Significance
Models have predicted that drier climates could reduce the amount of evolutionary history contained within ecological communities (i.e., phylogenetic diversity), but clear empirical evidence is still lacking. We report a drought-induced erosion of the phylogenetic diversity of grassland plant communities. During a long period of drier than normal growing seasons, many species, especially those with drought-intolerant functional traits, were lost from communities and contributed to the declines in phylogenetic diversity. Our study represents one of the first demonstrations that climate-induced loss of diversity may extend beyond the level of the species. However, our study also indicates the positive value of physically and biologically complex landscapes as refuges for maintaining biodiversity in a changing world.
Keywords: aridification, climate change, drought, evolutionary history, functional traits
Abstract
While climate change has already profoundly influenced biodiversity through local extinctions, range shifts, and altered interactions, its effects on the evolutionary history contained within sets of coexisting species—or phylogenetic community diversity—have yet to be documented. Phylogenetic community diversity may be a proxy for the diversity of functional strategies that can help sustain ecological systems in the face of disturbances. Under climatic warming, phylogenetic diversity may be especially vulnerable to decline in plant communities in warm, water-limited regions, as intensified water stress eliminates drought-intolerant species that may be relicts of past wetter climates and may be distantly related to coexisting species. Here, we document a 19-y decline of phylogenetic diversity in a grassland community as moisture became less abundant and predictable at a critical time of the year. This decline was strongest in native forbs, particularly those with high specific leaf area, a trait indicating drought sensitivity. This decline occurred at the small spatial scale where species interact, but the larger regional community has so far been buffered against loss of phylogenetic diversity by its high levels of physical and biotic heterogeneity.
Climate change is exerting increasingly strong effects on biodiversity at multiple organizational levels and spatial scales (1, 2). In warm and water-limited climates, further warming may cause plant species to disappear as their drought tolerances are exceeded (3–5). In colder environments, where warming relaxes the dominant limitation to growth, slow-growing resident plant species may be overtaken by faster growing ecological generalists (6), and changes in community diversity may be further shaped by dispersal and herbivory (7, 8). How these and other climate-driven changes to plant communities will affect phylogenetic diversity, or biodiversity at deeper levels in the evolutionary tree of life than the species (9, 10), is as yet largely unknown. Phylogenetic community diversity may indicate the diversity in functional strategies (11) that stabilizes communities in the face of environmental perturbations (12). Therefore, a better understanding of the effects of climate change on phylogenetic diversity is critical to forecasting the future of biodiversity and ecosystem services.
Climate change will likely alter phylogenetic diversity nonrandomly because climate tolerances tend to be shared among close relatives (13, 14). Plant phylogenetic diversity tends to be highest where mesic lineages dependent on mild temperatures and abundant water are found (15). Within water-limited regions, community phylogenetic diversity may decrease along gradients of increasing water limitation, reflecting the strong filter imposed by aridity against older lineages with conserved traits conferring drought intolerance (16). Declines in community phylogenetic diversity thus may be expected within water-limited climates as these become effectively drier and lose their mesic-adapted species (3, 17, 18). Indeed, using contemporary relationships of phylogenetic diversity to climate and projecting into a warmer future, a recent model predicted that phylogenetic diversity would decline in warm and water-limited southern Europe, while showing more complex trends in northern Europe (19). Another modeling study predicted declines in phylogenetic diversity of eucalypts in Australia (20). However, empirical evidence of climate-driven declines in phylogenetic diversity is scarce.
Here, we report a case of climate-driven decline in phylogenetic community diversity. Our 19-y study encompassed an episode of “precipitation whiplash,” a long sequence of drier than average growing seasons punctuated by a bout of extreme rainfall, as is increasingly expected in California and other semiarid regions (21, 22). Winter precipitation, cloud cover, and humidity declined over most of this period, while other climate variables did not change significantly (23). From 2000 to 2018, we recorded community composition at 80 sites in a 2,800-ha grassland landscape with high heterogeneity of soils and species composition and little extrinsic disturbance. Species richness and diversity at the community (5-m2 site) scale declined over this time period, regardless of grazing or fire history, soil type, or the abundance and diversity of exotic species (23, 24). Native forb species with high mean values of specific leaf area (SLA; leaf area/dry mass), a trait linked to drought intolerance, were disproportionately lost. Precipitation in winter (December 1 to March 1), when annuals are present as small seedlings, declined over most of the period and was a highly significant driver of the plant community changes (23, 24). The winter of 2016 to 2017 was exceptionally wet. Nonetheless, diversity did not rebound as it had in earlier wet years such as 2005 to 2006. Experimental and observational analyses indicated that the series of dry winters had led to elevated seedling mortality, thus depleting the seed bank and diminishing community resilience (24).
