Significance
Natural selection can drive evolution over short timescales. However, there is little understanding of which ecological factors are capable of driving rapid evolution and how rapid evolution alters allele frequencies across the genome. Here, we combine a field experiment with population genomic data from natural populations to assess whether and how microbiome composition drives rapid genomic evolution of host populations. We find that differences in microbiome composition cause divergence in allele frequencies genome-wide, including in genes previously associated with local adaptation. Moreover, we observed concordance between experimental and natural populations in terms of the direction of allele frequency change, suggesting that microbiome composition may be an agent of selection that drives adaptation in the wild.
Keywords: microbiome, rapid evolution, genomics of adaptation, Drosophila melanogaster
Abstract
Population genomic data has revealed patterns of genetic variation associated with adaptation in many taxa. Yet understanding the adaptive process that drives such patterns is challenging; it requires disentangling the ecological agents of selection, determining the relevant timescales over which evolution occurs, and elucidating the genetic architecture of adaptation. Doing so for the adaptation of hosts to their microbiome is of particular interest with growing recognition of the importance and complexity of host–microbe interactions. Here, we track the pace and genomic architecture of adaptation to an experimental microbiome manipulation in replicate populations of Drosophila melanogaster in field mesocosms. Shifts in microbiome composition altered population dynamics and led to divergence between treatments in allele frequencies, with regions showing strong divergence found on all chromosomes. Moreover, at divergent loci previously associated with adaptation across natural populations, we found that the more common allele in fly populations experimentally enriched for a certain microbial group was also more common in natural populations with high relative abundance of that microbial group. These results suggest that microbiomes may be an agent of selection that shapes the pattern and process of adaptation and, more broadly, that variation in a single ecological factor within a complex environment can drive rapid, polygenic adaptation over short timescales.
A growing number of studies have identified genes that contribute to adaptation (1–4), but the ecological mechanisms that drive evolution are rarely identified (5). Ecological factors often covary in nature, so disentangling the effects of putative agents of selection on changes in allele frequencies requires experimental manipulation. Patterns of intraspecific genomic variation in nature can be shaped by differences in founder populations, connectance between populations, and demography, complicating inferences of selection (6). Replicated selection experiments provide a way to test whether particular ecological mechanisms act as agents of selection and assess the genomic architecture of adaptation, both key challenges to understanding adaptation (2, 6–8). Yet, using selection experiments to identify mechanisms capable of driving rapid evolution in nature also presents methodological challenges; it is difficult to create both ecologically realistic (e.g., complex selective environment, population sizes allowed to vary across treatments) and evolutionarily realistic (e.g., sufficient standing genetic variation, multiple generations, selection agents similar to those in nature) conditions that allow experimental results to translate to populations in nature (5). Combining field selection experiments with population genomic data from both experimental and natural populations presents a powerful approach to determine whether and how particular agents of selection drive rapid evolution in the genome.
Many prominent theories in evolution suggest that species interactions are the primary mechanism that drives evolution and diversification (9–14). Yet, determining which species interactions actually drive evolution when selective landscapes are complex is crucial to understanding both the mechanisms and outcomes of adaptation (15–17). Outdoor experiments that manipulated specific species interactions have provided convincing evidence that competition and predation can act as agents of selection capable of driving rapid phenotypic evolution (18–21). Host–microbe interactions can be strong and there is evidence they can drive macroevolutionary patterns (22–26), but associated microorganisms have not been experimentally investigated as an agent capable of driving rapid host evolution (27, 28) except where symbiont evolution is tied to the host through vertical transmission (29, 30). Bacteria play a crucial role in the physiology, ecology, and evolution of animals even if they are not transmitted or acquired across generations (22, 31–34), and the composition of affiliated microbial communities can impact host performance and relative fitness (35). Moreover, patterns of intraspecific variation in microbiome composition that could have considerable effects on host physiology and performance have been described in a growing number of taxa (36–39). The amount of intraspecific variation in microbiome composition and its effects on host phenotypes have led to considerable speculation, but little data, on the important role the microbiome may play in host evolution (27, 28, 34, 40).
Drosophila melanogaster presents an excellent system in which to investigate whether microbiome composition acts as an agent that drives rapid host genomic adaptation. D. melanogaster populations vary in their microbiome composition in eastern North America, driven by latitudinal variation in the relative proportion of acetic acid bacteria (AAB) and lactic acid bacteria (LAB) (41). Inoculation experiments in the laboratory have demonstrated that LAB and AAB directly influence the functional traits of D. melanogaster including development rate, lipid storage, and starvation tolerance (42, 43). The influences of AAB and LAB on these traits are species-specific, but generally AAB speeds up development and decreases starvation resistance relative to LAB. D. melanogaster populations in eastern North America have long been a model for adaptation, as there are strong patterns of both phenotypic and genomic variation across latitudes that are presumed to be driven by temperature and photoperiod (44–48). Extensive genomic sequencing of natural populations has revealed thousands of independent SNPs that vary clinally and, hence, are likely involved in adaptation (46, 48). Finally, large D. melanogaster populations can be manipulated in replicated outdoor mesocosms, providing the opportunity to connect the wealth of genomic information about this species with an understanding of evolution in field contexts.
To test whether microbiome composition can drive rapid evolution, we introduced outbred populations of D. melanogaster into 14 individual 2 m × 2 m × 2 m outdoor experimental enclosures. We then applied 1 of 3 treatments to these populations as they evolved over a 45-d period: 1) Addition of the AAB species Acetobacter tropicalis to the food resource (At treatment), 2) addition of the LAB species Lactobacillus brevis to the food resource (Lb treatment), and 3) no microbial inoculation (No-Ad treatment). At and Lb strains were selected as representative AAB and LAB based on their different influences on D. melanogaster life history traits, with At-inoculated flies displaying faster development times and shorter periods of starvation resistance than Lb-inoculated flies (41). We used 16s rRNA sequencing and microbial culture to ascertain the efficacy of the treatments and tracked host population size in each replicate to determine whether treatments altered host population dynamics. We tested for rapid evolution in response to microbiome treatments by coupling whole genome data for each replicate with previously identified lists of putatively adaptive loci and examining whether microbiome treatments led to genomic divergence. In addition, we compared the direction of allele frequency change to determine whether differences between experimental treatments were similar to those observed in natural populations as a way of assessing the importance of microbial variation in driving adaptation across natural populations.
Results and Discussion
Efficacy of Shifting the Microbiome in an Outdoor Experiment.
