Significance
Living organisms depend on sensory systems to monitor changes in their surroundings and internal conditions. The physical and chemical signals detected by various sensory proteins must be properly processed and modulated in order to trigger prompt, precise, and coherent cellular responses. It is still puzzling at the molecular level how these input signals are perceived, transduced, and integrated into a finite number of signaling pathways inside the cells without information loss. Using dynamic crystallography and biochemistry, this work dissects the inner workings of a dual-sensor signaling protein that accomplishes a molecular logic OR in response to 2 distinct input signals. Findings provide structural insights into the operating mechanism of signal integration in modular signaling proteins.
Keywords: photoreceptor, signal integration, phosphorylation, molecular logic, dynamic crystallography
Abstract
Signal detection and integration by sensory proteins constitute the critical molecular events as living organisms respond to changes in a complex environment. Many sensory proteins adopt a modular architecture that integrates the perception of distinct chemical or physical signals and the generation of a biological response in the same protein molecule. Currently, how signal perception and integration are achieved in such a modular, often dimeric, framework remains elusive. Here, we report a dynamic crystallography study on the tandem sensor domains of a dual-sensor histidine kinase PPHK (phosphorylation-responsive photosensitive histidine kinase) that operates a molecular logic OR, by which the output kinase activity is modulated by a phosphorylation signal and a light signal. A joint analysis of ∼170 crystallographic datasets probing different signaling states shows remarkable dimer asymmetry as PPHK responds to the input signals and transitions from one state to the other. Supported by mutational data and structural analysis, these direct observations reveal the working mechanics of the molecular logic OR in PPHK, where the light-induced bending of a long signaling helix at the dimer interface is counteracted by the ligand-induced structural changes from a different sensor domain. We propose that the logic OR of PPHK, together with an upstream photoreceptor, implements a “long-pass” red light response distinct from those accomplished by classical phytochromes.
The ability to sense and integrate multiple environmental signals is critical for survival and adaptation of living organisms. At the molecular level, many modular signaling proteins, including photoreceptors and transmembrane chemoreceptors, are able to integrate signal perception and enzymatic response in the same protein framework (1–5). Yet, the underlying molecular mechanisms of allosteric regulation remain largely unknown. To study the molecular mechanism of signal integration in modular systems, we characterized a multidomain sensory histidine kinase from cyanobacterium Leptolyngbya sp. JSC-1 (6, 7), denoted PPHK (phosphorylation-responsive photosensitive histidine kinase). PPHK features 2 distinct sensor domains, where the N-terminal receiver (nREC) domain perceives a phosphorylation signal and the second bilin-binding GAF domain senses a light signal (Fig. 1A and SI Appendix, Fig. S1). As a photoreceptor, PPHK belongs to the phytochrome superfamily and undergoes reversible photoconversion between the green light-absorbing (Pg) state and the red light-absorbing (Pr) state (8, 9). The domain architecture of PPHK is highly representative of the sensory histidine kinase superfamily, including transmembrane chemoreceptors and mechanoreceptors (10) (Fig. 1A). Therefore, PPHK offers an excellent model system for structural dissection of the allosteric action in modular signaling proteins, where signals perceived by individual sensor domains are integrated to make coherent decisions (1, 3, 4).
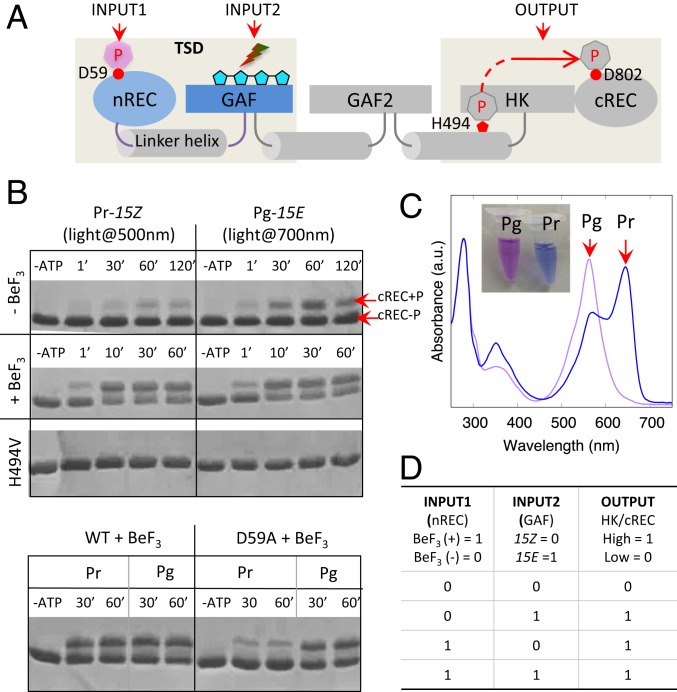
Fig. 1.
Signaling logic of dual-sensor PPHK. (A) Domain architecture of PPHK, in which modular domains are linked via long helices shown in cylinders. The nREC domain senses an upstream phosphorylation signal (INPUT1), and the bilin-binding GAF domain senses a red or green light signal (INPUT2). TSD denotes the truncated construct containing the tandem sensor domains (gray shade). Four linked pentagons represent the bilin chromophore (cyan). HK, histidine kinase. (B) Histidine kinase assays of wild-type (WT) and mutant PPHK in different signaling states. Control experiments with the single mutants (H494V and D59A) confirmed that Asp59 and His494 are the phosphorylation sites in the nREC domain and histidine kinase, respectively. ATP, adenosine 5′-triphosphate; BeF3, beryllofluride. (C) Absorption spectra show the reversible photoconversion between the Pg (λmax of ∼562 nm) and Pr (λmax of ∼644 nm) states, corresponding to purple and blue, respectively, in sample colors. (D) Truth table for the molecular logic “OR” gate. λmax, characteristic peak wavelength.
To interrogate the signaling logic of PPHK, we carried out PhosTag-based histidine kinase assays in 4 different input conditions (Fig. 1B). Specifically, we used beryllofluoride , a compound widely used for mimicking phosphorylation of the Asp residue, to introduce an input signal to the nREC domain (11–13). The output kinase activity of PPHK clearly responds to both and light signals. In the absence of , PPHK displays significantly higher kinase activity under red light (resulting in the Pg state) than under green light (resulting in the Pr state) (Fig. 1 B and C). In the presence of , however, the kinase activity is no longer light-dependent, although PPHK remains photoactive (Fig. 1B and SI Appendix, Fig. S2). In a negative control experiment, we made a single mutant, D59A, to disrupt the phosphorylation site, Asp59, which indeed abolishes the sensitivity and restores light-dependent responses in PPHK. Taken together, we demonstrate that PPHK is a dual-sensor histidine kinase regulated by both a chemical signal (phosphorylation) and a physical signal (light). If the output kinase activity is represented by the binary states 0 and 1 for low and high, respectively, PPHK accomplishes the logic OR gate (Fig. 1D).
