Significance
Phosphorylation is essential for the regulation and degradation of PHYTOCHROME-INTERACTING FACTORs (PIFs), key repressors of photomorphogenesis. Although the kinases responsible for PIF phosphorylation have been extensively studied, the phosphatases that dephosphorylate PIFs are largely unknown. Here, we reveal that mutations of FyPP1 and FyPP3, 2 catalytic subunits of PP6 phosphatases, promote seedling photomorphogenesis in the dark. FyPP1 and FyPP3 directly interact with and dephosphorylate PIF3 and PIF4, which may increase the activity of these proteins in the dark and stabilize PIF4 during the dark-to-light transition. The phosphatases identified in this study add a building block to the light signaling network.
Keywords: photomorphogenesis, PIF, PP6 phosphatase, phosphorylation, Arabidopsis
Abstract
The PHYTOCHROME-INTERACTING FACTORs (PIFs) play a central role in repressing photomorphogenesis, and phosphorylation mediates the stability of PIF proteins. Although the kinases responsible for PIF phosphorylation have been extensively studied, the phosphatases that dephosphorylate PIFs remain largely unknown. Here, we report that seedlings with mutations in FyPP1 and FyPP3, 2 genes encoding the catalytic subunits of protein phosphatase 6 (PP6), exhibited short hypocotyls and opened cotyledons in the dark, which resembled the photomorphogenic development of dark-grown pifq mutants. The hypocotyls of dark-grown sextuple mutant fypp1 fypp3 (f1 f3) pifq were shorter than those of parental mutants f1 f3 and pifq, indicating that PP6 phosphatases and PIFs function synergistically to repress photomorphogenesis in the dark. We showed that FyPPs directly interacted with PIF3 and PIF4, and PIF3 and PIF4 proteins exhibited mobility shifts in f1 f3 mutants, consistent with their hyperphosphorylation. Moreover, PIF4 was more rapidly degraded in f1 f3 mutants than in wild type after light exposure. Whole-genome transcriptomic analyses indicated that PP6 and PIFs coregulated many genes, and PP6 proteins may positively regulate PIF transcriptional activity. These data suggest that PP6 phosphatases may repress photomorphogenesis by controlling the stability and transcriptional activity of PIF proteins via regulating PIF phosphorylation.
Green plants utilize sunlight as the source of energy to provide both food and oxygen for most creatures on earth. Meanwhile, light regulates many processes of plant development (1). For example, Arabidopsis seedlings grown in the dark undergo skotomorphogenesis, which is featured by long hypocotyls, closed and yellow cotyledons, and apical hooks. After illumination, seedlings undergo photomorphogenesis, which is featured by inhibited hypocotyl elongation, open and expanded cotyledons, and disappearance of the apical hook. This latter process, which is also called de-etiolation, is regulated by the coordination of multiple photomorphogenic regulators (2). One of the most important partners are the transcription factors, PHYTOCHROME-INTERACTING FACTORs (PIFs), which repress photomorphogenesis in the dark (3, 4).
PIF proteins interact with phytochromes in a light-dependent manner: Red light converts the phytochromes into the active Pfr form, which can interact with PIF proteins, while far-red light converts phytochromes into the inactive Pr form, which reduces their interaction with PIF proteins (5, 6). During de-etiolation, light activates phytochromes, and the Pfr forms of phytochromes translocate from the cytosol into the nucleus and interact with PIF proteins to induce the phosphorylation and degradation of PIF proteins to initiate photomorphogenesis (7–9). For example, light can induce phosphorylation of PIF3 proteins at multiple sites, and phosphorylated PIF3 is then targeted for degradation by LIGHT RESPONSIVE BTB PROTEIN (LRB) and EIN3-BINDING F BOX PROTEIN (EBF) E3 ligases (10–12). Phosphorylation has also been shown to regulate the transcriptional activity of PIF4 and translocation of PIF7 in diurnal hypocotyl elongation and shade-induced stem elongation, respectively (13–15).
Several kinases have been identified that phosphorylate PIF proteins either in the dark and/or during the dark-to-light transition. Casein Kinase 2 (CK2) has been shown to phosphorylate PIF1 at multiple sites that promote light-induced degradation of PIF1 (16). The GSK3-like kinase BRASSINOSTEROID-INSENSITIVE 2 (BIN2) has been shown to phosphorylate PIF3 in the dark, and elevated activity of BIN2 destabilizes PIF3 in cop1 mutants (17). BIN2 has also been shown to regulate the phosphorylation and transcriptional activity of PIF4 in diurnal conditions (14). Recently, there is evidence showing that phytochromes, Photoregulatory Protein Kinases (PPKs), and MPK6 can also phosphorylate PIF3 proteins (18–20). In contrast to the extensive studies of PIF protein phosphorylation, dephosphorylation of PIF proteins is poorly understood. To our knowledge, the only known phosphatase acting on PIF proteins is TOPP4, which can dephosphorylate and stabilize PIF5 (21).
Ser/Thr phosphatases and Tyr phosphatases are major types of phosphatase in eukaryotes, and Ser/Thr phosphatases can be further divided into the Ser/Thr-specific phosphoprotein phosphatase (PPP) family and the Mg2+-dependent phosphoprotein phosphatase (PPM) family (22). In Arabidopsis, there are 26 PPP catalytic subunits subdivided into PP1, PP2A, PP4, PP5, PP6, and PP7 types (23). The catalytic subunits of PP6, FyPP1 (phytochrome-associated Ser/Thr protein phosphatase 1) and FyPP3, have been shown to physically interact with phytochromes A (phyA) and B (phyB), and regulate flowering time in Arabidopsis (24). Loss of function of both FyPP1 and FyPP3 in Arabidopsis has been shown to cause many developmental defects, including shorter roots, root meristem collapse, abnormal cotyledons, and altered leaf venation, which were shown to be due to elevated accumulation of phosphorylated PIN-FORMED (PIN) protein and abnormal auxin transport (25). It has also been shown that the PP6 holoenzyme containing FyPP1 and FyPP3 can directly dephosphorylate ABSCISIC ACID INSENSITIVE5 (ABI5) to regulate ABSCISIC ACID (ABA) signaling (26). Even though PP6 phosphatases were first identified as regulators of light signaling, the direct targets of PP6 in this process are still unknown.