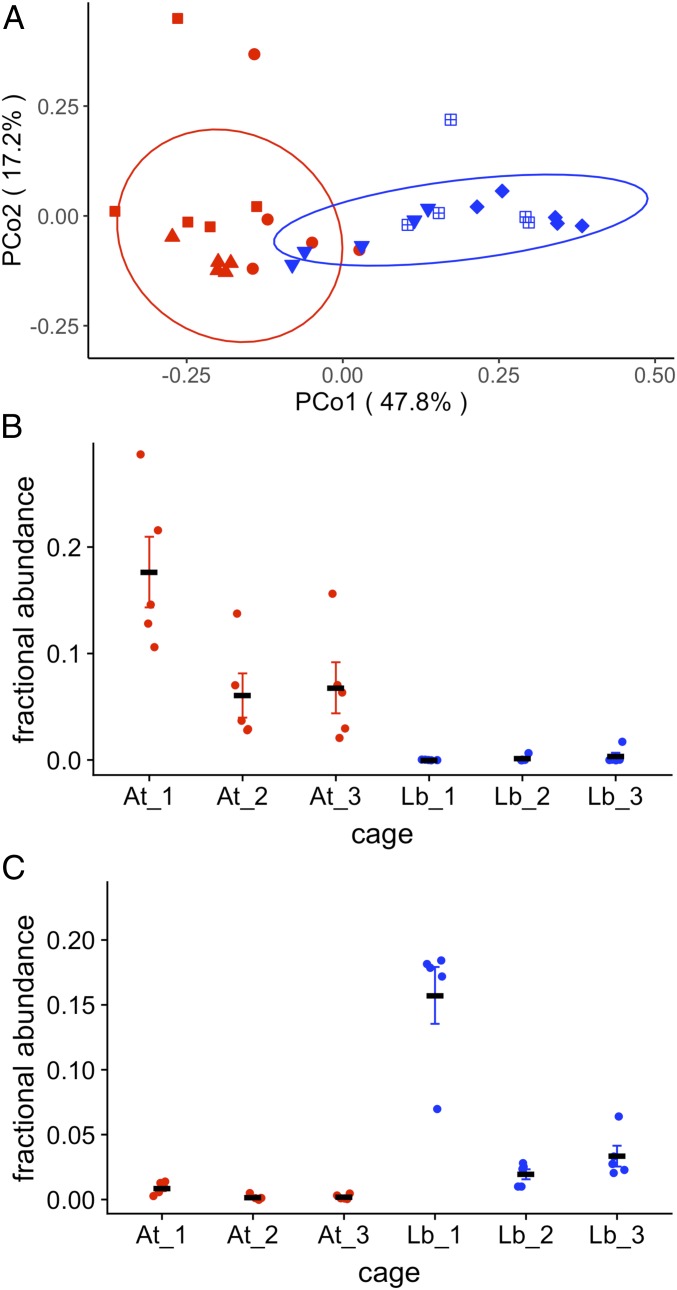
Microbial addition treatments shifted the overall microbiome composition of D. melanogaster populations (Bray Curtis F1,29 = 15.8, P < 0.001) (Fig. 1A; Unifrac metrics in SI Appendix, Fig. S1), the relative abundance of individual operational taxonomic units (OTUs), the abundance of colony forming units (CFUs), and the total abundance of microbes (SI Appendix, Figs. S2–S4). Microbiome composition in At and Lb cages became more similar over time (SI Appendix, Fig. S1), as expected if wild environmental microbiota established in the population in addition to the administered microorganisms. While the different treatments displayed substantial variation in the relative abundance of AAB and LAB, both microbial groups were present in the microbiome of all experimental populations (SI Appendix, Fig. S3). Sequencing the V4 region of the 16S rRNA gene demonstrated that microbiomes of D. melanogaster in At- and Lb-treated cages were enriched for OTUs with perfect identity to the 16s rRNA gene of At and Lb, respectively. Whole genome sequencing of randomly selected microbial colonies isolated from 1 At replicate revealed AAB with >99.9% whole-genome similarity to the added At strain, further supporting that inoculated strains were present in the microbiome (SI Appendix, Fig. S5). Wolbachia, an intracellular microbe common in D. melanogaster and many insect species (49), was present in all populations. Wolbachia increased in relative abundance during the experiment in flies from Lb replicates but not At replicates, consistent with the previously reported negative relationship between Wolbachia and Acetobacteraceae abundance (50, 51) (SI Appendix, Fig. S6B). Our experiment was conducted using a rich diet. Future work manipulating microbiomes on a variety of diets, which are known to influence the microbiome (36, 52, 53), could help disentangle the role of diet and microbiome in driving local adaptation. Overall, the differences in microbiome composition between the At and Lb treatments are modest compared to population-level differences in microbiome composition found across latitudes, where high-latitude locations have microbiomes dominated by LAB and microbiomes in low-latitude populations are dominated by AAB (41).
Fig. 1.
The effect of microbial additions on the gut microbiomes of D. melanogaster in the At and Lb treatments, measured by 16S rRNA marker gene analysis. (A) The effect of At and Lb treatments at the fourth week of the experiment on microbiome composition of pools of adult males collected from cages. (B and C) The relative abundance of AAB and LAB (respectively) in the microbiomes of D. melanogaster from each microbial addition replicate (plotted as means ± SEM).
The influences of distinct AAB and LAB on various D. melanogaster phenotypes are well characterized (42, 54–58). To confirm the previously reported phenotypic effects are also detectable in outbred D. melanogaster populations, we compared the larval development of individuals from the No-Ad experimental cages when monoassociated with At and Lb. Consistent with previous work, bacterial treatment significantly influenced larval development time: At led to ∼10% higher development rate than Lb (Z = −15.9, P < 0.001) (SI Appendix, Fig. S7). The effects of microbiome composition on host ecology presents a mechanism by which microbiomes may shape rapid evolution of host populations.
Influences of Microbiota Treatments on Host Ecology.
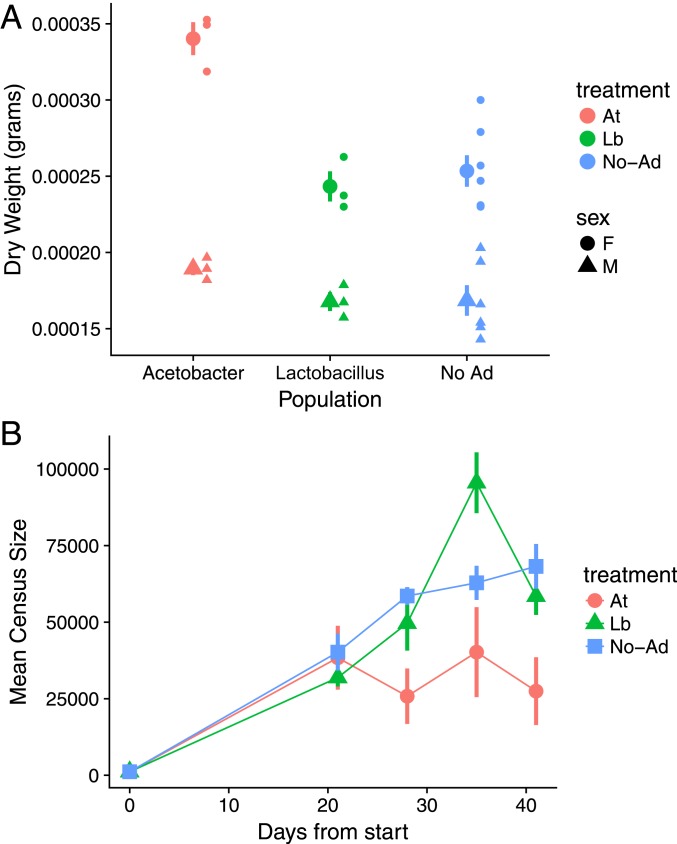
To determine whether microbiome communities alter the ecological characteristics of host populations in outdoor mesocosms and, hence, could plausibly shape host evolution, we measured 2 key ecological characteristics in field mesocosms: fly body mass and population size. Individuals collected directly from At treatment populations had 28% higher mass than those from Lb-treated populations (F2,19 = 13.81, P = 0.0002) (Fig. 2A). We also observed increased sexual dimorphism in At treatments in body size relative to the Lb- and No-Ad treatments (F2,19 = 5.73, P = 0.0113). In contrast, Lb replicates had significantly higher population sizes than At replicates (chisq = 14.86, df = 1, P = 0.0001) (Fig. 2B), suggesting that microbiome treatments influence the tradeoff between somatic and reproductive investment. The difference in population size demonstrates that shifts in the relative abundance of the D. melanogaster microbiota can significantly alter host population dynamics. Differences in population size associated with microbiome composition provides clear evidence to support previous assertions that natural population-level variation in the microbiota that has been observed across the animal kingdom (39, 41, 59, 60) may influence the population ecology of hosts bearing diverse communities of partners (28, 34, 61). Such patterns are established for hosts bearing obligate partners (62–64) or infected with microbial symbionts (65), but our data demonstrate that changes in the relative abundance of microbial taxa can shape host populations. These differences in body size and population dynamics, due to a presumed combination of ecological and evolutionary forces, demonstrate that modest shifts in microbiomes can alter host populations in outdoor settings, which bolsters the hypothesis that microbiomes could drive rapid evolution.
