Abstract
The WRKY gene family has never been identified in pigeonpea (Cajanus cajan). Therefore, objective of the present study was to identify the WRKY gene family in pigeonpea and characterize the Fusarium udum stress-responsive WRKY genes under normal, NaCl-stressed and Pseudomonas fluorescens OKC (a plant growth-promoting bacterial strain) treated conditions. The aim was to characterize the Fusarium udum stress-responsive WRKY genes under some commonly occurring field conditions. We identified 97 genes in the WRKY family of pigeonpea, using computational prediction method. The gene family was then classified into three groups through phylogenetic analysis of the homologous genes from the representative plant species. Among the 97 identified WRKY genes 35 were further classified as pathogen stress responsive genes. Functional validation of the 35 WRKY genes was done through generating transcriptional profiles of the genes from root tissues of pigeonpea plants under the influence of P. fluorescens OKC after 24 h of stress application (biotic: Fusarium udum, abiotic: NaCl). The entire experiment was conducted in two pigeonpea cultivars Asha (resistant to F. udum) and Bahar (susceptible to F. udum) and the results were concluded on the basis of transcriptional regulation of the WRKY genes in both the pigeonpea cultivars. The results revealed that among the 35 tentatively identified biotic stress responsive CcWRKY genes, 26 were highly F. udum responsive, 17 were better NaCl responsive compared to F. udum and 11 were dual responsive to both F. udum and NaCl. Application of OKC was able to enhance transcript accumulation of the individual CcWRKY genes to both the stresses when applied individually but not in combined challenge of the two stresses. The results thus indicated that CcWRKY genes play a vital role in the defense signaling against F. udum and some of the F. udum responsive CcWRKYs (at least 11 in pigeonpea) are also responsive to abiotic stresses such as NaCl. Further, plant beneficial microbes such as P. fluorescens OKC also help pegionpea to defend itself against the two stresses (F. udum and NaCl) through enhanced expression of the stress responsive CcWRKY genes when the stresses are applied individually.
Subject terms: Gene expression profiling, Gene expression
Introduction
The WRKY family of transcription factors (TFs) is one of the most important gene families in higher plants involved in biotic and abiotic stress responses1. As TFs, WRKY gene family acts with other components of the transcriptional machinery and plays a vital role in the plant response to both external and internal stimuli2,3. Identification and characterization of WRKY genes in Jatropha curcas revealed presence of 47 WRKY genes that showed responsiveness to one or more abiotic stresses4. Similarly, the Arabidopsis WRKY gene AtWRKY25 is induced in response to the bacterial pathogen Pseudomonas syringae and plays a role in the host defense5. Other Arabidopsis WRKY genes such as, AtWRKY3, AtWRKY4 and AtWRKY8 demonstrated to respond to the necrotrophic fungal pathogen Botrytis cinerea6,7. It is also well documented that a single gene may play a specific role at a varied level against a specific stress under different circumstances. WRKY proteins possess one or two unique DNA binding domains that consist of about 60 amino acid long peptide sequences including the highly conserved sequence WRKYGQK and a zinc-finger motif. WRKY family proteins have been classified into three groups (I-III) based on the WRKY domain numbers as well as structure of the zinc-finger motif. WRKY proteins in group I possess two WRKY domains along with a C2H2 like zinc-finger motif. Group II WRKY proteins possess a single WRKY domain along with a C2H2 like zinc-finger motif. Based on the type of conserved motifs group II can be further sub-divided into five different groups (IIa–IIe)8. Finally, group III WRKY proteins possess a single WRKY domain in addition to a C2HC type zinc-finger motif. Evolutionary studies indicated that the majority of WRKY gene members in groups I and II were emerged before the divergence of monocot and dicot plants, whereas group III genes appeared relatively later9.
WRKY genes have been identified and characterized in various plant species8–11, since the first cDNA of the WRKY gene SPF1 was cloned from sweet potato12. Different plant species carry different numbers of WRKY genes. For example, 72 WRKY family members have been identified in the Arabidopsis genome9, 102 in rice9, 55 in cucumber13, 119 in maize14, 59 in grapevine15, 104 in poplar16, etc. In addition to higher plants, WRKY-type genes were also reported in non-plant green alga and other eukaryotes17.
Pigeonpea (Cajanus cajan (L.) Millsp.) is a widely cultivated food legume in South Asia, Africa, the Caribbean and Latin America, where it is considered as an important source of protein in the human diet18. Fusarium udum is a very serious wilt pathogen for pigeonpea and the role of the pigeonpea WRKY genes against F. udum or any other pathogen stress is not yet investigated. Therefore, the aim of the present study is to (i) identify and validate the tentative WRKY TFs of pigeonpea that respond to F. udum challenge, (ii) considering the increasing areas of salinity in cultivable lands, the same set of WRKY genes was also evaluated for their responsiveness to NaCl stress either individually or in combination with F. udum and (iii) a plant growth-promoting rhizobacteria (PGPR) Pseudomonas fluorescens OKC, a potential plant stress reliever, was used to see its impact on the selected WRKY genes under both F. udum and NaCl stresses in pigeonpea.