In this study, we report that PP6 phosphatases repressed photomorphogenesis. PP6 directly interacted with PIF3 and PIF4 proteins, and regulated their phosphorylation status in the dark. Loss of PP6 function elevated light-induced PIF4 degradation. Transcriptome analysis of fypp1 fypp3 (f1 f3) double-mutant seedlings suggested that PP6 shared many downstream responsive genes with PIFs, and PP6 was required for PIF transcriptional activity in vivo. Our study indicates that PP6 functions as the phosphatase that dephosphorylates PIF3 and PIF4, and regulates photomorphogenesis in Arabidopsis.
Results
PP6 Repressed Photomorphogenesis.
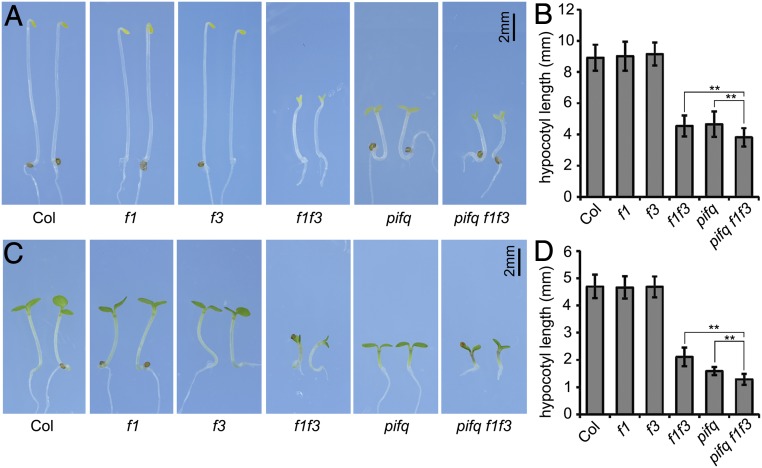
Previously, FyPP1- and FyPP3-containing PP6 phosphatases have been shown to be involved in light signaling (24). To better determine whether these PP6 phosphatases can regulate photomorphogenesis, we compared the phenotypes of wild-type (Columbia-0 [Col-0]), fypp1 (f1), fypp3 (f3), and the progeny of f1−/+ f3 (fypp1 allele is heterozygous, fypp3 allele is homozygous) in the dark. While both f1 and f3 single-mutant seedlings exhibited skotomorphogenic phenotypes similar to Col, the f1 f3 double-mutant seedlings from the progeny of f1−/+ f3 showed photomorphogenic phenotypes with short hypocotyls and open cotyledons (Fig. 1 A and B). Similarly, we found that under continuous red light, even though the hypocotyl lengths of f1 and f3 single mutants were comparable to those of Col, the hypocotyl lengths of f1 f3 double mutants were much shorter (Fig. 1 C and D). We generated transgenic plants with binary vectors FyPP1pro::GUS (GUS expression driven by the FyPP1 native promoter) or FyPP3pro::GUS to study the expression of FyPP genes (SI Appendix, Fig. S1 A and B). GUS staining showed that both FyPP1 and FyPP3 were expressed in hypocotyls and cotyledons of dark- and light-grown transgenic seedlings (SI Appendix, Fig. S1 C and D). Together, these data suggest that PP6 may play a negative role in regulation of photomorphogenesis.
Fig. 1.
PP6 negatively regulated photomorphogenesis. Phenotypes (A) and hypocotyl lengths (B) of wild-type (Col-0), f1, f3, f1 f3, pifq, and pifq f1 f3 mutants grown in the dark for 4 d. Phenotypes (C) and hypocotyl lengths (D) of Col-0, f1, f3, f1 f3, pifq, and pifq f1 f3 mutants grown under continuous red light (30 μmol⋅m−2⋅s−1) for 4 d. Data are shown as mean ± SD (n ≥ 20). **P < 0.01, as calculated by Student’s t test. The experiments were performed 3 times, with similar results each time.
PIFs are well-known photomorphogenic repressors, and pif1 pif3 pif4 pif5 (pifq) mutant seedlings show constitutive photomorphogenesis in the dark (3, 4). To investigate the genetic interactions between PIFs and FyPPs, we crossed pifq mutants with f1−/+ f3 mutants to obtain the sextuple mutant pifq f1−/+ f3 (SI Appendix, Fig. S2 A and B). Among the progeny of pifq f1−/+ f3, we found that the hypocotyl lengths of the homozygous pifq f1 f3 seedlings were shorter than those of both pifq and f1 f3 seedlings in the dark (Fig. 1 A and B) and under continuous red light (Fig. 1 C and D). These synergistic genetic interactions between FyPPs and PIFs are consistent with the hypothesis that PP6 phosphatases may function together with PIFs to repress hypocotyl elongation in photomorphogenesis.
PP6 Directly Interacted with PIF3 and PIF4.
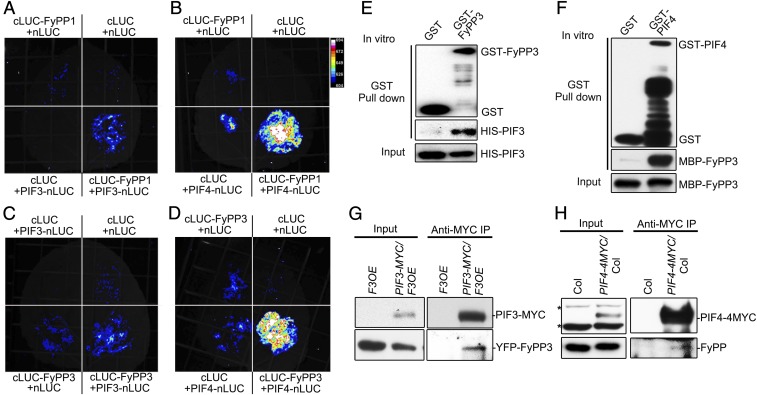
To reveal the functional relationship between PP6 and PIF proteins, we conducted several types of experiments to test whether the FyPP catalytic subunits of PP6 could interact with PIF3 and PIF4. Luciferase complementation imaging (LCI) assays showed that FyPPs could interact with PIF3 and PIF4 in tobacco leaves (Fig. 2 A–D). We also found that FyPPs could interact with PIF1 and PIF5 in vivo (SI Appendix, Fig. S3 A and B). In vitro pull-down assays showed that FyPP3 could directly interact with PIF3 and PIF4 (Fig. 2 E and F). Finally, we confirmed that FyPPs could associate with PIF3 and PIF4 in Arabidopsis seedlings by coimmunoprecipitation assays (Fig. 2 G and H). These data suggest that FyPPs are able to directly interact with PIF3 and PIF4 both in vitro and in vivo.
Fig. 2.