Fig. 2.
Population size and body mass of D. melanogaster populations from each microbial addition treatment. (A) The mean from each treatment at the end of experiment of the dry weight of D. melanogaster individuals of each sex from each replicate cage. (B) Host population size over the course of the experiment. In both graphs, values plotted are means ± SEM.
Microbiome Composition Shapes Host Genomic Evolution.
We assessed whether differences in the microbiome across At and Lb treatments shaped D. melanogaster evolution over the course of 5 host generations. Using a whole-genome pool-seq approach (66), we generated data on allele frequencies at 1,988,853 biallelic segregating sites after filtering (Materials and Methods and SI Appendix, Table S4) for the founder population and from each experimental replicate after 45 d of microbiome treatment. Given that our experiment was founded with a genetically diverse population with little linkage disequilibrium (67) and any divergent selection between treatments was limited to 5 overlapping generations, we did not expect substantial genome-wide divergence (68, 69). To assess any genome-wide divergence from the initial founding population, we calculated the mean FST statistic between the founder population and the 3 treatment populations, for subsets of 1,000 sites sampled randomly from across the genome (SI Appendix, Fig. S8). We also conducted a principal component analysis (PCA) of allele frequencies from all sampled populations to visualize divergence genome-wide (SI Appendix, Fig. S9). In both figures, we observe nonsignificant trends indicating that microbial addition treatments (both At and Lb) are associated with greater genome-wide divergence from the founder population than No-Ad over the relatively short duration of the experiment.
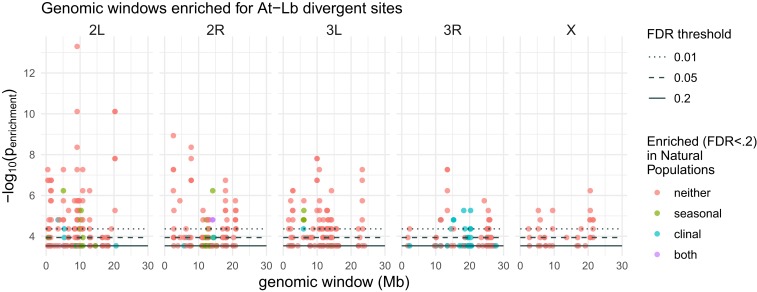
In addition to whole-genome analyses, we also assessed patterns of divergent selection between At and Lb populations at individual sites. Linkage disequilibrium decays over ∼200 bp in most regions of the D. melanogaster genome (67) and our founding populations contained substantial standing genetic variation, giving us considerable genomic resolution with which to detect selection. To assess divergent selection between treatments at each segregating site, we fit a generalized linear model to allele frequencies as a function of microbiome treatment. We found 297 sites diverged significantly between At and Lb treatments with false-discovery rate (FDR) < 0.05 and minimum effect size of 2% (SI Appendix, Table S1). These sites were located on all chromosomes and were found in or near 281 genes, indicating little linkage between the most divergent sites. As signal from individual sites can be confounded with technical and biological noise, we also conducted a region-based analysis to assess divergence between treatments in overlapping windows of 250 SNPs. We found 280 regions of significantly enhanced divergence (FDR ≤ 0.05) between At and Lb populations, with at least 23 such windows found on each of the 5 main chromosome arms (Fig. 3). The D. melanogaster genome contains several inversions that vary in frequency across natural populations in a way that is suggestive of adaptation (70), but we observed no enrichment for divergence of inversion frequencies associated with microbial treatment (based on marker sites) (SI Appendix, Table S2), meaning overall patterns of divergence were not driven by shifts in inversion frequencies. The patterns of divergence we observed across resolutions suggest that modest variation in microbiome composition can drive genomic divergence of host population when standing genetic variation is present. Moreover, the architecture of this divergence, with signatures of selection at many independent regions of the genome, fits with a polygenic model of adaptation, in which many genes contribute to adaptation (71), and suggest that the genomic basis of adaptation over very short timescales can be polygenic.
Fig. 3.
Genomic landscape of divergence between At and Lb populations. Local enrichment of divergent SNPs (divergence GLM P < 0.05 and effect size ≥ 2%) was calculated using a hypergeometric test in windows of 250 SNPs, tiled across the genome with 50 SNP shifts. Shown at the bottom is the −log10 of the enrichment P value for windows with FDR ≤ 0.2. Graphs are chromosomes, and the black dotted lines show the corresponding score thresholds for FDR < 0.01 and <0.05, in addition to the solid line for FDR < 0.2. Windows are colored according to whether they also show enrichment (FDR < 0.2) for sites that vary clinally and/or seasonally in natural populations.
Links Between Microbiome Manipulation and Changes in Allele Frequency in Nature.
Combining our experiment with population genomic data from nature allows us to test whether differences in microbiome composition alone are capable of driving divergence in allele frequencies at SNPs that vary across natural populations. Previous work has found predictable changes in allele frequency at many independent SNPs across seasons from spring to fall in North American orchard populations of D. melanogaster (72, 73). We found more overlap than expected by chance between SNPs that show significant differentiation between At and Lb treatments and SNPs that vary significantly across seasons, using multiple cutoffs for SNP significance (SI Appendix, Table S5). Notably, we did not find this same pattern of seasonal overlap with sites that showed differentiation between the No-Ad treatment and any other treatment, nor between subsets of No-Ad populations. Taken together, these results suggest that SNPs that diverged across At and Lb treatments are also involved in seasonal adaptation in wild D. melanogaster populations.