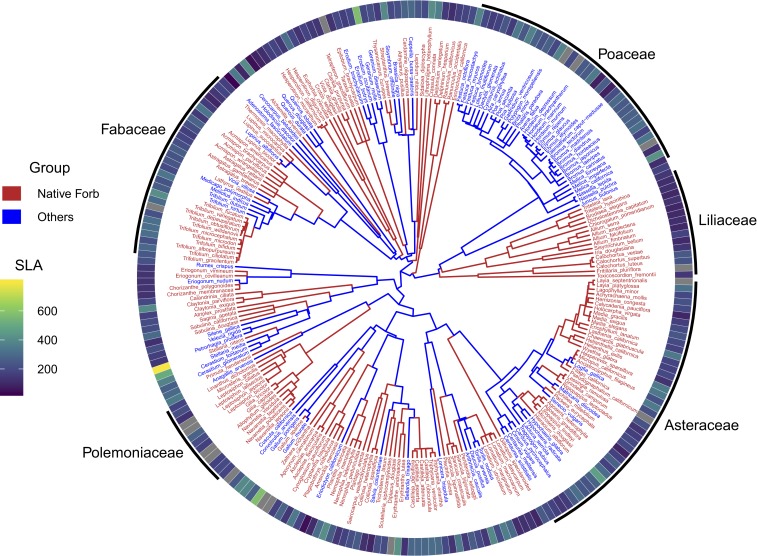
Given the already-observed community changes, we expected phylogenetic diversity to decline for 2 reasons: losses of native forbs relative to other species (many of which belong to the grass family, Poaceae; Fig. 1) and disproportionate loss of native forbs with high mean values of SLA, a trait that indicates drought intolerance and may be associated with older and less rapidly evolving lineages in the California flora (16, 25, 26). We calculated yearly phylogenetic diversity in these communities and analyzed its changes over the 19-y period in relation to spatial scale, precipitation patterns, and plant traits. We used phylogenetic mean pairwise distance (MPD) (27) and the phylogenetic component of phylogenetic community dissimilarity (PCDp) (28), which are mathematically independent of species-level richness (29) and dissimilarity (28), respectively. We tested whether phylogenetic diversity declined at the site (five 1-m2 quadrats combined), within-site (among five 1-m2 quadrats 10 m apart at each site), or regional (all sites combined) scale. We asked whether the decline in phylogenetic community diversity was related to precipitation, and whether it was associated with particular families or functional groups.
Fig. 1.
Phylogeny and SLA (square millimeters per gram) of study species (n = 248). Species names are colored according to status (native forbs and remaining species). The top 5 families are labeled.