PP6 physically interacted with PIF3 and PIF4. LCI assays showed that FyPP1 interacted with PIF3 (A) and PIF4 (B) and that FyPP3 interacted with PIF3 (C) and PIF4 (D) in tobacco leaves. Empty vectors (cLUC and nLUC only) were used as negative controls. GST-FyPP3, but not GST, interacted with HIS-PIF3 (E), and MBP-FyPP3 interacted with GST-PIF4, but not GST (F). Immunoblotting analyses were conducted with anti-GST, anti-HIS, and anti-MBP antibodies. (G) PIF3-MYC interacted with YFP-FyPP3 in vivo. Total proteins were extracted from F3OE and PIF3-MYC/F3OE seedlings grown in the dark for 3 d. Anti-MYC antibodies were used for immunoprecipitation. Pellets were analyzed with anti-MYC and anti-GFP antibodies. (H) PIF4-MYC interacted with endogenous FyPPs in vivo. Total proteins were extracted from 3-d-old dark-grown seedlings of Col-0 and PIF4-4MYC. Anti-MYC antibodies were used for immunoprecipitation. Pellets were analyzed with anti-MYC and anti-FyPP antibodies. Asterisks denote cross-reacting bands.
PP6 Regulated the Phosphorylation Status of PIF3 and PIF4.
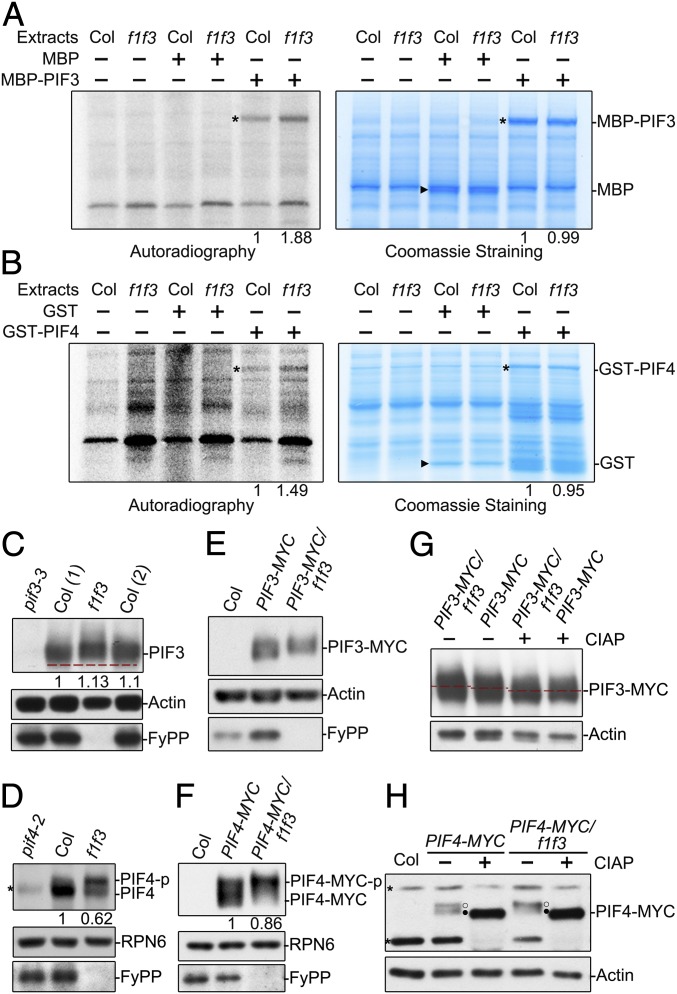
As FyPPs are catalytic subunits of PP6 phosphatases, we wanted to know whether FyPP-containing phosphatases could regulate PIF phosphorylation status. We first tested whether PP6 could directly dephosphorylate PIF3 and PIF4 by cell-free kinase assays using purified maltose binding protein (MBP)-PIF3 and glutathione S-transferase (GST)-PIF4 proteins from Escherichia coli as substrates and extracts from dark-grown Col and f1 f3 double-mutant seedlings as kinase sources. We observed that the signal of phosphorylated MBP-PIF3 or GST-PIF4, but not MBP or GST, was stronger in samples incubated with plant total protein extracts from f1 f3 mutant seedlings compared with Col seedlings (Fig. 3 A and B). These observations are consistent with the possibility that PP6 may directly mediate the dephosphorylation of PIF3 and PIF4.
Fig. 3.
PP6 regulated the phosphorylation status of PIF3 and PIF4 in the dark. Cell-free kinase assays showed that there were more phosphorylated MBP-PIF3 (A) and GST-PIF4 (B) proteins in total extracts of dark-grown f1 f3 seedlings than in total extracts of dark-grown Col-0 seedlings. (Left) [γ-32P] adenosine 5′-triphosphate autoradiography. (Right) Coomassie Blue staining. Asterisks denote the bands of GST-PIF4 or MBP-PIF3. Triangles denote the GST or MBP bands. The phosphorylated signals of recombinant PIF proteins incubated with Col were set as 1. Endogenous PIF3 proteins (C) and PIF4 proteins (D) from f1 f3 mutants migrated more slowly than those from Col-0 on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels. Immunoblotting analyses were performed with anti-PIF3, anti-PIF4, and anti-FyPP antibodies. The numbers in parentheses represents biological replicates. PIF3-MYC proteins (E) and PIF4-MYC proteins (F) from f1 f3 mutants migrated more slowly on SDS-PAGE gels than those from Col-0. Anti-MYC and anti-FyPP antibodies were used in the immunoblotting analyses. In C, D, and F, PIF3, PIF4, and PIF4-MYC protein levels were normalized to RPN6, and PIF protein levels in Col background were set as 1. The slow-migrating PIF3-MYC (G) and PIF4-MYC (H) were phosphorylated forms. Proteins extracted from dark-grown seedlings were incubated with (+) or without (−) alkaline phosphatase (CIAP) for 2 h before electrophoresis. Actin and RPN6 proteins were used as loading controls. Asterisks denote nonspecific bands. Open circles (○) represent the phosphorylated form, and filled circles (●) indicate the dephosphorylated form.