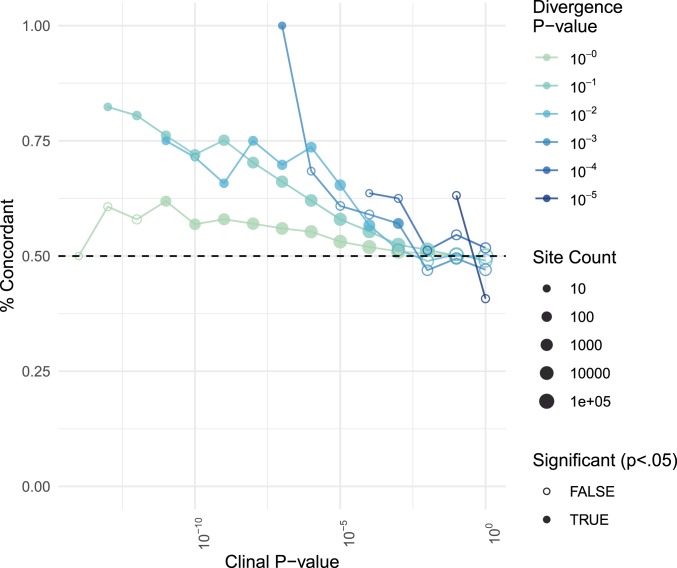
In addition to changes in allele frequency across time, population genomic sequencing of D. melanogaster populations along the east coast of North America has uncovered thousands of putatively adaptive sites that vary significantly (FDR < 0.05) in allele frequency with latitude (73), 15,399 of which were also segregating in our experimental populations. There is also variation in microbiome composition of D. melanogaster populations across latitudes, as high-latitude populations of D. melanogaster have LAB-enriched microbiomes and populations from lower latitudes have AAB-enriched microbiomes (41). We tested whether the allele that was more common in populations experimentally enriched for a microbial group was also more common in the natural clinal population that has a high relative abundance of the same microbial group, noting the caveat that At and Lb are individual strains and cannot represent the breadth of influence possible in wild flies bearing diverse AAB and LAB strains. We labeled sites as “directionally concordant” if the allele that was at higher frequencies in high-latitude populations compared to low-latitude populations was also the allele that was at higher frequencies in Lb populations compared to At populations. When we considered all ∼2 million variant sites, the percent of directionally concordant sites was 50.3%, indistinguishable from a null expectation. However, concordance rose significantly in subsets of sites with both strong divergence between microbial treatments and strong clinal variation (Fig. 4). For example, 70.7% were concordant among the 945 SNPs with At-Lb divergence pval < 0.05, effect size > 2%, and clinal P value <10−5, while 80.0% were concordant among the 35 SNPs with At-Lb divergence pval < 0.01, effect size > 2%, clinal P value <10−8. One-thousand rounds of randomly sampling sites matched to observed data for chromosome and allele frequency demonstrated that these concordance values are both significantly higher than expected by chance (P < 0.001 in both cases). In the latter case, the majority of the 35 SNPs are on chromosome arm 3R, yet are located in or near 32 different genes, several of which are known to play a role in local adaptation (72–74) (SI Appendix, Table S3). Although these high levels of concordance at top divergence sites may suggest long-range linkage, we did not find significantly elevated concordance in any of 7 large chromosomal inversions (SI Appendix, Table S2). The surprising concordance of the identity of AAB-associated and LAB-associated alleles in experimentally treated populations and natural clinal populations suggests microbiome composition may be a significant component of the fitness landscape and, hence, adaptation in natural populations.
Fig. 4.
Concordance of allelic divergence in natural and experimental populations. Concordance is calculated as the percent of sites in which the allele found at higher frequencies in natural high-latitude populations compared to low-latitude populations was also found at higher frequencies in experimental Lb populations compared to At populations. Each point refers to a distinct subset of sites, binned according to clinality (x axis) and At-Lb divergence (color); the number of sites examined is indicated by the size of the point. A dashed black line is drawn at the null expectation of 50% concordance. Solid-colored points represent site subsets in which concordance is significantly elevated compared to the shuffled null distribution.
Conclusion
Moving from documenting cases of rapid evolution to studying the driving mechanisms is crucial to understanding adaptation in natural populations (16). Microbiomes can influence nearly all aspects of host biology (27, 40, 75), and it has long been assumed that microbiomes are also an important factor at the population level (28, 76). Our manipulative experiment demonstrates that changes in the relative and total abundance of the D. melanogaster microbiome are sufficient to cause genomic divergence of host populations over only 5 generations. The magnitude of divergence was heterogeneous across the genome, but we uncovered regions of strong divergence on all chromosomes. Genomic patterns also illustrate that variation in microbiome composition is a sufficiently strong agent of selection to drive evolution at loci that exhibit putatively adaptive patterns across populations in nature. We detected concordance in the directionality of allelic change at these sites between our experiment and natural populations, which provides evidence that variation in microbiome composition is a substantial component of the fitness landscape. Overall, our results demonstrate that shifts in microbiome composition can be important drivers of ecological and evolutionary processes at the population level and that a single ecological factor within a complex environment can drive polygenic adaptation over short timescales.
Materials and Methods
Experimental Setup.
We constructed the founding D. melanogaster population for this experiment by crossing 150 wild-collected isofemale lines from Pennsylvania. Ten males and 10 females were taken from each line and combined into a single breeding cage. After 3 generations of mating and density-controlled rearing in favorable laboratory conditions, we introduced 500 females and 500 males of a single age cohort into each experimental cage on June 15, 2017. Subsamples of the founding population were collected on June 15th for initial genomic sequencing. Flies were in enclosures from June 15 to August 3, 2017, which, based on larval development rates in outdoor cages, allowed for ∼5 overlapping generations. Outdoor cages are 2 m × 2 m × 2 m enclosures constructed of fine mesh built around metal frames (BioQuip PO 1406C) (77, 78). Inside of these enclosures, we planted 1 peach tree and vegetative ground cover to provide shading and physically mimic the natural environment. Peaches were removed before ripening to prevent flies from feeding on them. Photographs of 8 quadrats within each cage were taken, and flies were counted to estimate population size at 5 time points during the experiment. We tested for effects of microbiome treatments on host population size using an LME with microbial treatment as a fixed effect and sample date as a random effect. Each cage was used as a statistical replicate, and our analysis was conducted on all census data after the initial population expansion (>day 21 of the experiment).
Microbial Treatments.
The experiment consisted of 3 treatments: diet supplemented with L. brevis DmCS_003 (Lb), diet supplemented with A. tropicalis DmCS_006 (At), and no bacterial addition (No-Ad). To prepare the bacterial inoculum, a 24- to 72-h culture of each species was centrifuged for 10 min at 15,000 × g and resuspended in PBS at OD600 = 0.1. Separately, 300 mL of modified Bloomington diet was prepared in a 1.5-lb aluminum loaf pan under standard laboratory conditions (nonsterile). Within 24 h of diet preparation, 2.2 mL of normalized bacteria were spread on the surface of the food inside of the loaf pan. The inoculated diets were covered for a 12- to 36-h incubation at 25 °C and transported to the outdoor experiment site 3 times each week. Pans were uncovered immediately after introduction to outdoor fly enclosures and placed on shelving units to protect from rain. Pans were left undisturbed for 2 to 3 d to allow for egg laying and then covered with mesh caps to permit larval development but exclude further egg laying. When adults started to eclose, pans were transferred to a small cage inside the larger cage and caps were removed to allow adults to emerge while preventing additional egg laying on pans where adults had already eclosed. We allowed 14–16 d (twice the time needed for the fastest developing eggs) for adults to eclose from the time pans were introduced before discarding them. The protocol for the No-Ad replicates mimicked the above but did not include any inoculation of the food. The diets provided were the only source of food available that was capable of supporting D. melanogaster development.
