Significance
Circadian clocks control various aspects of physiology. The circadian control of the response of T cells to antigen presentation, which is at the core of the specific immune response to pathogens, has remained unclear. Here, we show a role for circadian clocks in controlling the magnitude of the response of T cells to antigen presentation by dendritic cells. We demonstrate that the clock within T cells themselves is required for this circadian regulation, making these cells more or less prone to be activated according to time of day. Our study contributes to a better understanding of the role of circadian clocks in the adaptive immune response and opens a door to T cell-based therapies based on time of day.
Keywords: CD8 T cells, dendritic cells, vaccination, circadian clock, transcriptome
Abstract
Circadian variations of various aspects of the immune system have been described. However, the circadian control of T cells has been relatively unexplored. Here, we investigated the role of circadian clocks in regulating CD8 T cell response to antigen presentation by dendritic cells (DCs). The in vivo CD8 T cell response following vaccination with DCs loaded with the OVA257–264 peptide antigen (DC-OVA) leads to a higher expansion of OVA-specific T cells in response to vaccination done in the middle of the day, compared to other time points. This rhythm was dampened when DCs deficient for the essential clock gene Bmal1 were used and abolished in mice with a CD8 T cell-specific Bmal1 deletion. Thus, we assessed the circadian transcriptome of CD8 T cells and found an enrichment in the daytime of genes and pathways involved in T cell activation. Based on this, we investigated early T cell activation events. Three days postvaccination, we found higher T cell activation markers and related signaling pathways (including IRF4, mTOR, and AKT) after a vaccination done during the middle of the day compared to the middle of the night. Finally, the functional impact of the stronger daytime response was shown by a more efficient response to a bacterial challenge at this time of day. Altogether, these results suggest that the clock of CD8 T cells modulates the response to vaccination by shaping the transcriptional program of these cells and making them more prone to strong and efficient activation and proliferation according to the time of day.
Circadian clocks located in most tissues in mammals allow the adaptation to daily environmental variations (1). These clocks are composed of a set of clock genes (e.g., Clock, Bmal1, Period [Per1-3], Cryptochrome [Cry1-2]) involved in transcriptional–translational feedback loops (2). This molecular clock controls the expression of numerous clock-controlled genes in a rhythmic manner; depending on the tissue or cell studied, between 3% and 16% of genes are expressed with a circadian rhythm (3).
Whereas circadian rhythms of innate immune responses have been extensively characterized, the circadian regulation of the adaptive antigen (Ag)-specific immune responses has remained more elusive (4–6). CD4 T cells express clock genes (7, 8), and T cells rhythmically recirculate through the peripheral blood (9). The rhythm of T cell counts in the blood and lymph nodes depends on glucocorticoid and chemokine receptor rhythms (10, 11) and on adrenergic regulation as well as the clock intrinsic to T cells (12, 13).
Whether circadian clocks can regulate T cell functions (in particular, their response to Ag presentation by Ag-presenting cells, and their subsequent activation and expansion), and the mechanisms involved in this regulation, have remained relatively unexplored. Pioneer studies suggested a circadian control of the adaptive immune response (14, 15). Esquifino et al. (16) showed an effect of time of day on the proliferation of T and B cells after a stimulation with concanavalin or LPS, respectively. A rhythm of T cell proliferation in response to stimulation of the T cell receptor (TCR) was observed and was abolished in mice mutant for the Clock gene (17). Human CD4 T cells collected over a 24-h cycle and stimulated with phytohemagglutinin or PMA/ionomycin showed a rhythm of cytokine (e.g., IL-2 and IFNγ) secretion (7, 18).
Although previous studies have correlated the T cell trafficking rhythms with variations in the magnitude of immune response to immunization, Ag was delivered directly in mice, which means that variations in the response could be due to changes in Ag processing or presentation, antigen-presenting cell (APC) migration, or T cell response itself (12, 13). To be able to address the respective contributions of the APCs and the T cells and to define the mechanisms underlying the circadian variations of T cell response to Ag presentation, we have used a vaccination model where the Ag is presented by bone marrow-derived dendritic cells (BMDCs). Indeed, in contrast to immunization studies with soluble Ags, this system bypasses the steps anterior to Ag presentation to T cells (e.g., Ag take-up and processing, presentation at the surface of DCs). Hence, it allows focusing on the circadian regulation of the response to Ag presentation within T cells and following the rhythmicity of molecular events occurring within these cells. Using this model, we previously showed a day–night variation of the CD8 T cell response (17). Here, we used the same vaccination model to show that a circadian clock intrinsic to CD8 T cells controls the magnitude of their response to Ag presentation. Further, we uncover a rhythmicity in CD8 T cells of gene expression and pathways involved in T cell activation and proliferation and show that these pathways become activated more strongly upon daytime vaccination. The significance of this circadian regulation is indicated by a day–night difference in the control of an infectious bacterial challenge following vaccination.
Results
Circadian Variation of CD8 T Cell Response to DC-OVA Vaccination.
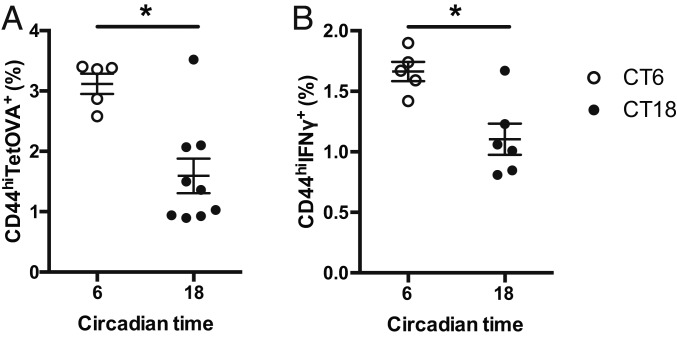
To confirm the circadian (endogenous) nature of the time-dependent variation observed in our previous study (17), we analyzed the CD8 T cell response induced by vaccination of C57BL/6J mice kept in constant darkness after entrainment to a 12-h light:12-h dark (LD) cycle. Mice were vaccinated with LPS-stimulated BMDCs loaded with the OVA257–264 peptide (DC-OVA) or without peptide (DC) as a negative control, either at circadian time (CT)6 (middle of subjective day) or CT18 (middle of subjective night). We confirmed that the level of Ag loading and the differentiation/activation of BMDCs were similar between time points (SI Appendix, Fig. S1 A and B). Seven days postvaccination, spleens were harvested and OVA-specific CD8 T cells were identified using Kb-OVA tetramer (tetOVA) staining. This tetramer recognizes TCRs that are specific for the OVA peptide in the context of the H-2Kb MHC molecule, and can therefore be used as a tool to identify the OVA-specific CD8 T cells. Mice vaccinated at CT6 had about 2 times more CD8 T cell expansion than mice vaccinated at CT18 (Fig. 1A and SI Appendix, Fig. S1 C and D). In parallel, a short ex vivo restimulation of splenic T cells with the OVA peptide was done to analyze their ability to produce cytokines following DC-OVA vaccination. Similarly to CD8 T cell expansion, the percentage of CD8+ CD44+ IFNγ+ cells was significantly higher after a vaccination at CT6 than at CT18 (Fig. 1B and SI Appendix, Fig. S1 E and F). These data show that the day–night difference in response to DC-OVA vaccination is endogenous and suggest the involvement of circadian clocks.
Fig. 1.
Circadian variation of CD8 T cell response to DC-OVA vaccination. BMDCs loaded with OVA peptide (DC-OVA) or unloaded BMDCs (DC) were injected i.v. at either CT6 or CT18 in WT mice. (A) Seven days postvaccination, spleens were harvested to assess the frequency of OVA-specific CD8+ T cells (TetOVA+CD44hi). (B) Ex vivo restimulation with the OVA peptide was done to assess the percentage of IFNγ-producing CD8+ T cells. Representative FACS plots are shown in SI Appendix, Fig. S1 C and D. Data are pooled from 2 independent experiments with similar results, n = 5 to 9 mice/CT. Data are shown for individual mice, as well as mean ± SEM. Unpaired t test, *P < 0.05. See SI Appendix, Statistical Details for more information on statistics.