By immunoblotting, we detected a lower shift in the mobilities of both endogenous PIF3 and PIF4 bands in dark-grown f1 f3 double-mutant seedlings compared with those from dark-grown Col seedlings (Fig. 3 C and D and SI Appendix, Fig. S4 A and B). Similarly, transgenically expressed PIF3-MYC or PIF4-MYC protein bands also exhibited mobility shifts in the f1 f3 double-mutant background compared with Col (Fig. 3 E and F). To further confirm that the mobility shifts of these bands were caused by phosphorylation, we used a calf intestinal alkaline phosphatase (CIAP) treatment. Without CIAP treatment, the movement of PIF3-MYC and PIF4-MYC proteins was slower in the f1 f3 background than in Col (Fig. 3 G and H); after CIAP treatment, these proteins moved faster in both the f1 f3 and Col backgrounds, and the mobility differences between the f1 f3 and Col backgrounds disappeared (Fig. 3 G and H). These results suggest that disruption of FyPP1 and FyPP3 in vivo will induce a mobility shift of PIF3 and PIF4 proteins, and the mobility shift is caused by overphosphorylation of PIF3 and PIF4 proteins.
PP6 Regulated the Stability of PIF4 Protein.
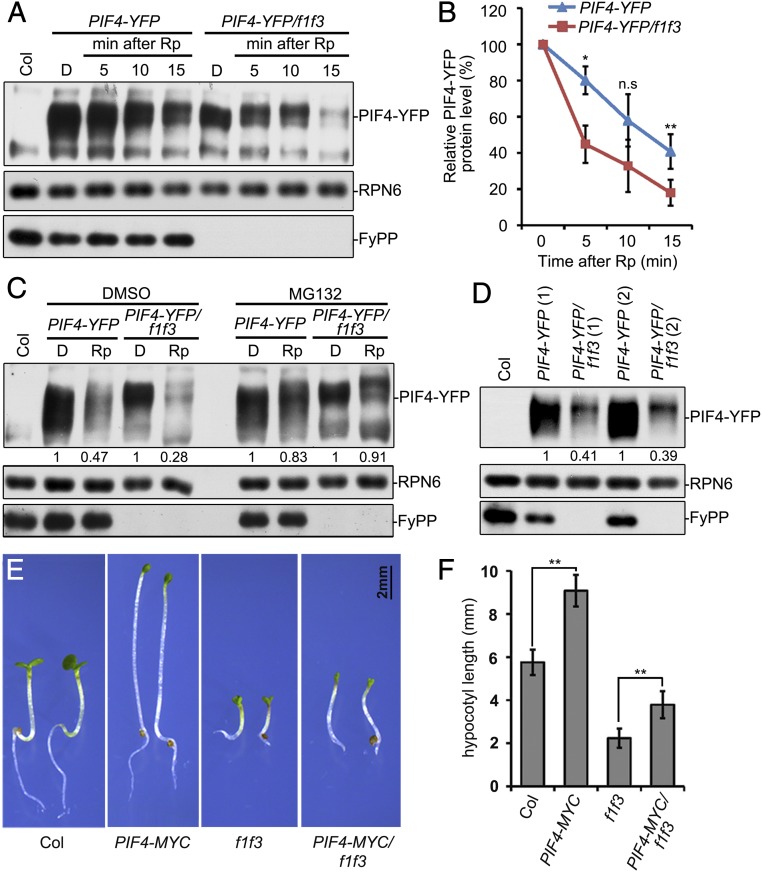
Phosphorylation of PIFs has been previously reported to correlate with protein stability or transcriptional activity (10, 14, 17). We therefore asked whether the regulation of PIF3 and PIF4 phosphorylation by PP6 had similar effects. We found that the levels of PIF4 proteins, but not PIF3, showed a slight decrease in f1 f3 double-mutant seedlings in the dark (Fig. 3 C and D). We further checked whether PP6 could regulate the stability of PIF3 and PIF4 proteins during de-etiolation, as the timely removal of PIF proteins is essential for de-etiolation and is coupled with phosphorylation (7, 8, 10, 11). Because the PIF4 messenger RNA (mRNA) levels were dramatically lower in f1 f3 mutant seedlings (SI Appendix, Fig. S5), PIF4-YFP was ectopically expressed in f1 f3 mutants (by crossing 35S::PIF4-YFP/Col plants with f1 f3 mutants) to investigate the regulation on protein level. We found that the light-induced degradation of PIF4-YFP, but not PIF3, was more rapid in f1 f3 mutant seedlings than in Col (Fig. 4 A and B and SI Appendix, Fig. S6A), and the rapid degradation of PIF4-YFP in f1 f3 could be inhibited by MG132 treatment (Fig. 4C). Similarly, we found that PIF4-YFP protein levels, but not PIF3-MYC protein levels, were significantly lower in f1 f3 than in Col under continuous red or white light (Fig. 4D and SI Appendix, Fig. S6 B–D). These data suggested a correlation between PP6-regulated phosphorylation and light-induced degradation of PIF4 proteins. We also found that the hypocotyls of f1 f3 double-mutant seedlings were significantly shorter than Col under continuous red light, while overexpression of PIF4 and PIF3 could partially rescue the hypocotyl lengths of f1 f3 double-mutant seedlings (Fig. 4 E and F and SI Appendix, Fig. S7 A and B). These data suggest that PP6 regulates light-triggered PIF4 degradation and PIF4-controlled hypocotyl elongation under continuous red light.
Fig. 4.
PP6 regulated the red-light–induced degradation of PIF4. Mutation of PP6 increased the red-light–induced degradation rate of PIF4-YFP as shown by immunoblotting (A) and statistical analyses (B). Seedlings were grown in the dark (D) for 4 d, and were then illuminated with a red-light pulse (Rp) of 3,000 μmol/m2 and kept in the dark for the indicated times before harvesting. Data in B are means of 3 independent biological replicates ± SD *P < 0.05 and **P < 0.01, as determined by Student’s t test. n.s, not significant. (C) MG132 reduced the degradation rate of PIF4-YFP in f1 f3 mutants during red-light irradiation. The 4-d-old dark-grown seedlings were incubated with dimethyl sulfoxide (DMSO) or 50 μM MG132, and were then given a red-light pulse and kept in the dark for 15 min. (D) PP6 facilitated PIF4 accumulation under continuous red light. The seedlings were grown under continuous red light (40 μmol⋅m−2⋅s−1) for 4 d. The numbers in parentheses represent biological replicates. In B, C, and D, the PIF4-YFP protein level was normalized to RPN6. The PIF4-YFP level in dark conditions (B and C) or Col background (D) was set as 100% or 1. Hypocotyl phenotypes (E) and lengths (F) of Col-0, PIF4-MYC, f1 f3, and PIF4-MYC/f1 f3 seedlings grown under continuous red light (30 μmol⋅m−2⋅s−1) for 4 d. Data are shown as mean ± SD (n ≥ 20). **P < 0.01, as calculated by Student’s t test. The experiments were performed 3 times, and similar results were obtained each time.