Results and Discussion
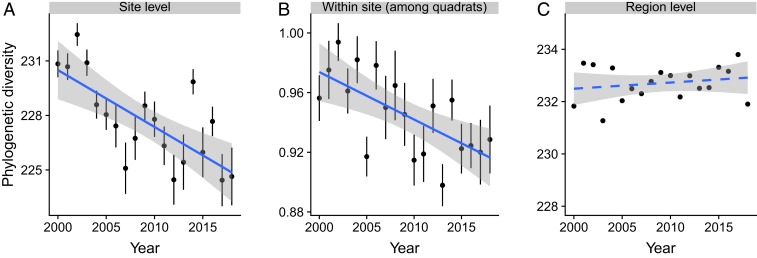
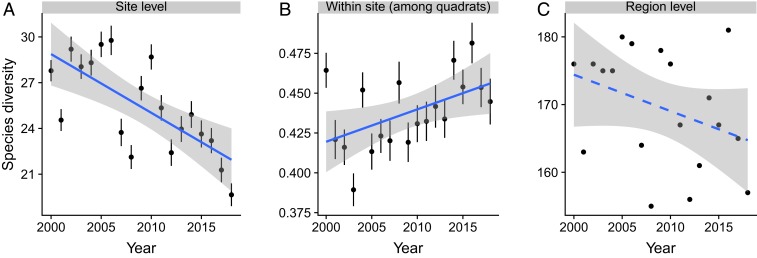
At the site scale, phylogenetic diversity (measured with MPD) declined over time with a rate of 0.317 million years per year (My/y; P < 0.001; Fig. 2A and SI Appendix, Table S1). Within sites, phylogenetic dissimilarity (measured with PCPp) among quadrats also declined (Fig. 2B and SI Appendix, Table S1), indicating that the 5 quadrats within a site tended to lose their unshared and distantly related species. Regionally, the MPD of species across all sites combined did not decline (Fig. 2C and SI Appendix, Table S1), indicating that the decline in site-level MPD resulted from losses of different species at different sites, and that the high biotic heterogeneity among the sites buffered regional MPD against decline. PCDp among sites within the region also did not diminish (SI Appendix, Table S1). Within-site, among-quadrat dissimilarity in species composition (i.e., within-site beta diversity), not investigated in previous studies, increased over time (Fig. 3B and SI Appendix, Table S2). This increase resulted from higher nestedness, or tendency of species-poor quadrats to support subsets of the flora of species-rich quadrats rather than to support unique species (i.e., turnover; SI Appendix, Table S2). Dissimilarity in species composition among sites within the region (i.e., among-site beta diversity) did not change (SI Appendix, Table S2).
Fig. 2.
Temporal trends in phylogenetic diversity at different spatial scales. (A) Site-level phylogenetic MPD (million years). (B) Within-site, among-quadrat PCDp (unitless). (C) Regional (all sites combined) MPD (million years). For each year, we plotted the mean and SE of phylogenetic diversity across all sites, which were then used to fit the regression lines. Solid lines indicate significant trends based on LMMS with all raw diversity values, while the dashed line indicates no significant changes over time. Test statistics are provided in SI Appendix, Table S1.
Fig. 3.
Temporal trends in taxonomic diversity at different spatial scales. (A) Site-level species richness. (B) Within-site, among-quadrat taxonomic diversity measured by Sorensen dissimilarity. (C) Regional (all sites) species richness. Solid lines indicate significant trends based on LMMS with all raw diversity values, while the dashed line indicates no significant changes over time. Test statistics are provided in SI Appendix, Table S2.
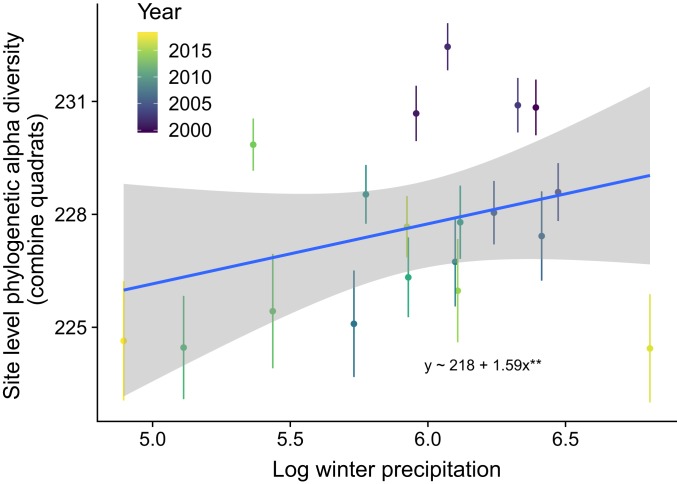
These changes in phylogenetic diversity (MPD) occurred in the course of declines in site-level species richness (Fig. 3A and SI Appendix, Table S2), as was documented in previous studies and linked statistically and experimentally to the effects of dry winters (23, 24). Site-scale MPD in this study was positively linked to winter rainfall (Fig. 4), consistent with earlier conclusions that dry winters were the main driver of loss of species richness in these communities (23, 24). As was previously found for species richness (23), the effects of winter rainfall were stronger in formerly grazed sites than those in ungrazed sites (P = 0.034; SI Appendix, Table S3). Although we observed a shift in the identity of dominant exotic grass species during the course of the study, this shift (which was unrelated to grazing history) was not significantly correlated with trends in site-level species richness or MPD.