The Dendritic Cell Clock Modulates CD8 T Cell Response to DC-OVA Vaccination.
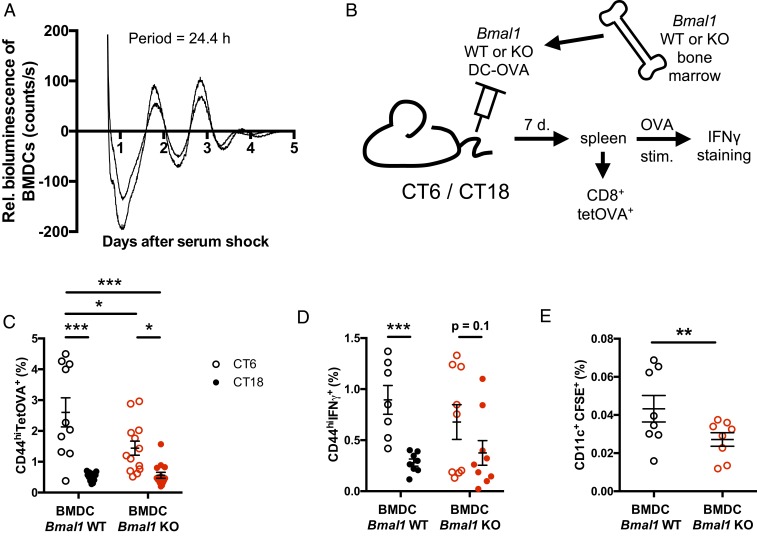
We prepared DCs from the bone marrow of PER2::Luciferase mice (19) and synchronized their clocks with a serum shock. Circadian rhythms of bioluminescence were observed for several days (Fig. 2A), confirming the presence of a functional circadian clock in these cells. Thus, we analyzed the role of the BMDC clock in the CD8 T cell response to DC-OVA vaccination. To do so, we generated BMDCs from Bmal1 knockout (KO) mice and wild-type (WT) littermates. We first looked at the Ag loading and the activation/differentiation levels of the Bmal1 WT vs. KO BMDCs and saw no difference between genotypes (SI Appendix, Fig. S2 A and B). We then vaccinated C57BL/6J WT mice at either CT6 or CT18 with Bmal1 WT or KO DC-OVA (or unloaded DCs) (Fig. 2B). Seven days postvaccination, CD8 T cell expansion was analyzed using Kb-OVA tetramer staining as well as cytokine production following ex vivo restimulation of T cells with OVA. The circadian variation of the CD8 T cell expansion in response to Bmal1 WT DC-OVA remained present when using Bmal1 KO BMDCs as Ag presenting cells for the vaccination (Fig. 2C and SI Appendix, Fig. S2C), showing that the DC clock is not essential for this time-dependent variation. However, the CD8 T cell expansion was significantly lower after a vaccination with Bmal1 KO BMDCs at CT6 compared to the vaccination with the WT BMDCs at the same time point (Fig. 2C and SI Appendix, Fig. S2C). In contrast, the variation observed in the percentage of the CD8+ CD44+ IFNγ+ cells in response to Bmal1 WT DC-OVA was lost in mice that received Bmal1 KO DC-OVA vaccination, although there was a statistical trend for a time-dependent difference (Fig. 2D and SI Appendix, Fig. S2D).
Fig. 2.
The dendritic cell clock modulates CD8 T cell response to DC-OVA vaccination. (A) Bioluminescence recording (baseline-subtracted data) of 2 preparations of BMDCs from PER2::Luc mouse bone marrow. The period of the rhythm is indicated within the graph. (B) Experimental protocol for vaccinations with Bmal1 WT vs. KO BMDCs. (C) Bmal1 WT vs. KO BMDCs loaded with OVA peptide were injected i.v. at CT6 or CT18 in WT mice. Seven days postvaccination, spleens were harvested to assess the frequency of OVA-specific CD8+ T cells (TetOVA+CD44hi). (D) Ex vivo restimulation with the OVA peptide was done to assess the percentage of IFNγ-producing CD8+ T cells. (E) CFSE-labeled WT and Bmal1−/− BMDCs were injected by i.v. at CT6 in B6.SJL mice. Four hours later, migration of DCs to the spleen was analyzed as CD11c+CFSE+ splenocytes. Representative FACS plots are shown in SI Appendix, Fig. S2. Data are pooled from 3 independent experiments with similar results, n = 9 to 14 mice/group for C and D, n = 8 mice/group for E. Data are shown for individual mice, as well as mean ± SEM. Two-way ANOVA with Bonferroni post hoc test where applicable (C: only an effect of time, so post hocs only between CTs, D: significant interaction, so all pairwise comparisons analyzed) or unpaired t test (E). *P < 0.05, **P < 0.01, ***P < 0.001. See SI Appendix, Statistical Details for more information on statistics.
Since the Ag loading and the activation/differentiation levels were unaffected in Bmal1 KO BMDCs, we hypothesized that the numbers of BMDCs delivered in the spleen after a DC-OVA vaccination might be lower for the KO cells. Consistent with this, in an in vivo migration assay, we noticed a reduction of the Bmal1 KO DC-OVA cells compared to WT DC-OVA in the spleen 4 h after the injection done at CT6 (Fig. 2E and SI Appendix, Fig. S2E), a time at which the T cell expansion difference between Bmal1 WT and KO DC-OVA is observed (Fig. 2C). Altogether, these data suggest a contribution of the clock gene Bmal1 in DCs to the regulation of the migration of the BMDCs to the spleen. However, they do not support a significant role for the DC clock in the circadian variation of the CD8 T cell response to vaccination subsequent to antigen uptake and presentation.
The CD8 T Cell Clock Is Essential for the Rhythmicity of the Response to DC-OVA Vaccination.
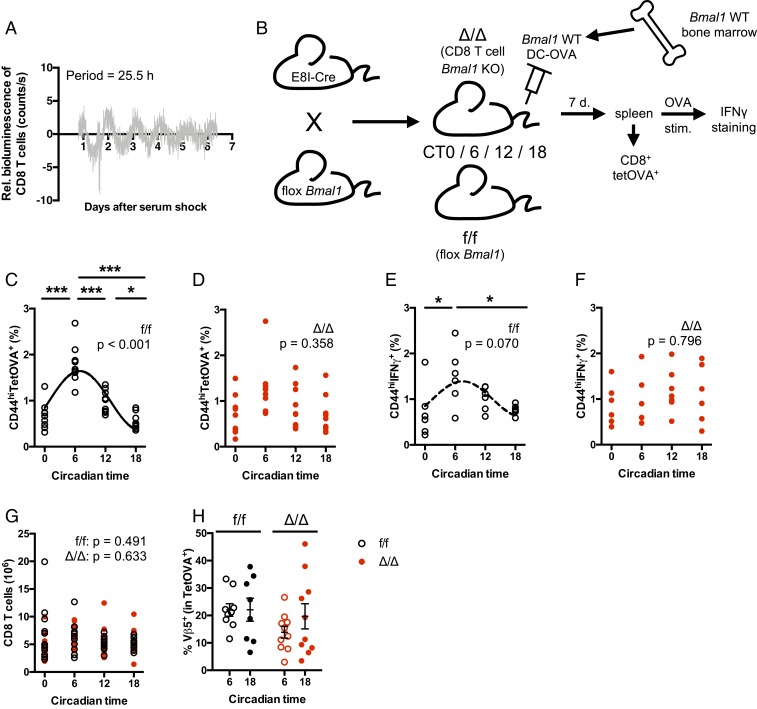
CD8 T cells were isolated from spleens of PER2::Luciferase mice and synchronized with a serum shock. Bioluminescence rhythms were observed, which, although of low amplitude, were robust enough to be observed for 5 to 6 cycles (Fig. 3A). These data showed that CD8 T cells harbor a circadian clock. To address the role of this clock in the CD8 T cell response to DC-OVA vaccination, we generated mice lacking clock function specifically in these cells. The Bmal1 gene was deleted in mature CD8 T cells by breeding E8I-Cre mice with Bmal1 flox/flox mice. We first ensured that Bmal1 gene expression (SI Appendix, Fig. S3A) and BMAL1 protein (SI Appendix, Fig. S3B) were efficiently and specifically suppressed in CD8 T cells (and not in CD4 T cells or other cells), and we confirmed that the deletion did not alter the general behavioral rhythmicity of the mice (SI Appendix, Fig. S3 C and D). We also checked that the proportion of DC-OVA cells migrating to the spleen after vaccination was similar between times of vaccination and between genotypes (SI Appendix, Fig. S3 E and F). We then vaccinated WT (f/f mice, no E8I-Cre transgene) and CD8 T cell-specific Bmal1 KO mice (Δ/Δ mice, both Bmal1 flox/flox and E8I-Cre constructs) at either CT0, 6, 12, or 18 with Bmal1 WT DC-OVA (or unloaded DCs) (Fig. 3B). Seven days postvaccination, CD8 T cell expansion was analyzed using Kb-OVA tetramer staining. The circadian variation of CD8 T cell expansion observed in the WT mice (Fig. 3C and SI Appendix, Fig. S4A) was abolished in the CD8 T cell-specific Bmal1 KO mice (Fig. 3D and SI Appendix, Fig. S4A), showing that the CD8 T cell clock is essential for the rhythmicity of the response of these cells to DC-OVA vaccination. Moreover, after a short ex vivo restimulation of the T cells with OVA, the rhythm of the percentage of CD8 T cells that secrete IFNγ (Fig. 3E and SI Appendix, Fig. S4B) was also abolished in the CD8 T cells Bmal1 KO mice (Fig. 3F and SI Appendix, Fig. S4B). CD8 T cell counts in the spleen did not vary across the day and were unaffected by Bmal1 gene deletion (Fig. 3G). Thus, the circadian rhythm of response to DC-OVA vaccination or the effect of Bmal1 deletion on this rhythm was not due to changes in CD8 T cell counts in the spleen. Moreover, we did not observe any difference in Vβ5 usage within the OVA-specific T cells, suggesting that a similar repertoire of TCR-expressing cells was recruited into the response (Fig. 3H and SI Appendix, Fig. S4C). These data indicated that circadian processes within CD8 T cells might make these cells more or less responsive according to the time of day.