In this work, we apply static and dynamic crystallography to investigate how the logic OR operation is achieved in this dual-sensor histidine kinase. We have determined the crystal structure of the tandem sensor domains of PPHK, which displays remarkable dimer asymmetry. We present a wealth of structural, biochemical, and mutational data to establish a mechanistic model that links the local and global structural changes to the kinase activation of the output domain. We show that the molecular logic is achieved in PPHK when phosphorylation-induced structural changes in one sensor domain counteract the light-induced allosteric effects from the other sensor domain in the helical spine.
Results
Crystal Structure of PPHK Reveals Remarkable Dimer Asymmetry.
The crystal structure of the 2 tandem sensor domains in PPHK (denoted PPHK-TSD) was determined at 1.95 Å resolution (ref. 14, Fig. 2A, and SI Appendix, Fig. S1 and Table S1). In comparison to the full-length protein, this truncated PPHK construct undergoes similar Pg/Pr photoconversion but renders a more stable Pr state (SI Appendix, Fig. S2 A and B). Single-crystal spectroscopy shows that PPHK-TSD is able to undergo Pg→Pr photoconversion in the crystalline state, although the extent of photolysis is much less compared with those in solution and the absorption spectrum of the Pg state is slightly blue-shifted in crystals (SI Appendix, Fig. S2C). PPHK forms a head-to-head parallel dimer, where the nREC and bilin-binding GAF domains juxtapose along a central helical spine at the dimer interface (Fig. 2A). The phycocyanobilin chromophore is covalently attached to Cys241 in the GAF domain and adopts the 15Ea configuration in the Pg state, confirmed by acid denaturation experiments (Figs. 2A and 3A and SI Appendix, Fig. S2B). The tandem sensor domains have minimal direct contacts with one another in contrast to the extensive dimer interface (buried surface area of 1,770 Å2), with long tethered linker helices spanning both domains. Two distinct sensory sites, namely, the phosphorylation site Asp59 in the nREC domain and the bilin chromophore in the GAF domain, are separated by ∼60 Å (SI Appendix, Fig. S1B).
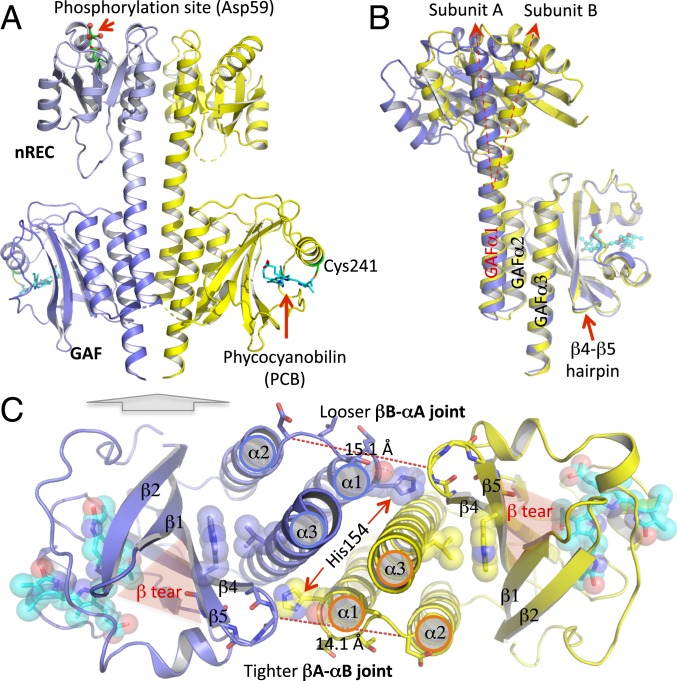
Fig. 2.
Crystal structure of the tandem sensor domains of PPHK reveals dimer asymmetry. (A) Ribbon diagram of the PPHK structure in a parallel dimer. In each subunit, the nREC (light blue) and GAF (blue) domains are connected via a long linker helix at the dimer interface. The phosphorylation site in the nREC domain is highlighted, where the acid side chain of Asp59 (green) forms hydrogen bonds with an Mg2+ ion in coordination with 3 water molecules (red sphere). The bilin chromophore (in cyan) embedded in the GAF domain is covalently attached to Cys241 (green). (B) Dimer asymmetry between subunits A (blue) and B (yellow) is evident in the diverging linker helices when the 2 monomer structures are aligned according to the GAF domain. (C) Bottom view of the PPHK dimer (indicated by a gray block arrow) illustrates the spatial organization of the GAFα1/α2/α3 helices at the dimer interface. Shaded red triangles highlight a unique feature, denoted “β-tear,” between strands β1 and β5 of the β-sheet in the GAF domain. The β4/β5 hairpin from one subunit is closely associated with the GAFα1/α2 turn from the partner subunit. The equivalent intersubunit distances (dashed red lines) show that such intersubunit contacts are not symmetrical, and the βA/αB joint renders shorter distances, and thus tighter interactions, than the βB/αA joint.
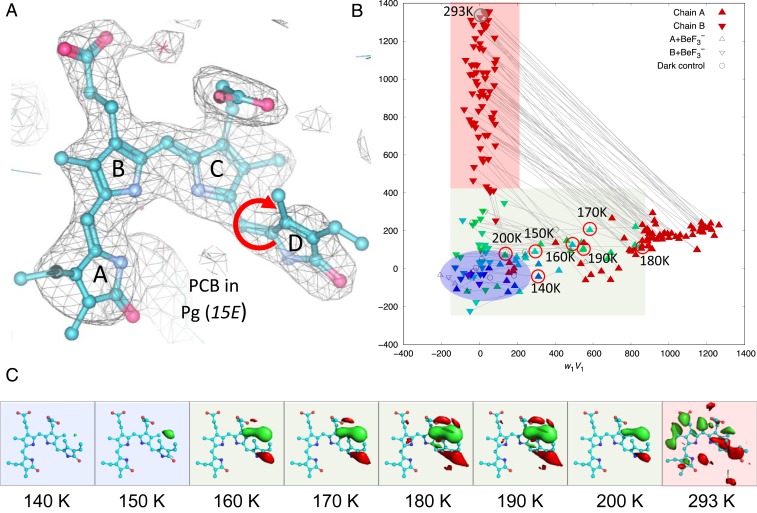
Fig. 3.
