Significance
Membrane proteins dwell in a sea of phospholipids that not only structurally stabilize the proteins by providing a hydrophobic environment for their transmembrane segments but also dynamically regulate protein function. While many cation channels are known to be regulated by phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2], relatively little is known about anion channel regulation by phosphoinositides. Using a combination of patch-clamp electrophysiology and atomistic molecular-dynamics simulations, we have identified several PI(4,5)P2 binding sites in ANO1 (TMEM16A), a Cl− channel that performs myriad physiological functions from epithelial fluid secretion to regulation of electrical excitability. These binding sites form a band at the cytosolic interface of the membrane that we propose constitute a network to dynamically regulate this highly allosteric protein.
Keywords: chloride channel, protein–lipid interaction, molecular dynamics, structure–function, phospholipid
Abstract
ANO1 (TMEM16A) is a Ca2+-activated Cl− channel that regulates diverse cellular functions including fluid secretion, neuronal excitability, and smooth muscle contraction. ANO1 is activated by elevation of cytosolic Ca2+ and modulated by phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2]. Here, we describe a closely concerted experimental and computational study, including electrophysiology, mutagenesis, functional assays, and extended sampling of lipid–protein interactions with molecular dynamics (MD) to characterize PI(4,5)P2 binding modes and sites on ANO1. ANO1 currents in excised inside-out patches activated by 270 nM Ca2+ at +100 mV are increased by exogenous PI(4,5)P2 with an EC50 = 1.24 µM. The effect of PI(4,5)P2 is dependent on membrane voltage and Ca2+ and is explained by a stabilization of the ANO1 Ca2+-bound open state. Unbiased atomistic MD simulations with 1.4 mol% PI(4,5)P2 in a phosphatidylcholine bilayer identified 8 binding sites with significant probability of binding PI(4,5)P2. Three of these sites captured 85% of all ANO1–PI(4,5)P2 interactions. Mutagenesis of basic amino acids near the membrane–cytosol interface found 3 regions of ANO1 critical for PI(4,5)P2 regulation that correspond to the same 3 sites identified by MD. PI(4,5)P2 is stabilized by hydrogen bonding between amino acid side chains and phosphate/hydroxyl groups on PI(4,5)P2. Binding of PI(4,5)P2 alters the position of the cytoplasmic extension of TM6, which plays a crucial role in ANO1 channel gating, and increases the accessibility of the inner vestibule to Cl− ions. We propose a model consisting of a network of 3 PI(4,5)P2 binding sites at the cytoplasmic face of the membrane allosterically regulating ANO1 channel gating.
Ca2+-activated Cl− channels (CaCCs) are jacks of all trades and masters of many. These ion channels facilitate the passive flow of Cl− across cell membranes in response to elevation of cytosolic Ca2+ (1). Although CaCCs are probably best known for driving fluid secretion across mammalian epithelia, they are intimately involved in manifold physiological functions in all eukaryotes. CaCCs mediate action potentials in algae, the fast block to polyspermy in Anuran eggs, and regulate functions as diverse as smooth muscle contraction, nociception, neuronal excitability, insulin secretion, and cell proliferation and migration in mammals (1–13). While there are several types of CaCCs, the so-called classical CaCCs are encoded by the ANO1 (TMEM16A) and ANO2 (TMEM16B) genes (14–16).
Activation of ANO1 in its physiological context typically begins with ligand binding to a G-protein–coupled receptor that activates phospholipase C (PLC) (17–20). PLC hydrolyzes phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] in the plasma membrane to produce diacylgycerol and inositol 1,4,5-trisphosphate [Ins(1,4,5)P3]. Ins(1,4,5)P3 binding to Ins(1,4,5)P3 receptors then triggers release of Ca2+ from internal endoplasmic reticulum stores and initiates store-operated Ca2+ entry. The resulting rise in cytosolic Ca2+ activates Cl− flux because of conformational changes induced by direct binding of Ca2+ to the ANO1 protein (21–25). Calmodulin is not required for channel activation (21, 26).
Structurally, ANO1 is a dimer with each subunit composed of 10 transmembrane (TM) segments. The Cl− selective pore of each subunit is surrounded by TMs 4 to 7 (22, 23, 27, 28), and each subunit has a Ca2+ binding site formed by amino acids E654, E702, E705, E734, and D738 in TMs 6 to 8 (22–25, 29). Channel gating involves conformational changes of TM6 (24, 25, 30).
ANO1 is also regulated by PI(4,5)P2 (31–35). Ta et al. (33) have reported that PI(4,5)P2 stimulates ANO1 currents in excised patches. We (32) and Tembo et al. (35) have shown that PI(4,5)P2 can prevent and rescue Ca2+-dependent rundown caused by spontaneous PI(4,5)P2 hydrolysis. In whole-cell recording, we also showed that reduction of cellular PI(4,5)P2 by the voltage-sensitive phosphatase Dr-VSP or by activation of G-protein–coupled receptors causes a reduction in ANO1 current (32). Pritchard et al. (34) show biochemical evidence that PI(4,5)P2 binds to ANO1, but they report that exogenous PI(4,5)P2 decreases endogenous Cl− currents thought to be encoded by ANO1 in inside-out patches of pulmonary artery cells, in contrast to the results with heterologously expressed ANO1 (32, 33).
PI(4,5)P2 binding sites typically have 2 or more positively charged amino acids with at least 1 Lys, and at least 1 aromatic residue (36–38). One well-known PI(4,5)P2 binding site is the pleckstrin homology (PH) domain, which binds PI(4,5)P2 in a pocket of basic amino acids that are noncontiguous in the primary sequence but are close in proximity in the folded protein (39). On the other hand, “electrostatic type” binding sites typified by the Clathrin Assembly Lymphoid Myeloid (CALM) domain are clusters of contiguous basic amino acids that interact with phospholipids relatively nonspecifically without forming a binding pocket (40). A wide variety of ion channels are regulated by PI(4,5)P2 (36–38, 41). In Kir channels, these basic amino acids are located near the interface between the cytosol and the bulk lipid bilayer where the polar headgroups of lipids typically reside (42–44).
The objective of this study was to understand the mechanisms of regulation of ANO1 by PI(4,5)P2 and identify amino acids that stabilize PI(4,5)P2 binding. We find that PI(4,5)P2 stimulates ANO1 currents in excised patches in a voltage- and Ca2+-regulated manner. Mutagenesis and molecular-dynamics (MD) simulations reveal that ANO1 has multiple PI(4,5)P2 binding sites. These data add to a growing body of knowledge showing that ANO1 is a highly allosteric protein that is gated by a network of interactions involving both Ca2+ and PI(4,5)P2.
Results
PI(4,5)P2 Is a Positive Regulator of ANO1 Currents.
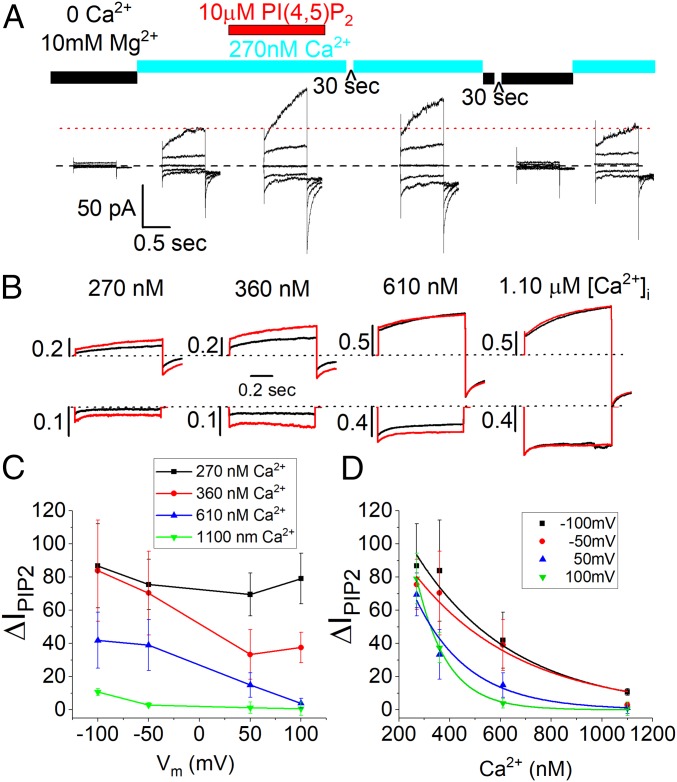
To determine the effect of PI(4,5)P2 on ANO1, we transiently expressed ANO1 in HEK293 cells and measured the effect of dioctanoyl phosphatidylinositol 4,5-bisphosphate [diC8-PI(4,5)P2], a water-soluble, short acyl chain PI(4,5)P2, on inside-out excised patches. To reduce variability caused by different amounts of endogenous PI(4,5)P2, we first removed PI(4,5)P2 from the membrane patch by excising it into a solution containing 10 mM Mg2+ and 0 Ca2+/5 mM EGTA. Mg2+ competes with ANO1 for binding to PI(4,5)P2 by electrostatically masking negative charges on PI(4,5)P2 (45). ANO1 currents were measured during voltage steps first in the presence of 270 nM Ca2+ (Iinitial) and then with 270 nM Ca2+ plus 10 µM diC8-PI(4,5)P2 (IPIP2) (Fig. 1A). The increase in current (ΔIPIP2) was calculated as (IPIP2/Iinitial − 1) × 100%. On average, application of diC8-PI(4,5)P2 at +100 mV increased the current 79.1 ± 15.1% (n = 8). The effect of diC8-PI(4,5)P2 was slowly reversible: The currents decreased in amplitude when the patch was exposed to 270 nM Ca2+ in the absence of diC8-PI(4,5)P2 and exposure to 10 mM Mg2+ reduced the current further to its initial level (Fig. 1A).
Fig. 1.
PI(4,5)P2 stimulates ANO1 current. (A) Current traces from a single inside-out excised patch exposed sequentially to solutions indicated above each set of traces. Each set of traces was obtained by voltage pulses from a holding potential of 0 mV to −100, −50, 0, 50, and 100 mV. (B) Representative current traces from different patches exposed to different Ca2+ concentrations. Black trace: control solution without PI(4,5)P2. Red trace: same solution with added 10 µM diC8-PI(4,5)P2. Top row: +100 mV. Bottom row: −100 mV. Scale bars represent current amplitudes normalized to the current obtained from the same patch exposed to 20 µM Ca2+ to evoke a maximal current. (C) Voltage dependence of PI(4,5)P2 stimulation of ANO1 current. Percent change in current amplitude caused by 10 µM diC8-PI(4,5)P2 (ΔIPIP2) is plotted vs. voltage for patches exposed to 270 nM (black squares), 360 nM (red circles), 610 nM (blue triangles), or 1.1 µM (green inverted triangles) free Ca2+. (D) Effect of free Ca2+ on stimulation of ANO1 current by diC8-PI(4,5)P2. Percent change in current amplitude is plotted vs. free Ca2+ concentration at −100 mV (black squares), −50 mV (red circles), 50 mV (blue triangle), and 100 mV (green inverted triangle).