Fig. 3.
The CD8 T cell clock is essential to the rhythmicity of the response to DC-OVA vaccination. (A) Bioluminescence recording (baseline-subtracted data) of CD8+ T cells purified from spleens of PER2::Luc mice. The period of the rhythm is indicated within the graph. (B) Experimental protocol for vaccinations of WT and CD8 T cell-specific Bmal1 KO mice. (C and D) BMDCs loaded with OVA257–264 peptide were injected by i.v. at either CT6 or CT18 in E8I-Cre+/− Bmal1f/f (Δ/Δ, D) and their littermates without Cre (f/f, C). Seven days postvaccination, spleens were harvested to assess the frequency of OVA-specific CD8+ T cells (TetOVA+CD44hi). (E and F) Ex vivo restimulation with the OVA peptide was done to assess the percentage of IFNγ-producing CD8+ T cells. (G) CD8 T cell counts in the spleen of Δ/Δ and f/f mice, 7 d after vaccination at different CTs. (H) Percentage of Vβ5+ cells among CD8+CD44hiTetOVA+ cells. Representative FACS plots are shown in SI Appendix, Fig. S4. Data are pooled from 3 (C and D) or 2 independent experiments (E–H) with similar results, n = 6 to 10 mice/group. Data are shown for individual mice, as well as mean ± SEM. In C–F, rhythmicity was assessed by cosine fits, with curves indicated when significant (C) or a trend (E, dotted line); no curve indicates nonsignificant cosine fit. The P value for the cosine fits is indicated within each graph. One-way (C–F) or 2-way (G and H) ANOVA, with Bonferroni post hoc test where applicable. *P < 0.05, ***P < 0.001. See SI Appendix, Statistical Details for more information on statistics.
Circadian Control of Pathways Involved in CD8 T Cell Activation.
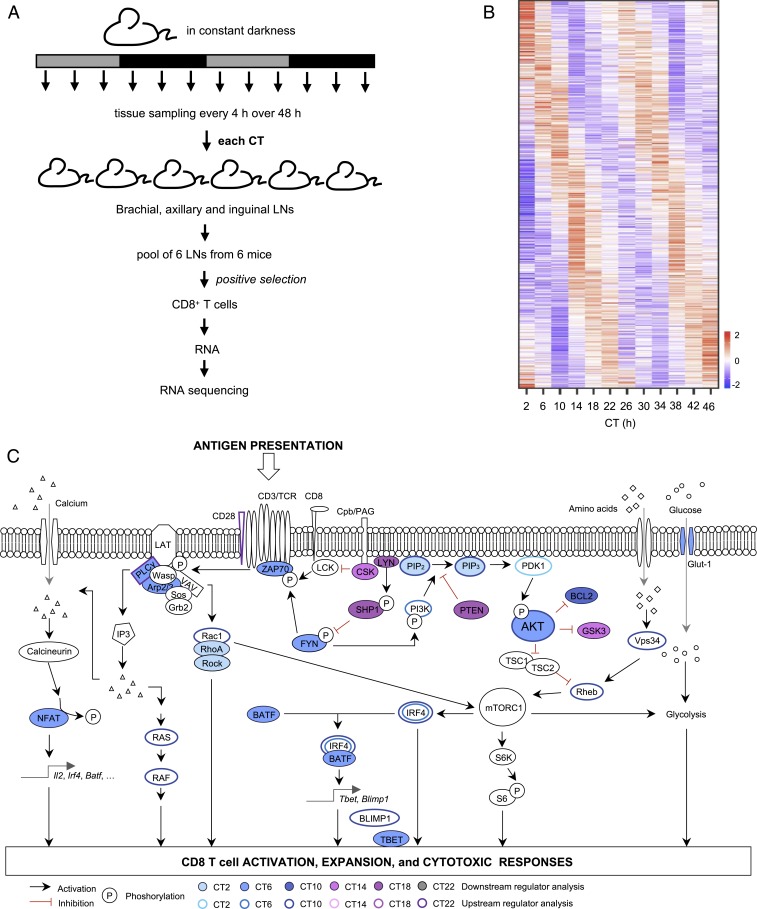
To address the T cell-intrinsic rhythms underlying the circadian variation of their response to Ag presentation, we decided to analyze gene expression rhythms before vaccination. We reasoned that since there is a difference of T cell response that depends on the time of vaccination, and since the clock intrinsic to CD8 T cells is necessary for this rhythm, there should be aspects of unstimulated CD8 T cells that present a circadian rhythm, making them more predisposed to be either activated or inhibited at certain times of the day. To address this, we characterized circadian rhythms in the genome-wide gene expression levels in CD8 T cells at steady state. We isolated CD8 T cells from lymph nodes harvested from mice kept in constant darkness after entrainment to a LD cycle, every 4 h over 48 h (Fig. 4A). The purity of the CD8 T cells was between 95.6% and 98.9% in all samples (SI Appendix, Fig. S5A). We extracted the RNA and performed RNA sequencing.
Fig. 4.
Circadian control of pathways involved in CD8 T cell activation. (A) Experimental protocol for the transcriptomic experiment. (B) Heat map of phase-sorted rhythmic transcripts in CD8+ T cells at steady state in C57BL/6J mice. CD8+ T cells were isolated by positive selection from lymph nodes harvested from mice every 4 h over 48 h in constant darkness. Rhythmic transcripts were determined in RNA sequencing data using RAIN with cutoff value set to FDR < 0.1. At this FDR cutoff, all uncorrected P values were below 0.006. (C) Schematic representation of the T cell receptor signaling pathways, and color coding of upstream and downstream regulators identified using GenExplain platform.