The Transcriptional Regulatory Activities of PIFs Were Dependent on PP6.
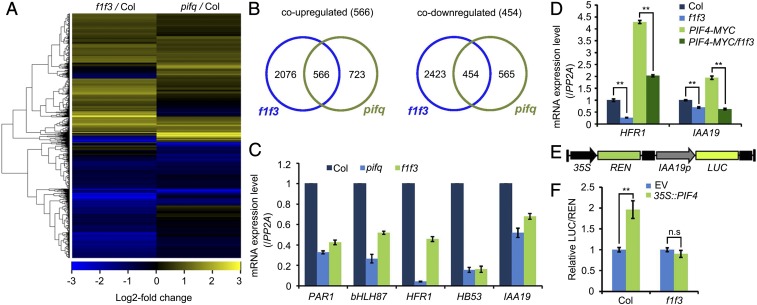
To examine the functional relationships between PP6 and PIFs in the regulation of photomorphogenesis, we compared the transcriptomes of 4-d-old, dark-grown, wild-type (Col-0), f1 f3 and pifq mutant seedlings by mRNA deep sequencing analysis. We identified 5,519 genes that were differentially expressed in dark-grown f1 f3 double-mutant seedlings compared with Col (Dataset S1). A heat map showed that the trends of changes in expression levels of many genes were similar between f1 f3 and pifq mutants (Fig. 5A). We identified 566 coup-regulated genes, which accounted for 43.9% (566 of 1,289) and 27% (566 of 2,642) of the total up-regulated genes in pifq and f1 f3, respectively, and we also identified 454 codown-regulated genes, which accounted for 44.6% (454 of 1,019) and 15.8% (454 of 2,877) of the total down-regulated genes in pifq and f1 f3, respectively (Fig. 5B).
Fig. 5.
PP6 mediated the PIF-regulated transcriptomic changes in the dark-grown seedlings. (A) Heat map of genes up- or down-regulated in f1 f3 and pifq mutants. Up to 44% of PIF-regulated genes were also regulated by PP6. (B) Venn diagrams showing the overlap of genes that were up- or down-regulated in f1 f3 and pifq mutants. (C) RT-qPCR validation of several genes that were coregulated in f1 f3 and pifq mutants. These genes were chosen for validation because they were bound and regulated by PIF4. (D) Relative expression levels of HFR1 and IAA19 in Col-0, f1 f3, PIF4-MYC, and PIF4-MYC/f1 f3 seedlings grown in the dark for 4 d. The expression of PP2A was used as an internal control. Data are mean ± SD (n = 3). **P < 0.01, as calculated by Student’s t test. (E) Structure of the IAA19p::LUC reporter. The positions of the 35S promoter, REN luciferase coding sequence (REN), IAA19 promoter (IAA19p, −2 kilobases to 0 base pair) and firefly luciferase coding sequence (LUC) are indicated. (F) Relative LUC activities (LUC/REN) in Col-0 and f1 f3 mesophyll protoplasts cotransformed with IAA19p::LUC (reporter) and 35S:PIF4 (effector). The empty vector (EV) was used as a reporter control. Data are shown as mean ± SD (n = 4). **P < 0.01, as calculated by Student’s t test. n.s, not significant.
We overlapped the genes that were coregulated in f1 f3 and pifq with previously published PIF4 chromatin immunoprecipitation-sequencing data (27). This analysis revealed that at least 266 genes were likely direct targets of PIF4 and were also regulated by PP6 (SI Appendix, Fig. S8 A and B and Dataset S2). We identified the biological processes in which these genes were involved using the functional annotation of DAVID (Database for Annotation, Visualization, and Integrated Discovery) (28) (SI Appendix, Fig. S8 C and D). Specifically, 186 genes whose promoters were bound by PIF4 and whose expression was decreased in both f1 f3 and pifq were enriched in regulation of transcription, oxidation-reduction process, far-red-light and red-light stimulus, regulation of growth, response to auxin, ethylene biosynthetic process, and brassinosteroid-mediated process (SI Appendix, Fig. S8D). Through quantitative reverse transcription polymerase chain reaction (RT-qPCR) analyses, we confirmed that the expression levels of several representative targets, PAR1, bHLH87, HFR1, HB53, and IAA19, were indeed decreased in f1 f3 double mutants (Fig. 5C).
More interestingly, we found that even though overexpression of PIF4 could significantly activate the expression of HFR1 and IAA19, PP6 mutations alleviated the activation (Fig. 5D), suggesting that PP6-mediated dephosphorylation of PIF4 may promote the transcriptional activation activity of PIF4 protein in vivo. To test whether transcriptional activity of PIF4 was influenced by PP6, we performed a dual-luciferase assay using mesophyll protoplasts prepared from Col and f1 f3 plants. We found that PIF4 could activate the expression of the IAA19pro::LUC reporter in Col but not in f1 f3 protoplasts (Fig. 5 E and F). These results indicate that dephosphorylation of PIF proteins by PP6 facilitates their transcriptional activity to repress photomorphogenesis in the dark.
Discussion
We present evidence that FyPP1 and FyPP3, 2 conserved catalytic subunits of PP6, are negative regulators of photomorphogenesis and light signal transduction. FyPP-null mutant seedlings showed shorter hypocotyl lengths and separated cotyledons in the dark, and also exhibited shorter hypocotyls under continuous red light (Fig. 1 A and C). Furthermore, among the 5,519 FyPP-regulated genes, light signaling pathway-related genes were significantly and strongly enriched (Dataset S3), suggesting that FyPPs may have a specific role in light signal transduction. Mutations of PIF1, PIF3, PIF4, and PIF5 in f1 f3 double-mutant seedlings further shortened the hypocotyl lengths in the dark, suggesting a functional convergence of PIFs and PP6 in repressing photomorphogenesis (Fig. 1). However, we observed that PP6 mutations did not further enhance the cotyledon separation of pifq mutant, probably because PP6 may mediate cotyledon development by factors other than PIFs. SAUR genes, which are auxin-responsive genes, have been shown to be the major factors regulating cotyledon opening (29, 30). We found that several SAURs were up-regulated to higher levels in pifq than those in f1 f3 (e.g., SAUR51) or down-regulated in f1 f3 (e.g., SAUR14, SAUR16) (Dataset S1), which could be part of the reason why f1 f3 did not further enhance the cotyledon separation phenotype of pifq mutant. Nonetheless, the transcriptomic comparison of f1 f3 and pifq demonstrated that PP6 shared many target genes with PIFs, and these genes were involved in light responses or related to cell elongation (SI Appendix, Fig. S8), further suggesting that FyPPs work in concert with PIFs to repress photomorphogenesis in vivo. However, the genes independently regulated by PIFs were also enriched in light response, suggesting that the FyPP-PIF regulation module only affected part of the light-responsive genes among PIF-regulated genes (SI Appendix, Fig. S9 A and B and Dataset S4). FyPPs independently regulated another 4,499 genes that were involved in general translation processes (SI Appendix, Fig. S9C and Dataset S4), suggesting that FyPPs also show broader function on plant development other than specifically regulating light signal transduction.
