic generation for reduced pigmentation when plants were placed under nutrient stress. Thus, 8 independent candidate ppo mutants were identified in the initial screen, and the presence of different deletion events in the targeted region of the Mapoly0021s0041 gene then confirmed for each of these lines (SI Appendix, Fig. S5). As the initial screen was visual, rather than an unbiased screening for mutation events, we confirmed the specificity of the ppo mutations by sequencing any predicted potential nontarget sites in the genome. No off-target genome sequence changes were found. The ppo mutants had reduced auronidin production but WT flavone production (Fig. 5), suggesting a specific role for Mapoly0021s0041 in auronidin biosynthesis and supporting the proposed aurone biosynthetic route to auronidins.
Discussion
The timing of the evolution of the flavonoid pathway is the subject of current debate (20). Most papers communicate that flavonoids are absent from algae and propose flavonoids arising during the evolution of the first land plants. Although genes for the initial steps of the phenylpropanoid pathway may be present in algae, and there is some evidence of various flavonoid groups (but not the pigmented anthocyanin nor aurone groups) at very low amounts in algae (40), there are no substantiated examples of the flavonoid-specific genes being present in algal genomes. What type of flavonoids the first land plants might have produced is not known. The UV-B–absorbing colorless flavones and flavonols are present in all major extant land plant groups examined, suggesting that they may have been present in the last common ancestor (8–11, 16). When it comes to pigmented flavonoids, anthocyanins are widely distributed in angiosperms and to some extent in gymnosperms. A few deoxyanthocyanidins (rather simple in structure) are reported to occur in nonseed plants, such as ferns and mosses (6, 41).
Anthocyanins serve 2 major functions in plants. In seed plants, they provide a diversity of colors vital for pollination and seed dispersal (14, 15). This color palette is based on several distinct structural elements of the anthocyanins, including various proportions of structurally different equilibrium forms of individual colors. The second function of anthocyanins is in protection against abiotic stress. Although the mechanism by which anthocyanins function is not resolved, they are known to directly absorb or distribute visible light and to have antioxidant activity (2, 3, 7, 8). In the liverwort gametophyte, auronidins obviously serve other functions besides participating in pollination and seed dispersal and (reported as riccionidin) have been found to be stress-induced (17–19). Thus, auronidins may be anthocyanin “replacements” in liverworts with regard to stress tolerance functions—despite the absence of the various structural forms typical of anthocyanins. Auronidins share some color attributes with anthocyanins in the orange to violet part of the light spectrum and are the only other flavonoid group to do so. However, in auronidins, the chromophore system is based on acid-base equilibrium forms and not on the different anthocyanin equilibrium forms. Also, the formation of the auronidin C- and D-rings based on the C9 skeleton is unique among the flavonoid groups. The chemical properties displayed by auronidins in vitro, and the presence of both cell wall-bound and soluble forms in planta, suggest unreported in planta functions that perhaps contribute to the remarkable ability of liverworts to survive in extreme environments.
More than 8,000 flavonoid structures were identified by 2006 (6), and more have been characterized since. We suggest that the development of and within the various flavonoid groups, as well as the regulation of the amounts of individual flavonoids in each plant, are in the evolutionary context of the various plant phyla driven by functional needs. R2R3MYB transcription factors are central regulators of flavonoid production (14, 17, 22, 23). Angiosperm R2R3MYB gene families are substantially larger than those of basal plants such as Marchantia (25). In Marchantia, a single R2R3MYB gene, MpMYB14, is the key regulator of auronidin production in response to stress triggers (17) (Fig. 5). Although these same stress triggers activate the R2R3MYB genes that induce anthocyanin production in angiosperms, additional R2R3MYB genes are required for the production of anthocyanins or other flavonoid groups in response to a variety of different developmental and environmental stimuli. As a consequence, we suggest that the progress of diversity and complexity of anthocyanin structures with their structurally different equilibrium forms, and associated increased complexity in pathway regulation, are advanced features in seed plants that facilitated communication with the biotic environment via their additional functions in pollination and seed dispersal. Thus, the biosynthesis of anthocyanins in the evolutionary context must be viewed from 2 directions, driven either by its proposed function in stress tolerance or its function in pollination and seed dispersal.