Using rhythmicity analysis incorporating nonparametric methods (RAIN), a nonparametric method to detect circadian rhythmicity in time series (20), we found that 5.9% (n = 786) of protein-coding transcripts show a significant circadian rhythm (false discovery rate [FDR] < 0.1) (Fig. 4B; full list of genes in Dataset S1). Among these rhythmically expressed genes were several core clock genes, such as Per2, Nr1d1, Rora, and Dbp, whose phase relationships are similar to those in other tissues. Other clock genes, such as Per1, Per3, Cry1, Cry2, and Bmal1, showed a hint of a rhythm but did not pass our stringent statistical cutoff for rhythmicity (SI Appendix, Fig. S5B). Gene ontology analyses revealed that gene categories related to immune responses and to catabolic processes were enriched among the rhythmic genes (SI Appendix, Fig. S6A). The phases of the rhythmic transcripts, as determined by cosinor analysis (Dataset S2), were nonuniformly distributed around the 24-h cycle (P < 0.01; Rao’s spacing test), with many transcripts peaking around the middle of the day and the middle of the night (SI Appendix, Fig. S6B).
To assess the pathway enrichment in the rhythmic gene list, we used the GenExplain platform (21). For the groups of rhythmic genes peaking at each of the different time points, we inferred the downstream pathways, as well as the upstream regulators and the transcription factors with enriched binding sites around the rhythmic genes. The analysis showed that many predicted rhythmic regulators are downstream of the TCR (Fig. 4C and Dataset S3). Interestingly, many regulators known to be related to the activation of TCR-dependent pathways were enriched at CT6 or CT10, the time at which DC-OVA immunization led to the highest T cell expansion (Figs. 1A and 3C). In particular, the enriched regulators and pathways included the TCR-associated ZAP70, as well as regulators of the AKT and mTOR pathways (including regulators of AKT PI3K and PDK1, and mTOR activator GTPases Rheb, Rac1, and RhoA). In contrast, regulators known to regulate TCR-dependent pathways negatively (e.g., PTEN and SHP-1) were enriched at CT14 and CT18 (Fig. 4C), i.e., when DC-OVA vaccination led to weaker T cell expansion (Fig. 3C). Further, there was an enrichment in the subjective day of genes with binding sites for transcription factors IRF4 and BLIMP1, which are known to be involved in T cell activation and proliferation (Fig. 4C and SI Appendix, Fig. S6C).
The Circadian Regulation of CD8 T Cell Response to Vaccination Occurs at the Early Stages of T Cell Activation.
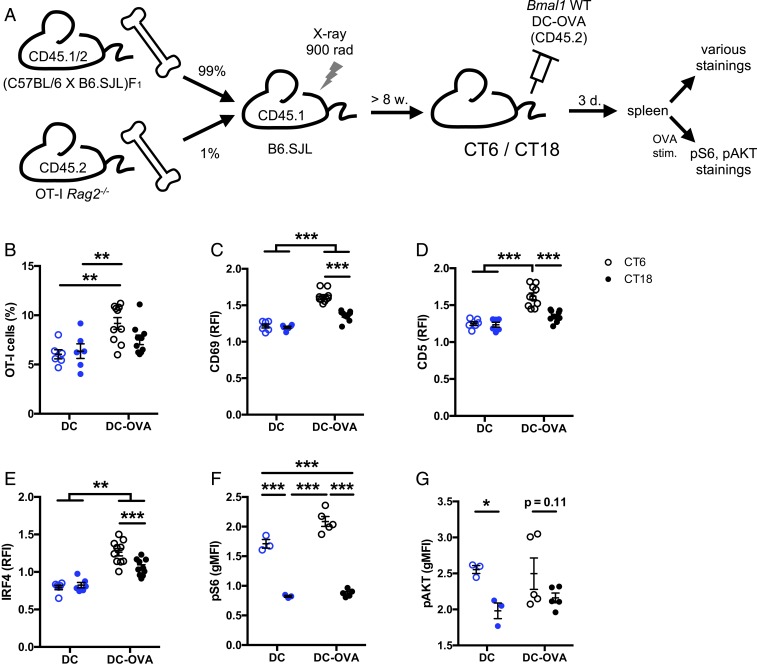
To assess whether the circadian variation of the CD8 T cell response to vaccination can be detected at early stages after vaccination, we generated mice in which we could follow the early response of the OVA-specific T cells bearing the OT-I TCR transgene. To do so, we used a mixed bone marrow chimera approach to increase the frequency of OVA-specific T cells, a prerequisite to follow the early response. B6.SJL mice were irradiated in order to eliminate hematopoietic cells expressing the congenic marker CD45.1. These mice were grafted with a mix of 1% bone marrow cells from OT-I Rag2−/− mice expressing the congenic marker CD45.2 and 99% bone marrow cells from F1 mice expressing both CD45.1 and CD45.2 (Fig. 5A). After bone marrow reconstitution, the mice were vaccinated at CT6 or CT18 with DC-OVA (or unloaded DCs), and several parameters were analyzed in the spleen at day 3 postvaccination (Fig. 5A, gating strategy in SI Appendix, Fig. S7A). At this early stage of the response, an expansion was already noticeable for mice vaccinated at CT6 (with significantly more OVA-specific OT-I cells in mice vaccinated with DC-OVA than with unloaded DCs), but not at CT18, suggesting that a time-dependent difference was already happening (Fig. 5B). There was no difference of apoptotic OVA-specific CD8 T cells, ruling out apoptosis as the mechanism behind the circadian rhythm (SI Appendix, Fig. S7 B and C). As expected, a significantly increased size of OVA-specific CD8 T cells and a down-regulation of surface CD8 were observed, consistent with cell activation after vaccination at both CTs (SI Appendix, Fig. S7 D and E). Given the importance of the metabolic regulation of T cells for their activation and proliferation, we tested the expression of CD71 and CD98, 2 metabolic markers involved in the T cell response. Neither CD71 nor CD98 presented a time-dependent level (SI Appendix, Fig. S8). However, as expected for activated T cells, we noted a significant increase of CD71 expression levels (SI Appendix, Fig. S8A).
Fig. 5.
The circadian variation of CD8 T cell response to vaccination occurs at the early stages of T cell response. (A) Experimental protocol. B6.SJL mice were irradiated followed by bone marrow graft (1% OT-I Rag2−/−, 99% F1). Eight weeks postgraft, BMDCs loaded with OVA peptide (DC-OVA) or unloaded (DC-LPS) were injected i.v. at either CT6 or CT18. Three days postvaccination, spleens were harvested to assess the response of OT-I CD8+ T cells. (B) Frequency of OT-I cells (CD45.1−CD45.2+) among CD8+ cells. Gating strategy is shown in SI Appendix, Fig. S7A. (C–G) Relative mean fluorescence intensity (ratio between OT-I [CD8+CD45.1−CD45.2+] cells and non OVA-specific CD8 T cells [CD8+CD45.1+CD45.2+] cells) for (C) CD69 and (D) CD5 surface stain, (E) IRF4 intracellular stain, and (F) phospho-S6 protein and (G) phospho-AKT after ex vivo restimulation with OVA. Representative FACS plots are shown in SI Appendix, Fig. S9. Data are pooled from 2 independent experiments with similar results, n = 6 to 10 mice/group. Two-way ANOVA with Bonferroni post hoc test where applicable. *P < 0.05, **P < 0.01, ***P < 0.001. For pAKT, since there was only an effect of time, post hocs were done only between CTs. See SI Appendix, Statistical Details for more information on statistics.
Interestingly, a higher activation profile of the OVA-specific OT-I T cells was observed after a vaccination done at CT6, compared to CT18. Indeed OVA-specific CD8 T cells expressed higher levels of CD69, CD5, and IRF4 following daytime vaccination (Fig. 5 C–E and SI Appendix, Fig. S9 A–C). CD69 and CD5 are 2 membrane glycoproteins well known to be up-regulated upon T cell activation (22). IRF4 (IFN regulatory factor 4) is a transcription factor critical for CD8 T cell expansion (23). Therefore, a higher expression of these proteins upon vaccination of the mice at CT6 in our experiments is consistent with a stronger T cell receptor activation as well as with our RNA-seq analyses (Fig. 4C and SI Appendix, Fig. S6C).