Light-induced local structural changes in the chromophore. (A) The 2Fo-Fc electron density map contoured at 1.2 σ shows that the bilin chromophore adopts the ZZEssa configuration in the Pg state. The circular red arrow marks the C15 = C16 double bond, where the photoisomerization occurs. (B) SVD of 112 difference (Fgreen/Fred) maps obtained at a cryogenic temperature and RT. The scatter plot of coefficient sets from the top 2 SVD components reveals that light-induced signals are temperature-dependent, and dimer asymmetry between subunits A (upward triangles) and B (downward triangles) further develops as the pump temperature rises from a low (blue) to high (green) cryogenic temperature and RT (red). (C) Fgreen/Fred difference densities (contoured at ±4 σ; green, positive density; red, negative density) show the rise and decay of the 15E/15Z photoisomerization signal as a function of temperature in subunit A. The difference map obtained by green-light illumination at RT (293 K) reveals an overall chromophore rotation in subunit B. Datasets corresponding to the featured maps in C are circled in B.
The PPHK structure shows remarkable dimer asymmetry evidenced by an angular displacement of ∼22° between the linker helices when the protomers are aligned according to the rigid GAF framework (Fig. 2B and SI Appendix, Fig. S3). These long linker helices display distinct local curvatures covarying along the helical spine, where the juxtaposed helical segments adjust to complement each other’s curvature (SI Appendix, Fig. S4A). Specifically, the linker helix in subunit B features one major bend near the middle, while its counterpart undulates twice, once in each sensor domain. The dimer asymmetry is further evident in the intersubunit joints, where the β4/β5 hairpin from the GAF core directly interacts with His154 from the C terminus of GAFα1 in the partner subunit (Fig. 2C and SI Appendix, Fig. S4B). In contrast to an extended loop conformation in subunit A, the GAFα1/α2 loop in B (denoted αB) is contracted, where Thr156 and Leu157 swap the side-chain dispositions relative to the β-hairpin of A (denoted βA) (Fig. 2C and SI Appendix, Fig. S4B). Both joints (denoted βA/αB joint and βB/αA joint, respectively) engage close Van der Waals (VDW) interactions with GAFα3, which constitutes a T-junction motif consisting of 3 key structural elements: β-hairpin, GAFα1/α2, and GAFα3 (Fig. 2C and SI Appendix, Figs. S4B and S5). Remarkably, many crystal structures of the GAF sensors also feature such a T-junction motif despite the distinct nature of their input signals (15–17) (SI Appendix, Fig. S5). We postulate that this conserved T-junction is important for signal coupling from the GAF core domain to the helical spine. Helix parameterization also reveals a shorter helical pitch in the nRECα5 segment of the linker helix in subunit B (SI Appendix, Fig. S6A). As the contracted βA/αB joint presses on GAFα1B on the other end, the linker helix in B is likely to endure compression at both ends, resulting in a large bend in the middle (SI Appendix, Fig. S4A). As shown later in this paper, perturbations to the T-junction by an input signal or mutation alter the dimer asymmetry, thereby affecting the kinase activity of PPHK.
The Chromophore Undergoes Light-Induced “Flip-and-Rotate” Motions.
To capture light-induced structural changes in the Pg→Pr pathway, we conducted temperature-scan (T-scan) experiments on photoactive PPHK-TSD crystals, where the Pg state serves as the reference or parent state (18) (SI Appendix, Fig. S7A). By collecting the light (denoted Fgreen) and reference (denoted Fred) datasets from the same crystal or from a pair of crystals, we calculated Fgreen/Fred difference maps to examine light-induced structural changes initiated at different temperatures ranging from 100 K to room temperature (RT; at 293 K) (Fig. 3C and SI Appendix, Figs. S7 and S8). Here, light and temperature are exploited as experimental variables to provoke and trap light-induced structural events along the reaction pathway. To resolve possible structural heterogeneity, we applied singular value decomposition (SVD) to a collection of difference maps or simulated-annealed omit maps (19) (Fig. 3B and SI Appendix, Fig. S10).
Remarkable difference signals appear in the Fgreen/Fred difference Fourier maps starting at 150 K, corresponding to flipping of ring D resulting from 15E→15Z photoisomerization about the C15 = C16 double bond of the bilin chromophore (Fig. 3). These signals are barely detectable at temperatures below 140 K, but quite prominent at 160 to 180 K before they spread to ring C. Between 180 to 200 K, the difference densities also appear in the helical spine, where strong difference signals propagate from the GAF domain to the nREC domain as the temperature rises (SI Appendix, Fig. S8). At RT, the entire chromophore is displaced with a clear overall rotation (Fig. 3 A and C and SI Appendix, Fig. S9A). The “flip-and-rotate” motions are evident in the top-ranked components in the SVD analysis of a collection of 224 difference maps masked around the chromophore (Fig. 3B and SI Appendix, Fig. S10). Therefore, they represent the most significant and distinct light-induced structural events. Based on their temperature dependence, we infer that the flip of ring D precedes the chromophore rotation in the Pg→Pr photoreaction pathway (SI Appendix, Fig. S11). It is noteworthy that the chromophore rotation signals are mostly found in subunit B, but not subunit A (Fig. 3 and SI Appendix, Fig. S10). We speculate that dimer asymmetry also contributes to kinetic trapping of the otherwise elusive intermediates, along with cryotemperatures and the crystal lattice (SI Appendix, Fig. S11). These crystallographic observations suggest that the “flip-and-rotate” model represents a common light signaling mechanism in both classical phytochromes and cyanobacteriochromes (20–22).
Light-Induced Global Structural Changes Amplify Dimer Asymmetry.
To examine light-induced global structural changes at RT, we jointly refined the light-induced Pr structure with the reference Pg structure against a representative Fgreen dataset using difference refinement (23–25) (SI Appendix, Table S1). Structural alignment according to the GAF domain reveals 2 major differences between the Pg and Pr structures. First, the linker helix in subunit B bends even further in the Pr state, resulting in a more asymmetrical helical spine (Fig. 4A and Movie S1). Second, 2 GAF domains undergo a small rotation relative to the helical spine but in the opposite directions, resulting in greater asymmetry between the intersubunit βA/αB and βB/αA joints (Fig. 4 B and C).
Fig. 4.