Notably, the molecular and biochemical data suggested that PP6 is a phosphatase of PIF3 and PIF4. The phosphorylation status of PIF3 and PIF4 proteins was altered in pp6-null mutants, and plant extracts containing PP6 could efficiently dephosphorylate PIF3 and PIF4 in vitro (Fig. 3 A and B), suggesting an important role of PP6 in regulating PIF phosphorylation. The ability to detect direct interactions of FyPPs with both PIF3 and PIF4 further implied the possibility that FyPPs directly catalyzed the dephosphorylation of PIF3 and PIF4 proteins (Fig. 2 E and F). FyPPs could also interact with PIF1 and PIF5 (SI Appendix, Fig. S3), indicating a more inclusive role of PP6 in regulating PIF protein phosphorylation. Moreover, it also has been shown that phosphorylation status will affect the activity of COP1, another key suppressor of photomorphogenesis (31). Even though the kinase of COP1 has been identified, the potential phosphatase(s) of COP1 is still unknown, and it will be interesting to investigate whether PP6 is also involved in the regulation of COP1 protein phosphorylation status or not.
PIF proteins have been shown to play a central role in light signaling (3–5). Following the light-induced activation of phytochromes, most PIF proteins are rapidly phosphorylated (7–9, 13). PIF protein phosphorylation regulates the stability and/or transcriptional activity of PIF proteins, and also plays an important role in promoting the degradation of phyB to attenuate light signaling (10, 11, 14). Interestingly, changes in PIF4, but not PIF3, phosphorylation decreased corresponding protein levels in f1 f3 double-mutant seedlings (Fig. 4 and SI Appendix, Fig. S6), suggesting a distinct effect of PP6-regulated phosphorylation on different PIF proteins. Also, there are both convergence and difference between light- and pp6-induced PIF phosphorylation. Both light- and pp6-induced PIF4 phosphorylation could regulate PIF4 protein level (this study and ref. 32). However, even though light-induced PIF3 phosphorylation can strongly induce the degradation of PIF3 protein (10), pp6-induced PIF3 phosphorylation did not affect its protein stability (SI Appendix, Fig. S6 A and B). We propose that FyPPs may regulate PIF3 phosphorylation at different sites than those induced by light, and other uncharacterized phosphatase(s) may be required to specifically regulate those light-induced phosphorylation sites on PIF3 protein. Furthermore, we noticed a more obvious mobility shift of PIF4 protein after red-light treatment even in f1 f3 mutant background (Fig. 4C), suggesting that there are other light-responsive factors to regulate PIF4 phosphorylation status together with FyPPs. The PP6 phosphatases counteract with light signal to maintain the dephosphorylation status and activity of PIF4 protein to negatively regulate light signal transduction (Figs. 4D and 5 C–F). As PIF4 protein is the key regulator in temperature signaling, and FyPPs can regulate PIF4 activity, we found that the response to warm temperature was significantly impaired in f1 f3 mutant compared with wild type (SI Appendix, Fig. S10), suggesting that the regulation of PIF4 by FyPPs is probably conserved across PIF4-involved biological processes.
Reversible phosphorylation is mainly dependent on the antagonistic role of kinases and phosphatases in eukaryotic organisms (33). Our data and previous studies demonstrate that the phosphorylation status of PIF proteins is strictly regulated by FyPP-containing PP6 phosphatases as well as numerous kinases in vivo. It will be interesting to study how light simultaneously regulates the activity of both phosphatases and kinases to fine-tune PIF protein phosphorylation and induce optimal plant development.
Methods
Plant Materials and Growth Conditions.
All Arabidopsis plant materials were in the Col-0 ecotype background. The 35S::PIF4-MYC (PIF4-MYC) transgenic lines were provided by Peter Quail, University of California, Berkeley, CA (34), and 35S::PIF4-YFP (PIF4-YFP) transgenic lines were provided by Mingyi Bai, Shandong University, Shandong, China (35). The f1, f3, f1−/+ f3 (25), and pifq (3) mutants and 2×35S::EYFP-FyPP3 (F3OE) (25) and 35S::PIF3-MYC (PIF3-MYC) (36) transgenic lines have been described previously. Because f1 f3 double mutants showed severe infertility, fertile f1−/+f3 heterozygous mutants were used to produce genetic materials. For generating pifq f1−/+ f3, PIF3-MYC/f1−/+ f3, PIF4-MYC/f1 f3−/+, and PIF4-YFP/f1−/+ f3 genetic materials, pifq, PIF3-MYC, PIF4-MYC, and PIF4-YFP plants were crossed with f1−/+ f3 mutants. For generating PIF3-MYC/F3OE, PIF3-MYC was crossed with F3OE transgenic lines. For generating 35S::PIF4-4×MYC/Col (PIF4-4MYC), Agrobacterium strain GV3101 carrying the binary plasmid pJIM19-PIF4-4×MYC was transformed into Col-0 plants.
For phenotypic observations, f1 f3, pifq f1 f3, PIF3-MYC/f1 f3, PIF4-MYC/f1 f3, and PIF4-YFP/f1 f3 homozygous seedlings were isolated from the progenies of f1−/+ f3, pifq f1−/+ f3, PIF3-MYC/f1−/+ f3, PIF4-MYC/f1 f3−/+, and PIF4-YFP/f1−/+ f3 plants. We identified homozygous f1 f3 through PCR on genomic DNA and RT-PCR. For biochemical assays, 3-d-old dark-grown homozygous seedlings of f1 f3, PIF3-MYC/f1 f3, PIF4-MYC/f1 f3, and PIF4-YFP/f1 f3 were isolated from the progenies of f1−/+ f3, PIF3-MYC/f1−/+ f3, PIF4-MYC/f1 f3−/+, and PIF4-YFP/f1−/+ f3 plants under green light, and then were grown on Murashige and Skoog (MS) medium in the dark for another 24 h before protein extraction. Wild-type Col-0, PIF3-MYC, PIF4-MYC, and PIF4-YFP were treated in the same way.