The similarity of the auronidin ring pattern, hydroxyl positions, and glycosylation pattern to that of aurones suggested an aurone precursor for auronidins. The phenotypes of the chi and chil mutants supported this proposed aurone biosynthetic route; the chi mutants lost production of flavanone-derived flavonoids (flavones) but not of auronidins, while the chil mutants had reduced production of both flavones and auronidins. The reduced flavone in chil plants suggests that CHIL has a similar role in promoting flavonoid production in Marchantia as it does in some angiosperms. The flavonoid route to auronidins was also supported by the genes identified as up-regulated by MpMYB14 coincident with auronidin production. These included the phenylpropanoid pathway enzyme PAL and the flavonoid pathway enzymes CHS and CHIL, as well as a candidate AUS PPO. Thus, the chemistry, chi/chil mutant, and RNA-seq analysis data show that auronidins are phenylpropanoid pigments and suggest a biosynthetic origin via aurones. However, a genetic mutation removing the activity of either CHS or a confirmed enzyme in the specific aurone/auronidin biosynthetic branch is required to rule out other possible biosynthetic routes. CHS is a challenging CRISPR/Cas9 target in Marchantia. Twenty-four putative CHS genes have been identified in the genome (25), including 9 of the 2,000 most highly expressed genes in the stressed WT RNA-seq data, with no single gene accounting for >25% of the total CHS transcript pool. The auronidin biosynthetic steps following chalcones are unknown; however, the candidate AUS PPO, Mapoly0021s0041, was coordinately expressed with auronidin production and the characterized flavonoid biosynthetic genes. In the top 2,000 genes in the stressed WT RNA-seq data, there are only 2 PPO candidates, with Mapoly0021s0041 accounting for 77% of the total PPO transcript pool, thus making it a suitable CRISPR target. Mapoly0021s0041 loss-of-function mutants (ppo lines) had greatly reduced production of both cell wall-bound auronidin and auronidin glycoside. Thus, Mapoly0021s0041 probably conducts a biosynthetic step before the cell wall attachment of auronidin (which presumably may involve polymerization), as the auronidin glycoside is thought to be vacuolar-located based on visual localization in cells, including the presence of anthocyanic vacuolar inclusion equivalents (24). It is unlikely that the reduced pigment amounts in the ppo plants were due to pathway feedback inhibition from the accumulation of unbound auronidin, as 35S:MYB14 plants accumulate large amounts of auronidin glycoside. The data thus support an aurone biosynthetic route. However, the possibility that Mapoly0021s0041 (which has an in silico localization prediction as “secretory”) or other MpPPO genes are involved in biosynthetic steps in addition to formation of the aurone, such as cell wall binding or polymerization, cannot be ruled out.
Aurones occur sporadically across the embryophyta, and PPOs with differing structures and activities have been adopted for aurone biosynthesis. Tran et al. (35) found that the size of the PPO gene family varied from 0 to 13 across 20 embryophyte genomes analyzed, which included those of the moss Physcomitrella patens and the lycophyte Selaginella moellendorffii. The same analysis found PPO genes across 5 green algae genomes, suggesting that PPO predates land colonization. In comparison, 63 candidate PPO genes were found in the Marchantia genome, with various predicted protein structures. These included several genes, like the candidate AUS, that are short sequences comparative to angiosperm PPOs. The large gene number and variation in protein structures suggests that the PPO gene family has undergone extensive duplication and neofunctionalization in Marchantia and could have roles in biosynthetic processes other than those typically expected for angiosperm PPOs. Whether this is a feature of liverworts in general awaits the completion of genome sequences for additional species.
Liverworts are often considered as the basal plant group with a sister relationship to all extant land plants. The discovery that riccionidin A is an auronidin means that there are now no validated reports of anthocyanins in liverworts. Thus, we suggest that the biosynthesis of anthocyanins arose after the occurrence of the last common ancestor of liverworts and seed plants.
Methods and Materials
Plant Material.
Plant material, growth conditions, and stress treatments of Marchantia polymorpha L. have been described previously (17). The WT and mpmyb14-1 lines have been described previously (17). CRISPR/Cas9-mutagenesis of the MpCHI, MpCHIL, and MpPPO genes was as described previously (17, 42), using the oligonucleotide guide sequences given in SI Appendix. Multiple independent knockout lines were generated, with results shown for representative lines (mpchi-1, mpchil-1, and mpppo-1).
Pigment Isolation and Analyses.