Results
Identification and phylogenetic analysis of pigeonpea WRKY genes
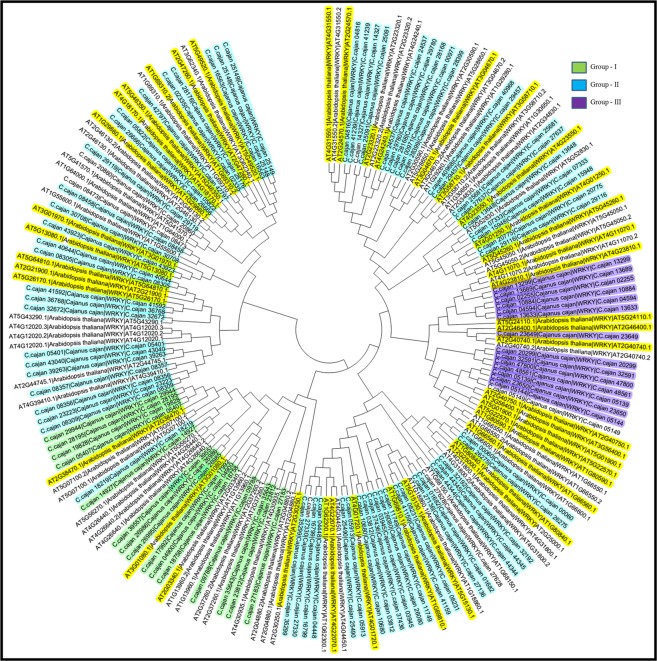
Sequence analysis of the conserved domains in pigeonpea led to identification of 97 WRKY genes (further defined as CcWRKY genes) (Supplementary Table S1). All putative 97 CcWRKY proteins in pigeonpea and 72 WRKY proteins in Arabidopsis thaliana were used to perform phylogenetic study to categorize and investigate the evolutionary relationships of the CcWRKY genes. Phylogenetic relationship study showed that the CcWRKY proteins in pigeonpea could be categorized into three groups, Group I–III, which are highly conserved in both monocots and eudicots8 (Fig. 1). A total of 11 CcWRKY proteins are placed in group I with two WRKY domains and C2H2-type of zinc-finger motif, 69 CcWRKY proteins that contained one WRKY domain and also C2H2-type zinc binding motif is placed in group II and the remaining 14 CcWRKY proteins with a single WRKY domain and C2HC type of zinc-binding motif is assigned to group III. Out of the 97 CcWRKY genes, 3 sequences (C.cajan_08356, C.cajan_08357, C.cajan_47800) were devoid of any WRKY domain and zinc binding motif. These exceptional WRKY proteins could represent pseudogenes or sequencing and/or assembly errors19,20.
Figure 1.
Phylogenetic tree of the WRKY proteins of pigeonpea (Cajanus cajan). The conserved WRKY domain sequences of pigeonpea WRKY proteins were used to construct multiple sequence alignments using Clustal W. Software MEGA was used to prepare the phylogenetic tree and neighbor-joining method was adopted by 1,000 bootstrap replications. The CcWRKY proteins of pigeonpea are classified into three groups, Group I–III, based on conserved WRKY domain sequences. Yellow colour indicates the Arabidopsis WRKY defense responsive genes used for selection of pigeonpea WRKY defense responsive genes.
In silico functional analysis of CcWRKY proteins
We retrieved a total of 97 protein sequences as members of the WRKY family in C. cajan. A homology search was carried out using BLAST in NCBI and TAIR databases, and all CcWRKY proteins showed sequence identity with WRKY proteins from Arabidopsis thaliana. In silico functional analysis showed that 56 CcWRKY proteins are similar to either biotic or abiotic stress-related AtWRKY proteins in A. thaliana and 35 of them are pathogen stress responsive (Supplementary Table S2). Thus, we assumed that a great number of the CcWRKY proteins are also involved in response to various biotic and abiotic stresses.
Motif analysis of CcWRKY Genes
In the CcWRKY family a total of 25 motifs were identified in different CcWRKY proteins (Supplementary Fig. SF1, Supplementary Table S3). Results from the MEME motif analysis showed that the CcWRKY proteins have variation in occurrence of motifs numbers (1–9), and also in the length of motifs (ranged from 7 to 95 amino acids long). However, 3 motifs out of the 25 namely motifs 1, 2 and 8 showed presence of the most conserved sequence WRKYGQK. The 22 remaining motifs were located outside the WRKY domain. Further, motif 1 is shared by 45 CcWRKYs belonging to all three groups while motif 2 is shared by 47 CcWRKYs belonging to group I and group II only and motif 8 is shared by 3 CcWRKY genes from group II (CcWRKY35, CcWRKY57, CcWRKY73) and 6 CcWRKY genes from group III (CcWRKY7, CcWRKY34, CcWRKY49, CcWRKY66, CcWRKY86, CcWRKY97). In addition, motifs 1, 3 and 4 are those which are shared by the members of all three groups while motifs 2, 5, 9 and 14 are shared by the members of group I and II only. Members of the group II and III are shared by motifs 8, 13, 15, 19 and 24 whereas, motifs 6, 7, 10, 12, 16, 20, 22 and 23 are found conserved only in the group II CcWRKY members and motifs 21 and 25 are conserved in the members of group III only. The motifs 1 and 2 showed similarity with AtWRKY4 which is involved in defense responses such as negative regulation of defense response to bacterium, regulation of defense response to fungus, regulation of transcription, DNA-templated, response to ethylene, jasmonic acid and salicylic acid21 while motif 8 showed similarity with AtWRKY1 which is linked to positive regulation of transcription, DNA-templated, and salicylic acid mediated signaling pathway22. All the 35 pathogen stress responsive CcWRKYs had one of the above mentioned motifs and therefore confirmed to be CcWRKYs. In the selected CcWRKYs, motif 24 (RGRHTCT) is found only in CcWRKYs 7, 34 and 35 that showed similarity with TOPLESS (TPL) and TOPLESS-related (TPR) proteins. In plants, TPL/TPR are co-repressors that regulate development, stress responses, and hormone signaling through interaction with small ethylene response factor-associated amphiphilic repression (EAR) motifs and normally found in diverse transcriptional repressors23 (Supplementary Table S3).