Seeds were surface-sterilized with 10% NaClO and 0.02% Triton X-100 for 10 min, and then were sown on solid MS medium and stratified in darkness for 3 d at 4 °C. For dark treatment, seeds were transferred to white-light chambers (80 μmol⋅m−2⋅s−1) for 12 h and then kept in a dark chamber maintained at 22 °C unless otherwise specified. For red-light treatment, seeds were irradiated with continuous white light for 4 h and then transferred to red light and maintained at 22 °C for 4 d. For biochemical assays, 1× MS medium (containing 4.3 g/L MS powder, 10 g/L sucrose, and 8 g/L agar [pH 5.7]) was used, and for phenotypic observations, 1/2× MS medium (containing 2.2 g/L MS powder, 3 g/L sucrose, and 8 g/L agar [pH 5.7]) was used. ImageJ software was used to calculate hypocotyl lengths. For MG132 treatment, 4-d-old dark-grown seedlings were treated with MG132 or dimethyl sulfoxide in liquid MS medium for the indicated times before harvesting.
Cell-Free Kinase Assays.
The cell-free kinase assays were performed as previously described (25). The bacterial strains expressing MBP-PIF3 and GST-PIF4 were obtained from previous research (17, 34).
In Vitro Pull-Down Assays.
The assays were essentially performed as previously described (12). Input and eluted proteins were analyzed by immunoblotting using anti-HIS (Sigma–Aldrich), anti-MBP (New England Biolabs), and anti-GST (Sigma–Aldrich) antibodies.
Coimmunoprecipitation Assays.
Four-day-old dark-grown F3OE and 35S::PIF3-MYC/F3OE (Col-0 and 35S::PIF4-4×MYC/Col) seedlings were harvested and ground into powder in liquid nitrogen. The assays were performed as previously described (34). Input and eluted proteins were analyzed by Western blot using anti-MYC (Sigma–Aldrich), anti-GFP (Abmart), and anti-FyPP (custom-made) antibodies.
RNA Extraction and RT-qPCR.
Total RNA was extracted from 4-d-old dark-grown seedlings using the RNeasy Plant Mini Kit (Qiagen), followed by RT using ReverTra Ace qPCR RT Master Mix (TOYOBO). RT-qPCR analysis was performed using the SYBR Premix Ex Taq Kit (TaKaRa) in a 7500 Fast Real-Time PCR System (Applied Biosystems). The primer sequences used for RT-qPCR are listed in SI Appendix, Table S1. The relative gene expression levels were normalized to the PP2A gene. Three biological repeats were used for the RT-qPCR experiments. Each sample was analyzed with 3 technical replicates.
Transcriptomic Analysis.
Total RNA was extracted from 4-d-old dark-grown Col, pifq, and f1 f3 seedlings using the RNeasy Plant Mini Kit. Deep sequencing of mRNA was performed using an Illumina HiSeq 2500 system (Illumina), generating more than 20 million paired-end reads of 150 base pairs per sample. Two biological repeats of each sample were prepared for RNA-sequencing analyses. The reads were viewed by fastqc (https://www.bioinformatics.babraham.ac.uk/projects/fastqc) to assess quality and trimmed by Seqtk (https://github.com/lh3/seqtk). Next, trimmed reads were mapped to the Arabidopsis TAIR10 genome using the splice site-guided HISAT2 alignment software with default parameters (37). Numbers of reads per gene were determined by the Python package HTSeq (38). Differential expression analysis was performed with the DESeq2 R package (39). Genes were considered differentially expressed when the adjusted P value was ≤0.05 and the absolute fold change was ≥1.5. Functional enrichment analysis was applied by the DAVID functional annotation clustering tool (28). Heat maps were generated by the heat map function in the gplots R package (40). The RNA-sequencing data have been deposited in the Gene Expression Omnibus under accession number GSE128498.
Plant Total Protein Extraction and Immunoblot Analysis.
Arabidopsis seedlings were ground into powder in liquid nitrogen. Total proteins were extracted with denaturing buffer (8 M urea, 100 mM NaH2PO4, 100 mM Tris⋅HCl [pH 8.0], 1 mM phenylmethylsulfonyl fluoride [PMSF], 1× protease inhibitor, and 100 μM MG132) and cleared by centrifugation. Protein supernatants were mixed with 5× sodium dodecyl sulfate (SDS) loading buffer and subsequently boiled for 10 min. Immunoblot assays were performed as previously described (34). Immunoblotting analyses were performed with anti-MYC, anti-PIF3 (34), anti-PIF4 (Agrisera), anti-RPN6 (41), anti-Actin (Sigma–Aldrich), and anti-FyPP antibodies. ImageJ software was used for protein quantification. Proteins of interest were normalized to the loading control.
LCI Assays.
The LCI assays were performed as previously described (25, 26, 42).
Alkaline Phosphatase Treatment.
Total proteins were extracted from 4-d-old dark-grown seedlings with extraction buffer (100 mM Tris⋅HCl [pH 6.8], 20% glycerol, 5% SDS, 20 mM dithiothreitol, 1 mM PMSF, 1× protease inhibitor, and 100 μM MG132). The extracts were boiled for 3 min and cleared by centrifugation. Total protein supernatants were diluted 20-fold into dephosphorylation buffer (Roche) containing 1 mM PMSF, 1× protease inhibitor, and 50 μM MG132, and then incubated with or without 400 units/mL Alkaline Phosphatase (Roche) at 37 °C for 2 h. The reaction mixtures were terminated by adding 5× SDS loading buffer and boiling at 99 °C for 10 min. Immunoblotting analyses were performed with anti-MYC and anti-RPN6 antibodies.
Transient Gene Expression Assays.
Protoplast isolation and polyethylene glycol transformation were performed as previously described (43). Arabidopsis Col-0 and f1 f3 were grown under a 12-h light/12-h dark photoperiod for 3 wk. Isolated mesophyll protoplasts (2 × 104) were transfected with 20 μg of DNA (empty vector or 35S::PIF4 effector and pGreenII0800-IAA19p::LUC reporter). After overnight incubation in the dark, protoplasts were harvested by centrifugation and lysed in passive lysis buffer (Promega). Firefly and Renilla luciferase activities were measured using a SpectraMax i3x reader (Molecular Devices) and Dual-Luciferase Reporter kit (Promega).