Flavone glycoside and auronidin quantification were done as described previously (17). All analyses were performed using at least 3 biological replicates. Pigments 1 and 2 were extracted from freeze-dried M. polymorpha 35S:MYB14 tissue with methanol/water/trifluoroacetic acid (TFA) (80:20:0.5, vol/vol). The pigments were purified using liquid-liquid extraction against ethyl acetate (monitored with an Agilent 1160 analytical high-performance liquid chromatography [HPLC] system), followed by preparative HPLC. Full details of the extraction and HPLC procedures are provided in SI Appendix. Structural determination used homonuclear and heteronuclear 2D nuclear magnetic resonance (NMR) and high-resolution electrospray ionization (ESI+) mass spectrometry. NMR spectroscopy was done with a Bruker Ascend 850-MHz spectrometer equipped with a TCl CryoProbe at 850.13 MHz for 1H and 213.77 MHz for 13C. Mass spectrometry was done using a JEOL JMS-T100LC-AccuTOF mass spectrometer with ESI+ and time-of-flight separation. Instrumentation and analytical procedures are detailed in Fig. 1, SI Appendix, Table S1, and SI Appendix. UV/visible-light absorption spectra were recorded using a Biochrom Libra S32 PC spectrophotometer.
Data and Materials Availability.
All data underlying the study are available in the paper or SI Appendix.
Supplementary Material
Acknowledgments
We thank Bjarte Holmelid for the high-resolution mass spectra recordings; Sarah Cordiner for assistance with HPLC; Takayuki Kohchi and colleagues for pMpGE010; and Nigel Perry and John Bowman for helpful discussions on the research. Financial support was provided by the Research Council of Norway through the Norwegian NMR Platform (NNP-226244/F50) and The Marsden Fund of New Zealand (Grants PAF1302 and PAF1701).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912741116/-/DCSupplemental.
References
- 1.Björn L. O., Widell S., Wang T., Evolution of UV-B regulation and protection in plants. Adv. Space Res. 30, 1557–1562 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Davies K., Albert N., Zhou Y., Schwinn K., Functions of flavonoid and betalain pigments in abiotic stress tolerance in plants. Annu. Plant Rev. 1, 1–41 (2018). [Google Scholar]
- 3.Landi M., Tattini M., Gould K. S., Multiple functional roles of anthocyanins in plant-environment interactions. Environ. Exp. Bot. 119, 4–17 (2015). [Google Scholar]
- 4.Weng J. K., The evolutionary paths towards complexity: A metabolic perspective. New Phytol. 201, 1141–1149 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Banks J. A., et al. , The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332, 960–963 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersen Ø. M., Markham K. R., Eds., The Flavonoids: Chemistry, Biochemistry and Applications (CRC Press, Boca Raton, FL, 2006), p. 1237. [Google Scholar]
- 7.Agati G., Azzarello E., Pollastri S., Tattini M., Flavonoids as antioxidants in plants: Location and functional significance. Plant Sci. 196, 67–76 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Agati G., et al. , Functional roles of flavonoids in photoprotection: New evidence, lessons from the past. Plant Physiol. Biochem. 72, 35–45 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Clayton W. A., et al. , UVR8-mediated induction of flavonoid biosynthesis for UVB tolerance is conserved between the liverwort Marchantia polymorpha and flowering plants. Plant J. 96, 503–517 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Pollastri S., Tattini M., Flavonols: Old compounds for old roles. Ann. Bot. 108, 1225–1233 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolf L., Rizzini L., Stracke R., Ulm R., Rensing S. A., The molecular and physiological responses of Physcomitrella patens to ultraviolet-B radiation. Plant Physiol. 153, 1123–1134 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin R., Ulm R., How plants cope with UV-B: From perception to response. Curr. Opin. Plant Biol. 37, 42–48 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Andersen Ø. M., Jordheim M., “The anthocyanins” in The Flavonoids: Chemistry, Biochemistry, and Applications, Andersen Ø. M., Markham K. R., Eds. (CRC Press, Boca Raton, FL, 2006), pp. 471–552. [Google Scholar]
- 14.Davies K. M., Albert N. W., Schwinn K. E., From landing lights to mimicry: The molecular regulation of flower colouration and mechanisms for pigmentation patterning. Funct. Plant Biol. 39, 619–638 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Valenta K., Nevo O., Martel C., Chapman C. A., Plant attractants: Integrating insights from pollination and seed dispersal ecology. Evol. Ecol. 31, 249–267 (2017). [Google Scholar]
- 16.Asakawa Y., Ludwiczuk A., Nagashima F., “Chemical constituents of bryophytes: Bio- and chemical diversity, biological activity, and chemosystematics” in Progress in the Chemistry of Organic Natural Products, Kinghorn A. D., Falk H., Kobayashi J., eds. (Springer, Vienna, 2013), Vol. 95, pp. 1–796. [DOI] [PubMed] [Google Scholar]
- 17.Albert N. W., et al. , Genetic analysis of the liverwort Marchantia polymorpha reveals that R2R3MYB activation of flavonoid production in response to abiotic stress is an ancient character in land plants. New Phytol. 218, 554–566 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Kunz S., Burkhardt G., Becker H., Riccionidins A and B, anthocyanidins from the cell walls of the liverwort Ricciocarpos natans. Phytochemistry 35, 233–235 (1993). [Google Scholar]