Light-induced global structural changes in protein. (A) Superposition of the Pg structure (A in blue, B in yellow) and light-induced Pr structure (green) shows that the linker helix in subunit B is bent further in the Pr state, while the counterpart helix in subunit A becomes straighter, resulting in the more asymmetrical dimer structure. Black dashed arrows mark the axial positions of the linker helices in the GAF domains, while red arrows show the overall directions of helical bending. (B) Difference distance matrix between the Pg and Pr dimer structures examines global structural changes in the protein backbone. Blue and red circles highlight a pair of equivalent intersubunit interactions that exhibit opposing structural changes in subunits A (blue triangle) and B (yellow triangle). Specifically, the negative distance changes (red) associated with the tightened βA/αB joint contrasts with the positive distance changes (blue) in a “loosened” βB/αA contact. The color bar shows the amplitude of distance change. (C) Dimer structure shown in a bottom-up view from A reveals a small rotation in both subunits relative to the helical spine. Green arrows indicate the amplitudes (at 5× amplification) and directions of light-induced motions. Large shaded arrows show the collective motions in subunits A and B as they rotate in opposite directions. Shaded bars highlight the β-tear features of the GAF core.
To examine what leads to these global structural changes, we discovered a remarkable structure feature in the central β-sheet conserved among the bilin-binding GAF proteins. Each of the β2 and β5 strands contains an extra residue, giving rise to an irregular hydrogen-bonding pattern in the core β-sheet, with a typical 2-1-5-4-3 topology (SI Appendix, Fig. S12). As these creased β-strands are directed toward different directions, the β-sheet seems to be torn between the β1 and β5 strands, resulting in a β-tear that is spatially flanked by 2 sets of highly conserved residues, namely, LWG in β5 and YR/KF in β1 (Fig. 2C and SI Appendix, Fig. S12D). On one side is the T-junction, where the β4/β5 hairpin directly interacts with the helical spine (SI Appendix, Figs. S4B and S12D). On the other side is the YR/KF motif, where Tyr182 is adjacent to ring D of the chromophore (SI Appendix, Fig. S9B). In the Pr structure, this β-tear further develops in subunit B, while joining up in subunit A (SI Appendix, Fig. S13).
We postulate that the β-tear, together with the T-junction, bridges the chromophore activity to the helical spine, which directly accounts for further bending of the helical spine in response to green light (SI Appendix, Fig. S9C). On one hand, the rigid βA/αB joint exerts a shear force on the β-tear in subunit A, resulting in a tense protein pocket that apparently hinders the chromophore rotation (SI Appendix, Fig. S9B). On the other hand, the restricted chromophore relaxation following 15E/15Z photoisomerization in subunit A would apply a counterforce on GAFα1B, which compresses the βA/αB joint, thus further deforming the linker helix (Fig. 4A). This conserved T-junction structure thereby converts a local signal into a global signal without large-amplitude motions.
Phosphorylation of Asp59 Counteracts Light-Induced Structural Changes.
To address signal integration, we probed the phosphorylation-induced signals by soaking the PPHK-TSD crystals in a crystallization buffer containing (SI Appendix, Tables S1 and S2). We determined the -bound PPHK structure at 2.44 Å resolution ref. 26 and (Fig. 5A). Upon binding, the side chain of Tyr60 moves away from Asp59 to accommodate , which is further stabilized by a highly conserved Thr89 (Fig. 5A). Concomitantly, the nRECβ4/α4 loop undergoes major rearrangements as the backbone structure is displaced by as much as 4.8 Å; meanwhile, the bulky Tyr108 retreats from the dimer interface to fill a space vacated by Thr89. We must point out that although both phosphorylation sites are accessible in the crystal lattice, only subunit A undergoes large protein structural changes, with strong -binding signals at the phosphorylation site in comparison to subunit B (SI Appendix, Fig. S14). The binding was only detectable in B at a much higher concentration of . Such asymmetrical responses to a phosphorylation signal are also manifested in the top-ranked SVD components when >300 simulated-annealing omit maps around the phosphorylation site were jointly analyzed (SI Appendix, Fig. S14A).
Fig. 5.
Ligand-induced changes counteract light-induced structural changes. (A, Left) Superposition of the PPHK-TSD structures in the absence (blue/yellow) and presence of (magenta) reveals large structural changes in subunit A, which extend from the phosphorylation site to the dimer interface, while minimal changes are observed in subunit B. (A, Right) Close-up view shows ligand-induced motions in the conserved residues (Thr89 and Tyr108), along with a displaced loop and nRECα4. (B, Left) Side-by-side comparisons of light-induced motions (green arrows) versus -induced changes (magenta arrows) at the dimer interface. (B, Right) Cartoon illustration shows that the -induced motions in nRECα4A clash with the light-induced bending of nRECα5B. The reference Pg structure is shown in blue (subunit A) and yellow (subunit B).
In the absence of , nRECα4 is overwound by an additional ∼4° per residue compared with nRECα5 (SI Appendix, Fig. S6C). This helical overwinding arises from a tight nRECβ4/α4 turn stabilized by short hydrogen bonds involving Thr89. Upon binding, Thr89 moves toward as the nRECβ4/α4 turn is transformed to an extended loop forming new hydrogen bonds with Tyr108 relocated between nRECα4 and β5 (Fig. 5A). As the relaxed nRECα4A tilts toward the dimer interface, it inevitably hinders the motion of nRECα5B, thereby preventing further bending of the linker helix in subunit B (Fig. 5B). Meanwhile, nRECα5A is further separated from nRECα4A by the retracted bulky side chain of Tyr108, as evidenced by an increase of ∼10° in their interhelix angle (SI Appendix, Fig. S4C). Collectively, these -induced motions convert a local phosphorylation signal into the global structural changes that directly conflict with the light-induced allosteric effects (Fig. 5B). Given the conservation of the Thr89/Tyr108 coupling, these results also shed light on the general signaling mechanism of response regulators (27).
Mechanism of Signal Integration.
In the PPHK structure, the long linker helices at the dimer interface are asymmetrically tethered in a left-handed manner (SI Appendix, Fig. S4A). This twisted helical spine is likely to predicate different structural responses between the 2 chemically identical subunits, as observed by dynamic crystallography (Fig. 6). Each sensor domain in this modular architecture employs a unique coupling mechanism to trigger the global structural responses (Fig. 6 and SI Appendix, Fig. S15). We propose that the GAF sensor module uses the T-junction structure to bridge the light-induced photoisomerization to the helical spine, while the nREC domain relays a phosphorylation signal to the dimer interface via the Thr89/Tyr108 coupling (Fig. 5A). It is the collective allosteric action of all input signals on the helical spine that governs the outcome in the effector histidine kinase domain (Fig. 6). In this work, we show that the light-induced effect is overridden by -induced tilting of nRECα4A at the dimer interface (Fig. 5 B and C). In other words, nRECα4A acts as a limit switch that counteracts the light responses in the helical spine. As a result, the molecular logic OR gate is achieved in PPHK.