The Effect of PI(4,5)P2 Is Regulated by Voltage and Ca2+.
The effect of diC8-PI(4,5)P2 was modulated by both Ca2+ and voltage (Fig. 1 B–D). At all voltages, ΔIPIP2 was greatest at lower Ca2+ concentrations (Fig. 1D). With 270 nM Ca2+, ΔIPIP2 at +100 mV was 79.1%, while with 1.1 µM Ca2+ ΔIPIP2 was <10%. Moreover, at all Ca2+ concentrations except the lowest [Ca2+] tested (270 nM), current stimulation by PI(4,5)P2 decreased with depolarization (Fig. 1C). For example, with 360 nM Ca2+ at −100 mV ΔIPIP2 was 83.8 ± 30%, but at +100 mV ΔIPIP2 was only 37.5 ± 9.1%. The IC50 for Ca2+ suppression of the PI(4,5)P2 effect was determined by fitting the data in Fig. 1D to exponential equations. The estimated IC50 for attenuation of the diC8-PI(4,5)P2 stimulatory effect was 390 nM Ca2+ at 100 mV and 656 nM Ca2+ at −100 mV (Fig. 1D). Because ANO1 has a lower affinity for Ca2+ at hyperpolarized potentials (46), the observation that PI(4,5)P2 has a larger effect at hyperpolarized potentials and at lower Ca2+ concentrations indicates that the lipid stabilizes the open, Ca2+-liganded state of the channel. At saturating Ca2+ concentration, PI(4,5)P2 has no additional effect.
The Effect of PI(4,5)P2 Is Specific and Moderate Affinity.
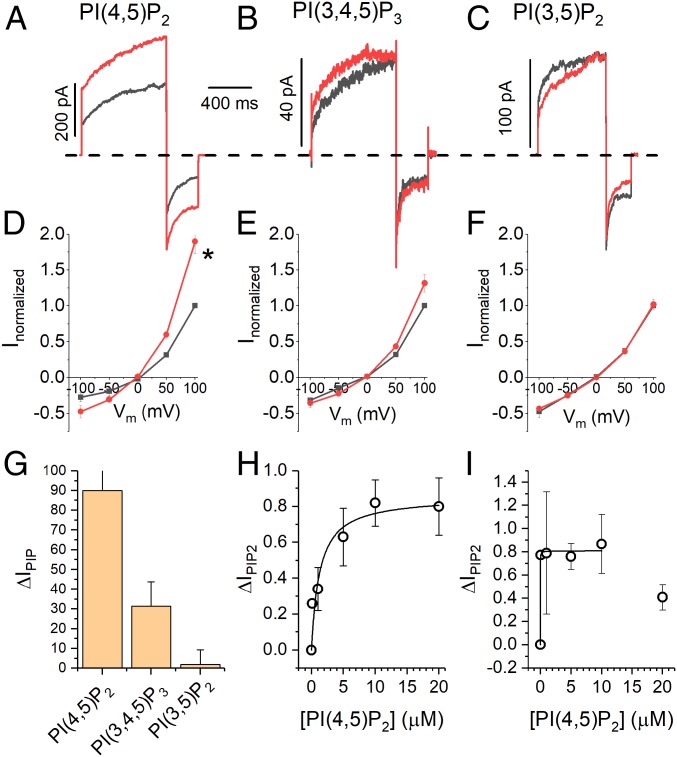
We compared the effects of 2 other physiologically important phosphoinositides (dioctanoyl phosphatidylinositol 3,4,5-trisphosphate [diC8-PI(3,4,5)P3] and dioctanoyl phosphatidylinositol 3,5-bisphosphate [diC8-PI(3,5)P2]) (47, 48) on ANO1 currents (Fig. 2). diC8-PI(3,4,5)P3, which has one additional phosphate at the 3-position, was less than half as efficacious at 10 µM as diC8-PI(4,5)P2: ANO1 current increased 31 ± 12% (Fig. 2 B, E, and G). diC8-PI(3,5)P2, which has a phosphate in the 3-position instead of the 4-position had no effect at 10 µM (Fig. 2 C, F, and G). Thus, the effect of PI(4,5)P2 on ANO1 exhibits specificity.
Fig. 2.
PI(4,5)P2 effect is selective and moderately high affinity. (A–C) Representative current traces at +100 mV with 270 nM Ca2+ in the absence (black) and presence (red) of 10 µM (A) diC8-PI(4,5)P2, (B) diC8-PI(3,4,5)P3, and (C) diC8-PI(3,5)P2. (D–F) Average ANO1 current–voltage relationships in the absence of phosphoinositides (black squares) and in the presence of 10 µM (D) diC8-PI(4,5)P2, (E) diC8-PI(3,4,5)P3, and (F) diC8-PI(3,5)P2. (G) Summary of stimulatory effect of phosphoinositides on ANO1 current at 100 mV with 270 nM Ca2+. (H and I) Concentration–response curve for diC8-PI(4,5)P2 on ANO1 current with 270 nM Ca2+at (H) 100 mV and (I) −100 mV.
To estimate the apparent EC50 for PI(4,5)P2, we measured ΔIPIP2 in response to different diC8-PI(4,5)P2 concentrations. The data, fitted to the Michaelis–Menton equation, gave an EC50 = 1.24 µM (Fig. 2H) at 100 mV with 270 nM Ca2+. This falls within the range for the effect of PI(4,5)P2 on other ion channels (0.12 to 4.6 µM) (49–51). At −100 mV, inward currents were more sensitive to diC8-PI(4,5)P2 with EC50 < 0.1 µM (Fig. 2I).
Selection of Potential PI(4,5)P2 Regulatory Sites.
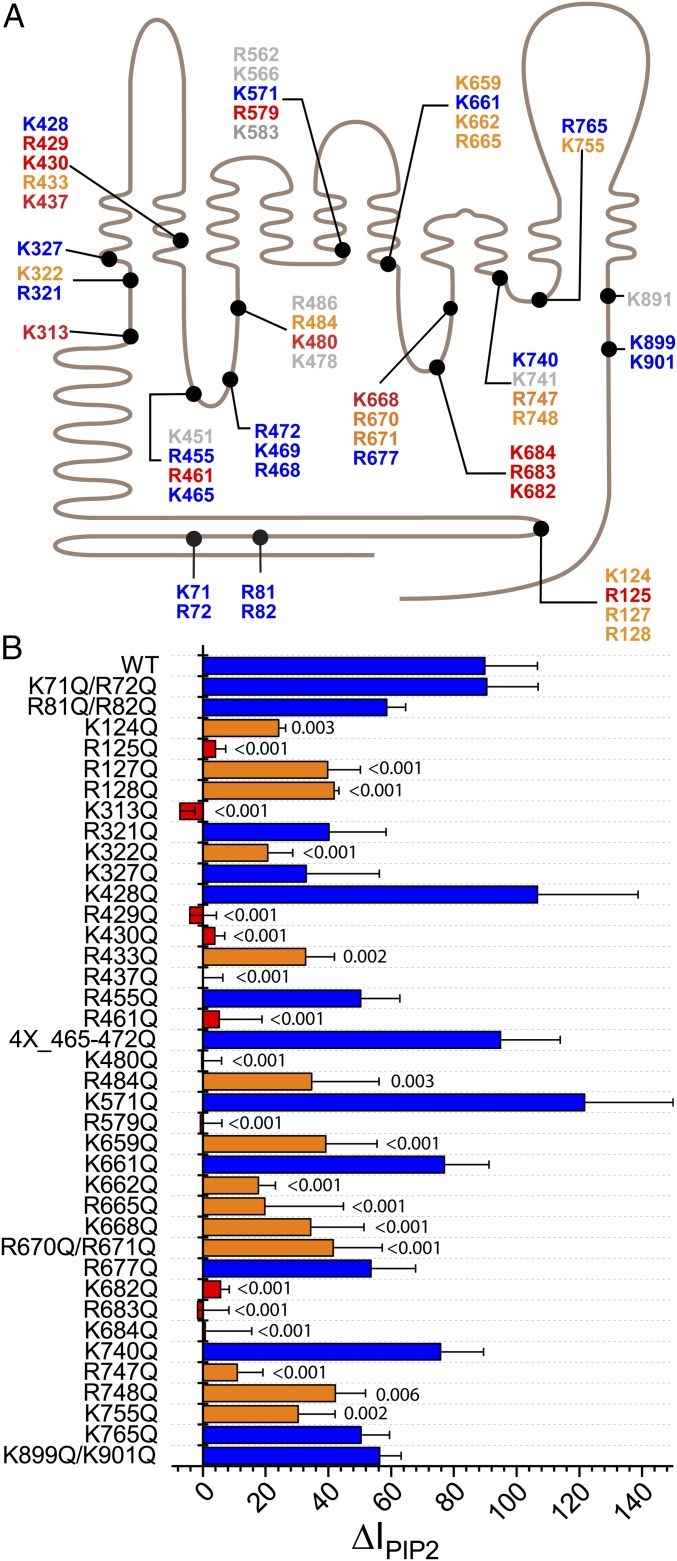
To identify amino acids that might play a role in PI(4,5)P2 regulation of ANO1, we mutagenized amino acids singly or in groups and tested whether stimulation of ANO1 currents in excised patches by diC8-PI(4,5)P2 was altered. We focused on basic amino acids because known PI(4,5)P2 binding sites contain 2 or more positively charged amino acids (38, 52). There are 62 Lys and 60 Arg residues in mouse ANO1. To narrow candidate residues for mutagenesis, we identified Lys and Arg residues located within 10 Å of the cytoplasmic membrane interface. To this list, we added amino acids in the cytoplasmic N terminus: K71, R72, R81, and R82 because they are predicted to constitute a phospholipid binding site by BHSEARCH (53), and K124, R125, R127, and R128 because they align partly with a region that ostensibly forms a PI(4,5)P2 binding site (54, 55) in the ANO1 paralog ANO6 (amino acids 93 to 100) (54, 55) (Fig. 3A).
Fig. 3.