Since our transcriptomic analyses had suggested a daytime activation of many components of the AKT and mTOR pathways (Fig. 4C), we then looked at these pathways. Splenocytes of the same mice were cultured and restimulated with the OVA peptide and then stained for the phosphorylated form of S6, a target of mTOR: a higher phosphorylation was observed after a vaccination at CT6, compared to CT18 (Fig. 5F and SI Appendix, Fig. S9D). A similar time-dependent difference appears to exist for AKT phosphorylation, although the difference reached statistical significance only for the unloaded DC-vaccinated mice, and not with the DC-OVA–vaccinated mice (Fig. 5G and SI Appendix, Fig. S9E).
Interestingly, the CT6/CT18 difference in S6 and AKT phosphorylation following OVA stimulation of splenocytes from the vaccinated chimeras was observed both in the DC-OVA–vaccinated mice and for the unloaded DCs (which have not encountered the Ag) (Fig. 5 F and G and SI Appendix, Fig. S9 D and E). This suggested that even before Ag presentation, T cells in the subjective day (CT6) are more primed for TCR-dependent AKT and mTOR pathway activation than T cells in the subjective night (CT18). Therefore, we harvested spleens of OT-I mice at CT6 and CT18, mixed them 1:1 with splenocytes from B6.SJL mice, and stimulated them ex vivo with OVA. A CT6/CT18 difference in both S6 and AKT phosphorylation was observed in the OVA-responsive OT-I cells, but not in the B6.SJL cells, mirroring what was found in spleens of chimeric mice (Fig. 6 and SI Appendix, Fig. S10).
Fig. 6.
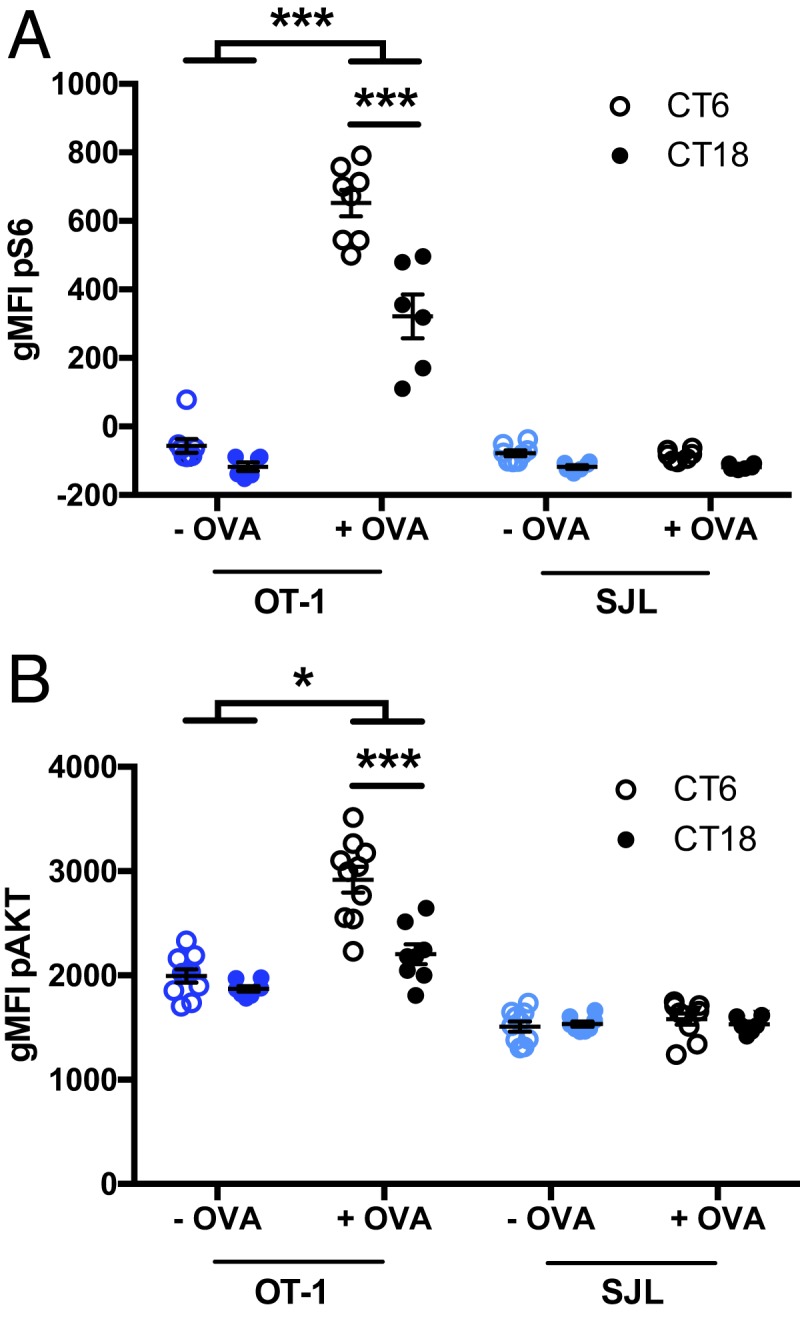
The AKT and mTOR pathways are more sensitive to T cell receptor stimulation at CT6 than CT18. Splenocytes were collected from OT-I Rag2−/− mice and from B6.SJL mice at CT6 or CT18 and mixed 1:1. Cells were stimulated with OVA for 30 min to assess the phosphorylation of S6 (A) and for 60 min to assess the phosphorylation of AKT (B). The analysis was done for OT-I and B6.SJL cells independently. Representative FACS plots are shown in SI Appendix, Fig. S10. Data are pooled from 2 mice per time point with at least 3 replicates per group and are representative of 4 and 2 experiments with similar results, for pS6 and pAKT, respectively. Two-way ANOVA (for each cell type independently), with Bonferroni post hoc test where applicable. *P < 0.05, ***P < 0.001; there was no interaction or effect of time or treatment for B6.SJL cells. See SI Appendix, Statistical Details for more information on statistics.
Altogether, our transcriptomic, in vivo vaccination and ex vivo stimulation results show that the circadian variation observed in CD8 T cell response to DC-OVA can be observed early after vaccination, and involves factors associated with TCR-dependent T cell activation such as IRF4, AKT, and mTOR, pathways which are primed to respond more efficiently in naïve CD8 T cells in the daytime, compared to nighttime.
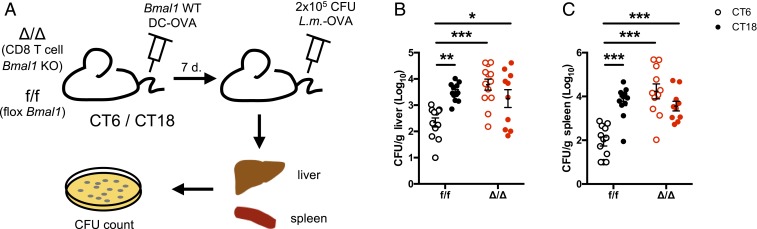
The CD8 T Cell Clock Underlies a Day–Night Difference in the Control of an Infectious Challenge following DC-OVA Vaccination.
To confirm the functional impact of the CD8 T cell clock on the ability to control an infectious challenge, 7 d after DC-OVA (or unloaded DCs) vaccination, CD8 T cell-specific Bmal1 KO mice and WT littermates were challenged with a lethal dose of OVA-expressing Listeria monocytogenes (L.m.-OVA) (Fig. 7A). Three days later, the level of infection was assessed by quantifying the bacterial load in the liver and the spleen of infected mice. WT mice had fewer bacteria in the liver and the spleen after a DC-OVA vaccination at CT6 compared to CT18 (Fig. 7 B and C). The circadian difference was abolished in the CD8 T cell-specific Bmal1 KO mice (Fig. 7 B and C). This is in agreement with the stronger CD8 T cell response after a vaccination at CT6 compared to CT18, and thus, a better protection against L.m.-OVA in the former case. Thus, the CD8 T cell clock controls the circadian variation of the ability of the CD8 T cell to respond and fight against an infectious challenge.
Fig. 7.