Fig. 6.
Signal integration in modular signaling proteins. PPHK features a typical “beans-on-the-stalk” architecture, where the sensor (input) and effector (output) domains are attached to a helical spine, where long linker helices (cylinders) are tethered at the dimer interface. The helical tethering may result in 2 kinds of asymmetry (marked by double-headed arrows): the dimer asymmetry across the dimer interface and the state asymmetry (the R vs. T state) between the adjacent domains. The sensor and effector domains are often separated by lipid bilayers in transmembrane receptors that share similar architectures. The purple arrow and red 3-way arrows represent the “molecular levers” in PPHK that facilitate the signal coupling from each sensory site to the helical spine, where global structural changes are reconciled via concerted helical bending, unwinding, and/or compression. The helical spine thus functions as a signaling hub, where various input signals are integrated to affect the same effector domain in the same protein framework. HK, histidine kinase; P, phosphorylation signal.
To test this model, we carried out site-directed mutagenesis at several key positions in full-length PPHK (Fig. 7). We made a single mutant Y108A to examine the role of Tyr108 in the nREC domain (Fig. 7 A and B). Y108A does not affect the Pg/Pr photoconversion (Fig. 7D), but this single mutation abrogates the -induced response in PPHK and significantly weakens the light dependence of the kinase activity (Fig. 7E). We also note that the indole ring of Trp266 in the highly conserved LWG motif engages close VDW interactions with a hydrophobic residue (Leu299) in GAFα3 of the GAF domain (Fig. 4C). We made 2 single mutations at this position, W266A and W266L. W266L seems to be stuck in the Pg state, while W266A is still photoactive (Fig. 7D). As a result, the kinase activity of W266L is no longer light-dependent and constitutively high (Fig. 7E).
Fig. 7.
Mutational studies on key residues for signal coupling of full-length PPHK. (A) Sequence logos in the regions of interest for the nREC (Left) and bilin-binding GAF (Right) domains. Residues are numbered according to PPHK. The nREC logo was generated based on the sequence alignment of 31 unique receiver domain structures from the Protein Data Bank. The LWG logo was generated from the sequence alignment of >40 representative bilin-binding photoreceptors, including plant and bacterial phytochromes and cyanobacteriochromes. (B) Locations of Tyr108 and Trp266 (green spheres) in the PPHK structure. The indole ring of Trp266 is packed against GAFα3. (C) Positions of His154 and Trp266 with respect to the helical spine viewed from the C terminus of the PPHK dimer structure. The side chain of His154 points to the β4/β5 hairpin and GAFα3 from the partner subunit. The indole ring of Trp266 is packed against a hydrophobic residue on GAFα3. (D) Photoconversion of wild-type (WT) and mutant PPHK in solution. Spectra colored in purple were measured after illumination with 10 min of red light (700 ± 20 nm), and those in blue were measured following illumination with 10 min of green light (500 ± 20 nm). (E) HK assays of WT and mutants in different signaling states. Pg and Pr correspond to the red and green light conditions under which the reaction was conducted. Samples were taken at 2 time points (2 and 10 or 17 min) after adding adenosine 5′-triphosphate (ATP) to a reaction mix. The reaction without ATP serves as a control.
To examine the role of the T-junction, we made a single mutant H154A to disrupt the intersubunit joints between the β4/β5 hairpin and GAFα1 (Fig. 7C). Remarkably, H154A completely abolishes the kinase activity of PPHK in the absence of . Adding to the reaction mix partially recovers the histidine kinase activity (Fig. 7E). Not surprisingly, the photoactivity of PPHK is minimally affected by H154A since His154 is located far away (>30 Å) from the chromophore pocket (Fig. 7D). We postulate that the His154→Ala substitution makes the intersubunit βA/αB and βB/αA joints more asymmetrical, given the twisted or coiled helical spine (Fig. 4). The βA/αB joint on one side of the dimer engages closer backbone interactions, while the βB/αA joint loosens up on the other side. The helical spine is further bent to such an extent that the histidine kinase activity is no longer detectable.
Taken together, we advance a model to elucidate how PPHK operates the molecular logic “OR” gate in response to 2 distinct input signals: here, a phosphorylation signal and a light signal. In a modular architecture, long linker helices at the dimer interface are tethered to form a helical spine, to which distinct sensor (input) and effector (output) domains are attached (Fig. 6). Given the minimal contacts between the modular domains, such helical tethering leads to the dimer asymmetry across the dimer interface and the state asymmetry (the R vs. T state) between the adjacent domains. The resulting twisted helical spine acts as a signaling hub. This hub takes an input as an individual sensor domain tweaks or deforms the helical spine. As global protein structural changes are reconciled via helical bending, unwinding, and/or compression (Fig. 5 and SI Appendix, Fig. S6), different input signals are integrated in the shared spine to ultimately alter the same effector domain. Similar scaffolds are found in the transmembrane receptors, where the sensor and effector domains are separated by lipid bilayers (Fig. 6). PPHK thus offers a prototype for studying how modular signaling proteins make autonomous decisions within the same molecule.
To understand what in vivo function would be gained by PPHK, we examined its genome neighborhood in JSC-1, and found a Cph1-like phytochrome gene immediately upstream of PPHK (SI Appendix, Fig. S16). We speculate that the nREC domain of PPHK receives a phosphoryl group from this upstream protein, which features 3 N-terminal photosensory domains (namely, PAS, GAF, and PHY) typical of classical phytochromes conferring the photoconversion between the red-light–absorbing (Pr) state and far-red-light–absorbing (Pfr) state (8). We and others have shown that the output histidine kinase is more active in the Pr state than the Pfr state in bacterial phytochromes (18, 28, 29). Upon far-red light, this upstream phytochrome is likely to form the Pr state, resulting in higher histidine kinase activity that promotes the nREC phosphorylation in PPHK. Upon red light, formation of the Pfr state in the upstream phytochrome would reduce the nREC phosphorylation, while PPHK forms the Pg state. Given the molecular logic OR (Fig. 1), we postulate that the dual-sensor PPHK works in tandem with the upstream phytochrome to implement a “long-pass” photoreceptor, where the output kinase activity is promoted by both red and far-red light. This long-pass light response is distinct from that rendered by a canonical phytochrome, which is activated by far-red light and deactivated by red light (SI Appendix, Fig. S16).
Discussion
Structural Asymmetry.