Amino acids involved in mediating the effect PI(4,5)P2 on ANO1. (A) Schematic of 51 basic amino acids that were considered potential PI(4,5)P2 interacting residues. ANO1 is represented as a line with 10 transmembrane helices. Extracellular is upward. The length of the line is scaled to the amino acid sequence. Colors indicate effect of mutagenesis on stimulation of ANO1 current by PI(4,5)P2 corresponding to B: blue, response was like WT; red, PI(4,5)P2 effect was essentially abolished; orange, PI(4,5)P2 effect was significantly reduced; gray, not tested. (B) Effects of mutation of basic amino acids in ANO1 on the stimulation of ANO1 current in inside-out patches by PI(4,5)P2. Ten micromolar PI(4,5)P2 stimulates WT ANO1 current by ΔIPIP2 = 89.9%. Error bars are ±SEM; n = 3 to 13 patches per mutation. Numbers above bars show statistical P calculated by one-way ANOVA with Fisher LSD post hoc analysis for difference between means. Blue bars: P > 0.05. Orange bars: P < 0.05 and >50% reduction in response to PI(4,5)P2. Red bars: P < 0.01 and >90% reduction in response to PI(4,5)P2.
Mutagenesis to Identify PI(4,5)P2 Regulatory Sites.
We neutralized the charge on selected Arg and Lys residues by replacement with Gln. Surprisingly, more than 20 mutants were found to have a statistically significant reduction in response to diC8-PI(4,5)P2 compared to wild type (WT). For WT ANO1, ΔIPIP2 = 89.9 ± 16.8% (n = 13) in this set of experiments. ΔIPIP2 for “significant” mutants was <40% (P < 0.05; Fig. 3B, orange and red bars). Eleven amino acids were considered “critical” for PI(4,5)P2 regulation of ANO1 because their mutation reduced ΔIPIP2 to <10% (Fig. 3B, red bars).
These critical residues define 3 locations in the ANO1 structure that correspond to pockets of surface electronegativity (Fig. 4). In the following description, the number in parentheses is ΔIPIP2 when this amino acid is substituted with Gln (WT = 89.9%). Site A is near the dimer interface and is defined by critical amino acids R429 (−5%), K430 (4%), and R437 (0%) in TM2 and K313 (−8%) preceding TM1. R433 (36%), located one helix turn from K430, also contributes to this site. Site B is located at the cytoplasmic end of TM6, which plays a central role in ANO1 gating. It is defined by K682 (6%), R683 (−2%), and K684 (1%). Four nearby amino acids also significantly reduce the effect of PI(4,5)P2: K659 (44%), K662 (20%), K665 (23%), and K668 (42%). Site C is located in the short intracellular loop between TM2 and TM3 that forms one side of the Cl− ion permeation pathway. It is defined by R461 (10%), K480 (0%), and R484 (48%).
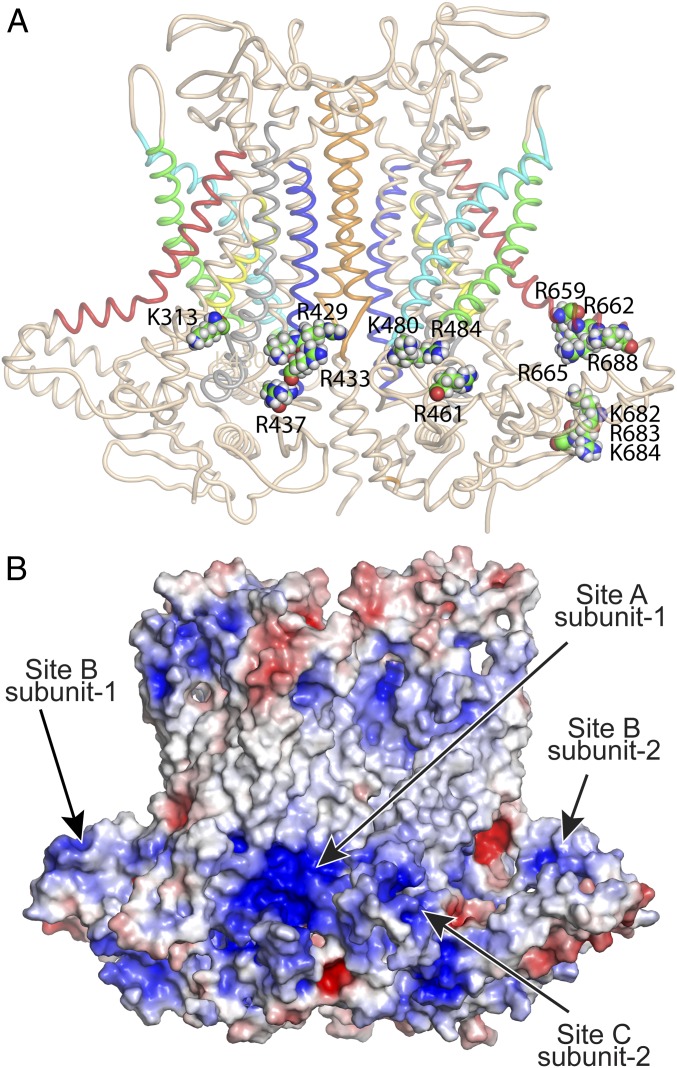
Fig. 4.
Location of mutations that are critical for the stimulatory effect of PI(4,5)P2 on ANO1. (A) Cartoon representation of ANO1 with critical amino acids as space-fill labeled. For clarity, site A (K313, R429, K430, R433, and R437) is shown only in the left subunit; site B (K659, R662, R665, R668, R682, R683, and K684) and site C (R461, K480, and R484) are shown only in the right subunit. Helices are colored: blue (TM2), cyan (TM3), green (TM4), red (TM6), yellow (TM7), and orange (TM10). (B) Electrostatic surface of ANO1 calculated by APBS Electrostatics in PyMOL. Red (+5 kT/e); blue (−5 kT/e).
Microscopic Characterization of PI(4,5)P2–ANO1 Interaction.
In order to gain more insight into the binding of PI(4,5)P2 to ANO1, we performed MD simulations using the highly mobile membrane mimetic model (HMMM) (54, 56). The HMMM model was introduced to accelerate lipid diffusion in atomistic MD simulations in order to obtain significantly enhanced sampling of interaction of lipid headgroups with proteins within simulation timescales currently achievable. In conventional membrane models, lipids with long acyl chains have a relatively low lateral diffusion constant (∼8 × 10−8 cm2⋅s−1). Therefore, during a 500-ns simulation, lipids do not diffuse far enough to allow sufficient mixing and sampling of the protein surface. The HMMM model accelerates lipid lateral diffusion by shortening lipid acyl tails to 6 carbons and filling the membrane core with a liquid phase (Fig. 5A). The HMMM model has been used successfully to study the diffusion and domain formation of lipids and membrane-associated proteins, pore formation by peptides, hydrophobic matching of transmembrane helices, and membrane-associated assembly of coagulation factors (56–64). The purpose of the model is to provide a more flexible and mobile environment that allows for rapid rearrangement and displacement of the lipid headgroups, thereby facilitating phenomena that might be inaccessible with conventional membrane models due to the inherently slow dynamics of the lipids.
Fig. 5.
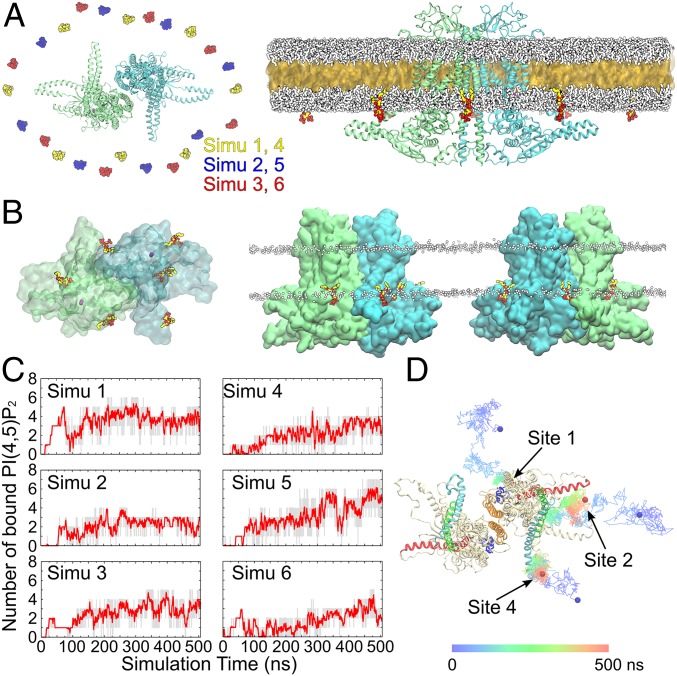
Spontaneous PI(4,5)P2 binding captured in MD simulations. (A, Left) Top view showing the initial placement of 8 C6-PI(4,5)P2 molecules in the inner leaflet of the bilayer surrounding the channel. Six independent simulations were performed with the C6-PI(4,5)P2 molecules placed in different initial positions, as shown by different colors. The 2 subunits of ANO1 are shown in green and blue. (A, Right) The initial HMMM simulation system viewed from the membrane. A large fraction of the acyl tails of the membrane-forming lipids is replaced by a liquid organic phase (yellow surface representation). The C6-PC molecules are shown in white. The C6-PI(4,5)P2 molecules are colored (oxygen, red; phosphorus, tan; carbon, yellow). Ions and water molecules are not shown for clarity. (B) Top and side views showing the positions of the C6-PI(4,5)P2 molecules at the end of one representative HMMM simulation (simu 5). Six C6-PI(4,5)P2 molecules were directly coordinated by the charged/polar residues of the channel. Ca2+ ions are shown as purple spheres. The membrane position is demarcated by the phosphorus atoms (white spheres) of the C6-PC lipids. (C) Number of bound C6-PI(4,5)P2 molecules (gray trace) and the moving average (red; bin = 20) plotted vs. simulation time for the 6 simulations. (D) Representative trajectories showing the binding of C6-PI(4,5)P2 to sites 1, 2, and 4. The colored lines illustrate the position of the 4′-phosphate of the C6-PI(4,5)P2 over the 500-ns simulations at 100-ps intervals. The initial and final positions of the 4′-phosphate are shown as blue and red spheres, respectively.
To examine the interaction of ANO1 with PI(4,5)P2, 8 C6-PI(4,5)P2 [short-tailed PI(4,5)P2 lipid] molecules were added to the inner leaflet of a C6-PC (short-tailed PC lipid) bilayer evenly surrounding ANO1 (Fig. 5A) at the beginning of the simulation. C6-PI(4,5)P2 constitutes ∼1.4 mol% of the total lipids in the inner leaflet. To diminish any bias introduced by the initial placement, the position of the C6-PI(4,5)P2 molecules were shifted in each of the 6 independent simulations such that each simulation began with different C6-PI(4,5)P2 starting positions. Within 50 ns, the C6-PI(4,5)P2 molecules diffused around the protein and interacted with it. The number of bound C6-PI(4,5)P2 molecules reached a plateau after ∼300 ns with an average of 4.5 C6-PI(4,5)P2 molecules bound per dimer (range, 3 to 6) in each simulation at the end of 500 ns (Fig. 5 B and C). Representative trajectories of C6-PI(4,5)P2 interacting with ANO1 are shown in Fig. 5D. At the end of the 6 HMMM simulations with C6-PI(4,5)P2, all subunits had at least one C6-PI(4,5)P2 molecule bound: 2 subunits had site 1 occupied only, 6 subunits had site 2 and/or 4 occupied, and 4 subunits had site 1 plus site 2 and/or 4 occupied.