Protein structure analysis of CcWRKY
The protein structures of CcWRKYs were further analyzed and the results showed that 84 CcWRKYs contained the highly conserved sequence WRKYGQK whereas proteins of 5 CcWRKYs (CcWRKY21, CcWRKY22, CcWRKY23, CcWRKY24 and CcWRKY57) consist of the most common variant sequence WRKYGKK. Proteins of four CcWRKYs (CcWRKY19, CcWRKY34, CcWRKY35 and CcWRKY84) consist of the less common variant sequence WRKYGEK. CcWRKY50 protein consists of four and CcWRKY13 consists of one amino acid variation in their WRKY domains. In three CcWRKY genes the WRKY domain was not found (Supplementary Fig. SF2).
Physico-chemical characterization of CcWRKY proteins
Amino acid residues calculated from the primary sequences of the CcWRKY proteins ranged from 66 to 678 (Supplementary Table S4). Isoelectric points of all CcWRKYs ranged between 4.74 and 10.09 and fifty percent of the proteins are acidic in nature (pI < 7) and fifty percent showed alkaline nature (pI > 7). Molecular weight of the proteins was between 7935.92 and 73688.00 Da. Almost all proteins except seven have instability index greater than 40, which indicated that the proteins are unstable in nature. The aliphatic index of all the proteins is significantly high which may be regarded as a positive factor for increased thermostability of globular proteins24. Lower range of GRAVY indicated that the proteins have better interaction with water.
Transcript profiling of CcWRKY genes against biotic and abiotic stresses
In silico functional analysis of the 97 pigeonpea WRKY genes showed that 35 are pathogen stress responsive genes. Quantification analyses of the CcWRKY gene transcripts in response to the biotic (Fusarium udum WSP-V2) and abiotic (NaCl) challenges revealed differential expression patterns of the 35 CcWRKY genes (Fig. 2, Supplementary Tables S5 and S6). Details of expression profiles of the genes in the two pigeonpea cultivars are described with the internal control Actin as both the internal controls (Actin and β-tubulin) provided similar trends of expression profiles of the selected genes and expression profiles of the individual genes were statistically not different between the two internal controls.
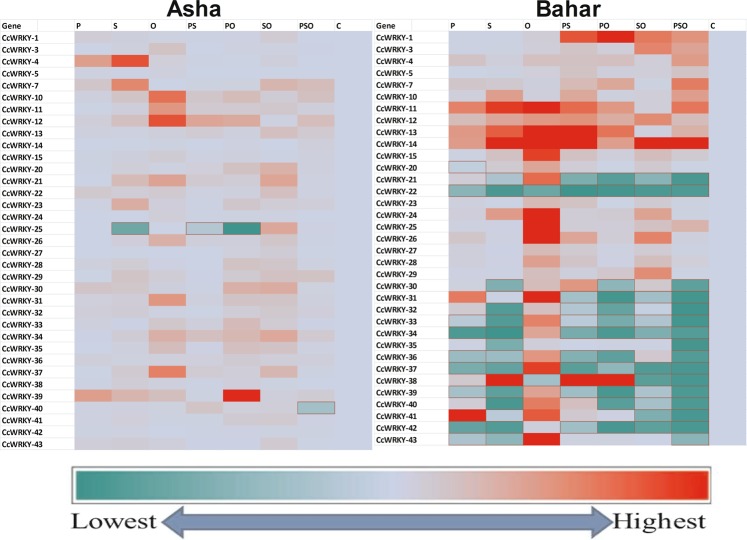
Figure 2.
Heat maps of expression profiles of 35 selected CcWRKY genes in the pigeonpea cultivars Asha and Bahar in different treatments: P (Pathogen Fusarium udum), S (Salt-NaCl), O (Pseudomonas fluorescens OKC), PS (Pathogen + Salt), PO (Pathogen + Pseudomonas), SO (Salt + Pseudomonas), PSO (Pathogen + Salt + Pseudomonas) and C (Control). The colour scale indicates the expression value (dark red indicate higher expression value, darker green indicates lower gene expression values).
Transcript profiling of the CcWRKY genes in response to Fusarium udum in resistant and susceptible cultivars
Transcript profiling of the WRKY genes was done in both the F. udum resistant and susceptible pigeonpea cultivars ‘Asha’ and ‘Bahar’, respectively, to observe the cultivar-specific response to F. udum challenge. In pigeonpea, all 35 CcWRKY genes did not respond equally to F. udum stress in the two cultivars. In the resistant cultivar ‘Asha’ all 35 CcWRKY’s were up-regulated but in different folds under the pathogen stress, viz., CcWRKYs 3, 5, 11, 15, 20, 21, 23, 24, 25, 26, 27, 29, 34, 35, 37, 38, 40, and 42 were up-regulated by 1–5 fold, whereas CcWRKYs 10, 13, 28, 31, 33, 36, and 41 by 5–10 fold, CcWRKYs 1, 12, 32, and 43 by 10–15 fold, CcWRKYs7 and 22 by 15–20 fold, CcWRKY30 by 20 fold while two CcWRKYs 4 and 39 by more than 50 fold. However, 9 CcWRKYs (CcWRKYs 3, 14, 21, 24, 25, 26, 27, 29, and 35) were least responsive to F. udum. These CcWRKYs were up-regulated minimally (0.5–1 fold). At the same time CcWRKYs in the susceptible cultivar ‘Bahar’ revealed that 9 CcWRKYs (CcWRKYs 20, 22, 33, 34, 36, 37, 39, 42, and 43) were down-regulated and only 26 CcWRKYs were up-regulated. Among the up-regulated ones 21 CcWRKYs (CcWRKYs 1, 3, 4, 5, 7, 10, 12, 15, 21, 23, 24, 25, 26, 27, 28, 29, 30, 32, 35, 38, and 40) were up-regulated by 1–5 fold, 2 (CcWRKYs 12, and 13) by 5–10 fold, 2 (CcWRKYs 11, and 31) by 10–15 fold and one (CcWRKY41) by 29 fold under F. udum stress (Fig. 2, Supplementary Tables S5 and S6). It was clearly observed that not all 35 selected CcWRKYs responded to F. udum in the susceptible cultivar ‘Bahar’ unlike in the case of the resistant cultivar ‘Asha’.