Structural asymmetry has been an emerging theme among signaling proteins, where individual domains of distinct function are connected by long helices (17, 30–32). Such asymmetry is distinct from noncrystallography symmetry or the structural variations due to crystal packing. While the former concerns structural differences in the core regions and interfaces, the latter primarily arises from thermal fluctuation or flexibility of the protein surface features. Despite the importance of structural asymmetry in enzyme regulation and signaling, the mechanistic understanding has been largely conceptual and lacking in structural details due to technical difficulties in biophysical interrogation (32, 33). In this work, we apply dynamic crystallography to provoke, probe, and dissect the dimer asymmetry as PPHK transitions from one signaling state to the other.
If the “relaxed” or R state denotes a protein conformation prone to structural changes upon stimulation and the “tense” or T state denotes the rigid counterpart, the nREC domains of subunits A and B are thus assigned to the R and T states, respectively, because subunit A allows more extensive protein structural changes upon ligand binding. The state assignment, however, is reversed for the GAF domain, where the “tense” protein framework in subunit A apparently hinders the chromophore rotation. For the same reason, strong local signals arising from ring D flipping have been observed in subunit A due to kinetic trapping (Fig. 3C), in contrast to the “relaxed” protein framework in subunit B, which allows the chromophore rotation at RT (Fig. 3C and SI Appendix, Fig. S9A). Here, PPHK features 2 distinct types of asymmetry: the dimer asymmetry between 2 chemically identical subunits as well as the state asymmetry between adjacent domains in the same subunit (Fig. 6). In the head-to-head dimer structure, these asymmetries are orthogonal but intertwined, thereby rendering distinct states via tethering.
It is noteworthy that dimer asymmetry is rarely seen in the crystal structures of single-domain or shorter constructs (34–36) but is often self-evident in the structures of multiple modular domains (30, 32, 37), where the long flexible linker helices are tethered in the shared scaffold. The α4/α5/α5 dimer interface featured by the nREC domain is quite common among response regulators (11, 27, 38), as is the 6-helix bundle for the GAF sensor proteins (15, 17, 36, 38). In PPHK, the slightly deformed helical spine likely results from simultaneous accommodation of these conserved dimer interfaces (SI Appendix, Fig. S4A). Here, deformation of the helical spine arms PPHK like a mousetrap ready for a trigger. Due to dimer asymmetry, each protomer may proceed differently along the reaction trajectory (SI Appendix, Fig. S11). GAF-A in the rigid T state seems to render deeper energy wells such that the signals associated with ring D flipping are sufficiently populated for trapping (Fig. 3B). On the other hand, GAF-B in the R state allows the reaction to advance further along the Pg→Pr pathway (Fig. 3C and SI Appendix, Fig. S10). Dimer asymmetry ultimately alters the kinase activity as the helical spine extends to the effector domain. Our PPHK data suggest that the greater the dimer asymmetry in the tandem sensor domains, the lower is the kinase activity. Similar phenomena observed in other modular signaling proteins (30, 32, 37) suggest a tug-of-war in dimer symmetry between the sensor and effector domains.
The Helical Spine as a Signaling Hub.
A long linker helix of 40 residues is a recurring motif among modular signaling proteins. Such a “signaling helix” often contains a heptad sequence, suggesting a coiled-coil conformation (39). With a weak heptad pattern, the linker helices in PPHK feature Glu and Gln directly involved in helical tethering (SI Appendix, Fig. S15 B and C). Given the spring-like mechanics of long helices, their juxtapositions at the dimer interface would impose certain torsional strains on the helical spine. Strategic positioning of Gln300/301 at the staggered junctions between the overlapping helices also suggests a role in signal coupling between the sensor and effector domains (Fig. 6 and SI Appendix, Fig. S15B).
In a modular system, each input signal is entered into the signaling hub via a unique mechanism (10, 30). In PPHK, a phosphorylation signal is relayed to the helical spine via the Thr89/Tyr108 coupling, and the GAF domain is likely to transduce a light signal to the helical spine via the conserved T-junction (Fig. 6 and SI Appendix, Fig. S15A). This T-junction motif is found in other GAF proteins, including the classical phytochromes, oxygen sensor proteins, and human phosphodiesterases despite their distinct nature of input signals (15–17, 40, 41) (SI Appendix, Fig. S5). Although helical supercoiling has been proposed to account for the light action in a chimeric blue-light photoreceptor, YF1 (30, 42), it was not clear what light-induced structural events trigger such a change in supercoiling, given the limited resolution afforded by X-ray solution scattering. In YF1, Trp103 makes direct VDW contacts with the Jα-helix (Protein Data Bank ID code 4GCZ) (30, 43) in a similar disposition of Trp266 in PPHK. Thus, the conserved Trp103 may serve as a molecular lever in light signaling of the LOV (light-oxygen-voltage) proteins. It is plausible that these subtle local signals exerted by individual molecular levers are further integrated or amplified as the tethered helices adjust themselves via bending, compression, and winding in the signaling hub.
Motion, Force, and Structure Rigidity in Protein Mechanics.
To dissect the inner workings of a signaling protein, it is important to establish what structural events drive the next, and how various forms of energy are introduced and released during signal transduction. Such information can be derived from the time sequence and/or temperature dependence of distinct reaction intermediates (18, 44–47). Using the T-scan as an example, the higher the temperature required to provoke a structural signal, the more advanced is the corresponding event along the reaction pathway (18, 48, 49). Our T-scan data on PPHK suggest that the strain energy arising from the primary photoisomerization event (140 to 170 K) drives the helical motions at the dimer interface (180 to 190 K) before the chromophore is allowed to undergo further relaxation with a rotational motion at 293 K (Fig. 3C and SI Appendix, Fig. S8).
The forces that drive protein motions are difficult to measure experimentally. However, they can be deduced from the changes they effect. To this end, helix parameterization offers a very useful tool for analyzing the nature and extent of helical distortion or deformation segment by segment. In a previous study, we demonstrated that subtle changes in helical winding or torsion represent a critical event in rhodopsin signaling (50). Changes in helical pitch, diameter, and interhelix angles inform both the direction and amplitude of forces associated with the strain energy stored in distorted or deformed helices (SI Appendix, Fig. S6).
In addition to motion and force, protein rigidity plays an important role in long-range signaling. A rigid structure would sustain and transmit force without large displacements. In this sense, the T state presents structural rigidity essential for lossless signaling over a long distance, although it is less prone to changes compared with the R state. In this work, we calculate the root-mean-square deviation distance matrix from a collection of perturbed structures to identify rigid elements and/or close associations between different elements (SI Appendix, Fig. S3). Such rigid frameworks provide unbiased references for structural alignment to reveal subtle changes that are otherwise concealed by random variations from structure to structure (51). We posit that protein motions, force, and structural rigidity are 3 integral parts that constitute the working mechanics of a signaling protein.