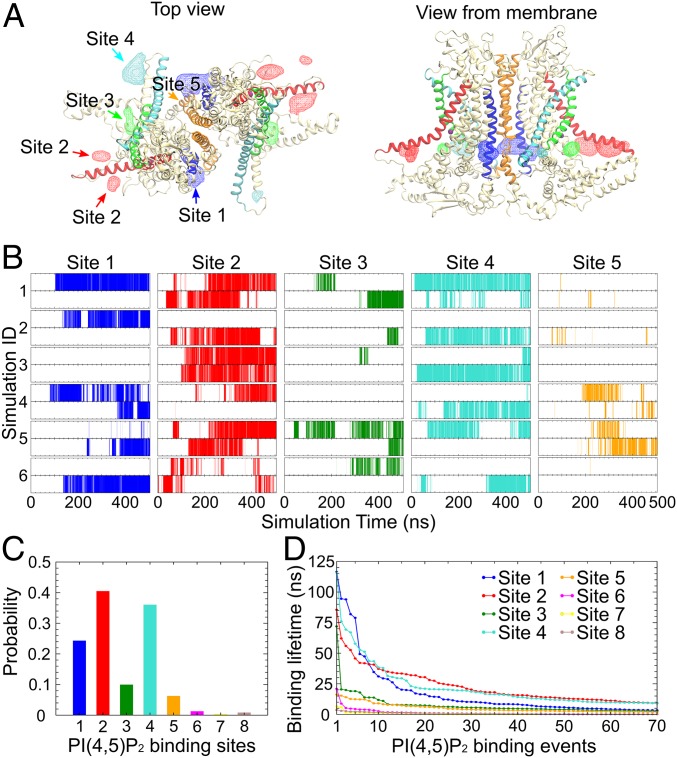
By calculating the density of the inositol groups over the simulation trajectories and the interactions between the phosphate/hydroxyl groups of the C6-PI(4,5)P2 and ANO1, 8 sites on the cytoplasmic surface of ANO1 were found to be visited by the C6-PI(4,5)P2 molecules. Sites 1 to 5 (labeled according to their clockwise location) involve basic and aromatic residues at the membrane–cytoplasm interface around the ends of transmembrane helices TM1, TM2, TM3, TM4, TM6, and TM10 (Fig. 6A). C6-PI(4,5)P2 binding to these sites was observed in multiple independent trajectories and for both subunits of ANO1 (Fig. 6B), revealing several specific and stable interactions between the C6-PI(4,5)P2 and the protein. Sites 6 to 8, which bind the C6-PI(4,5)P2 only transiently, mainly involve residues from the N-terminal region of ANO1, including the short α-helices α0a and α0b and the loop preceding them, and the loop between Nβ1 and Nα1. Among the 8 sites captured in the simulations, sites 1, 2, and 4 showed the highest occupancies (Fig. 6C). These 3 sites represent 84.3% of all of the binding events observed. C6-PI(4,5)P2 headgroups are well coordinated in these binding sites, resulting in residence lifetimes as long as 116.5 ns (Fig. 6D). Because C6-PI(4,5)P2 binding to sites 1, 2, and 4 was more robust than binding to the other sites, we focused on these 3 sites for further analysis.
Fig. 6.
PI(4,5)P2 binding at specific sites on ANO1. (A) Volumetric map of inositol occupancy extracted from the last 200 ns of the HMMM simulations is shown as colored wireframe contoured at isovalue 0.05 overlaid on the protein structure. Analysis combines results from the 6 simulations. Each local map is colored with the same color used for the nearby transmembrane helix that is involved in C6-PI(4,5)P2 binding (TM2, blue; TM3, cyan; TM4, green; TM6, red; and TM10, orange). (B) C6-PI(4,5)P2 occupancy at sites 1 to 5 in each subunit (Upper: subunit 1; Lower: subunit 2) during the 6 independent C6-PI(4,5)P2-binding simulations. C6-PI(4,5)P2 binding is indicated by the vertical lines at the corresponding time point. (C) The probability of C6-PI(4,5)P2 occupancy for each site over the 6 C6-PI(4,5)P2-binding simulations. C6-PI(4,5)P2 binding was observed mainly in sites 1, 2, and 4. (D) Dwell time of the top 70 binding events in each of the 8 binding sites, sorted in descending order. Only sites 1, 2, and 4 exhibit significant dwell times.
Insight into the Functional Roles of Specific Key Amino Acids.
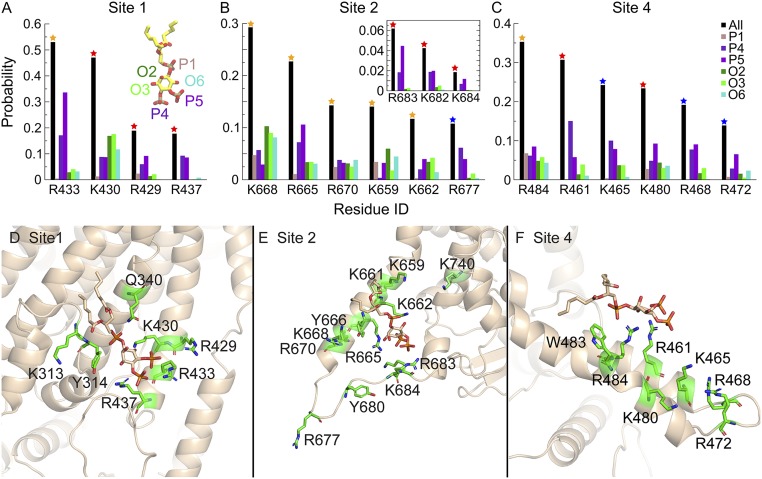
In site 1, there are 5 basic residues (R433, K430, R429, R437, and K313; Fig. 7 A and D), which stabilize the binding of C6-PI(4,5)P2 by coordinating the inositol ring and its phosphates. All of these residues are experimentally verified to strongly affect PI(4,5)P2 binding (Fig. 3B). Of the 4 residues that coordinate C6-PI(4,5)P2 for >10% of the total residence time in site 1, K430 shows strong preference for interaction with the hydroxyl groups on the inositol ring, while the other 3 basic residues mainly coordinate 4′- and 5′-phosphate groups. R437 is located closer to the cytoplasm than the other residues and interacts almost exclusively with the 4′- and 5′-phosphate groups. This feature suggests that R437 might be crucial for specific recognition of PI(4,5)P2. Experimentally, the R437Q mutation totally abolishes the PI(4,5)P2-induced effect. Similarly, in site 2 (Fig. 7 B and E), all of the residues facing the cytoplasm including those in the cytoplasmic loop connecting TM6 and TM7 (R677, R683, K682, and K684) and R665 in TM6 are mainly involved in coordinating the 4′- and 5′-phosphate groups. The other residues in TM6 show less discrimination and also interact with the 1′-phosphate and hydroxyl groups on the inositol ring. In site 4 (Fig. 7 C and F), all major interacting residues show preference for 4′- and 5′-phosphate groups except for R484, which indiscriminately coordinates all of the functional groups on the inositol ring.
Fig. 7.
PI(4,5)P2 coordination in major binding sites. (A–C) The probability of C6-PI(4,5)P2 headgroup coordination by the key basic residues in sites 1, 2, and 4 is shown as black bars. Residues that coordinate C6-PI(4,5)P2 for >10% of the total binding time in each site are shown [other residues that affect PI(4,5)P2 binding experimentally are also shown in the Inset]. The color of the star on each bar represents the experimental results in Fig. 3B, where the mutation of the residues affects PI(4,5)P2 binding to different degrees. The coordination probability for each functional group on the inositol ring (1′-phosphate, 4′-phosphate, 5′-phosphate, 2′-hydroxyl, 3′-hydroxyl, and 6′-hydroxyl) is shown individually. (D–F) Coordination of C6-PI(4,5)P2 in sites 1, 2, and 4. Amino acids that interact with each binding site are shown as stick representation in green. PI(4,5)P2 is shown in stick representation in tan.
Specificity of Phospholipid Binding.
To evaluate the specificity of PI(4,5)P2 binding, we measured the effect of PtdSer on PI(4,5)P2 binding by performing 3 350-ns HMMM simulations with a mixture 8 C6-PI(4,5)P2 and 8 C6-PtdSer molecules with each simulation having a different initial placement of the lipids. On average, the number of bound C6-PI(4,5)P2 molecules over the trajectories was reduced ∼20% from 2.73 ± 1.28 to 2.21 ± 1.45 (per monomer), which was not statistically significant. In these simulations, C6-PI(4,5)P2 interacted with 42 residues. We analyzed whether the C6-PI(4,5)P2 coordination probability was different for these 42 residues in simulations with and without C6-PtdSer and found that they were not statistically different (one-way ANOVA, P > 0.1). In contrast, C6-PtdSer binding to these sites was significantly different from C6-PI(4,5)P2 in the same simulations (P < 0.001). In these simulations, the C6-PtdSer headgroup only interacted with 7 residues with significant probability (>0.01). Two residues are in site 1 (R429: 0.09; R433: 0.06), 1 in site 2 (R670: 0.02), 2 in site 4 (R461: 0.02, K465: 0.01), and 2 others near site 4 (R486: 0.04, K469: 0.01). These data suggest that, although PtdSer may also bind to these sites, PI(4,5)P2 has a higher probability of interaction.
Potential Conformational Changes Induced by PI(4,5)P2.
To determine whether binding of PI(4,5)P2 was influenced by full-length acyl chains, the short-tailed lipid molecules [C6-PI(4,5)P2 and C6-PC] used in the initial probing of lipid–protein interactions were converted back to full-length lipids [palmitoyl-oleoyl-PI(4,5)P2 and palmitoyl-oleoyl-PC] at the end of the HMMM simulations, and the resulting full systems were subjected to an additional equilibrium simulation of 600 ns each. During these simulations with full-length lipid, each bound PI(4,5)P2 molecule remained coordinated around its binding site, suggesting that these sites stably bind full-length PI(4,5)P2. In the HMMM simulations, the C6-PI(4,5)P2 molecules were more likely to dissociate from their binding sites and sample more binding events.