The CD8 T cell clock controls a day–night difference in the control of an infectious challenge following DC-OVA vaccination. (A) Experimental protocol. BMDCs loaded with OVA peptide were injected i.v. at either CT6 or CT18 in E8I-Cre+/− Bmal1f/f (Δ/Δ) and their littermates without Cre (f/f). Seven days postvaccination, mice were challenged with 2 × 105 CFU of L. monocytogenes-OVA i.v. at ZT8. (B) Livers and (C) spleens were harvested to assess bacterial load. Data are pooled from 3 independent experiments with similar results, n = 10 to 13 mice/group. Two-way ANOVA with Bonferroni test where applicable. *P < 0.05, **P < 0.01, ***P < 0.001. See SI Appendix, Statistical Details for more information on statistics.
Discussion
Here we reported that the response of CD8 T cells to vaccination with dendritic cells loaded with an antigen follows a circadian rhythm. This rhythm of a T cell functional response requires an intact clock within the CD8 T cells themselves, as does the control of a bacterial challenge. Analysis of the CD8 T cell transcriptome revealed an enrichment of genes and pathways involved in T cell activation in the daytime, i.e., the time of higher response to DC-OVA vaccination. Moreover, we showed that the CD8 T cell clock controls the magnitude of the CD8 T cell response at the early stage of the response to vaccination, with a day–night difference of key molecular players of TCR-dependent signaling pathways such as IRF4 and mTOR.
Our results add to a few recent studies that have documented an impact of the circadian system in the adaptive immune response and have addressed the respective roles of clocks in antigen presenting cells (e.g., DCs) and T cells. Hopwood et al. (24) showed a contribution of the DC clock to the Th1/Th2 balance in the response to parasitic worm infection. In our experiments, the DC clock was not essential to the circadian variation of the CD8 T cell response to vaccination. However, our data suggest that the DC clock acts on the migration of the DCs to the spleen after an i.v. injection. Hemmers and Rudensky (8) reported that T cell-specific deletion of Bmal1 had little effect on the T cell response in experimental autoimmune encephalomyelitis (EAE; a mouse model of multiple sclerosis) and antiviral and antibacterial T cell responses. However, Druzd et al. (13) also studied T cell-specific Bmal1 KO mice in the EAE model and found a difference in the clinical score, depending on the time of treatment, and a lack of such a day–night difference upon T cell clock ablation. Nguyen et al. (25) showed increased IFNγ+ T cells in the spleen after L. monocytogenes infection at zeitgeber time (ZT)8, compared to ZT0, a time-dependent difference that was not lost in myeloid-specific KO mice. Here, we showed an essential role of the clock intrinsic to CD8 T cells in their response to vaccination, as well as the time-dependent difference in the response to an infectious challenge with L. monocytogenes.
Discrepancies between studies could be due to the times selected for the assays, as the Bmal1 deletion may have an impact at some times of day and not at others. One example of this can be found in our data: in Figs. 3 and 7, the effects of Bmal1 deletion are seen for mice vaccinated at CT6 but not for mice vaccinated at CT18. Similarly, in Druzd et al.’s report (13), the effects of Bmal1 KO on EAE clinical score are observed for the ZT8-immunized mice but not for the ZT20-immunized mice. Using a similar KO, Hemmers and Rudensky did not observe an effect, but only 1 time point (not specified) was used (8). Similarly, an apparent absence of rhythm could sometimes be due to the selection of time points not at peak and trough of the response, making it important to test more than 2 time points. For example, in Fig. 3 of the present report, among the 4 time points tested, CT6 vaccination yielded a higher response than at any other time point. Had we chosen other CTs than CT6 and 18 for our 2 time point experiments, we would have missed the time-dependent variation. In this regard, it is interesting to note that Nguyen et al. (25) tested the cytokine response to L. monocytogenes infection at ZT0 and ZT8, and found a highest response when done at ZT8, consistent with the T cell responses in our model, whereas Hemmers and Rudensky did not see a difference in response to infection with this same bacterium done at ZT2 and ZT14 (8), which are the shoulders of our T cell response rhythm (Fig. 3).
The data from our group (the current study) and others (13) showing effects of T cell-specific Bmal1 gene KO on T cell functions suggest that a clock within these cells controls their functions. However, this view has been challenged by the relatively low amplitude of clock gene transcript rhythms in mouse or human T cells (our data of SI Appendix, Fig. S5 and refs. 7 and 8). However, despite this, sustained rhythms (over at least several days) were found using bioluminescent reporters in isolated human CD4+ (7) or mouse CD8+ T cells (our work), supporting endogenous and cell-autonomous clock function within T cells.
Recently, 2 groups showed a positive correlation between the circadian variation of the T and B cell counts in secondary lymphoid organs and the circadian variation of the efficacy of these cells in response to immunization with NP-CGG or with MOG35–55 peptide (12, 13). Also, Keller et al. (26) observed a rhythm of the percentage of T cells (using CD90.2 as a marker) in mouse spleens (although this was due to rhythmic total splenocytes, as the absolute counts of T cells did not present a rhythm). However, in our study, we can exclude the role of the proportion of T cells in secondary lymphoid organs. Indeed, based on the study published by Druzd et al. (13), the proportion of CD8 T cells at ZT5 and ZT17 in secondary lymphoid organs is similar. These 2 time points are close to the time points where we saw a difference in the circadian variation of the CD8 T cell response to vaccination. Moreover, we did not find a circadian variation of CD8 T cell numbers in the spleen (our vaccination model relies on CD8 T cells and Ag presentation occurs in the spleen).
Our previous work suggested a role of the clock in controlling T cell activation at the level of pathways downstream of the TCR (17). Indeed, TCR triggering showed a circadian rhythm of the proliferative response of T cells, whereas such a rhythm was not seen (CD8 T cells) or had a much lower amplitude (CD4 T cells) upon PMA/ionomycin stimulation, which activates more downstream pathways. Consistent with this, ZAP70 transcript and protein levels were rhythmic in lymph nodes, with a peak at CT8 (17), a time point close to that where we showed a higher CD8 T cell response to vaccination. A circadian regulation of TCR-dependent signaling pathways is supported by our CD8 T cell transcriptome data. We searched for pathways which can explain the set of rhythmic genes (upstream regulators) or could be differentially regulated by them (downstream pathways). Our analysis suggests an activation in the subjective day of TCR-dependent pathways and metabolic and cell proliferation regulators related to T cell response. In the daytime, the TCR-associated kinase ZAP70 was among the regulators identified in our analysis, which is consistent with our previous report (17). Also identified in the analysis, we found the transcription factor IRF4, which is known to be required for the CD8 T cell expansion after a viral and bacterial challenge as well as the maintenance of effector functions (23). Many components of the mTOR and AKT pathways were also inferred from the set of genes with peak expression in the subjective day. This included kinases upstream of AKT, PI3K, and PDK1, as well as the direct activator of mTOR, the GTPase Rheb. The activation of the mTOR pathway is critical to the switch of T cell metabolism to aerobic glycolysis, which is a hallmark of activated T cells (27). Consistent with this, the pathway analysis revealed GLUT-1, the main glucose transporter of activated T cells, among the regulators in the daytime. Interestingly, nighttime pathways showed many factors known to down-regulate TCR-dependent signaling and T cell activation/proliferation, such as SHP-1 and CSK (negatively acting on TCR proximal signaling) and PTEN (negatively regulating the PI3K–PIP3 pathway).
The transcriptomic data are in agreement with our in vivo experiments, with a higher activation profile of the CD8 T cells after a vaccination at CT6 compared to CT18, 3 d postvaccination, including higher expression of IRF4, and phosphorylation of mTOR target S6 and of AKT. Actually, even in nonvaccinated OT-I mice, the stimulation of OT-I cells with OVA peptide led to a higher S6 and AKT phosphorylation when splenocytes were harvested at CT6 than at CT18. Although additional studies will be required to uncover all of the mechanisms involved in the circadian regulation of CD8 T cells, our transcriptomic analysis and our vaccination and ex vivo stimulation assays all converge to show that the transcriptional program of CD8 T cells is shaped to make them more prone to respond strongly and efficiently to Ag presentation in the daytime, and to tone down this response at night.