Fig. 4.
Positive relationship between annual winter (December 1 to March 1) precipitation (millimeters) and site-level MPD. Winter precipitation of 2017 was much higher than that of other years. We considered it as an outlier, and thus did not fit a nonlinear model. **P < 0.01.
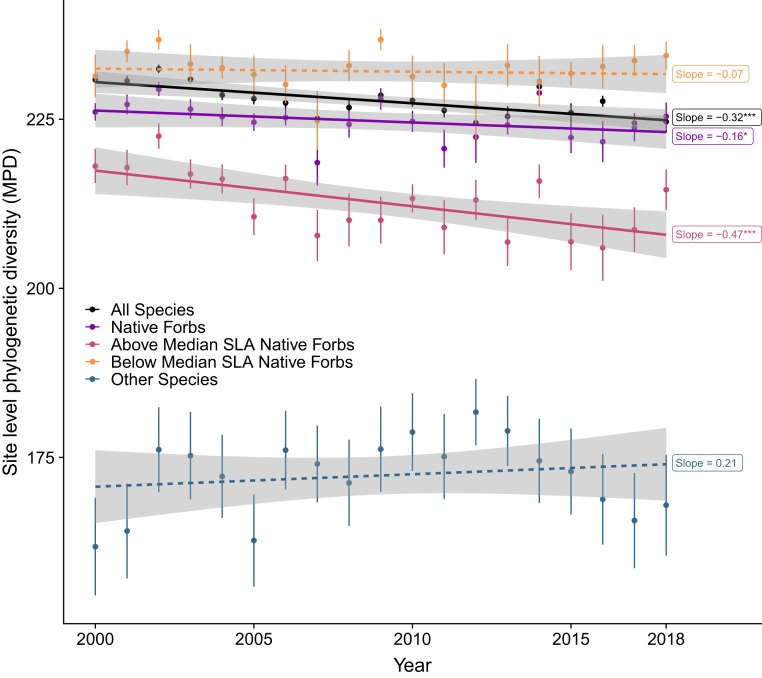
Because species richness declined in all groups of species (SI Appendix, Fig. S1), the within-site total branch lengths of all species, native forbs, and other species all declined over time, with rates of 21 My/y, 19.8 My/y, and 3.06 My/y, respectively (all P < 0.001; SI Appendix, Fig. S1). Along with all species, within-site MPD of native forbs also declined significantly, with a rate of 0.16 My/y, while the remaining species did not (Fig. 5).
Fig. 5.
Temporal trends in site-level MPD for different groups of species. Native forbs, especially those having higher than median SLA within sites, showed a significant trend of declines in phylogenetic diversity. Species with lower than median seed mass within sites showed a similar pattern as species with higher than median SLA; we did not plot this subgroup to avoid overlapping. “Other species” refers to species that are not native forbs. *P < 0.05; ***P < 0.001.
Dividing native forbs into those with mean values of SLA above and below the median SLA value for each site, the decline in site-level MPD was found only in species with above-median SLA (slope = −0.47 My/y, P < 0.001; Fig. 5). Also, the decline in site-level MPD was positively correlated with the decline in the community mean SLA of native forbs (Pearson’s ρ = 0.233, P = 0.037). When considering other functional traits (seed mass, leaf dry matter content, and plant height), only seed mass showed a pattern similar to SLA, in which only the native forbs with lower than median seed mass declined in site-level MPD (slope = −0.46 My/y, P = 0.007). High SLA and small seeds are both part of the fast-growing, drought-intolerant strategy, and are correlated in our dataset (log10-transformed seed mass and SLA: ρ = −0.31, P < 0.001). The removal of any family with >5 native forb species from the analysis did not reverse the decline in site-level MPD. Together, these results suggest that the decline in MPD was caused by the loss of native forbs, especially those with the correlated traits of high SLA and low-seed mass relative to their sites, and that these vulnerable species were well distributed across families as well as across the study region.