Quantification of Microbial Communities from Experimental Treatments.
For culture-dependent analysis, 5 pools of 5 male flies were collected from each treated outdoor cage and homogenized in a microcentrifuge tube containing 125 μL of mMRS (modified De Man, Rogosa, Sharpe agar) medium. Homogenates were dilution plated onto mMRS and grown at 30 °C under ambient and restricted oxygen conditions. Tan- or copper-colored colonies were classified as AABs, and white or yellow colonies were classified as LABs. One milliliter of the same homogenate was pelleted for DNA extraction via the QuickDNA Fecal/Soil Microbe kit (Zymo Research, D6011) and analyzed by culture-independent analysis as described below. Pairwise comparisons between absolute CFU abundances were determined by a Dunn test.
We used 16S rRNA marker genes of pooled whole-body flies to survey the microbial community associated with the pooled fly homogenates. From each DNA extraction, the V4 region of the 16S rRNA gene was amplified as described previously, except using a HiSeq 2500 at the Brigham Young University DNA sequencing center (79). Sequence variants were clustered and assigned to the sequencing data using QIIME 2 (80, 81). After taxonomic assignment, sequences identified as Wolbachia, which were present in every sample, were removed (Wolbachia are analyzed separately in SI Appendix, Fig. S6), and the OTU tables were rarefied to balance sequence depth with sample retention (OUT table available as Dataset S1). The single OTUs with perfect matches to the At and Lb genomes were identified using BLASTn (82). Tests for significant differences in microbial beta-diversity (Bray-Curtis, weighted Unifrac, unweighted Unifrac) were performed in R using PERMANOVA (83). Differences in taxonomic abundance were assessed using ANCOM, which uses relative abundances to assess differences in community composition (84). Figures were created using ggplot2 (85).
Measuring Body Size and Development Rate.
At the conclusion of the experiment, we sampled adult individuals from all cages. To determine adult mass content of cage-caught individuals, we took pools of 5 individuals of each sex, dried them at 55 °C for 24 h, weighed them, and divided the total weight by 5 to obtain average individual mass. Body size data (dry weight) were analyzed using a ANOVA with microbial treatment and sex as fixed effects with cage used as the unit of replication.
We collected eggs from each No-Ad cage to determine the effect of monoassociation with At and Lb on development rate. To rear in monoassociation, fly eggs were collected within 24 h of deposition, bleached twice for 150 s each, rinsed thrice in sterile H2O, transferred to a sterile diet at a target density of 30 to 60 eggs per vial, and inoculated with a PBS-washed overnight culture of either bacterial species, normalized to OD600 = 0.1 (86). The period of larval development was determined by counting the number of empty pupae in each vial 3 times each day (at 1, 6, and 11 h into the daily light cycle) until all flies had eclosed or until no flies eclosed in 3 consecutive time periods, whichever came first. Bacteria-dependent differences in D. melanogaster development were analyzed using Cox mixed survival models in R. Development rate was calculated as the inverse time to eclosion. Significant differences between treatments were determined by a Cox proportional hazards model, analyzed separately for each bacterial inoculation, and are reported as different letters over the symbols. Summary statistics were also calculated by ANOVA.
Genomic Sequencing.
We sequenced pools composed of 120 males and 80 females collected from each cage at the end of the study. We extracted the DNA and prepared libraries using ∼500-bp fragments for whole-genome sequencing using KAPA Hyper Prep kit. Libraries were multiplexed with dual-indexing and sequenced on multiple lanes of an Illumina NovaSeq (6 samples on each lane) system with 150-bp paired-end reads. Reads were checked for quality using FastQC. Adapters were trimmed with Skewer (87) and reads with a quality score <20 were removed, and overlapping read pairs were merged with PEAR (88). We aligned reads to a reference genome composed of the D. melanogaster reference sequence (v5) (89), the L. brevis, and the A. tropicalis genomes using BWA (90), then removed duplicate reads with Picard tools (91) and realigned remaining reads around indels with GATK’s IndelRealigner (92). For logistical reasons, the 16 samples included in this study were multiplexed and sequenced with other samples in 2 batches run on separate days. The first batch of samples (n = 12) was sequenced in the same lanes as multiplexed human genome samples, and we detected trace numbers of human reads, likely due to index switching that can happen on Illumina HiSeq platforms (93). We removed any reads that mapped to the human genome (version hg19) using bbmap (94) and excluded from our analysis any D. melanogaster sites to which these putatively human reads mapped.
After mapping and QC, we retained an average of 83 M mapped reads per sample at an average coverage (mosdepth; ref. 95) of 109× of the D. melanogaster autosomes (range 92–133×) and average coverage 92× on the X chromosome. We then used PoPoolation2 (96) to obtain allele counts at segregating sites, discarding bases with quality <20. To be included for down stream analysis, we required SNPs to be biallelic with 1 of the 2 alleles matching the reference allele, and we excluded SNPs overlapping any called indels, SNPs with less than 10 mapped reads containing the minor allele (an allele frequency of ∼0.5% across all samples), and SNPs with min and max read depths less than 50 or greater than 250, respectively. Since the timescale of our experiment was too short to expect any true signal from new mutations arising during the 5 generations of evolution, we additionally filtered out any SNPs with allele frequencies <1% in either sample from the founder population. SNPs within repeat regions as defined by University of California, Santa Cruz RepeatMasker (97) were excluded. Finally, we examined a larger panel of 112 samples all founded from the same starting population (of which the 16 samples included in this study were a subset) that were sequenced in 2 separate sets of lanes and excluded any SNPs that showed distinct allele frequency ranges across sets. This yielded at dataset of ∼2 million SNPs. A full table of the number of sites excluded due to different filters is presented in SI Appendix, Table S4.
PCA and Fst Analyses.
Allele frequencies at each segregating site for each sample were used to conduct a PCA using the R function prcmp with scale = TRUE, and the first 2 PCs were plotted to examine genome-wide divergence across samples visually. To obtain a more quantitative account of the divergence of populations under each treatment from the founder population, a bootstrap-Fst analysis was conducted with 1,000 rounds. In each round, 1,000 sites were randomly selected from across the genome, and Fst was calculated at each site between the average allele frequency in the 2 founder samples and allele frequencies averaged within treatment groups (3 of the 8 No-Ad samples were randomly averaged for each round to match the number of At and Lb samples). Fst values for each round were averaged across the 1,000 sites for each treatment, and the resulting distributions were plotted as boxplots.
SNP Divergence Analysis.