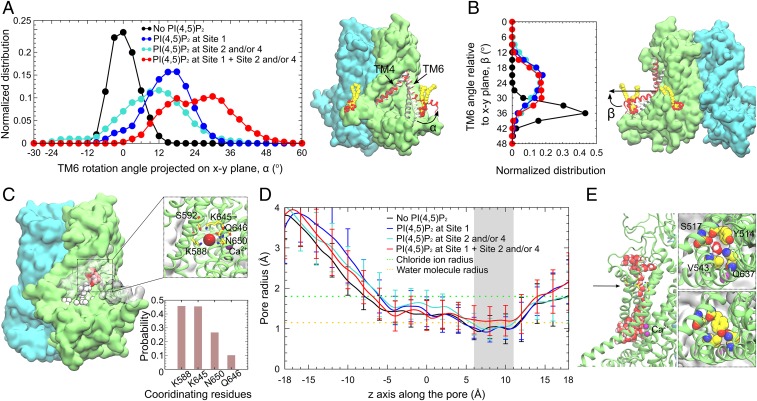
We compared the conformation of the PI(4,5)P2-bound ANO1 with its conformation in a control simulation performed in the absence of PI(4,5)P2. The most significant change captured in the simulations with PI(4,5)P2 bound is a rotation of the cytoplasmic half of TM6, which forms one side of the channel pore and plays a key role in channel gating (Fig. 8 A and B). The position of the cytoplasmic portion of TM6 is described by 2 angles. α is the angle of TM6 projected onto the x–y plane with the position in the cryogenic electron microscopy (cryo-EM) structure defined as 0°. β is the angle of TM6 relative to the x–y plane. In the absence of PI(4,5)P2, the cytoplasmic end of TM6 fluctuates only slightly around its initial position (peak α = 0°, peak β = 36°). When any site is occupied at sites 2/4, or single occupancy at site 1, there is a significant shift of TM6 away from the pore (peak α = 12° to 18°, peak β = 22°). When site 1 plus site 2 and/or 4 are occupied, there is an even more dramatic rotation of the cytoplasmic end of TM6 away from the pore (peak α = 18° to 32°, peak β = 22°).
Fig. 8.
Conformational changes induced by PI(4,5)P2 binding. (A) Rotation angle of the cytoplasmic portion of TM6 with respect to its position in the cryo-EM structure, projected onto the x–y plane. (A, Left) Distribution of the rotation angle normalized over the 600-ns full-length lipid simulations for each palmitoyl-oleoyl PI(4,5)P2 occupancy state. The cytoplasmic portion of TM6 fluctuates around its initial position (0°) in the absence of PI(4,5)P2 (black). Single occupancy of site 1 (blue), or single/double occupancy of sites 2 and 4 (cyan) shifts the rotation angle to positive values (cytoplasmic portion of TM6 away from the pore). Multiple occupancy of PI(4,5)P2 at sites 1 plus 2 and/or 4 (red) rotates TM6 most dramatically away from the pore. (A, Right) Snapshots of TM6 in the multiply occupied state (red), the PI(4,5)P2-free state (black), and the cryo-EM structure (white). Bound PI(4,5)P2 is shown in van der Waals (vdW) representation. (B, Left) Distribution of the angle between the TM6 cytoplasmic portion and the x–y plane. (B, Right) Snapshot showing the angle relative to the membrane in the multiple-occupied state (red), the PI(4,5)P2-free state (black), and the cryo-EM structure (white). (C) Time series snapshots showing the spontaneous penetration of Cl− ions (vdW spheres, white-to-red over time) in the multiply occupied state. Coordination of the Cl− ion deeply binding inside the pore is shown in the Inset, with the probability of coordination for all of the binding events shown below. Cl− and Ca2+ ions are shown as red and purple spheres, respectively. (D) Average radius of the ion conduction pore calculated using HOLE (86) illustrates a bottleneck between 6 and 11 Å (gray shading). The bottleneck in the multiply occupied state dilates ∼30% compared to other states. Orange dotted line: radius of water molecule (1.15 Å). Green dotted line: radius of Cl− ion (1.8 Å). (E, Left) Hydration of the ion conduction pore when the bottleneck (black arrow) dilates in the multiply occupied state. (E, Right) Top views of the pore showing the amino acids forming the bottleneck. The side-chain orientation of Y514 affects the pore radius and hydration.
When PI(4,5)P2 was bound, penetration of Cl− ions into the pore was frequently observed (Fig. 8C). Especially in the case of the multioccupied simulations (sites 1 and 2/4), deep penetration of Cl− occurred ∼25% of the total time. The major Cl−-coordinating amino acids were K588, K645, N650, and Q646. S592 also participated when Cl− entered more deeply. Although we had hoped to observe Cl− ions transiting the entire conduction pathway, this did not happen in the timeframe of our simulations. However, these simulations were performed with no applied voltage. Without driving force for Cl− movement, ion conduction may be too slow to capture in these 600-ns simulations. Furthermore, the voltage-dependent gating of ANO1 was not activated, so the conditions for the protein entering a conducting state may be suboptimal. The cryo-EM model of ANO1 we used for these simulations is a nonconducting state with a constriction in the pore between 6 and 11 Å formed by the bulky residue Y514 and 3 nearby residues (S517, V543, and Q637). When multiple PI(4,5)P2 sites are occupied (site 1 and 2/4), we observed that the conduction pathway dilated ∼30% to a radius of 1.2 Å (Fig. 8 D and E), which is slightly greater than the radius of a water molecule (1.15 Å), but is smaller than a Cl− ion (1.8 Å). Thus, forces that stabilize this nonconducting conformation are apparently sufficiently strong to prevent pore opening in the timeframe of these simulations.
Discussion
Correspondence between Mutagenesis and MD Predictions.
In this study, we used mutagenesis and computational modeling to identify lipid-binding sites in ANO1 that are important in regulation of the channel by PI(4,5)P2. Sites 1, 2, and 4 predicted by MD simulations to capture 84% of all interactions between ANO1 and PI(4,5)P2 correspond topographically to sites A, B, and C determined by mutagenesis. In further commentary, these sites are referred to as sites A/1, B/2, and C/4.
Both the mutagenesis and computational approaches have their limitations. Mutagenesis experiments can identify sites that are important in functionally mediating the effect of PI(4,5)P2 on ANO1, but this approach cannot easily distinguish between amino acids that coordinate PI(4,5)P2 binding, stabilize the structure of the binding site, or allosterically couple PI(4,5)P2 binding to channel gating or ion permeation. Furthermore, it should be noted that amino acids critical for the PI(4,5)P2 effect were identified under very specific recording conditions of 270 nM Ca2+ and 100 mV. Given the sensitivity of the PI(4,5)P2 effect to both voltage and Ca2+, it is entirely possible that under different conditions where ANO1 occupies different conformational states, a different complement of amino acids may be involved in mediating PI(4,5)P2 binding and effect. The MD simulations provide direct information about amino acids that are energetically favorable for binding PI(4,5)P2, but suffer from incomplete knowledge about native ANO1 and membrane structure in living cells. Furthermore, the MD simulations were not performed under identical conditions as the live-cell patch-clamp experiments. For example, no voltage was applied during the MD simulations and there was no free Ca2+ present in the system. While the correspondence between the functional live-cell experiments and the MD simulations is not perfect, the global agreement is remarkable. This provides a high level of confidence that these sites are involved in PI(4,5)P2 binding.
Site A/1 is the most robust site we have identified with all of the same amino acids identified in mutagenesis and MD (Table 1). Mutation of a single amino acid (R429, K430, R437, or K313) in this site nearly abolishes the effect of PI(4,5)P2. The MD simulation shows the headgroup of PI(4,5)P2 is well coordinated by a pocket formed by R429, K430, and R437 on one side and K313, Y314, and Q340 on the other sides (Fig. 7D). The site fulfils the requirements for a canonical PI(4,5)P2 binding site by having at least one Lys (K430) and one aromatic amino acid (Y314).
Table 1.
Comparison of amino acids in PI(4,5)P2 sites identified by MD simulation and mutagenesis
| MD simulation | PI(4,5)P2 effect | |
| Site | Probability | Mutant/WT ×100 |
| Site A/1 | ||
| R433 | 0.52 | 36 |
| K430 | 0.47 | 4 |
| R429 | 0.18 | −5 |
| R437 | 0.17 | 0 |
| K313 | 0.07 | −8 |
| Site B/2 | ||
| K668 | 0.29 | 42 |
| R665 | 0.22 | 23 |
| R670 | 0.15 | 46 |
| K659 | 0.15 | 44 |
| K662 | 0.13 | 20 |
| R677 | 0.12 | 60 |
| K740 | 0.06 | 84 |
| R683 | 0.06 | −2 |
| K661 | 0.05 | 86 |
| K682 | 0.04 | 6 |
| K684 | 0.02 | 1 |
| Site C/4 | ||
| R484 | 0.35 | 48 |
| R461 | 0.31 | 10 |
| K465 | 0.25 | 106 |
| K480 | 0.23 | 0 |
| R468 | 0.20 | 100 |
| R472 | 0.14 | 100 |
| Others | ||
| R125 | N terminus | 4 |
| R579 | Structural ? | −1 |
| R747 | Structural ? | 12 |
MD probability is probability of the amino acid coordinating C6-PI(4,5)P2 within that site (Fig. 7). PI(4,5)P2 effect is expressed as percentage of effect on WT ANO1.
A close agreement also exists between the MD and functional studies for site B/2 (Table 1). By both MD and mutagenesis, site B/2 is composed of at least 6 basic amino acids, 3 of which (R683, K682, K684) are essential for the PI(4,5)P2 effect. Mutation of each of the other amino acids has a partial effect, but this agrees with the modest probability of each of these amino acids coordinating PI(4,5)P2. Site B/2 also resembles a bona fide PI(4,5)P2 binding pocket (aromatic Y666 and K684) (Fig. 7E).
Site C/4 has both Lys and aromatic residues but does not form a pocket and may be an electrostatic CALM-type PI(4,5)P2 binding site (Fig. 7F). Mutation of K480 abolishes the response to PI(4,5)P2, and mutation of R461 and R484 have a significant effect. However, mutation of the other 3 basic residues identified by MD has no effect.
In addition to these 3 major sites, mutagenesis identified several amino acids outside these sites. We suspect that some of these amino acids may play structural or allosteric roles. R579 makes a hydrogen bond with E568 in the loop between TM4 and TM5, which are important in forming the conduction pathway. R747 forms hydrogen bonds with L693 and E694 (L693:O-K747:NE and E694:O-K747:N), which lock the cytoplasmic ends of TM7 and TM8. This may be important in maintaining a proper conformation of site B/2 (K682, R683, and R684) to interact with PI(4,5)P2. Mutation of R125 and its adjacent amino acids in the N terminus significantly affect PI(4,5)P2 binding, but interpretation is hampered because the N terminus is not modeled well in the cryo-EM structures (see below).