In summary, we observed a decline in the phylogenetic diversity of local communities that arose, in large part, from the loss of native forbs relative to other species, many of which were exotic grasses. Indeed, native forbs were entirely lost from the 3 sites at which the declines in phylogenetic diversity were strongest. The other principal cause of declining phylogenetic diversity was the relative loss of native forb species that were less drought-tolerant than their immediate neighbors, as evidenced by having higher values of SLA and lower seed mass than the site-level medians. This high-SLA and low-seed mass subgroup at each site was responsible for the loss of site-scale phylogenetic diversity within native forbs. Because sites varied considerably in their species composition and their median SLA and seed mass, the identities of the lost species were highly variable among sites, and local losses did not scale up to produce a decline in the phylogenetic diversity of the entire set of sites.
Our study illustrates that climate change in water-limited regions may erode not only the species richness of plant communities but the amount of evolutionary history they contain. Such a pattern of loss may become widespread because climatic warming is associated with increasing scarcity and unpredictability of growing-season soil moisture in many regions of the world (30, 31). While models have predicted declining phylogenetic diversity for plant and animal assemblages in water-limited regions (19), our study provides some of the first empirical support.
Phylogenetic diversity has been proposed as a useful proxy for diversity in both measured and unmeasured functional traits (11). Declining phylogenetic diversity thus suggests the potential for concomitant loss of diversity in functional traits and resource-use strategies (11). Recent work in our system supports this suggestion (32); our study communities have lost some of their functional diversity, as measured by the multivariate dispersion in a suite of correlated traits (SLA, carbon/nitrogen [C/N] ratio, leaf dry matter content, plant height, and seed mass; all have significant phylogenetic signal; SI Appendix, Table S4). Just as for phylogenetic diversity, drought-induced loss of species with high SLA and low-seed mass species appears to drive this loss of functional diversity (32), although other recent evidence suggests that root and seedling traits are also important (33). In any case, our study clearly establishes that evolutionary, trait-based, and species-based measures of community diversity are all vulnerable to erosion under climatic change.
Although losses of phylogenetic and functional diversity may undermine community functioning and resilience (11, 34), we have detected 2 potentially mitigating mechanisms in our study system. The first is functional redundancy, or the existence of multiple species with similar functional traits (34). During the wet winter of 2016 to 2017, while measures of community diversity did not recover from their long drought-induced declines, community mean values of SLA did recover, indicating that surviving high-SLA species increased strongly in abundance (24). The existence of multiple drought-intolerant, high-SLA species constitutes a form of functional redundancy that could help stabilize community properties, such as soil microbial activity (35), in the face of increased rainfall variability. The second mitigating mechanism is spatial heterogeneity. We found no declines in phylogenetic diversity at the scale of the whole landscape (i.e., all sites combined), despite the substantial declines observed at the local-community scale. As long as the pronounced physical heterogeneity and biotic heterogeneity of the landscape are not eroded by pollution, invasion, or other alterations, they will serve to slow larger scale losses and maintain sources for possible future recovery of declining species.
Materials and Methods
Study Site.
Our study is a continuation of work (23, 24) taking place at the University of California McLaughlin Reserve, a 2,776-ha facility at an elevation of 366 to 914 m in the Inner North Coast Range (N 38°52′, W 122°26′), largely surrounded by >105 ha of publicly owned wildlands. The climate is Mediterranean, with mean temperatures of 8 °C in January and 25 °C in July, and mean annual precipitation (all rainfall) of 620 mm. Substrates include fertile soils derived from volcanic and sedimentary rocks and infertile, Mg-rich, and nutrient-poor soils derived from serpentine rock. Grasslands consist mainly of annuals that germinate in fall (September to November) shortly after rains begin, are present as seedlings during winter (December to February), and flower in spring (March to May) except for <10 species that flower in summer. The most abundant species on fertile soils are ∼10 species of exotic (Eurasian) annual grasses, and the majority of other species are native and exotic annual forbs (nongrasses), with natives being most prevalent on the infertile serpentine soils where exotic grasses are sparse.
Prior analyses (23, 24, 32) found that (1) total species richness, and richness of nearly all functional groups, declined over the study period, with native annual forb richness declining fastest, and no functional group increased substantially in richness or cover; (2) community mean SLA declined as high-SLA species “blinked out” more frequently and “blinked in” less frequently over time than low-SLA species, and community mean values of the other traits did not change; (3) the loss of high-SLA species drove a loss of multivariate trait-based functional diversity; (4) soils, grazing and fire histories, and levels of exotic species cover did not qualitatively influence these trends; (5) winter (December to February) rainfall and winter and spring (March to May) humidity and cloud cover declined over most of the study period, while no other climatic variables showed significant time trends; (6) time series models indicated that declining winter rainfall was the driver of declining richness and mean SLA, although the positive influence of winter rainfall on diversity weakened over time; and (7) experimental evidence supported drought-induced seedling mortality as the main mechanism underlying these changes.