Dynamic Crystallography.
Dynamic crystallography aims to provoke and probe structural changes as a function of an experimental variable, which could be time, temperature, ligand, or light conditions among others (18, 33, 52). However, the functional relevant structural changes are often so subtle in the crystal lattice that they cannot be readily distinguished from thermal fluctuations. We have shown that it is entirely possible to extract small signals by joint analysis of a large dynamic crystallography dataset using SVD (18, 19, 51). No matter how small the amplitudes, signals shall be consistent and reproducible, and therefore self-evident, in the SVD analysis. Furthermore, the scatter plots between the SVD coefficient sets are highly informative for establishing the origins and correlations of these signals (SI Appendix, Fig. S10B).
This work demonstrates that the crystal lattice, temperature, and dimer asymmetry can be exploited by dynamic crystallography. Generally speaking, a reaction initiated in crystals renders a more rugged energy landscape compared with the same reaction in solution (SI Appendix, Fig. S11). In addition, the reaction temperature and trapping temperature can be used to alter the rate constants of a reaction, thereby providing a floodgate to populate certain transient intermediates. As discussed before, the different reaction trajectories between protomers due to dimer asymmetry offer additional means for capturing intermediates.
Protein crystals have an average solvent content of ∼50%. The corresponding protein concentration in the crystal lattice (∼300 mg/mL) is comparable to the local macromolecular concentrations inside a bacterial cell (∼200 to 320 mg/mL). This suggests that it is entirely possible to provoke functionally relevant structure dynamics in crystals. However, large-amplitude structural changes, often associated with formation of the final products, are less likely to occur in protein crystals due to lattice constraints. In addition, the achievable extent of photolysis is rather limited in a photoactive crystal due to its high optical density, which impedes full light penetration. As a result, the structural changes captured by dynamic crystallography may not fully represent protein dynamics in solution. Nevertheless, these small-amplitude signals are extremely informative from a mechanistic perspective because they provide unparalleled insights into the nature and trend of the structural events leading up to a full development in solution. As shown in this work, the ligand-induced and light-induced structural changes allow us to examine the interplay between 2 distinct sensor domains and to arrive at a model for signal integration. In this regard, rather than a caveat, the lattice hindrance offers a kinetic trap for capturing structural events otherwise too elusive or transient to detect by solution-based methods.
Methods
A PhosTag-based method (Wako Chemicals) was used to assay the histidine kinase activity of both wild-type and mutant full-length PPHK. All X-ray diffraction datasets from the truncated PPHK-TSD crystals were collected at the Life Science Consortium Access Team Sector 21 of the Advanced Photon Source, Argonne National Laboratory. We carried out joint analysis of the dynamic crystallography datasets using dynamiX (19).
Supplementary Material
Acknowledgments
We thank Prof. D. Bryant (The Pennsylvania State University) for providing the genomic DNA of Leptolyngbya sp. strain JSC-1. We thank the Life Science Consortium Access Team (LS-CAT) staff for support in X-ray data collection. Use of LS-CAT Sector 21 is supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor under Grant 085P1000817. Use of the Advanced Photon Source is supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-06CH11357. This work was supported by grants from the NIH (Grant R01EY024363 to X.Y.), Chicago Biomedical Consortium (Grant CBC C-086 to X.Y.) and the University of Illinois at Chicago (to X.Y.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The coordinates of the structures reported have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 6OAP, 6OB8, and 6OAQ).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1910855116/-/DCSupplemental.
References
- 1.Capra E. J., Laub M. T., Evolution of two-component signal transduction systems. Annu. Rev. Microbiol. 66, 325–347 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Groves J. T., Kuriyan J., Molecular mechanisms in signal transduction at the membrane. Nat. Struct. Mol. Biol. 17, 659–665 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laub M. T., Keeping signals straight: How cells process information and make decisions. PLoS Biol. 14, e1002519 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lim W. A., The modular logic of signaling proteins: Building allosteric switches from simple binding domains. Curr. Opin. Struct. Biol. 12, 61–68 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Möglich A., Yang X., Ayers R. A., Moffat K., Structure and function of plant photoreceptors. Annu. Rev. Plant Biol. 61, 21–47 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Gan F., et al. , Extensive remodeling of a cyanobacterial photosynthetic apparatus in far-red light. Science 345, 1312–1317 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Rockwell N. C., Martin S. S., Gan F., Bryant D. A., Lagarias J. C., NpR3784 is the prototype for a distinctive group of red/green cyanobacteriochromes using alternative Phe residues for photoproduct tuning. Photochem. Photobiol. Sci. 14, 258–269 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Rockwell N. C., Su Y.-S., Lagarias J. C., Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 57, 837–858 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rockwell N. C., Lagarias J. C., Phytochrome diversification in cyanobacteria and eukaryotic algae. Curr. Opin. Plant Biol. 37, 87–93 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhate M. P., Molnar K. S., Goulian M., DeGrado W. F., Signal transduction in histidine kinases: Insights from new structures. Structure 23, 981–994 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bachhawat P., Stock A. M., Crystal structures of the receiver domain of the response regulator PhoP from Escherichia coli in the absence and presence of the phosphoryl analog beryllofluoride. J. Bacteriol. 189, 5987–5995 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casino P., Rubio V., Marina A., Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell 139, 325–336 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Wemmer D. E., Kern D., Beryllofluoride binding mimics phosphorylation of aspartate in response regulators. J. Bacteriol. 187, 8229–8230 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shin H., Ren Z., Zeng X., Bandara S., Yang X., Crystal structure of a dual sensor histidine kinase in the green light absorbing Pg state. Protein Data Bank. http://www.rcsb.org/pdb/explore/explore.do?structureId=6OAP. Deposited 12 June 2019.