Role of the N Terminus.
It has been reported that ANO6, a paralog of ANO1 with 54% sequence similarity, is regulated by PI(4,5)P2 binding to the N terminus (54, 55). Ye et al. (55) showed that mutation of amino acids K87, K88, K95, R96, K97, and R98 decreases the Ca2+ sensitivity and reduces the ability of exogenously applied PI(4,5)P2 to restore ANO6 current after rundown. ANO6 residues 95-KRKR-98 align in PROMAL3D with ANO1 residues 124-KRFRR-128, and alignment of the ANO1 (5oyb) and ANO6 (6qp6) structures shows these sequences are similarly located in the protein structure. Although we find that mutation of these residues in ANO1 decreases the responsiveness to PI(4,5)P2, the N terminus is not well resolved in the ANO1 structure, and amino acids 1 to 116 and 131 to 164 are not modeled. Thus, it remains unclear how these amino acids contribute to ANO1 regulation. Also, the role of these amino acids in ANO6 remains in question because Aoun et al. (54) showed that deletion or mutagenesis of this site in ANO6 had no effect on PI(4,5)P2 binding. Rather, they propose that amino acids 302-KKQPLDLIRK-311 in the proximal N terminus of ANO6 are important for PI(4,5)P2 binding. This sequence aligns well with 313-KKQPLDLIRK-322 in ANO1, and we find that mutation of K313 in ANO1 significantly reduces the stimulatory effect of PI(4,5)P2 on ANO1 current. K313 is located in a reentrant membrane loop just before TM1 in both ANO1 and ANO6 structures and is a crucial residue in site A/1.
A Network of PI(4,5)P2 Binding Sites.
A major question raised by this study is, “Why does ANO1 have multiple PI(4,5)P2 binding sites and what are their functions?” Most studies on PI(4,5)P2–protein interaction generally assume that the effect of PI(4,5)P2 is mediated by a single binding site. However, there are several examples of proteins interacting with PI(4,5)P2 in different ways. For example, X-ray crystallography, coarse-grained MD, and reconstitution in artificial membranes have demonstrated that the Arf GTPase and its GEF Brag2 form a complex that binds to the membrane via multiple PI(4,5)P2 interaction sites (65). There is also some mutational and biochemical evidence for multiple PI(4,5)P2 binding sites on profilin-1, gelsolin, TRPV1, EAG1, and Kir2.1 (66–70).
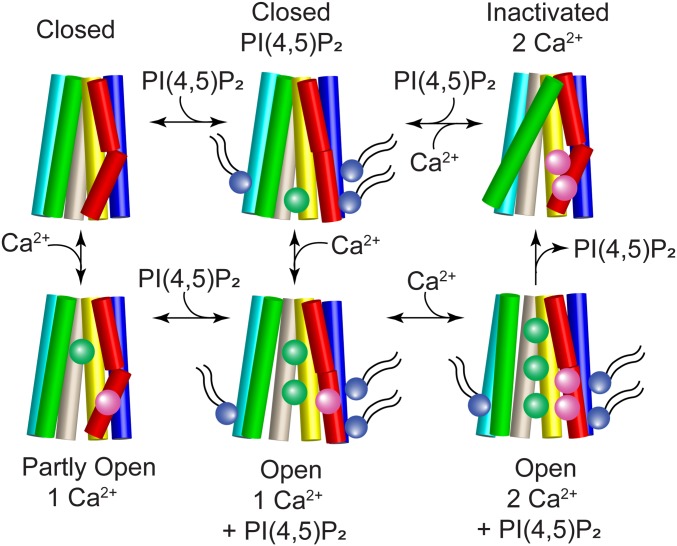
We anticipate that the 3 PI(4,5)P2 binding sites in ANO1 may have different and/or interacting effects. Although considerable work will be required to dissect these functional consequences, at this point in time we propose a general model for ANO1 gating that involves the binding of Ca2+ and PI(4,5)P2 to regulate this complex channel (Fig. 9). One corollary conclusion of this model is that the cryo-EM structures of ANO1 that have been published (22, 23) represent the inactivated state because the channel does not have PI(4,5)P2 bound.
Fig. 9.
Cartoon model of ANO1 gating. TM2 (blue), TM3 (cyan), TM4 (green), TM5 (wheat), TM6 (red), and TM7 (yellow) are shown as cylinders. The pore is formed by TM4–TM7 and Ca2+ (magenta spheres) binds to residues in TM6 and TM7. When PI(4,5)P2 (purple sphere with tails) binds to the cytoplasmic ends of TM2 (site A/1), TM6 (site B/2), and TM3 (site C/4), the cytoplasmic end of TM6 swings away from the pore to ultimately open the cytoplasmic vestibule to Cl− (green spheres). Top row: ANO1 is closed in the absence of Ca2+ and inactivated when 2 Ca2+ ions are bound without PI(4,5)P2. Bottom row: the channel partly opens when one Ca2+ binds without PI(4,5)P2 but full channel opening requires both Ca2+ and PI(4,5)P2.
The locations of the 3 sites in the protein may provide some insights into their possible function. Key residues forming site A/1 are located in TM1, TM2, and the loop between α0a and α0b. Because TM1 and TM2 are tightly packed against TM7 and TM8 that harbor part of the Ca2+ binding site, PI(4,5)P2 binding to site A/1 could have widespread allosteric effects on Ca2+ sensitivity and could be responsible for stabilizing the Ca2+-bound open state. Site B/2 is formed by amino acids at the cytoplasmic end of TM6 and intracellular loop 3 (ICL3) connecting TM6 and TM7. TM6 plays a crucial role in ANO1 gating (24, 25, 30) so that its conformational changes could alter channel gating. Also, TM6 and TM7 harbor 3 amino acids that form the binding site for Ca2+, so PI(4,5)P2 binding to this site could also affect Ca2+-dependent gating. Site C/4 is located at the cytoplasmic end of TM3 in the loop connecting TM3 and TM2. TM3 is tightly packed with TM4 that lines the conduction pathway, so binding of PI(4,5)P2 to site C/4 could modify the ion conduction pathway.
Methods
Cell Culture and Transfection.
HEK-293 cells (ATCC) were maintained in modified DMEM with 10% FBS, 100 U/mL penicillin G, and 100 µg/mL streptomycin, and transiently transfected with mTMEM16A (Uniprot Q8BHY3). HEK293 cells were authenticated by short tandem repeat profiling and were tested for myoplasm contamination. TMEM16A was tagged on the C terminus with EGFP. PCR-based mutagenesis was used to generate single amino acid mutations. Mutations were verified by sequencing.
Phosphatidylinositols and Inositol Phosphates.
Phosphatidylinositols were purchased from Echelon Research Laboratories. Unless noted otherwise, the synthetic lipids used in these experiments have C8 saturated fatty acid chains. At concentrations used here, these short chain phosphoinositides are likely monodisperse in solution. It has been estimated that the critical micelle concentration for lipids with phosphorylated inositol headgroups is >3 mM (71). Stock solutions of 10 mM PI(4,5)P2 were made in deionized H2O and stored at −20 °C. Working solutions were made fresh immediately before the experiment.
Preparation of the ANO1 Structure for Simulation.
The cryo-EM structure of mouse TMEM16A (Protein Data Bank ID 5OYB) at 3.75-Å resolution (72) was used as the starting structure for the MD simulations. Nomenclature of α-helices is from ref. 72. Because amino acids 1 to 116 (N terminus), 131 to 164 (N terminus), 260 to 266 (N terminus), 467 to 487 (TM2–TM3 linker), 669 to 682 (TM6–TM7 linker), and 911 to end (C terminus) are unstructured in the cryo-EM model, we used a model in which the TM2–TM3 and TM6–TM7 linkers and N-terminal residues 131 to 164 and 260 to 266 were added using SuperLooper2 (73) and subjected to energy minimization. The 2 Ca2+ ions bound in each of the 2 subunits were preserved for all of the simulations. The pKa of each ionizable residue was estimated using PROPKA (74, 75), and default protonation states were assigned based on the pKa analysis. Missing hydrogen atoms were added using PSFGEN in VMD (76). Internal water molecules were placed in energetically favorable positions within the protein using DOWSER (77, 78).
Simulation System Setup.
The ANO1 protein was first embedded in a palmitoyl-oleolyl-PC bilayer generated using the CHARMM-GUI membrane builder (79). In each of the 6 independent simulation systems (Simu 1 to Simu 6), 8 PI(4,5)P2 molecules were evenly placed around the protein in the inner leaflet of the membrane [∼1.4% PI(4,5)P2] (Fig. 5A). PI(4,5)P2 molecules were parameterized using CHARMM36 parameters. Considering the possible protonation states of PI(4,5)P2, 2 variations of PI(4,5)P2 headgroups were used in the simulations: Systems 1 to 3 used palmitoyl-oleoyl-phosphatidylinositol-(4,5)-bisphosphate with protonation on P4, while systems 4 to 6 used the palmitoyl-oleoyl-phosphatidylinositol-(4,5)-bisphosphate with protonation on P5. The initial positions of the PI(4,5)P2 molecules were at least 30 Å away from any atom of the channel. The membrane was then converted to an HMMM model using in-house scripts. This model replaces a portion of the membrane hydrophobic core by a more fluid representation using simple carbon solvent ethane (SCSE), while using short-tailed lipids to maintain full description of the headgroups and the initial part of the tails. The membrane/protein systems were fully solvated with TIP3P water (80) and buffered in 150 mM NaCl to keep the system neutral. The resulting systems consisting of ∼575,000 atoms were contained in a 244 × 180 × 141-Å3 simulation box.
To evaluate the specificity of PI(4,5)P2 binding to the channel, an additional set of simulations (3 trajectories, 350 ns each) were performed with 8 PtdSer molecules evenly placed around the protein in the inner leaflet of the membrane, in addition to the 8 PI(4,5)P2 molecules. Each simulation of mixed PI(4,5)P2 and PtdSer has a different initial placement of the lipids. The simulation setup was otherwise the same as described above.
Simulation Protocols.
MD simulations were carried out with NAMD2.12 (81) using CHARMM36m force field (82) and a time step of 2 fs. Periodic boundary conditions were used throughout the simulations. To evaluate long-range electrostatic interactions without truncation, the particle mesh Ewald method (83) was used. A smoothing function was employed for short-range nonbonded van der Waals forces starting at a distance of 10 Å with a cutoff of 12 Å. Bonded interactions and short-range nonbonded interactions were calculated every 2 fs. Pairs of atoms whose interactions were evaluated were searched and updated every 20 fs. A cutoff (13.5 Å) slightly longer than the nonbonded cutoff was applied to search for interacting atom pairs. Simulation systems were subjected to Langevin dynamics and the Nosé–Hoover Langevin piston method (84, 85) to maintain constant pressure (P = 1 atm) and temperature (T = 310 K) (NPT ensemble). To allow the simulation systems to fluctuate in all dimensions, in pressure control, a constant ratio was used for both HMMM and full-length simulations. This parameter keeps the x:y ratio of the unit cell constant rather than keeping the dimensions of the unit cell constant, thus allowing the surface area of the membrane to fluctuate during the simulation.