Transcript profiling of the CcWRKY genes in response to NaCl stress
Transcript profiling of the WRKY genes was done in both the F. udum resistant and susceptible pigeonpea cultivars ‘Asha’ and ‘Bahar’, respectively, to observe the cultivar-specific response to NaCl challenge. Transcript profiling of all 35 pathogen responsive CcWRKYs in the two pigeonpea cultivars revealed that, in ‘Asha’ 34 genes were up-regulated viz., CcWRKYs 5, 14, 24, 27, 33, and 42 by 1–5 fold, CcWRKYs 1, 3, 11, 13, 15, 20, 26, 35, 36, 40, and 41 by 5–10 fold, CcWRKYs 22, 28, 31, 32, 34, 38, and 43 by 10–15 fold, CcWRKYs 30, and 37 by 15–20 fold, CcWRKYs 10, 12, and 29 by 20–25 fold, CcWRKY21 by 25–30 fold, CcWRKY39 by 35 fold, CcWRKY23 by 41 fold, and CcWRKYs 4 and 7 by more than 50 fold. Only one gene (CcWRKY25) was down-regulated. In contrast, 15 CcWRKY genes viz., CcWRKYs 21, 22, 30, 31, 32, 33, 34, 35, 36, 37, 39, 40, 41, 42, and 43 were down-regulated in ‘Bahar’ whereas the rest of the 20 CcWRKY genes were up-regulated. Very few CcWRKY genes viz., CcWRKYs 11, 13, 14, 24, 25, and 38 in ‘Bahar’ showed higher transcript accumulation than ‘Asha’ whereas rest of the genes in ‘Bahar’ showed lower transcript accumulation compared to ‘Asha’. Interestingly, all selected 35 CcWRKYs are presumed to be F. udum responsive, but 17 of those CcWRKYs (CcWRKYs 7, 10, 11, 12, 15, 20, 21, 23, 26, 28, 29, 31, 34, 35, 37, 38 and 40) were observed to be better responsive to NaCl compared to F. udum in the resistant cultivar ‘Asha’. (Fig. 2, Supplementary Tables S5 and S6).
Transcript profiling of the CcWRKY genes in Pseudomonas fluorescens OKC treatment
Transcript profiling of the WRKY genes was done in both the F. udum resistant and susceptible pigeonpea cultivars ‘Asha’ and ‘Bahar’, respectively, to observe the cultivar-specific response to P. fluorescens OKC treatment. All 35 CcWRKY genes were up-regulated in ‘Asha’ whereas only 33 CcWRKY genes in ‘Bahar’ (CcWRKYs 22 and 38 were down-regulated) in the P. fluorescens OKC treatment. Interestingly, 5 CcWRKYs (CcWRKYs 10, 11, 12, 31, and 37) in ‘Asha’ and 3 CcWRKYs (CcWRKYs 24, 26, and 43) in ‘Bahar’ showed more than 50 fold up-regulation. The results indicated that OKC induce the expression of all 35 selected CcWRKYs (Fig. 2, Supplementary Tables S5 and S6).
Transcript profiling of the CcWRKY genes in response to combined stresses of Fusarium udum and NaCl
Transcript profiling of the WRKY genes was done in both the F. udum resistant and susceptible pigeonpea cultivars ‘Asha’ and ‘Bahar’, respectively, to observe the cultivar-specific response to the combined challenges of F. udum and NaCl. In most of the earlier studies WRKY genes were analyzed only either in biotic or abiotic stress conditions. In the present study we analyzed the combined effect of the pathogen and NaCl stress in a single treatment on the CcWRKY genes. Transcript accumulation of the CcWRKY genes showed differential patterns in both ‘Asha’ and ‘Bahar’ cultivars. In ‘Asha’ only CcWRKY25 was down-regulated but the rest of the CcWRKY genes were up-regulated with highest fold change increase in CcWRKY12. Similarly, CcWRKY genes in ‘Bahar’ showed down-regulation of 10 CcWRKYs (CcWRKYs 21, 22, 31, 32, 33, 34, 36, 37, 39, and 42) compared to only one in ‘Asha’. Interestingly, 5 CcWRKYs (CcWRKYs 30, 35, 40, 41, and 43) which were down-regulated in the individual NaCl treatment were slightly up-regulated in the combined application of the pathogen and NaCl with the highest fold change in CcWRKY30. Additionally, out of the 17 CcWRKYs that were observed to be better NaCl responsive compared to F. udum in the single application of NaCl (as listed in 2.6.2), 11 (CcWRKYs 1, 10, 11, 12, 13, 14, 20, 26, 38, 40 and 43) were identified to be dual responsive following combined application of both the stresses. This indicates dual role of these 11 CcWRKYs in addressing both F. udum and NaCl challenges (Fig. 2, Supplementary Tables S5 and S6).