Data.
Grassland community composition was recorded beginning in 2000 at 80 highly heterogeneous sites widely dispersed around the reserve, with about half on serpentine and half on nonserpentine soils (23). Each site consisted of 5 permanently marked 1-m2 quadrats evenly spaced on a 40-m transect. Species composition was sampled annually in April and June; presence (2000 to 2018) and visually estimated maximum cover (2006 to 2018) were recorded for each species.
In 2010, species-specific mean values of SLA, plant height, seed mass, leaf dry matter content, and foliar C/N ratio were measured using standard protocols on 10 adult individuals per species on each soil type. Using these single-time measurements, community mean trait values were computed for each site in each year, both weighted by cover (2006 to 2018) and unweighted (2000 to 2018). In the present study, we found that all of these traits showed significant phylogenetic signal (SI Appendix, Table S4).
Data on annual and quarterly values of minimum, maximum, and mean precipitation were obtained from the Knoxville Creek weather station of the Western Regional Climate Center, near the center of the study landscape.
Response Measurements.
We standardized species names using the taxonomic name resolution service (36) and derived a phylogeny from an updated subset of the Open Tree of Life (37), which has been shown to produce robust estimates of phylogenetic community diversity (38). We calculated site-scale and regional-scale values of phylogenetic MPD (27), a metric that is independent of species richness (29). Note that MPD is essentially the same as phylogenetic species variation (39); the only difference is that phylogenetic species variation is scaled to have a range of 0 to 1. We also calculated within-site (among-quadrat) and among-site (within-region) dissimilarity or beta diversity using phylogenetic community dissimilarity (PCD), a metric of the variance in a hypothetical trait among species in one community that can be predicted by the values of species in another community (28). PCD can be partitioned into a nonphylogenetic component reflecting shared species (PCDc; highly correlated with Sorenson beta diversity) and a phylogenetic component reflecting evolutionary relationships of unshared species (PCDp). For each site and year, we calculated mean pairwise PCDp of all unique combinations of quadrats. At the regional level, we calculated the mean pairwise PCDp of all unique combinations of sites.
To compare trends in phylogenetic diversity with those in species-level diversity, we also calculated species (taxonomic) diversity at the same scales (i.e., species richness at the site and regional scales, and species-based dissimilarity [beta diversity] at the within-site [among-quadrat] and among-site [within-region] scales). For dissimilarities, we used pairwise values of the Sorensen index, and further partitioned it into its turnover and nestedness components (40).
Statistical Analysis.
To test for time trends of within-site dissimilarity and site-level MPD, we used linear mixed models (LMMs). We regressed average pairwise PCDp between quadrats within each site and MPD of each site against year, with site as a random term for the intercept, and accounted for potential temporal autocorrelations with first-order autoregressive residual structure. For within-region dissimilarity and region-level MPD, we used ordinary least squares linear models. For each year, we calculated average pairwise PCDp between sites and regional MPD based on species from all sites. We then regressed among-site PCDp (1 value per year) and region-level MPD against year.
To test potential drivers of declining site-level MPD, we looked at the following variables based on previous studies (23) in the same community: winter precipitation, grazing history (grazed or not), and soil types (serpentine or nonserpentine). We also examined 3 common exotic grasses (Avena fatua, Taeniatherum caput-medusae, and Bromus hordeaceus) that were recently found to have shifted in abundance over the study period. We used the first principal component of the occurrence values of these 3 species, which explained 71% of total variation. We used LMMs to study the effects of these variables on site-level MPD. We first fitted a full model with log-transformed winter precipitation, grazing history, soil type, first axis of the 3 exotic species, and all 2-way interactions between continuous variables and categorical variables as fixed terms; site was included as random terms for intercept and slopes of continuous variables. We then conducted model selections to derive the final model, which included log-transformed winter precipitation, grazing history, and their interactions as fixed terms, and site as a random term for intercept. To test whether declining winter precipitation drove changes in site-scale MPD across all sites (grazed and ungrazed), we regressed MPD against log-transformed winter precipitation in LMMs with site as random terms for the intercept and the slope.