HMMM Simulations.
Because the HMMM representation simplifies the lipid bilayer by making the core more fluid, grid forces were applied on the carbon atoms affected by the conversion from the full-length membrane to the HMMM model to restrain the HMMM lipids, in order to resemble the full-length membrane while still allowing lipid molecules to fluctuate. The target atoms affected by the grid forces included carbon atoms in the solvent SCSE molecules and the terminal carbons (C26 and C36) of the C6-PC and C6-PI(4,5)P2 lipids. The aim was to make the positions (z coordinates) and flexibility of these target atoms resemble their counterparts in full-length membrane. To realize this, 3D grids with 1-Å spacing were specified in the simulation box to define the potential to be applied. Different potential values were then assigned to each grid point depending on the z position within the membrane (centered at z = 0). The grid potential is defined by the equation
where k = 0.025 kcal⋅mol−1⋅Å−2, zc = 10 Å, and denotes the z position of the grid point. For each target atom within the grid, the force was computed by a tricubic interpolation of the potential from the surrounding grid values based on the z position of the atom. The target atoms thus experience zero or close-to-zero force when they are in the region comparable to that of their counterparts in the full-length membrane (z within ±10 Å); increasing force is experienced when they are diffusing toward the headgroup/bulk-solvent region (z beyond ±10 Å). Since only the terminal carbon atoms (C26 and C36) in the short-tailed lipids are affected by the grid forces, this method provides the majority part of the molecules, especially the headgroups, with considerable flexibility in all dimensions during the simulation.
To verify that the HMMM simulations were providing a reasonable model of bilayer, we compared the dynamics of the phospholipids in a simulation of TMEM16A in full-length membrane (600 ns, no restraint) with the HMMM model used here (500 ns). The fluctuations of the choline-nitrogen atoms or phosphate atoms of PC headgroups in the z axis were used as metrics. In the HMMM simulations, the z positions of the choline nitrogens within 10 Å of the protein fluctuate up to 10 to 10.5 Å in each direction, and phosphorus atoms can fluctuate 9 to 9.5 Å. The capacity for fluctuation of the headgroup components shows that the model used in this study provides sufficient freedom for the HMMM membrane mimetics to behave similarly to a full-length membrane (57, 64).
The 6 HMMM simulation systems were first energy-minimized for 10,000 steps followed by a 1-ns relaxation MD, during which the heavy atoms from the protein were positionally restrained (k = 1 kcal⋅mol−1⋅Å−2) to allow the de novo added structures to relax. Then a 500-ns equilibrium simulation was performed for each system with the Cα atoms of the transmembrane region (estimated using the PPM server) slightly restrained (k = 0.1 kcal⋅mol−1⋅Å−2). Our published data showed that using SCSE solvent can greatly improve the behavior of transmembrane proteins within the HMMM model (57). SCSE showed substantially less solvent intercalation into transmembrane proteins compared to DCLE and SCSM solvents in absence of restraint on the proteins (64). Nevertheless, to prevent any potential disruption of the transmembrane structure, we applied a slight restraint on the C-alpha atoms of the transmembrane region of TMEM16A. The rest of the protein (side chains of the transmembrane region and the backbone atoms and side chains of the cytoplasmic and extracellular regions) was free to move. Such restraint is not likely to affect the sampling of lipid–protein interactions because the transmembrane region of the channel does not move much during unrestrained simulations. We compared the dynamics of the protein in the HMMM simulations with the ones in normal membranes without restraint. The transmembrane region is stable and does not move much for either simulation (average heavy-atom rmsd was 1.57 Å for HMMM vs. 2.00 Å for full-length). The rmsd for the transmembrane region in full-length membranes over the measured timescale (600 ns) is only ∼0.43 Å larger than the HMMM simulations (500 ns). In comparison, the average heavy atom rmsd for the entire protein is 7.01 Å for HMMM simulations and 8.67 Å for full-length simulations, meaning that most of the flexibility of the protein originates from the dynamics of the cytoplasmic and extracellular domains. Based on this observation, we conclude that the slight restraint on the protein will not affect results of the simulations.
Full-Length Simulations.
To investigate protein conformational changes upon PI(4,5)P2 binding, the C6-PI(4,5)P2-bound HMMM systems were converted back to full-length phospholipids, followed by an additional 600-ns simulation without any restraints on the protein. A control simulation in the absence of PI(4,5)P2 molecules was also performed using the same protocol.
Analysis of Lipid–Protein Interactions.
To characterize the interaction of lipid headgroups with potential binding sites on the protein, occupancy maps of the inositol group (and its phosphate groups) over the trajectories were calculated using the Volmap plugin in VMD (68). To determine the residues at those sites that stabilize PI(4,5)P2 binding, the polar interactions between the residues (side-chain nitrogen/oxygen atoms) and the PI(4,5)P2 inositol ring (phosphate/hydroxyl groups) in the binding site were calculated for each frame of the trajectory using a distance cutoff of 4 Å for phosphorus atoms of the phosphates and 3.5 Å for oxygen atoms of the hydroxyls.
Statistical Analysis.
Data were analyzed using Origin 2017 SR2. Error bars are SEM. Statistics are described in each figure.
Acknowledgments
Research reported in this publication was supported by the National Eye Institute, National Institute of Arthritis and Musculoskeletal Diseases, and National Institute of General Medical Sciences of the National Institutes of Health under Awards R01EY114852 (to H.C.H.), R01AR067786 (to H.C.H.), P41-GM104601 (to E.T.), and R01-GM123455 (to E.T.). Ninety-five percent of this research was financed with federal money and 5% from nongovernmental sources. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We also acknowledge computing resources provided by Blue Waters at National Center for Supercomputing Applications, and Extreme Science and Engineering Discovery Environment (Grant MCA06N060 to E.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Hartzell C., Putzier I., Arreola J., Calcium-activated chloride channels. Annu. Rev. Physiol. 67, 719–758 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Fromm J., Lautner S., Electrical signals and their physiological significance in plants. Plant Cell Environ. 30, 249–257 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Wozniak K. L., Phelps W. A., Tembo M., Lee M. T., Carlson A. E., The TMEM16A channel mediates the fast polyspermy block in Xenopus laevis. J. Gen. Physiol. 150, 1249–1259 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pedemonte N., Galietta L. J., Structure and function of TMEM16 proteins (anoctamins). Physiol. Rev. 94, 419–459 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Duvvuri U., et al. , TMEM16A induces MAPK and contributes directly to tumorigenesis and cancer progression. Cancer Res. 72, 3270–3281 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frings S., Reuter D., Kleene S. J., Neuronal Ca2+-activated Cl− channels—homing in on an elusive channel species. Prog. Neurobiol. 60, 247–289 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Malysz J., et al. , Conditional genetic deletion of Ano1 in interstitial cells of Cajal impairs Ca2+ transients and slow waves in adult mouse small intestine. Am. J. Physiol. Gastrointest. Liver Physiol. 312, G228–G245 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Catalán M. A., et al. , A fluid secretion pathway unmasked by acinar-specific Tmem16A gene ablation in the adult mouse salivary gland. Proc. Natl. Acad. Sci. U.S.A. 112, 2263–2268 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duran C., Hartzell H. C., Physiological roles and diseases of tmem16/anoctamin proteins: Are they all chloride channels? Acta Pharmacol. Sin. 32, 685–692 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bill A., Alex Gaither L., The mechanistic role of the calcium-activated chloride channel ANO1 in tumor growth and signaling. Adv. Exp. Med. Biol. 966, 1–14 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Oh U., Jung J., Cellular functions of TMEM16/anoctamin. Pflugers Arch. 468, 443–453 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berg J., Yang H., Jan L. Y., Ca2+-activated Cl− channels at a glance. J. Cell Sci. 125, 1367–1371 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crottès D., et al. , TMEM16A controls EGF-induced calcium signaling implicated in pancreatic cancer prognosis. Proc. Natl. Acad. Sci. U.S.A. 116, 13026–13035 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schroeder B. C., Cheng T., Jan Y. N., Jan L. Y., Expression cloning of TMEM16A as a calcium-activated chloride channel subunit. Cell 134, 1019–1029 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caputo A., et al. , TMEM16A, a membrane protein associated with calcium-dependent chloride channel activity. Science 322, 590–594 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Yang Y. D., et al. , TMEM16A confers receptor-activated calcium-dependent chloride conductance. Nature 455, 1210–1215 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Concepcion A. R., Feske S., Regulation of epithelial ion transport in exocrine glands by store-operated Ca2+ entry. Cell Calcium 63, 53–59 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Romanenko V. G., et al. , Tmem16A encodes the Ca2+-activated Cl− channel in mouse submandibular salivary gland acinar cells. J. Biol. Chem. 285, 12990–13001 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ambudkar I. S., Ong H. L., Liu X., Bandyopadhyay B. C., Cheng K. T., TRPC1: The link between functionally distinct store-operated calcium channels. Cell Calcium 42, 213–223 (2007). Correction in: Cell Calcium.44, 427 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Sung T. S., et al. , The cells and conductance mediating cholinergic neurotransmission in the murine proximal stomach. J. Physiol. 596, 1549–1574 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu K., Zhu J., Qu Z., Cui Y. Y., Hartzell H. C., Activation of the Ano1 (TMEM16A) chloride channel by calcium is not mediated by calmodulin. J. Gen. Physiol. 143, 253–267 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paulino C., et al. , Structural basis for anion conduction in the calcium-activated chloride channel TMEM16A. Elife 6, e26232 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dang S., et al. , Cryo-EM structures of the TMEM16A calcium-activated chloride channel. Nature 552, 426–429 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu K., Duran C., Qu Z., Cui Y. Y., Hartzell H. C., Explaining calcium-dependent gating of anoctamin-1 chloride channels requires a revised topology. Circ. Res. 110, 990–999 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tien J., et al. , A comprehensive search for calcium binding sites critical for TMEM16A calcium-activated chloride channel activity. Elife 3, e02772 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Terashima H., Picollo A., Accardi A., Purified TMEM16A is sufficient to form Ca2+-activated Cl− channels. Proc. Natl. Acad. Sci. U.S.A. 110, 19354–19359 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim N. K., Lam A. K., Dutzler R., Independent activation of ion conduction pores in the double-barreled calcium-activated chloride channel TMEM16A. J. Gen. Physiol. 