Transcript profiling of the CcWRKY genes in response to Fusarium udum and Pseudomonas fluorescens OKC
Transcript profiling of the WRKY genes was done in both the F. udum resistant and susceptible pigeonpea cultivars ‘Asha’ and ‘Bahar’, respectively, to observe the cultivar-specific response to P. fluorescens OKC treatment followed by F. udum challenge. In the resistant cultivar ‘Asha’ only CcWRKY25 was down-regulated while all other CcWRKY genes were up-regulated. CcWRKY25 also did not show true responsiveness to F. udum without OKC. Interestingly, most CcWRKYs which did not respond well to F. udum challenge without OKC were highly up-regulated in presence of OKC. CcWRKYs 3, 10, 11, 12, 13, 14, 15, 20, 21, 23, 24, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, and 42 showed more fold changes compared to pathogen stress alone without OKC treatment with highest fold change in CcWRKY39 while other CcWRKY genes namely CcWRKYs 1, 4, 5, 7, 22, and 43 showed lower fold changes. In contrast, in the susceptible cultivar ‘Bahar’ differential fold changes were observed compared to the ‘Asha’ cultivar. CcWRKYs 1, 3, 5, 7, 12, 13, 15, 20, 24, 25, 29, and 38 showed higher fold changes than pathogen stress alone while other CcWRKYs 11, 14, 23, 27, 28, 35, and 41 showed lower fold changes and CcWRKYs 21, 30, 31, 32, and 40, were down-regulated. There were some CcWRKY genes (CcWRKYs 22, 33, 34, 36, 37, 39, and 42) that were down-regulated in both pathogen and pathogen with OKC treatments in ‘Bahar’ cultivar. The results indicated minimal role of the selected CcWRKYs in the F. udum susceptible pigeonpea cultivar ‘Bahar’ except CcWRKYs 11, 13, and 14 where the role of OKC is established in sensitizing the genes (Fig. 2, Supplementary Tables S5 and S6).
Transcript profiling of the CcWRKY genes in response to NaCl and Pseudomonas fluorescens OKC
Transcript profiling of the WRKY genes was done in both the F. udum resistant and susceptible pigeonpea cultivars ‘Asha’ and ‘Bahar’, respectively, to observe the cultivar-specific response to P. fluorescens OKC treatment followed by NaCl challenge. In ‘Asha’, 22 CcWRKY genes viz., CcWRKYs 1, 11, 13, 20, 21, 22, 24, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 40, 41, and 43 were up-regulated under NaCl response in which higher fold changes were observed when OKC applied as seed priming. Interestingly CcWRKY25 that was down-regulated in NaCl treatment was up-regulated in NaCl treatment combined with OKC treatment. Some other CcWRKYs 3, 4, 5, 7, 10, 12, 14, 15, 23, 38, 39, and 42 that were up-regulated under NaCl stress were also up-regulated in the combined treatment with OKC but relatively in lower folds compared to NaCl treatment alone. In ‘Bahar’ also a similar trend was seen in OKC treatments as in ‘Asha’. The CcWRKY genes (CcWRKYs 1, 3, 4, 5, 12, 15, 16, 23, 25, 26, 27, 28, and 29) that were up-regulated in NaCl treatment without OKC found increase in fold changes under NaCl treatment combined with OKC. Some CcWRKYs (CcWRKYs 30, 35, 36, and 43) that were down-regulated in NaCl treatment alone were up-regulated in NaCl treatment combined with OKC. In CcWRKYs 7, 10, 11, 13, 14, and 24 less fold change was observed during their up-regulation in NaCl treatment with OKC compared to NaCl treatment alone. There were other CcWRKYs (CcWRKYs 21, 22, 31, 32, 33, 34, 37, 39, 40, 41, and 42) which were down-regulated in both NaCl alone and NaCl with OKC treatment indicating their non-involvement in NaCl stress mediation. Only CcWRKY38 which was up-regulated in NaCl treatment without OKC was down-regulated in NaCl with OKC treatment (Fig. 2, Supplementary Tables S5 and S6).
Transcript profiling of the CcWRKY genes in response to combined inoculation of NaCl and Fusarium udum with OKC
Transcript profiling of the WRKY genes was done in both the F. udum resistant and susceptible pigeonpea cultivars ‘Asha’ and ‘Bahar’, respectively, to observe the cultivar-specific response to P. fluorescens OKC treatment followed by combined challenges of F. udum and NaCl. In ‘Asha’ all CcWRKY genes were up-regulated in different folds except CcWRKY40. Among those, 11 CcWRKY genes (CcWRKYs 7, 10, 14, 15, 22, 23, 28, 29, 30, 32, and 39) were up-regulated in higher folds whereas 7 CcWRKYs (CcWRKYs 11, 12, 21, 26, 33, 34, and 37) up-regulated in lower folds in presence of OKC. CcWRKY25 that was down-regulated without OKC was up-regulated in presence of OKC whereas CcWRKY40 was down-regulated. Similarly, in ‘Bahar’ only 4 CcWRKY genes i.e. CcWRKYs 3, 4, 7, and 25 showed higher fold change in presence of OKC while in 4 (CcWRKYs 1, 12, 13, and 26) the fold change was lowered. CcWRKYs 21, 22, 31, 32, 33, 34, 36, 37, 39, and 42 were down-regulated with or without OKC while CcWRKYs 30, 35, 38, 40, and 43 were down-regulated in presence of OKC (Fig. 2, Supplementary Tables S5 and S6). The results clearly indicated cross-talk between the F. udum and NaCl challenges.