Altogether, our work suggests that the CD8 T cell clock modulates the magnitude of the CD8 T cell response to vaccination by controlling the early T cell activation and the rhythmicity of signaling pathways mediating the effects of TCR triggering on T cell activation, proliferation, and acquisition of effector functions. This has an impact on the capacity of T cells to fight a bacterial infection. This study joins others in the description of the circadian control of the adaptive immune response. We selected a vaccination model using antigen-presenting cells already loaded with an Ag which allows bypassing the steps of Ag take-up and processing, and to focus on T cell-intrinsic functions. DC-based vaccination is already used as a therapeutic approach to activate tumor-specific T cells (28). Our study allows understanding of how CD8 T cell functional responses to Ag presentation are shaped by circadian clocks and bears promise to the improvement of therapies based on T cell responses.
Materials and Methods
Mice.
Animal use was in accordance with the guidelines of the Canadian Council of Animal Care and was approved by the Douglas Institute Facility Animal Care Committee. Details about the mouse strains are available in SI Appendix, Supplemental Materials and Methods.
Flow Cytometry, Bioluminescence Recordings, BMDC Migration Assays, Quantitative PCR, and Immunoblotting.
Details about these procedures are available in SI Appendix, Supplemental Materials and Methods.
DC-OVA Vaccination.
BMDCs were generated from bone marrow cells cultured in 6-well plates with complete RPMI medium 1640 supplemented with GM-CSF (500 U/mL, Invitrogen) and IL-4 (supernatant of P-815IL4 cells, prepared in-house) at days 0, 2, 3, and 6. Maturation of BMDCs was induced with 1 µg/mL LPS (Millipore Sigma) at day 6 followed by an incubation overnight with 2 µg/mL OVA257–264 peptide (SIINFEKL) (Midwest Biotech). At day 7, nonadherent BMDCs were collected and isolated using 14.7% Histodenz gradient. BMDC differentiation and activation was confirmed based on the expression of CD11c, I-Ab, Kb, and CD86. BMDC loading with the OVA257–264 peptide was assessed based on the expression of Kb-OVA. The BMDCs were collected prior to the first time point of vaccination and kept in complete RPMI medium supplemented with 10% FBS on ice with gentle shaking in order to keep the cells alive. Before the first time point of vaccination, we assessed the activation level (using antibodies against IAb, CD86, and Kb) and the peptide loading level (using an antibody against Kb-OVA) (29) of the BMDCs by flow cytometry. To control and confirm that the cells delivered by vaccination were similar throughout the experiment, we repeated the activation and loading phenotyping right after the last vaccination. Before each vaccination, we also controlled for mortality using Trypan blue to count the cells to adjust to the right amount. Note that all BMDCs used in the experiments are LPS activated as described above, but to avoid overloading the figures, they are labeled as DC-OVA or DC (unloaded DCs).
Mice were entrained to a LD cycle for at least 2 wk and then put in constant darkness (DD) for 3 d. During the second day in DD, mice were injected i.v. with 1.25 × 106 OVA-loaded and LPS-stimulated BMDCs (DC-OVA) or unloaded BMDCs as controls (DC) under dim red light at the indicated circadian times (CT0/CT12 are the times of lights on/off in the prior LD cycle). Depending on the experiments, the first time point of injection varied, to control for effect of order; the results were identical irrespective of order. After the third day in DD, mice were put back in LD until the end of the experiment, to prevent them from free-running and to be at different endogenous circadian times on the day of killing. Our previous report (17) showed that what matters for the T cell response rhythm is the time of vaccination on day 0, hence using DD for the day of vaccinations.
Spleens were harvested 7 d post DC-OVA vaccination at the same time points as for vaccination and processed as described (17). In short, splenocytes were stained with Kb-OVA tetramer and for CD8 and CD44 and analyzed by flow cytometry. In parallel, splenocytes were restimulated ex vivo with 2 µg/mL OVA peptide and 10 µg/mL brefeldin A for 6 h, fixed, and stained for IFNγ, followed by surface staining for CD8 and CD44. In some experiments, an antibody for the Vβ5 chain was also used.
L. monocytogenes-OVA Challenge.
Seven days post DC-OVA vaccination, mice were injected i.v. with a lethal dose (2 × 105 colony-forming units [CFU]) of L.m.-OVA bacteria at ZT8. Spleen and liver were harvested 3 d later and the colony-forming units were analyzed to determine the bacterial load in those organs (30).
RNA Sequencing.
Brachial, axillary, and inguinal lymph nodes (LNs) from C57BL/6J mice were collected during the second and the third days in DD (every 4 h over 48 h, starting at CT2). For each of the 12 time points, LNs from 6 mice were pooled. CD8 T cells of each pool of cells were isolated using EasySep mouse CD8a positive selection kit (STEMCELL Technologies, catalog no. 18953), and the percentage of CD8 T cells was checked by flow cytometry before and after isolation. Total RNA was extracted using TRIzol reagent (Invitrogen) followed by cleanup on RNeasy MinElute Cleanup Kit (Qiagen). LNs were used for this experiment because in our preliminary tests, the purity of the CD8 T cell isolation was better using LNs than spleens. In any case, since CD8 T cells are recirculating among secondary lymphoid organs, circadian gene expression in any of these tissues is informative for CD8 T cell function irrespective of their location in the peripheral immune system.
RNA sequencing was performed by the Genomic Platform of Institut de recherche en immunologie et en cancérologie (IRIC, Montreal). RNA integrity number (RIN) was determined using a Bioanalyzer Nano, and was above 8 for all samples. NextSeq High Output 2 × 75 pb was used, with a coverage of 67 million paired-end reads per sample. RNA-seq data were deposited in the Gene Expression Omnibus database (accession no. GSE128995) (31). To reduce noise due to low expression levels, only protein-coding transcripts with >0 fragments per kilobase per million fragments mapped (FPKM) at all time points were used for subsequent analyses (n = 13,351 transcripts).
Data were analyzed in R v3.5.1 (32). RAIN (20), a well-validated and established method for the detection of rhythms in time series with moderate numbers of time points (e.g., 12, as in our study), was used to determine circadian rhythmicity of protein-coding transcripts in the RNA-seq dataset. P values were corrected for multiple testing using the Benjamini–Hochberg method (FDR < 0.1 considered statistically significant; at this cutoff, all uncorrected P values were below 0.006). To determine the phase of the transcripts identified as rhythmic using RAIN, linearized cosinor analysis was performed using R with the following Eq. (33):
In this model, yjk is the expression level of gene k at time point j, ak is the fitted average expression (mesor), bk and ck are the cosinor coefficients, tj is the time after lights off at time point j (in hours), and εijk is the residual variance. The cosinor coefficients bk and ck were used to compute the phase and amplitude of each transcript as described in Refinetti (34).
Gene Ontology analysis to find enriched biological processes within the whole list of rhythmic transcripts was performed with the WEB-based GEne SeT AnaLysis Toolkit (WebGestalt) using default settings and the protein-coding genome as the background list (35).
The phases of the rhythmic transcripts determined by cosinor analysis were used to group them into 4-h bins based on their phase (CT2, 6, 10, 14, 18, 22). On the lists of rhythmic transcripts peaking in each of these time bins, we performed pathway analyses using the GeneXplain platform, with mouse Immune Parameters profile, using DNA-binding motifs of the TRANSFAC library (promoter positions −1,000 to +100) and signaling pathway information of the TRANSPATH database (http://genexplain.com) (21). For downstream regulators, we searched for Common Effector with TRANSPATH, and for pathways and master regulators upstream of the rhythmic genes, we did Enriched Upstream Analysis using TRANSFAC and TRANSPATH.
Bone Marrow Transplantation and Ex Vivo Stimulations.