- 19.Snell K. R. S., et al. , Quantifying the metabolic cost to an Antarctic liverwort of responding to an abrupt increase in UVB radiation exposure. Glob. Change Biol. 15, 2563–2573 (2009). [Google Scholar]
- 20.de Vries J., de Vries S., Slamovits C. H., Rose L. E., Archibald J. M., How embryophytic is the biosynthesis of phenylpropanoids and their derivatives in streptophyte algae? Plant Cell Physiol. 58, 934–945 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Cheynier V., Comte G., Davies K. M., Lattanzio V., Martens S., Plant phenolics: Recent advances on their biosynthesis, genetics, and ecophysiology. Plant Physiol. Biochem. 72, 1–20 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Xu W., Dubos C., Lepiniec L., Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 20, 176–185 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Lloyd A., et al. , Advances in the MYB–bHLH–WD repeat (MBW) pigment regulatory model: Addition of a WRKY factor and co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol. 58, 1431–1441 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kubo H., et al. , Biosynthesis of riccionidins and marchantins is regulated by R2R3-MYB transcription factors in Marchantia polymorpha. J. Plant Res. 131, 849–864 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Bowman J. L., et al. , Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 171, 287–304.e15 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Kumara N. T. R. N., Lim A., Lim C. M., Petra M. I., Ekanayake P., Recent progress and utilization of natural pigments in dye-sensitized solar cells: A review. Renew. Sustain. Energy Rev. 78, 301–317 (2017). [Google Scholar]
- 27.Demirayak S., Yurttas L., Gundogdu-Karaburun N., Karaburun A. C., Kayagil I., Synthesis and anti-cancer activity evaluation of new aurone derivatives. J. Enzyme Inhib. Med. Chem. 30, 816–825 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Markham K. R., Porter L. J., Production of an aurone by bryophytes in the reproductive phase. Phytochemistry 17, 159–160 (1978). [Google Scholar]
- 29.Boucherle B., Peuchmaur M., Boumendjel A., Haudecoeur R., Occurrences, biosynthesis and properties of aurones as high-end evolutionary products. Phytochemistry 142, 92–111 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Molitor C., Mauracher S. G., Rompel A., Aurone synthase is a catechol oxidase with hydroxylase activity and provides insights into the mechanism of plant polyphenol oxidases. Proc. Natl. Acad. Sci. U.S.A. 113, E1806–E1815 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakayama T., et al. , Aureusidin synthase: A polyphenol oxidase homolog responsible for flower coloration. Science 290, 1163–1166 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Ono E., et al. , Localization of a flavonoid biosynthetic polyphenol oxidase in vacuoles. Plant J. 45, 133–143 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Kaintz C., et al. , Cloning and functional expression in E. coli of a polyphenol oxidase transcript from Coreopsis grandiflora involved in aurone formation. FEBS Lett. 588, 3417–3426 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Molitor C., et al. , Latent and active aurone synthase from petals of C. grandiflora: A polyphenol oxidase with unique characteristics. Planta 242, 519–537 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tran L. T., Taylor J. S., Constabel C. P., The polyphenol oxidase gene family in land plants: Lineage-specific duplication and expansion. BMC Genomics 13, 395 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng A. X., et al. , Identification of chalcone isomerase in the basal land plants reveals an ancient evolution of enzymatic cyclization activity for synthesis of flavonoids. New Phytol. 217, 909–924 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Jiang W., et al. , Role of a chalcone isomerase-like protein in flavonoid biosynthesis in Arabidopsis thaliana. J. Exp. Bot. 66, 7165–7179 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morita Y., et al. , A chalcone isomerase-like protein enhances flavonoid production and flower pigmentation. Plant J. 78, 294–304 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Ban Z., et al. , Noncatalytic chalcone isomerase-fold proteins in Humulus lupulus are auxiliary components in prenylated flavonoid biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 115, E5223–E5232 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goiris K., et al. , Detection of flavonoids in microalgae from different evolutionary lineages. J. Phycol. 50, 483–492 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Cohen M. F., Meziane T., Tsuchiya M., Yamasaki H., Feeding deterrence of Azolla in relation to deoxyanthocyanin and fatty acid composition. Aquat. Bot. 74, 181–187 (2002). [Google Scholar]
- 42.Sugano S. S., et al. , Efficient CRISPR/Cas9-based genome editing and its application to conditional genetic analysis in Marchantia polymorpha. PLoS One 13, e0205117 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.