To examine which groups of species were responsible for the changes in site-scale MPD, we repeated the site-scale model using (1) native forbs only, (2) native forbs above the median SLA for each site, (3) native forbs below the median SLA for each site [with both (2) and (3) paralleling the analysis of SLA and species loss in a study by Harrison et al. (23)], and (4) all other species. We also divided native forbs into above and below the median value of the other traits (height, seed mass, and leaf dry matter content, but not foliar C/N ratio given its relatively weak phylogenetic signal; SI Appendix, Table S4) and tested which groups’ site-scale MPDs have declined. A small fraction of species did not have SLA and seed mass measurements. Removing these species did not qualitatively change our results of the above analyses. We thus presented phylogenetic diversity results based on all species in the main text. To test whether any plant families were particularly influential, we repeated the model of site-scale MPD after sequentially removing each family with >5 species.
Data and Materials Availability.
All data used in this study have been deposited on figshare (DOI: 10.6084/m9.figshare.9747455).
Supplementary Material
Acknowledgments
Logistical support by C. Koehler, P. Aigner, and their staff at the University of California McLaughlin Reserve made this project possible. M. LaForgia assisted with database development and management. Valuable discussions and comments were provided by Anthony R. Ives, Marc W. Cadotte, and the members of the laboratory of Ben Baiser. The work was supported by National Science Foundation OPUS Award DEB-1748610 (to S.H.).
Footnotes
The authors declare no conflict of interest.
Data deposition: All data used in this study have been deposited on figshare (DOI: 10.6084/m9.figshare.9747455).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912247116/-/DCSupplemental.
References
- 1.Parmesan C., Ecological and evolutionary responses to recent climate change. Annu. Rev. Ecol. Evol. Syst. 37, 637–669 (2006). [Google Scholar]
- 2.Walther G.-R., Community and ecosystem responses to recent climate change. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 2019–2024 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrison S., Damschen E. I., Grace J. B., Ecological contingency in the effects of climatic warming on forest herb communities. Proc. Natl. Acad. Sci. U.S.A. 107, 19362–19367 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pauli H., et al. , Recent plant diversity changes on Europe’s mountain summits. Science 336, 353–355 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Slingsby J. A., et al. , Intensifying postfire weather and biological invasion drive species loss in a Mediterranean-type biodiversity hotspot. Proc. Natl. Acad. Sci. U.S.A. 114, 4697–4702 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gottfried M., et al. , Continent-wide response of mountain vegetation to climate change. Nat. Clim. Chang. 2, 111–115 (2012). [Google Scholar]
- 7.Kaarlejärvi E., Eskelinen A., Olofsson J., Herbivory prevents positive responses of lowland plants to warmer and more fertile conditions at high altitudes. Funct. Ecol. 27, 1244–1253 (2013). [Google Scholar]
- 8.Kaarlejärvi E., Eskelinen A., Olofsson J., Herbivores rescue diversity in warming tundra by modulating trait-dependent species losses and gains. Nat. Commun. 8, 419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nee S., May R. M., Extinction and the loss of evolutionary history. Science 278, 692–694 (1997). [DOI] [PubMed] [Google Scholar]
- 10.Mace G. M., Gittleman J. L., Purvis A., Preserving the tree of life. Science 300, 1707–1709 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Tucker C. M., Davies T. J., Cadotte M. W., Pearse W. D., On the relationship between phylogenetic diversity and trait diversity. Ecology 99, 1473–1479 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Ives A. R., Carpenter S. R., Stability and diversity of ecosystems. Science 317, 58–62 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Willis C. G., Ruhfel B., Primack R. B., Miller-Rushing A. J., Davis C. C., Phylogenetic patterns of species loss in Thoreau’s woods are driven by climate change. Proc. Natl. Acad. Sci. U.S.A. 105, 17029–17033 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis C. C., Willis C. G., Primack R. B., Miller-Rushing A. J., The importance of phylogeny to the study of phenological response to global climate change. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 3201–3213 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wiens J. J., Donoghue M. J., Historical biogeography, ecology and species richness. Trends Ecol. Evol. 19, 639–644 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Anacker B. L., Harrison S. P., Historical and ecological controls on phylogenetic diversity in Californian plant communities. Am. Nat. 180, 257–269 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Lavergne S., Molina J., Debussche M., Fingerprints of environmental change on the rare Mediterranean flora: A 115-year study. Glob. Change Biol. 12, 1466–1478 (2006). [Google Scholar]