- 15.Pandit J., Forman M. D., Fennell K. F., Dillman K. S., Menniti F. S., Mechanism for the allosteric regulation of phosphodiesterase 2A deduced from the X-ray structure of a near full-length construct. Proc. Natl. Acad. Sci. U.S.A. 106, 18225–18230 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang H., Robinson H., Ke H., Conformation changes, N-terminal involvement, and cGMP signal relay in the phosphodiesterase-5 GAF domain. J. Biol. Chem. 285, 38149–38156 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang X., Kuk J., Moffat K., Crystal structure of Pseudomonas aeruginosa bacteriophytochrome: Photoconversion and signal transduction. Proc. Natl. Acad. Sci. U.S.A. 105, 14715–14720 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang X., Ren Z., Kuk J., Moffat K., Temperature-scan cryocrystallography reveals reaction intermediates in bacteriophytochrome. Nature 479, 428–432 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ren Z., et al. , Resolution of structural heterogeneity in dynamic crystallography. Acta Crystallogr. D Biol. Crystallogr. 69, 946–959 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang X., Kuk J., Moffat K., Conformational differences between the Pfr and Pr states in Pseudomonas aeruginosa bacteriophytochrome. Proc. Natl. Acad. Sci. U.S.A. 106, 15639–15644 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burgie E. S., Zhang J., Vierstra R. D., Crystal structure of deinococcus phytochrome in the photoactivated state reveals a cascade of structural rearrangements during photoconversion. Structure 24, 448–457 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Schmidt A., et al. , Structural snapshot of a bacterial phytochrome in its functional intermediate state. Nat. Commun. 9, 4912 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Terwilliger T. C., Berendzen J., Difference refinement: Obtaining differences between two related structures. Acta Crystallogr. D Biol. Crystallogr. 51, 609–618 (1995). [DOI] [PubMed] [Google Scholar]
- 24.Terwilliger T. C., Berendzen J., Bayesian difference refinement. Acta Crystallogr. D Biol. Crystallogr. 52, 1004–1011 (1996). [DOI] [PubMed] [Google Scholar]
- 25.Shin H., Ren Z., Zeng X., Bandara S., Yang X., Crystal structure of a dual sensor histidine kinase in green light illuminated state. Protein Data Bank. http://www.rcsb.org/pdb/explore/explore.do?structureId=6OB8. Deposited 31 July 2019.
- 26.Shin H., Ren Z., Zeng X., Bandara S., Yang X., Crystal structure of a dual sensor histidine kinase in BeF3-bound state. Protein Data Bank. http://www.rcsb.org/pdb/explore/explore.do?structureId=6OAQ. Deposited 12 June 2019.
- 27.Bourret R. B., Receiver domain structure and function in response regulator proteins. Curr. Opin. Microbiol. 13, 142–149 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giraud E., et al. , A new type of bacteriophytochrome acts in tandem with a classical bacteriophytochrome to control the antennae synthesis in Rhodopseudomonas palustris. J. Biol. Chem. 280, 32389–32397 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Psakis G., et al. , Signaling kinetics of cyanobacterial phytochrome Cph1, a light regulated histidine kinase. Biochemistry 50, 6178–6188 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Diensthuber R. P., Bommer M., Gleichmann T., Möglich A., Full-length structure of a sensor histidine kinase pinpoints coaxial coiled coils as signal transducers and modulators. Structure 21, 1127–1136 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Gourinchas G., et al. , Long-range allosteric signaling in red light-regulated diguanylyl cyclases. Sci. Adv. 3, e1602498 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neiditch M. B., et al. , Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell 126, 1095–1108 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim T. H., et al. , The role of dimer asymmetry and protomer dynamics in enzyme catalysis. Science 355, eaag2355 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Burgie E. S., Walker J. M., Phillips G. N. Jr, Vierstra R. D., A photo-labile thioether linkage to phycoviolobilin provides the foundation for the blue/green photocycles in DXCF-cyanobacteriochromes. Structure 21, 88–97 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Essen L.-O., Mailliet J., Hughes J., The structure of a complete phytochrome sensory module in the Pr ground state. Proc. Natl. Acad. Sci. U.S.A. 105, 14709–14714 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wagner J. R., Brunzelle J. S., Forest K. T., Vierstra R. D., A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature 438, 325–331 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Gourinchas G., Heintz U., Winkler A., Asymmetric activation mechanism of a homodimeric red light-regulated photoreceptor. eLife 7, e34815 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang X., Zeng X., Moffat K., Yang X., Structure of the response regulator RPA3017 involved in red-light signaling in Rhodopseudomonas palustris. Acta Crystallogr. F Struct. Biol. Commun. 71, 1215–1222 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anantharaman V., Balaji S., Aravind L., The signaling helix: A common functional theme in diverse signaling proteins. Biol. Direct 1, 25 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burgie E. S., Bussell A. N., Walker J. M., Dubiel K., Vierstra R. D., Crystal structure of the photosensing module from a red/far-red light-absorbing plant phytochrome. Proc. Natl. Acad. Sci. U.S.A. 111, 10179–10184 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cho H. Y., Cho H. J., Kim M. H., Kang B. S., Blockage of the channel to heme by the E87 side chain in the GAF domain of Mycobacterium tuberculosis DosS confers the unique sensitivity of DosS to oxygen. FEBS Lett. 585, 1873–1878 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Berntsson O., et al. , Sequential conformational transitions and α-helical supercoiling regulate a sensor histidine kinase. Nat. Commun. 8, 284 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herrou J., Crosson S., Function, structure and mechanism of bacterial photosensory LOV proteins. Nat. Rev. Microbiol. 9, 713–723 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jung Y. O., et al. , Volume-conserving trans-cis isomerization pathways in photoactive yellow protein visualized by picosecond X-ray crystallography. Nat. Chem. 5, 212–220 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ren Z., et al. , A molecular movie at 1.8 A resolution displays the photocycle of photoactive yellow protein, a eubacterial blue-light receptor, from nanoseconds to seconds. Biochemistry 40, 13788–13801 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Ren Z., Šrajer V., Knapp J. E., Royer W. E. Jr, Cooperative macromolecular device revealed by meta-analysis of static and time-resolved structures. Proc. Natl. Acad. Sci. U.S.A. 109, 107–112 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schotte F., et al. , Watching a signaling protein function in real time via 100-ps time-resolved Laue crystallography. Proc. Natl. Acad. Sci. U.S.A. 109, 19256–19261 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bandara S., et al. , Photoactivation mechanism of a carotenoid-based photoreceptor. Proc. Natl. Acad. Sci. U.S.A. 114, 6286–6291 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zeng X., et al. , Dynamic crystallography reveals early signalling events in ultraviolet photoreceptor UVR8. Nat. Plants 1, 14006 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ren Z., Ren P. X., Balusu R., Yang X., Transmembrane helices tilt, bend, slide, torque, and unwind between functional states of rhodopsin. Sci. Rep. 6, 34129 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ren Z., Molecular events during translocation and proofreading extracted from 200 static structures of DNA polymerase. Nucleic Acids Res. 44, 7457–7474 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tenboer J., et al. , Time-resolved serial crystallography captures high-resolution intermediates of photoactive yellow protein. Science 346, 1242–1246 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.