To find SNPs that changed in associated with microbial treatment, we used the R function glm to fit a generalized linear model (GLM) to the allele frequencies at each SNP to test for significant associations between allele frequency and treatment. GLMs were fit using a quasibinomial error structure, as this reduces the rates of false positives relative to other significance testing protocols in genomic data (98), and to account for sampling of chromosomes, all allele frequencies were first scaled to counts out of Neffective, where n is the number of individuals sampled from the population (200 for all samples), rd is the true read depth, and Neffective = (72, 99, 100). We identified outliers as sites with significant divergence between At and Lb samples at an FDR < 0.05 (101), and a mean difference in allele frequency (effect size) of 2%, as this was approximately the average difference in allele frequency between treatments for all SNPs.
Window-Based Divergence Analysis.
To identify local regions of enhanced divergence, we first identified putatively diverged sites between At and Lb treatments using a relaxed GLM cutoff of P < 0.05 and an effect size threshold of 2% (n = 81,492 sites). Then, a hypergeometric test was conducted (with R function “phyper”) to assess enrichment of these sites in windows of 250 consecutive SNPs, with 50-SNP step-size between windows. Enriched windows were identified as those with enrichment FDR < 0.2, which resulted in a minimum of 22 putatively diverged sites in each enriched window. The same process was used to separately identify windows enriched for sites with clinal GLM P value <0.05 and seasonal GLM P value <0.05.
Seasonal Enrichment Analysis.
We used a hypergeometric test to determine whether sites that were divergent between treatments were enriched among sites previously found to vary over seasonal time in populations from eastern North America (73). From the 1,372,676 sites assayed in both the seasonal analysis and our experiment, nsea putatively seasonal sites were first identified using various GLM cutoffs (P < 0.1, P < 0.05, P < 0.01, P < 0.005, P < 0.001). Then, for each pair of treatments, ndiv putatively diverged sites between treatments were identified using the same GLM cutoff and an effect size threshold of 2%, and the number of overlapping sites nboth was calculated.
Test for Directional Concordance with Clinality.
SNPs that vary across the North American latitudinal cline may reflect local adaptation (72–74, 102), and represent potential sources of adaptation to microbiome composition, which is 1 of many factors known to vary along this cline. Although we do not expect extensive overlap between SNPs that vary predictably along the cline and SNPs that vary predictably between treatments in our experiment (due to different segregating sites, different nonmicrobiome-related selective pressures, and different timescales of adaptation), we did predict that the subset of SNPs that are strongly predictable in both cases should be “oriented” in the same direction: i.e., an allele strongly associated with natural clinal populations harboring more AAB should also be the allele associated with experimental populations experimentally enriched for AAB (here, the At treatment). As such, we used an existing genomic dataset on clinal variation (72, 73) to see if the SNPs that showed both 1) divergence between microbial treatments in our experiment, and 2) divergence between natural clinal populations, were more likely to be “directionally concordant” than other SNPs. We first collected P values and coefficients for each SNP in our dataset from our generalized linear model of allele frequency divergence between treatments (pAt-Lb and coefAt-Lb), and P values and coefficients from a previously conducted generalized linear model of allele frequency divergence across the cline (pcline and coefcline). The models were oriented such that a positive coefAt-Lb indicated that the frequency of the alternate allele was higher in Lb samples than At samples, while a positive coefcline indicated that the frequency of the alternate allele was higher in high-latitude (LAB-enriched) populations than low-latitude (AAB-enriched) populations. We assigned each SNP to 2 bins: an At-Lb divergence bin equal to the integer nearest −log10(pAt-Lb), and a clinality bin equal to the integer nearest −log10(pcline). We then examined the intersection of each At-Lb bin and each clinality bin and recorded the percent of SNPs where the sign of coefAt-Lb matched the sign of coefcline, which we termed “directional concordance.” Finally, we shuffled the bin labels across SNPs 500 times (maintaining the same bin pairs) and remeasured directional concordance values to obtain a P value for each true concordance value.
Tests for Enrichment at Inversions.
We identified breakpoints (103) and segregating marker sites (104) associated with 7 large chromosomal inversions. To test for enrichment of divergence between At and Lb samples at marker sites for each inversion, we first assigned every segregating site a divergence score equal to −log10 of the P value from the GLM analysis of per-site divergence. We then recorded the percent of times (of 1,000 replicates) that an equally sized random set of sites had a mean divergence score higher than the markers of a particular inversion. Similarly, to test for enrichment of At-Lb divergence at sites within each inversion, we recorded the percent of times (of 1,000 replicates) that a randomly selected set of 1,000 sites from outside an inversion had a mean divergence score higher than a randomly selected set of 1,000 sites from inside an inversion. Finally, to test for enrichment of clinal concordance within each inversion, we recorded the percent of times (of 1,000 replicates) that a randomly selected set of 1,000 sites from outside an inversion had a concordance rate higher than a randomly selected set of 1,000 sites from inside an inversion.
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the National Center for Biotechnology Information (NCBI Bioproject IDs PRJNA562479 and PRJNA550209).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1907787116/-/DCSupplemental.