- 18.Pfeifer-Meister L., et al. , Climate change alters plant biogeography in Mediterranean prairies along the West Coast, USA. Glob. Change Biol. 22, 845–855 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Thuiller W., et al. , Consequences of climate change on the tree of life in Europe. Nature 470, 531–534 (2011). [DOI] [PubMed] [Google Scholar]
- 20.González-Orozco C. E., et al. , Phylogenetic approaches reveal biodiversity threats under climate change. Nat. Clim. Chang. 6, 1110–1114 (2016). [Google Scholar]
- 21.Wang S.-Y. S., Yoon J.-H., Becker E., Gillies R., California from drought to deluge. Nat. Clim. Chang. 7, 465–468 (2017). [Google Scholar]
- 22.Swain D. L., Langenbrunner B., Neelin J. D., Hall A., Increasing precipitation volatility in twenty-first-century California. Nat. Clim. Chang. 8, 427–433 (2018). [Google Scholar]
- 23.Harrison S. P., Gornish E. S., Copeland S., Climate-driven diversity loss in a grassland community. Proc. Natl. Acad. Sci. U.S.A. 112, 8672–8677 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrison S. P., LaForgia M. L., Latimer A. M., Climate-driven diversity change in annual grasslands: Drought plus deluge does not equal normal. Glob. Change Biol. 24, 1782–1792 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Raven P. H., Axelrod D. I., Origin and Relationships of the California Flora (University of California Press, 1978). [Google Scholar]
- 26.Ackerly D. D., Community assembly, niche conservatism, and adaptive evolution in changing environments. Int. J. Plant Sci. 164 (suppl. 3), S165–S184 (2003). [Google Scholar]
- 27.Webb C. O., Ackerly D. D., Kembel S. W., Phylocom: Software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics 24, 2098–2100 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Ives A. R., Helmus M. R., Phylogenetic metrics of community similarity. Am. Nat. 176, E128–E142 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Miller E. T., Farine D. R., Trisos C. H., Phylogenetic community structure metrics and null models: A review with new methods and software. Ecography 40, 461–477 (2017). [Google Scholar]
- 30.Cook B. I., Ault T. R., Smerdon J. E., Unprecedented 21st century drought risk in the American Southwest and Central Plains. Sci. Adv. 1, e1400082 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang J., Yu H., Guan X., Wang G., Guo R., Accelerated dryland expansion under climate change. Nat. Clim. Chang. 6, 166–171 (2016). [Google Scholar]
- 32.Miller J. E. D., Li D., LaForgia M., Harrison S., Functional diversity is a passenger but not driver of drought-related plant diversity losses in annual grasslands. J. Ecol. 107, 2033–2039 (2019). [Google Scholar]
- 33.Harrison S., LaForgia M., Seedling traits predict drought-induced mortality linked to diversity loss. Proc. Natl. Acad. Sci. U.S.A. 116, 5576–5581 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCann K. S., The diversity-stability debate. Nature 405, 228–233 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Gravuer K., Eskelinen A., Nutrient and rainfall additions shift phylogenetically estimated traits of soil microbial communities. Front. Microbiol. 8, 1271 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boyle B., et al. , The taxonomic name resolution service: An online tool for automated standardization of plant names. BMC Bioinformatics 14, 16 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith S. A., Brown J. W., Constructing a broadly inclusive seed plant phylogeny. Am. J. Bot. 105, 302–314 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Li D., et al. , For common community phylogenetic analyses, go ahead and use synthesis phylogenies. Ecology, e02788 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Helmus M. R., Bland T. J., Williams C. K., Ives A. R., Phylogenetic measures of biodiversity. Am. Nat. 169, E68–E83 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Baselga A., Partitioning the turnover and nestedness components of beta diversity. Glob. Ecol. Biogeogr. 19, 134–143 (2010). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.