Discussion
Cells regulate gene expression through control of the transcriptional mechanism. WRKY proteins are also considered as transcriptional regulators. Proper annotation of genes of a gene family is important and essentially required for their functional studies. We identified and categorized 97 gene sequences as members of the WRKY family in pigeonpea (Cajanus cajan) and in silico functional analysis was done using BLAST as a tool to search the NCBI and TAIR databases. Results indicated that majority of the WRKY proteins in pigeonpea showed similarity with biotic and abiotic stress-related WRKY proteins of Arabidopsis thaliana. It made us to propose that CcWRKY proteins are also involved in resistance activities against various biotic and abiotic stresses as in A. thaliana.
The phylogenetic relationship study used in the present work allowed division of the pigeonpea WRKY genes into the three previously reported groups9. Though the WRKYGQK sequence was found highly conserved among pigeonpea WRKY proteins (Supplementary Fig. SF2), variations are observed in 9 CcWRKY genes. Previous studies also showed that GmWRKY6 and GmWRKY21 in soybean have the variant sequence WRKYGKK rather than the conserved WRKYGQK motif sequence25. Minor variations at this particular region was also reported earlier in Arabidopsis, sunflower, barley, rice, tobacco, and canola19,26–30. Variation in the conserved protein sequences may affect adversely resulting in termination or decreasing the capacity of WRKY proteins in binding with the signature W-box element3. It is quite possible that the WRKY proteins without the conserved WRKYGQK motif may bind to a different site in the target genes28 and play a different role27. For example, soybean WRKY protein GmWRKY6 and tobacco WRKY protein NtWRKY12 can bind to WK-box (TTTTCCAC) instead of binding to the canonical W-box element25,28. In the present study also, we observed that some CcWRKYs showed variations in the zinc finger motif in a similar way with the three VvWRKY proteins of Vitis vinifera31. However, the functionality of such variations in the zinc-finger like motif remains to be elucidated. In addition to the conserved WRKY domain, other motifs which are observed as conserved sequence could be important for different functions of the WRKY proteins. Up-regulation of the CcWRKY7 gene more than 50 fold under NaCl stress might be due to the presence of the extra motif 24 in this gene.
In field conditions, crop plants face variety of biotic and abiotic stresses and thereby remain under continuous threat. Plants adapt to these kinds of situations through reprogramming of their inherent metabolic pathways via differential expression of genes. WRKY gene family can respond to these situations and act as activators or repressors of certain genes of the crop species. In the current study, transcript accumulation patterns of 35 CcWRKY genes under the two stresses (F. udum and NaCl) with or without application of Pseudomonas fluorescens OKC in two pigeonpea cultivars revealed distinct variations in different treatments. Responsiveness of these selected WRKY genes to F. udum challenge in the resistant cultivar ‘Asha’ confirms their selection and validates their functionality. Significant up-regulation of most of the 35 selected biotic stress-responsive CcWRKY genes in the resistant cultivar ‘Asha’ endorses their responsiveness to F. udum stress and thereby, contributing to resisting progress of the pathogen in pigeonpea. Further, relatively high responsiveness of 4 CcWRKYs (CcWRKYs 12, 13, 31 and 41) to F. udum stress in both the cultivars can be attributed to their involvement in the initial response by pigeonpea to F. udum. Interestingly, 4 CcWRKYs (CcWRKYs 15, 21, 24 and 36) that did not respond highly to F. udum stress, responded very strongly when the plants were bio-primed with the P. fluorescens OKC strain. Significance of PGPR strains in reducing NaCl stress in pulses had been recorded earlier32,33. However, the mechanisms underlying thereof particularly the role of WRKY transcription factors have not been worked out so far. The results thus demonstrate significance of application of PGPR strains such as OKC in pigeonpea for enhancing resistance against biotic stresses such as F. udum through enhanced activation of CcWRKYs. Additionally, the role of OKC in enhancing transcript accumulation in some other CcWRKYs (CcWRKYs 25, 30 and 35) which were otherwise responded lowly to the lone NaCl stress indicated its worth in enhancing salt tolerance in pigeonpea as well. However, the role of OKC was not very evident in enhancing transcript levels of the selected CcWRKYs in combined application of both the stresses (F. udum and NaCl) compared to its ability to enhance transcript levels of the same CcWRKYs under the same stresses when applied individually. The variations may be attributed to cross-talk of the signaling cascades operative under F. udum as well as NaCl stresses in pigeonpea. Additionally, the use of microbial consortium may be another option to address dual stresses of different kinds34.
Although, different WRKY genes respond to different stresses some of the WRKY genes are also reported to respond to combined biotic and abiotic stresses35. In the current study, all 35 CcWRKY genes showed differential expression under salt stress, where the expression of all the genes were up-regulated except CcWRKY25 in ‘Asha’ compared to ‘Bahar’ in which only 25 CcWRKYs genes were up-regulated and rest 10 were down-regulated. This variation in expression of the same genes in the susceptible cultivar ‘Bahar’ may be attributed to lack or absence of some common factors (genes) that govern both biotic and abiotic stresses in the resistant pigeonpea cultivar ‘Asha’. However, it was interesting to note that among the 35 selected biotic stress-responsive CcWRKY genes 26 were responsive to F. udum, 17 were NaCl responsive and most significantly 11 were dual responsive to both F. udum and NaCl stresses in both the pigeonpea cultivars. This shows the role of the 11 CcWRKY genes in mitigating both biotic and abiotic stresses probably in most of the cultivated pigeonpea cultivars. Differential expression patterns of the same WRKY genes were also recorded in 54 OsWRKY from rice and 26 GmWRKY from soybean under abiotic stresses36. Further, it is quite likely that the highly induced CcWRKY genes might also play an important role against other abiotic stresses as it was shown in case of BcWRKY46 which is a cold-inducible gene from Pak-choi also enhances tolerance to salt and dehydration stresses in transgenic tobacco37 and in 23 CsWRKY genes from Cucumis sativus that expressed differentially in response to at least one abiotic stress such as cold, drought, or salinity13. From the current study, it can be concluded that the 35 biotic stress responsive CcWRKY genes identified in pigeonpea, based on the homologous genes from the representative plant species A. thaliana, responded well to F. udum stress and thereby confirm their selection as biotic stress responsive CcWRKYs. However, among the selected 35 CcWRKYs, 26 responded very highly to F. udum, 17 responded highly to NaCl compared to F. udum and 11 were dual responsive to both F. udum and NaCl. The study thus signifies the role of the biotic stress responsive CcWRKYs in mitigating F. udum and NaCl stresses in pigeonpea.