References
- 1.Jones F. C., et al. ; Broad Institute Genome Sequencing Platform & Whole Genome Assembly Team , The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484, 55–61 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gompert Z., et al. , Experimental evidence for ecological selection on genome variation in the wild. Ecol. Lett. 17, 369–379 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradley D., et al. , Evolution of flower color pattern through selection on regulatory small RNAs. Science 358, 925–928 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Miller S. E., Roesti M., Schluter D., A single interacting species leads to widespread parallel evolution of the stickleback genome. Curr. Biol. 29, 530–537.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrett R. D. H., et al. , Linking a mutation to survival in wild mice. Science 363, 499–504 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Savolainen O., Lascoux M., Merilä J., Ecological genomics of local adaptation. Nat. Rev. Genet. 14, 807–820 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Stapley J., et al. , Adaptation genomics: The next generation. Trends Ecol. Evol. 25, 705–712 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Long A., Liti G., Luptak A., Tenaillon O., Elucidating the molecular architecture of adaptation via evolve and resequence experiments. Nat. Rev. Genet. 16, 567–582 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobzhansky T., Evolution in the tropics. Am. Sci. 38, 209–221 (1950). [Google Scholar]
- 10.Ehrlich P. R., Raven P. H., Butterflies and plants: A study in coevolution. Evolution 18, 586–608 (1964). [Google Scholar]
- 11.Valen V. L., A new evolutionary law. Evol. Theory 1, 1–30 (1973). [Google Scholar]
- 12.Berenbaum M., Feeny P., Toxicity of angular furanocoumarins to swallowtail butterflies: Escalation in a coevolutionary arms race? Science 212, 927–929 (1981). [DOI] [PubMed] [Google Scholar]
- 13.Schluter D., The Ecology of Adaptive Radiation (OUP Oxford, 2000). [Google Scholar]
- 14.Harmon L. J., et al. , Detecting the macroevolutionary signal of species interactions. J. Evol. Biol. 32, 769–782 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Benkman C. W., Biotic interaction strength and the intensity of selection. Ecol. Lett. 16, 1054–1060 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Nosil P., et al. , Natural selection and the predictability of evolution in Timema stick insects. Science 359, 765–770 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Rudman S. M., et al. , What genomic data can reveal about eco-evolutionary dynamics. Nat. Ecol. Evol. 2, 9–15 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Endler J. A., Natural Selection in the Wild (Princeton University Press, 1986). [Google Scholar]
- 19.Reznick D. A., Bryga H., Endler J. A., Experimentally induced life-history evolution in a natural population. Nature 346, 357–359 (1990). [Google Scholar]
- 20.Schluter D., Experimental evidence that competition promotes divergence in adaptive radiation. Science 266, 798–801 (1994). [DOI] [PubMed] [Google Scholar]
- 21.Reznick D. N., Losos J., Travis J., From low to high gear: There has been a paradigm shift in our understanding of evolution. Ecol. Lett. 22, 233–244 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Ley R. E., et al. , Evolution of mammals and their gut microbes. Science 320, 1647–1651 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brooks A. W., Kohl K. D., Brucker R. M., van Opstal E. J., Bordenstein S. R., Phylosymbiosis: Relationships and functional effects of microbial communities across host evolutionary history. PLoS Biol. 14, e2000225 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Groussin M., et al. , Unraveling the processes shaping mammalian gut microbiomes over evolutionary time. Nat. Commun. 8, 14319 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaulke C. A., et al. , Ecophylogenetics clarifies the evolutionary association between mammals and their gut microbiota. MBio 9, e01348-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharpton T. J., Role of the gut microbiome in vertebrate evolution. mSystems 3, e00174-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shapira M., Gut microbiotas and host evolution: Scaling up symbiosis. Trends Ecol. Evol. 31, 539–549 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Macke E., Tasiemski A., Massol F., Callens M., Decaestecker E., Life history and eco‐evolutionary dynamics in light of the gut microbiota. Oikos 126, 508–531 (2017). [Google Scholar]
- 29.Moran N. A., Baumann P., Bacterial endosymbionts in animals. Curr. Opin. Microbiol. 3, 270–275 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Douglas A. E., How multi-partner endosymbioses function. Nat. Rev. Microbiol. 14, 731–743 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Turnbaugh P. J., et al. , An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Semova I., et al. , Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microbe 12, 277–288 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tremaroli V., Bäckhed F., Functional interactions between the gut microbiota and host metabolism. Nature 489, 242–249 (2012). [DOI] [PubMed] [Google Scholar]
- 34.McFall-Ngai M., et al. , Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. U.S.A. 110, 3229–3236 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gould A. L., et al. , Microbiome interactions shape host fitness. Proc. Natl. Acad. Sci. U.S.A. 115, E11951–E11960 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bolnick D. I., et al. , Individuals’ diet diversity influences gut microbial diversity in two freshwater fish (threespine stickleback and Eurasian perch). Ecol. Lett. 17, 979–987 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sevellec M., et al. , Microbiome investigation in the ecological speciation context of lake whitefish (Coregonus clupeaformis) using next-generation sequencing. J. Evol. Biol. 27, 1029–1046 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Wang J., et al. , Analysis of intestinal microbiota in hybrid house mice reveals evolutionary divergence in a vertebrate hologenome. Nat. Commun. 6, 6440 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kohl K. D., Varner J., Wilkening J. L., Dearing M. D., Gut microbial communities of American pikas (Ochotona princeps): Evidence for phylosymbiosis and adaptations to novel diets. J. Anim. Ecol. 87, 323–330 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Rosenberg E., Zilber-Rosenberg I., Microbes drive evolution of animals and plants: The hologenome concept. MBio 7, e01395 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walters A. W., et al. , The microbiota influences the Drosophila melanogaster life history strategy. 10.1101/471540 (16 November 2018). [DOI] [PMC free article] [PubMed]
- 42.Chaston J. M., Newell P. D., Douglas A. E., Metagenome-wide association of microbial determinants of host phenotype in Drosophila melanogaster. MBio 5, e01631-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Judd A. M., et al. , Bacterial methionine metabolism genes influence Drosophila melanogaster starvation resistance. Appl. Environ. Microbiol. 84, e00662-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmidt P. S., Matzkin L., Ippolito M., Eanes W. F., Geographic variation in diapause incidence, life-history traits, and climatic adaptation in Drosophila melanogaster. Evolution 59, 1721–1732 (2005). [PubMed] [Google Scholar]
- 45.Schmidt P. S., Paaby A. B., Reproductive diapause and life-history clines in North American populations of Drosophila melanogaster. Evolution 62, 1204–1215 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Fabian D. K., et al. , Genome-wide patterns of latitudinal differentiation among populations of Drosophila melanogaster from North America. Mol. Ecol. 21, 4748–4769 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paaby A. B., Bergland A. O., Behrman E. L., Schmidt P. S., A highly pleiotropic amino acid polymorphism in the Drosophila insulin receptor contributes to life-history adaptation. Evolution 68, 3395–3409 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Machado H. E., et al. , Comparative population genomics of latitudinal variation in Drosophila simulans and Drosophila melanogaster. Mol. Ecol. 25, 723–740 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clark M. E., Anderson C. L., Cande J., Karr T. L., Widespread prevalence of wolbachia in laboratory stocks and the implications for Drosophila research. Genetics 170, 1667–1675 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simhadri R. K., et al. , The gut commensal microbiome of Drosophila melanogaster is modified by the endosymbiont Wolbachia. MSphere 2, e00287-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moghadam N. N., et al. , Strong responses of Drosophila melanogaster microbiota to developmental temperature. Fly (Austin) 12, 1–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Turnbaugh P. J., et al. , The effect of diet on the human gut microbiome: A metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 1, 6ra14 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muegge B. D., et al. , Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332, 970–974 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Storelli G., et al. , Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab. 14, 403–414 (2011). [DOI] [PubMed] [Google Scholar]