Irradiated B6.SJL host mice (CD45.1) received 5 × 106 bone marrow cells (1% from OT-I Rag2−/− mice [CD45.2], 99% from [C57BL/6 × B6.JSL]F1 mice [CD45.1/CD45.2]). After reconstitution, DC-OVA or DC-LPS vaccination was done at CT6 or CT18. Three days postvaccination at the same time points as vaccination, spleens were harvested and splenocytes stained and analyzed by flow cytometry. To assess phosphorylated S6 and AKT, splenocytes were restimulated with OVA ex vivo. In other experiments, OT-I Rag2−/− and B6.SJL splenocytes (1:1) were stimulated ex vivo with OVA and stained for CD8, CD45.1, CD45.2, pS6, and pAKT. See SI Appendix, Supplemental Materials and Methods for details.
Statistical Analysis.
Statistical analyses were done with GraphPad Prism. Circadian variation was tested by fitting a cosine wave equation y = B + (A * cos (2 * π * (x − Ps)/24)) on data, where B is the baseline, A is the amplitude, Ps is the phase shift, with a fixed 24-h period; significance was determined by F-test. Statistical analysis for differences between time points and genotypes was done using 2-way ANOVA with Bonferroni post hoc test where applicable: all pairwise comparisons were performed when there was a significant interaction of the factors, whereas when there was only an effect for 1 of the factors, only pairwise comparisons for this factor were performed. Student’s t tests were used for experiments comparing only 2 time points or groups. A statistically significant difference was assumed when P < 0.05. Complete statistical information can be found in SI Appendix, Statistical Details.
Supplementary Material
Acknowledgments
The authors thank the members of the N.C. and N.L. laboratories for helpful discussions, technical help, and materials. We are grateful to Dr. Florian Storch for Bmal1 KO mice, Drs. Michel Tremblay and Kelly Pike for the E8I-Cre mice, Dr. Simona Stäger and Aymeric Fabié for the (C57BL/6 × B6.JSL)F1 mice, the Douglas Institute animal care staff (Ève-Marie Charbonneau, Geneviève Hamel, Ève Saint-Pierre Lussier), and Martine Dupuis and Tanya Koch for technical support (for flow cytometry and irradiator platform, respectively). This study was supported by a grant from the Canadian Institute of Health Research (MOP-119322, to N.C. and N.L.), a graduate scholarship from the University of Montreal (to C.C.N.), and a postdoctoral fellowships from the Fonds de Recherche du Québec–Santé (to L.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: RNA-seq data were deposited in the Gene Expression Omnibus database (accession no. GSE128995).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1905080116/-/DCSupplemental.
References
- 1.Dibner C., Schibler U., Albrecht U., The mammalian circadian timing system: Organization and coordination of central and peripheral clocks. Annu. Rev. Physiol. 72, 517–549 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Takahashi J. S., Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 18, 164–179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang R., Lahens N. F., Ballance H. I., Hughes M. E., Hogenesch J. B., A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc. Natl. Acad. Sci. U.S.A. 111, 16219–16224 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Curtis A. M., Bellet M. M., Sassone-Corsi P., O’Neill L. A., Circadian clock proteins and immunity. Immunity 40, 178–186 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Labrecque N., Cermakian N., Circadian clocks in the immune system. J. Biol. Rhythms 30, 277–290 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Scheiermann C., Gibbs J., Ince L., Loudon A., Clocking in to immunity. Nat. Rev. Immunol. 18, 423–437 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Bollinger T., et al. , Circadian clocks in mouse and human CD4+ T cells. PLoS One 6, e29801 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hemmers S., Rudensky A. Y., The cell-intrinsic circadian clock is dispensable for lymphocyte differentiation and function. Cell Rep. 11, 1339–1349 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Born J., Lange T., Hansen K., Mölle M., Fehm H. L., Effects of sleep and circadian rhythm on human circulating immune cells. J. Immunol. 158, 4454–4464 (1997). [PubMed] [Google Scholar]
- 10.Shimba A, et al. (2018) Glucocorticoids drive diurnal oscillations in T cell distribution and responses by inducing interleukin-7 receptor and CXCR4. Immunity 48, 286–298.e6. [DOI] [PubMed] [Google Scholar]
- 11.Dimitrov S., et al. , Cortisol and epinephrine control opposing circadian rhythms in T cell subsets. Blood 113, 5134–5143 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki K., Hayano Y., Nakai A., Furuta F., Noda M., Adrenergic control of the adaptive immune response by diurnal lymphocyte recirculation through lymph nodes. J. Exp. Med. 213, 2567–2574 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Druzd D., et al. , Lymphocyte circadian clocks control lymph node trafficking and adaptive immune responses. Immunity 46, 120–132 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernandes G., Halberg F., Yunis E. J., Good R. A., Circadian rhythmic plaque-forming cell response of spleens from mice immunized with SRBC. J. Immunol. 117, 962–966 (1976). [PubMed] [Google Scholar]
- 15.Kaplan M. S., et al. , Circadian rhythm of stimulated lymphocyte blastogenesis. A 24 hour cycle in the mixed leukocyte culture reaction and with SKSD stimulation. J. Allergy Clin. Immunol. 58, 180–189 (1976). [DOI] [PubMed] [Google Scholar]
- 16.Esquifino A. I., Selgas L., Arce A., Maggiore V. D., Cardinali D. P., Twenty-four-hour rhythms in immune responses in rat submaxillary lymph nodes and spleen: Effect of cyclosporine. Brain Behav. Immun. 10, 92–102 (1996). [DOI] [PubMed] [Google Scholar]
- 17.Fortier E. E., et al. , Circadian variation of the response of T cells to antigen. J. Immunol. 187, 6291–6300 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Cuesta M., Boudreau P., Dubeau-Laramée G., Cermakian N., Boivin D. B., Simulated night shift disrupts circadian rhythms of immune functions in humans. J. Immunol. 196, 2466–2475 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Yoo S. H., et al. , PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. U.S.A. 101, 5339–5346 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thaben P. F., Westermark P. O., Detecting rhythms in time series with RAIN. J. Biol. Rhythms 29, 391–400 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matys V., et al. , TRANSFAC and its module TRANSCompel: Transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 34, D108–D110 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang N., Bevan M. J., CD8(+) T cells: Foot soldiers of the immune system. Immunity 35, 161–168 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Man K., et al. , The transcription factor IRF4 is essential for TCR affinity-mediated metabolic programming and clonal expansion of T cells. Nat. Immunol. 14, 1155–1165 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Hopwood T. W., et al. , The circadian regulator BMAL1 programmes responses to parasitic worm infection via a dendritic cell clock. Sci. Rep. 8, 3782 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nguyen K. D., et al. , Circadian gene Bmal1 regulates diurnal oscillations of Ly6C(hi) inflammatory monocytes. Science 341, 1483–1488 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keller M., et al. , A circadian clock in macrophages controls inflammatory immune responses. Proc. Natl. Acad. Sci. U.S.A. 106, 21407–21412 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pollizzi K. N., Powell J. D., Regulation of T cells by mTOR: The known knowns and the known unknowns. Trends Immunol. 36, 13–20 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palucka K., Banchereau J., Dendritic-cell-based therapeutic cancer vaccines. Immunity 39, 38–48 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Porgador A., Yewdell J. W., Deng Y., Bennink J. R., Germain R. N., Localization, quantitation, and in situ detection of specific peptide-MHC class I complexes using a monoclonal antibody. Immunity 6, 715–726 (1997). [DOI] [PubMed] [Google Scholar]
- 30.Mathieu M., et al. , CD40-activated B cells can efficiently prime antigen-specific naïve CD8+ T cells to generate effector but not memory T cells. PLoS One 7, e30139 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cermakian N., Labrecque N., Nobis C. C., Dubeau Laramée G., The circadian transcriptome of mouse CD8+ T cells. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE128995. Deposited 28 March 2019.
- 32.Ihaka R. G. R., R: A language for data analysis and graphics. J. Comput. Graph. Stat. 5, 299–314 (1996). [Google Scholar]
- 33.Cornelissen G., Cosinor-based rhythmometry. Theor. Biol. Med. Model. 11, 16 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Refinetti R., Absence of circadian and photoperiodic conservation of energy expenditure in three rodent species. J. Comp. Physiol. B 177, 309–318 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Wang J., Vasaikar S., Shi Z., Greer M., Zhang B., WebGestalt 2017: A more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 45, W130–W137 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.