Materials and Methods
Plant materials, pathogen and bio-agent culture
Pigeonpea varieties ‘Asha’ (Fusarium wilt resistant) and ‘Bahar’ (Fusarium wilt susceptible) and the wilt pathogen Fusarium udum strain WSP-V2 were obtained from Indian Institute of Pulses Research, Kanpur, Uttar Pradesh, India. Pseudomonas fluorescens OKC (Accession numbers JN128891) was used as a PGPR and biocontrol agent having antagonistic properties against F. udum.
Biopriming and stress inoculation
Seeds of cultivars ‘Asha’ and ‘Bahar’ were bio-primed with Pseudomonas fluorescens OKC according to Yadav et al.38. Briefly, cell suspension of OKC was adjusted to 1.6 × 108 cfu mL−1 before seed biopriming. Seed were bio-primed for 2 h, dried under shed at room temperature and the primed seeds were sown in sterilized garden soil mixed with 10% vermiculite. Five replicates of each treatment were maintained. After 21 days of sowing, plants were inoculated with conidial suspension (2 × 107 conidia/mL) of Fusarium udum and 200 mM NaCl either alone or in combination. Conidial suspensions and NaCl solutions were prepared in sterilized distilled water. Root samples were collected 24 h after pathogen and salt inoculation for quantification of transcripts of target genes by quantitative RT-PCR. The treatments applied were: (i) Fusarium udum, (ii) NaCl (200 mM), (iii) Fusarium udum + NaCl and (iv) untreated control. One set of all 4 treatments were maintained separately without seed bio-priming with OKC.
RNA isolation, cDNA synthesis and qRT-PCR analysis
Total RNA was extracted and purified from 200 mg of homogenized root tissue thrice from each sample with some modifications and quantified using a Nano-Drop 2000 UV Vis spectrophotometer (Thermo Scientific, Wilmington, DE, USA)39. Approximately 3 μg total RNA was treated using RNase-free DNase I at 37 °C to remove the remaining genomic DNA. cDNA was prepared using oligo (dT) primers and reverse transcriptase enzyme according to Sambrook and Russell40. cDNAs were used as templates for semi-quantitative and quantitative RT-PCR.
Sequence analysis and classification of CcWRKY genes
Cajanus cajan WRKY genomic and protein sequences were retrieved from Plant Transcription Factor database41 and analysis were completed to confirm the presence of the WRKY domain using the SMART program42. Additionally, primary structure analysis of CcWRKY proteins such as length, molecular weight, isoelectric point, total number of atoms extinction coefficients, instability index, aliphatic index, grand average of hydropathicity was completed using the ExPasy website (http://au.expasy.org/tools/pi_tool.html), whereas protein homology study of WRKY proteins was done using the Basic Local Alignment Search Tool (BLAST). Since detailed study of WRKY family was done in Arabidopsis8, phylogenetic tree was constructed based on the alignment of WRKY domains from pigeonpea and Arabidopsis to evaluate the phylogenetic relationships and classified them into different groups8.
Motif analysis and phylogenetic relationship
WRKY proteins were also used to detect the conserved motifs using MEME (http://meme.nbcr.net/meme/cgibin/meme.cgi) with parameters such as number of repetitions: any; maximum number of motifs: 25; and the optimum motif widths: 6–200 amino acid residues. Software MEGA was used to prepare the phylogenetic tree and neighbor-joining method was adopted by 1,000 bootstrap replications8. Functional analysis of motifs was done through NCBI protein blast search within the protein databank of plant.
Primer designing and transcript profiling of CcWRKY genes
cDNA sequences of the candidate CcWRKY genes were retrieved from GenBank and primers were designed by using the online software Primer-343 (Supplementary Table S7). In order to prevent off-target amplification, the most homologous sequences of the WRKY TFs were identified through BLAST search in the NCBI database. Sequence alignment was done using Clustal Omega, and the software suggested primers were compared to the alignments to select the primers with the least homology to off target sequences. Transcript profiling of the selected CcWRKY transcription factors was carried out using qRT-PCR according to the modified protocol of Marone et al.44 in three technical replicates per sample. Eva Green SYBR Green Supermix Kit (Bio-Rad) was used in a iQ5 Real-Time PCR Detection System (Bio-Rad Laboratories, Munchen, Germany) for qRT-PCR. We used gene-specific primers at a final concentration of 10pmol/µl and transcript levels of each mRNA were determined and normalized with the level of internal control. The PCR condition involved denaturation at 95 °C for 2 min, 40 repeats at 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s. Actin (Accession No. XM_020367686.1) and β-tubulin (Accession No. XM_020353202.1) were used as internal controls. Data normalization was done with mean CT values of the endogenous genes Actin and β-tubulin using the 2−ΔΔCT method45. Fold accumulation of transcripts was compared by using the mean of the CT values of the three technical replicates from each five biological replications with control.