- 55.Shin S. C., et al. , Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 334, 670–674 (2011). [DOI] [PubMed] [Google Scholar]
- 56.Newell P. D., Douglas A. E., Interspecies interactions determine the impact of the gut microbiota on nutrient allocation in Drosophila melanogaster. Appl. Environ. Microbiol. 80, 788–796 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Keebaugh E. S., Yamada R., Obadia B., Ludington W. B., Ja W. W., Microbial quantity impacts Drosophila nutrition, development, and lifespan. iScience 4, 247–259 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Obadia B., Keebaugh E. S., Yamada R., Ludington W. B., Ja W. W., Diet influences host-microbiota associations in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 115, E4547–E4548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sullam K. E., et al. , Divergence across diet, time and populations rules out parallel evolution in the gut microbiomes of Trinidadian guppies. ISME J. 9, 1508–1522 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sevellec M., Derome N., Bernatchez L., Holobionts and ecological speciation: The intestinal microbiota of lake whitefish species pairs. Microbiome 6, 47 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trevelline B. K., Fontaine S. S., Hartup B. K., Kohl K. D., Conservation biology needs a microbial renaissance: A call for the consideration of host-associated microbiota in wildlife management practices. Proc. Biol. Sci. 286, 20182448 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Buchner P., Endosymbiose der Tiere mit Pflanzlichen Mikroorganismen (Springer-Verlag, 1953). [Google Scholar]
- 63.Hongoh Y., et al. , Complete genome of the uncultured Termite Group 1 bacteria in a single host protist cell. Proc. Natl. Acad. Sci. U.S.A. 105, 5555–5560 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Feldhaar H., Bacterial symbionts as mediators of ecologically important traits of insect hosts. Ecol. Entomol. 36, 533–543 (2011). [Google Scholar]
- 65.Asiimwe P., Kelly S. E., Hunter M. S., Symbiont infection affects whitefly dynamics in the field. Basic Appl. Ecol. 15, 507–515 (2014). [Google Scholar]
- 66.Schlötterer C., Tobler R., Kofler R., Nolte V., Sequencing pools of individuals - mining genome-wide polymorphism data without big funding. Nat. Rev. Genet. 15, 749–763 (2014). [DOI] [PubMed] [Google Scholar]
- 67.Langley C. H., et al. , Genomic variation in natural populations of Drosophila melanogaster. Genetics 192, 533–598 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Messer P. W., Petrov D. A., Population genomics of rapid adaptation by soft selective sweeps. Trends Ecol. Evol. 28, 659–669 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Messer P. W., Ellner S. P., Hairston N. G. Jr, Can population genetics adapt to rapid evolution? Trends Genet. 32, 408–418 (2016). [DOI] [PubMed] [Google Scholar]
- 70.Kapun M., Fabian D. K., Goudet J., Flatt T., Genomic evidence for adaptive inversion clines in Drosophila melanogaster. Mol. Biol. Evol. 33, 1317–1336 (2016). [DOI] [PubMed] [Google Scholar]
- 71.Boyle E. A., Li Y. I., Pritchard J. K., An expanded view of complex traits: From polygenic to omnigenic. Cell 169, 1177–1186 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bergland A. O., Behrman E. L., O’Brien K. R., Schmidt P. S., Petrov D. A., Genomic evidence of rapid and stable adaptive oscillations over seasonal time scales in Drosophila. PLoS Genet. 10, e1004775 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Machado H.E., et al. , Broad geographic sampling reveals predictable and pervasive seasonal adaptation in Drosophila. 10.1101/337543 (5 June 2018). [DOI]
- 74.Schmidt P. S., et al. , An amino acid polymorphism in the couch potato gene forms the basis for climatic adaptation in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 105, 16207–16211 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sharon G., et al. , Commensal bacteria play a role in mating preference of Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 107, 20051–20056 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zilber-Rosenberg I., Rosenberg E., Role of microorganisms in the evolution of animals and plants: The hologenome theory of evolution. FEMS Microbiol. Rev. 32, 723–735 (2008). [DOI] [PubMed] [Google Scholar]
- 77.Rajpurohit S., et al. , Adaptive dynamics of cuticular hydrocarbons in Drosophila. J. Evol. Biol. 30, 66–80 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rajpurohit S., et al. , Spatiotemporal dynamics and genome-wide association genome-wide association analysis of desiccation tolerance in Drosophila melanogaster. Mol. Ecol. 27, 3525–3540 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kozich J. J., Westcott S. L., Baxter N. T., Highlander S. K., Schloss P. D., Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Caporaso J. G., et al. , QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bolyen E., et al. , QIIME 2: Reproducible, interactive, scalable, and extensible microbiome data science. PeerJ. Preprints 6, e27295v2 (3 December 2018). [Google Scholar]
- 82.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 83.Oksanen J., et al. , vegan: Community Ecology Package. 2015 (R Package Version:2–2, 2015). https://CRAN.R-project.org/package=vegan. Accessed 21 November 2018.
- 84.Mandal S., et al. , Analysis of composition of microbiomes: A novel method for studying microbial composition. Microb. Ecol. Health Dis. 26, 27663 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wickham H., Ggplot2: Elegant Graphics for Data Analysis (Springer Publishing Company, Incorporated, ed. 2, 2009). [Google Scholar]
- 86.Koyle M. L., et al. , Rearing the fruit fly Drosophila melanogaster under axenic and gnotobiotic conditions. J. Vis. Exp., 10.3791/54219 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jiang H., Lei R., Ding S.-W., Zhu S., Skewer: A fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics 15, 182 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang J., Kobert K., Flouri T., Stamatakis A., PEAR: A fast and accurate Illumina paired-end reAd mergeR. Bioinformatics 30, 614–620 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hoskins R. A., et al. , Sequence finishing and mapping of Drosophila melanogaster heterochromatin. Science 316, 1625–1628 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Broad Institute , Picard tools. (2018). broadinstitute.github.io/picard/. Accessed 15 October 2018.
- 92.Van der Auwera G. A., et al. , From FastQ data to high confidence variant calls: The genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinformatics 43, 11.10.1–11.10.33 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Costello M., et al. , Characterization and remediation of sample index swaps by non-redundant dual indexing on massively parallel sequencing platforms. BMC Genomics 19, 332 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bushnell B., “BBMap: A fast, accurate, splice-aware aligner” (Lawrence Berkeley National Laboratory, Berkeley, CA, 2014). https://www.osti.gov/biblio/1241166-bbmap-fast-accurate-splice-aware-aligner. Accessed 10 July 2019.
- 95.Pedersen B. S., Quinlan A. R., Mosdepth: Quick coverage calculation for genomes and exomes. Bioinformatics 34, 867–868 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kofler R., Pandey R. V., Schlötterer C., PoPoolation2: Identifying differentiation between populations using sequencing of pooled DNA samples (Pool-Seq). Bioinformatics 27, 3435–3436 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kuhn R. M., Haussler D., Kent W. J., The UCSC genome browser and associated tools. Brief. Bioinform. 14, 144–161 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wiberg R. A. W., Gaggiotti O. E., Morrissey M. B., Ritchie M. G., Identifying consistent allele frequency differences in studies of stratified populations. Methods Ecol. Evol. 8, 1899–1909 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kolaczkowski B., Kern A. D., Holloway A. K., Begun D. J., Genomic differentiation between temperate and tropical Australian populations of Drosophila melanogaster. Genetics 187, 245–260 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Feder A. F., Petrov D. A., Bergland A. O., LDx: Estimation of linkage disequilibrium from high-throughput pooled resequencing data. PLoS One 7, e48588 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Benjamini Y., Yekutieli D., The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 29, 1165–1188 (2001). [Google Scholar]
- 102.Bergland A. O., Tobler R., González J., Schmidt P., Petrov D., Secondary contact and local adaptation contribute to genome-wide patterns of clinal variation in Drosophila melanogaster. Mol. Ecol. 25, 1157–1174 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Corbett-Detig R. B., Hartl D. L., Population genomics of inversion polymorphisms in Drosophila melanogaster. PLoS Genet. 8, e1003056 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kapun M., van Schalkwyk H., McAllister B., Flatt T., Schlötterer C., Inference of chromosomal inversion dynamics from Pool-Seq data in natural and laboratory populations of Drosophila melanogaster. Mol. Ecol. 23, 1813–1827 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.