Supplementary Material
Acknowledgments
We thank Dr. Peter Quail (University of California, Berkeley) for providing the 35S::PIF4-MYC transgenic seeds and Dr. Mingyi Bai (Shandong University) for providing the 35S::PIF4-YFP transgenic seeds. We thank Dr. Jian Li (Peking University) for providing technical assistance with cell-free kinase assays and Dr. Junjie Ling (Peking University) for providing bacterial strains expressing MBP-PIF3. This work was supported by grants from the National Key R&D Program of China (Grant 2017YFA0503800), the National Nature Science Foundation of China (Grants 31671256 and 31621001), the Fundamental Research Funds for the Central Universities of China (2014PY056), and the National Key Laboratory of Crop Genetic Improvement Self-Research Program (Grant ZW13A0401).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE128498).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1907540116/-/DCSupplemental.
References
- 1.Chen M., Chory J., Fankhauser C., Light signal transduction in higher plants. Annu. Rev. Genet. 38, 87–117 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Dong J., Terzaghi W., Deng X. W., Chen H., Multiple photomorphogenic repressors work in concert to regulate Arabidopsis seedling development. Plant Signal. Behav. 10, e1011934 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leivar P., et al. , Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 18, 1815–1823 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shin J., et al. , Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc. Natl. Acad. Sci. U.S.A. 106, 7660–7665 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ni M., Tepperman J. M., Quail P. H., PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95, 657–667 (1998). [DOI] [PubMed] [Google Scholar]
- 6.Ni M., Tepperman J. M., Quail P. H., Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature 400, 781–784 (1999). [DOI] [PubMed] [Google Scholar]
- 7.Al-Sady B., Ni W., Kircher S., Schäfer E., Quail P. H., Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23, 439–446 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Shen Y., Khanna R., Carle C. M., Quail P. H., Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol. 145, 1043–1051 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen H., et al. , Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell 20, 1586–1602 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ni W., et al. , Multisite light-induced phosphorylation of the transcription factor PIF3 is necessary for both its rapid degradation and concomitant negative feedback modulation of photoreceptor phyB levels in Arabidopsis. Plant Cell 25, 2679–2698 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ni W., et al. , A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science 344, 1160–1164 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong J., et al. , Light-dependent degradation of PIF3 by SCFEBF1/2 promotes a photomorphogenic response in Arabidopsis. Curr. Biol. 27, 2420–2430.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li L., et al. , Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 26, 785–790 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernardo-García S., et al. , BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes Dev. 28, 1681–1694 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang X., et al. , Shade-induced nuclear localization of PIF7 is regulated by phosphorylation and 14-3-3 proteins in Arabidopsis. eLife 7, e31636 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bu Q., et al. , Phosphorylation by CK2 enhances the rapid light-induced degradation of phytochrome interacting factor 1 in Arabidopsis. J. Biol. Chem. 286, 12066–12074 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ling J. J., Li J., Zhu D., Deng X. W., Noncanonical role of Arabidopsis COP1/SPA complex in repressing BIN2-mediated PIF3 phosphorylation and degradation in darkness. Proc. Natl. Acad. Sci. U.S.A. 114, 3539–3544 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shin A. Y., et al. , Evidence that phytochrome functions as a protein kinase in plant light signalling. Nat. Commun. 7, 11545 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ni W., et al. , PPKs mediate direct signal transfer from phytochrome photoreceptors to transcription factor PIF3. Nat. Commun. 8, 15236 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xin X., et al. , Arabidopsis MKK10-MPK6 mediates red-light-regulated opening of seedling cotyledons through phosphorylation of PIF3. J. Exp. Bot. 69, 423–439 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yue J., et al. , TOPP4 regulates the stability of PHYTOCHROME INTERACTING FACTOR5 during photomorphogenesis in Arabidopsis. Plant Physiol. 170, 1381–1397 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kerk D., Templeton G., Moorhead G. B., Evolutionary radiation pattern of novel protein phosphatases revealed by analysis of protein data from the completely sequenced genomes of humans, green algae, and higher plants. Plant Physiol. 146, 351–367 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farkas I., Dombrádi V., Miskei M., Szabados L., Koncz C., Arabidopsis PPP family of serine/threonine phosphatases. Trends Plant Sci. 12, 169–176 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Kim D. H., et al. , A phytochrome-associated protein phosphatase 2A modulates light signals in flowering time control in Arabidopsis. Plant Cell 14, 3043–3056 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dai M., et al. , A PP6-type phosphatase holoenzyme directly regulates PIN phosphorylation and auxin efflux in Arabidopsis. Plant Cell 24, 2497–2514 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dai M., et al. , The PP6 phosphatase regulates ABI5 phosphorylation and abscisic acid signaling in Arabidopsis. Plant Cell 25, 517–534 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oh E., Zhu J. Y., Wang Z. Y., Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14, 802–809 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang W., Sherman B. T., Lempicki R. A., Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Sun N., et al. , Arabidopsis SAURs are critical for differential light regulation of the development of various organs. Proc. Natl. Acad. Sci. U.S.A. 113, 6071–6076 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong J., et al. , The transcription factors TCP4 and PIF3 antagonistically regulate organ-specific light induction of SAUR genes to modulate cotyledon opening during de-etiolation in Arabidopsis. Plant Cell 31, 1155–1170 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin F., et al. , Phosphorylation and negative regulation of CONSTITUTIVELY PHOTOMORPHOGENIC 1 by PINOID in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 114, 6617–6622 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lorrain S., Allen T., Duek P. D., Whitelam G. C., Fankhauser C., Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53, 312–323 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Terol J., Bargues M., Carrasco P., Pérez-Alonso M., Paricio N., Molecular characterization and evolution of the protein phosphatase 2A B’ regulatory subunit family in plants. Plant Physiol. 129, 808–822 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dong J., et al. , Arabidopsis DE-ETIOLATED1 represses photomorphogenesis by positively regulating phytochrome-interacting factors in the dark. Plant Cell 26, 3630–3645 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bai M. Y., et al. , Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 14, 810–817 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim J., et al. , Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 15, 2399–2407 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim D., Langmead B., Salzberg S. L., HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anders S., Pyl P. T., Huber W., HTSeq–A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warnes G. R., et al. , gplots: Various R Programming Tools for Plotting Data. R Package, Version 3.0.1. https://cran.r-project.org/web/packages/gplots/index.html. Accessed 7 September 2019.
- 41.Kwok S. F., Staub J. M., Deng X. W., Characterization of two subunits of Arabidopsis 19S proteasome regulatory complex and its possible interaction with the COP9 complex. J. Mol. Biol. 285, 85–95 (1999). [DOI] [PubMed] [Google Scholar]
- 42.Chen H., et al. , Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146, 368–376 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoo S. D., Cho Y. H., Sheen J., Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.