148, 375–392 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeng G., Aggarwal M., Yu W. P., Chen T. Y., Independent activation of distinct pores in dimeric TMEM16A channels. J. Gen. Physiol. 148, 393–404 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paulino C., Kalienkova V., Lam A. K. M., Neldner Y., Dutzler R., Activation mechanism of the calcium-activated chloride channel TMEM16A revealed by cryo-EM. Nature 552, 421–425 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Peters C. J., et al. , The sixth transmembrane segment is a major gating component of the TMEM16A calcium-activated chloride channel. Neuron 97, 1063–1077.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian Y., et al. , Control of TMEM16A by INO-4995 and other inositolphosphates. Br. J. Pharmacol. 168, 253–265 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Jesús-Pérez J. J., et al. , Phosphatidylinositol 4,5-bisphosphate, cholesterol, and fatty acids modulate the calcium-activated chloride channel TMEM16A (ANO1). Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1863, 299–312 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ta C. M., Acheson K. E., Rorsman N. J. G., Jongkind R. C., Tammaro P., Contrasting effects of phosphatidylinositol 4,5-bisphosphate on cloned TMEM16A and TMEM16B channels. Br. J. Pharmacol. 174, 2984–2999 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pritchard H. A., Leblanc N., Albert A. P., Greenwood I. A., Inhibitory role of phosphatidylinositol 4,5-bisphosphate on TMEM16A-encoded calcium-activated chloride channels in rat pulmonary artery. Br. J. Pharmacol. 171, 4311–4321 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tembo M., Wozniak K. L., Bainbridge R. E., Carlson A. E., Phosphatidylinositol 4,5-bisphosphate (PIP2) and Ca2+ are both required to open the Cl− channel TMEM16A. J. Biol. Chem. 294, 12556–12564 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suh B. C., Hille B., PIP2 is a necessary cofactor for ion channel function: How and why? Annu. Rev. Biophys. 37, 175–195 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hansen S. B., Lipid agonism: The PIP2 paradigm of ligand-gated ion channels. Biochim. Biophys. Acta 1851, 620–628 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Logothetis D. E., et al. , Phosphoinositide control of membrane protein function: A frontier led by studies on ion channels. Annu. Rev. Physiol. 77, 81–104 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jian X., et al. , Molecular basis for cooperative binding of anionic phospholipids to the PH domain of the Arf GAP ASAP1. Structure 23, 1977–1988 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ford M. G., et al. , Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science 291, 1051–1055 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Hille B., Dickson E. J., Kruse M., Vivas O., Suh B. C., Phosphoinositides regulate ion channels. Biochim. Biophys. Acta 1851, 844–856 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hansen S. B., Tao X., MacKinnon R., Structural basis of PIP2 activation of the classical inward rectifier K+ channel Kir2.2. Nature 477, 495–498 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang W., Whorton M. R., MacKinnon R., Quantitative analysis of mammalian GIRK2 channel regulation by G proteins, the signaling lipid PIP2 and Na+ in a reconstituted system. Elife 3, e03671 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whorton M. R., MacKinnon R., Crystal structure of the mammalian GIRK2 K+ channel and gating regulation by G proteins, PIP2, and sodium. Cell 147, 199–208 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suh B. C., Hille B., Electrostatic interaction of internal Mg2+ with membrane PIP2 Seen with KCNQ K+ channels. J. Gen. Physiol. 130, 241–256 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiao Q., et al. , Voltage- and calcium-dependent gating of TMEM16A/Ano1 chloride channels are physically coupled by the first intracellular loop. Proc. Natl. Acad. Sci. U.S.A. 108, 8891–8896 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Craene J. O., Bertazzi D. L., Bär S., Friant S., Phosphoinositides, major actors in membrane trafficking and lipid signaling pathways. Int. J. Mol. Sci. 18, E634 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Riehle R. D., Cornea S., Degterev A., Role of phosphatidylinositol 3,4,5-trisphosphate in cell signaling. Adv. Exp. Med. Biol. 991, 105–139 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Cabanos C., Wang M., Han X., Hansen S. B., A soluble fluorescent binding assay reveals PIP2 antagonism of TREK-1 channels. Cell Rep. 20, 1287–1294 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Y., et al. , KCNE1 enhances phosphatidylinositol 4,5-bisphosphate (PIP2) sensitivity of IKs to modulate channel activity. Proc. Natl. Acad. Sci. U.S.A. 108, 9095–9100 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lopes C. M., et al. , Alterations in conserved Kir channel-PIP2 interactions underlie channelopathies. Neuron 34, 933–944 (2002). [DOI] [PubMed] [Google Scholar]
- 52.Rosenhouse-Dantsker A., Logothetis D. E., Molecular characteristics of phosphoinositide binding. Pflugers Arch. 455, 45–53 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Brzeska H., Guag J., Remmert K., Chacko S., Korn E. D., An experimentally based computer search identifies unstructured membrane-binding sites in proteins: Application to class I myosins, PAKS, and CARMIL. J. Biol. Chem. 285, 5738–5747 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aoun J., et al. , Anoctamin 6 contributes to Cl− secretion in accessory cholera enterotoxin (Ace)-stimulated diarrhea: An essential role for phosphatidylinositol 4,5-bisphosphate (PIP2) signaling in cholera. J. Biol. Chem. 291, 26816–26836 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ye W., et al. , Phosphatidylinositol-(4,5)-bisphosphate regulates calcium gating of small-conductance cation channel TMEM16F. Proc. Natl. Acad. Sci. U.S.A. 115, E1667–E1674 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vermaas J. V., et al. , Efficient exploration of membrane-associated phenomena at atomic resolution. J. Membr. Biol. 248, 563–582 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vermaas J. V., Pogorelov T. V., Tajkhorshid E., Extension of the highly mobile membrane mimetic to transmembrane systems through customized in silico solvents. J. Phys. Chem. B 121, 3764–3776 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baylon J. L., et al. , Atomic-level description of protein-lipid interactions using an accelerated membrane model. Biochim. Biophys. Acta 1858, 1573–1583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qi Y., et al. , CHARMM-GUI HMMM builder for membrane simulations with the highly mobile membrane-mimetic model. Biophys. J. 109, 2012–2022 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pogorelov T. V., Vermaas J. V., Arcario M. J., Tajkhorshid E., Partitioning of amino acids into a model membrane: Capturing the interface. J. Phys. Chem. B 118, 1481–1492 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ohkubo Y. Z., Pogorelov T. V., Arcario M. J., Christensen G. A., Tajkhorshid E., Accelerating membrane insertion of peripheral proteins with a novel membrane mimetic model. Biophys. J. 102, 2130–2139 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tietjen G. T., et al. , Coupling X-ray reflectivity and in silico binding to yield dynamics of membrane recognition by Tim1. Biophys. J. 113, 1505–1519 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Muller M. P., Wang Y., Morrissey J. H., Tajkhorshid E., Lipid specificity of the membrane binding domain of coagulation factor X. J. Thromb. Haemost. 15, 2005–2016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vermaas J. V., Tajkhorshid E., Differential membrane binding mechanics of synaptotagmin isoforms observed in atomic detail. Biochemistry 56, 281–293 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Karandur D., Nawrotek A., Kuriyan J., Cherfils J., Multiple interactions between an Arf/GEF complex and charged lipids determine activation kinetics on the membrane. Proc. Natl. Acad. Sci. U.S.A. 114, 11416–11421 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Morales-Lázaro S. L., Lemus L., Rosenbaum T., Regulation of thermoTRPs by lipids. Temperature (Austin) 4, 24–40 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rohacs T., Phosphoinositide regulation of TRPV1 revisited. Pflugers Arch. 467, 1851–1869 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Delgado-Ramírez M., López-Izquierdo A., Rodríguez-Menchaca A. A., Dual regulation of hEAG1 channels by phosphatidylinositol 4,5-bisphosphate. Biochem. Biophys. Res. Commun. 503, 2531–2535 (2018). [DOI] [PubMed] [Google Scholar]
- 69.Lee S. J., et al. , Secondary anionic phospholipid binding site and gating mechanism in Kir2.1 inward rectifier channels. Nat. Commun. 4, 2786 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.D’Avanzo N., Lee S. J., Cheng W. W., Nichols C. G., Energetics and location of phosphoinositide binding in human Kir2.1 channels. J. Biol. Chem. 288, 16726–16737 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Campbell R. B., Liu F., Ross A. H., Allosteric activation of PTEN phosphatase by phosphatidylinositol 4,5-bisphosphate. J. Biol. Chem. 278, 33617–33620 (2003). [DOI] [PubMed] [Google Scholar]
- 72.Brunner J. D., Schenck S., Dutzler R., Structural basis for phospholipid scrambling in the TMEM16 family. Curr. Opin. Struct. Biol. 39, 61–70 (2016). [DOI] [PubMed] [Google Scholar]
- 73.Hildebrand P. W., et al. , SuperLooper—a prediction server for the modeling of loops in globular and membrane proteins. Nucleic Acids Res. 37, W571–W574 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Olsson M. H., Søndergaard C. R., Rostkowski M., Jensen J. H., PROPKA3: Consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 7, 525–537 (2011). [DOI] [PubMed] [Google Scholar]
- 75.Rostkowski M., Olsson M. H., Søndergaard C. R., Jensen J. H., Graphical analysis of pH-dependent properties of proteins predicted using PROPKA. BMC Struct. Biol. 11, 6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Humphrey W., Dalke A., Schulten K., VMD: Visual molecular dynamics. J. Mol. Graph. 14, 33–38, 27–38 (1996). [DOI] [PubMed] [Google Scholar]
- 77.Zhang L., Hermans J., Hydrophilicity of cavities in proteins. Proteins 24, 433–438 (1996). [DOI] [PubMed] [Google Scholar]
- 78.Morozenko A., Leontyev I. V., Stuchebrukhov A. A., Dipole moment and binding energy of water in proteins from crystallographic analysis. J. Chem. Theory Comput. 10, 4618–4623 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu E. L., et al. , CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J. Comput. Chem. 35, 1997–2004 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L., Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983). [Google Scholar]
- 81.Phillips J. C., et al. , Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Klauda J. B., et al. , Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J. Phys. Chem. B 114, 7830–7843 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Darden T., York D., Pedersen L., Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1998). [Google Scholar]
- 84.Nose S., A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 81, 511–519 (1984). [Google Scholar]
- 85.Hoover W. G., Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A Gen. Phys. 31, 1695–1697 (1985). [DOI] [PubMed] [Google Scholar]
- 86.Smart O. S., Neduvelil J. G., Wang X., Wallace B. A., Sansom M. S., HOLE: A program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360, 376 (1996). [DOI] [PubMed] [Google Scholar]