Conclusions
WRKY genes family is known to be responsive to both biotic and abiotic stresses. We analyzed the transcript accumulation patterns of 35 biotic stress responsive CcWRKY genes against biotic (Fusarium udum) and abiotic (NaCl) stresses alone and in combination treatments in the F. udum resistant and susceptible pigeonpea cultivars ‘Asha’ and ‘Bahar’. A comparative expression profile of highly and lowly up-regulated CcWRKYs is presented in Supplementary Table S8 based on their responses in the resistant cultivar ‘Asha’ under the two individual stresses of F. udum and NaCl. We observed that in ‘Asha’, most of the selected CcWRKYs were up-regulated in all treatments except CcWRKY25 which was down-regulated. Similarly, CcWRKY40 was up-regulated in all treatments except in two treatments viz., F. udum and NaCl + OKC. However, differential expression patterns of CcWRKY genes were observed in ‘Bahar’ compared to ‘Asha’. In ‘Bahar’ eighteen CcWRKY genes (CcWRKYs 1, 2, 4, 5, 7, 10, 11, 12, 13, 14, 15, 23, 24, 25, 26, 27, 28,and 29) were up-regulated in all treatments while rest of the CcWRKY genes (CcWRKYs 20, 21, 30, 31, 32, 33, 34, 36, 37, 38, 39, 40, 41, 42, and 43) were differentially expressed in different treatments either lowly up-regulated or down-regulated. Interestingly, CcWRKY22 was down-regulated in all treatments in ‘Bahar’. To conclude, 26 CcWRKYs were observed to be highly F. udum responsive and 9 least responsive, and among the 35 CcWRKY genes selected 17 are better NaCl responsive compared to F. udum and 11 are dual responsive to both F. udum and NaCl. The PGPR strain P. fluorescens OKC proved to play a role in stimulating CcWRKY-mediated stress responsiveness in pigeonpea in individual stresses of F. udum and NaCl compared to their combined stresses.
Supplementary information
Author Contributions
B.K.S., V.K.G. and R.K.M. planned the experiments, G.K., R.B. and A.S. conducted the experiments, G.K., V.K.G., H.B.S., R.K.M. and B.K.S. wrote the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-50696-x.
References
- 1.Cormack RS, et al. Leucine zipper containing WRKY proteins widen the spectrum of immediate early elicitor-induced WRKY transcription factors in parsley. Biochim. Biophys. Acta. 2002;1576:92–100. doi: 10.1016/S0167-4781(02)00298-1. [DOI] [PubMed] [Google Scholar]
- 2.Eulgem T, Somssich IE. Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 2007;10:366–371. doi: 10.1016/j.pbi.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 3.Ciolkowski I, Wanke D, Birkenbihl RP, Somssich IE. Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol. Biol. 2008;68:81–92. doi: 10.1007/s11103-008-9353-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiong W, et al. Genome-wide analysis of the WRKY gene family in physic nut (Jatropha curcas L.) Gene. 2013;524:124–132. doi: 10.1016/j.gene.2013.04.047. [DOI] [PubMed] [Google Scholar]
- 5.Zheng, Z., Mosher, S. L., Fan, B., Klessig, D. F. & Chen, Z. Functional analysis of Arabidopsis WRKY25 transcription factor in plant defense against Pseudomonas syringae. BMC Plant Biol, 10.1186/1471-2229-7-2 (2007). [DOI] [PMC free article] [PubMed]
- 6.Lai, Z. Roles of Arabidopsis WRKY3 and WRKY4 transcription factors in plant responses to pathogens. BMC Plant Biol, 10.1186/1471-2229-8-68 (2008). [DOI] [PMC free article] [PubMed]
- 7.Chen L, Zhang L, Yu D. Wounding-induced WRKY8 is involved in basal defense in Arabidopsis. Mol. Plant-Microbe Interact. 2010;23:558–565. doi: 10.1094/MPMI-23-5-0558. [DOI] [PubMed] [Google Scholar]
- 8.Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/S1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 9.Wu KL, Guo ZJ, Wang HH, Li J. The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res. 2005;12:9–26. doi: 10.1093/dnares/12.1.9. [DOI] [PubMed] [Google Scholar]
- 10.Li HL, Zhang LB, Guo D, Li CZ, Peng SQ. Identification and expression profiles of the WRKY transcription factor family in Ricinus communis. Gene. 2012;503:248–253. doi: 10.1016/j.gene.2012.04.069. [DOI] [PubMed] [Google Scholar]
- 11.Tripathi, P. et al. The WRKY transcription factor family in Brachypodium distachyon. BMC Genomics, 10.1186/1471-2164-13-270 (2012). [DOI] [PMC free article] [PubMed]
- 12.Ishiguro S, Nakamura K. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 50 upstream regions of genes coding for sporamin and beta amylase from sweet potato. Mol. Gen. Genet. 1994;244:563–571. doi: 10.1007/BF00282746. [DOI] [PubMed] [Google Scholar]
- 13.Ling, J. et al. Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genomics, 10.1186/1471-2164-12-471 (2011). [DOI] [PMC free article] [PubMed]
- 14.Wei KF, Chen J, Chen YF, Wu LJ, Xie DX. Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in Maize. DNA Res. 2012;19:153–164. doi: 10.1093/dnares/dsr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang LN, et al. Genome-wide identification of WRKY family genes and their response to cold stress in Vitis vinifera. BMC Plant Biol. 2014;14:103. doi: 10.1186/1471-2229-14-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He H, et al. Genome-wide survey and characterization of the WRKY gene family in Populus trichocarpa. Plant Cell Rep. 2012;31:1199–1217. doi: 10.1007/s00299-012-1241-0. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Wang L. The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol. Biol. 2005;5:1–12. doi: 10.1186/1471-2148-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kassa MT, et al. Genetic patterns of domestication in pigeonpea (Cajanus cajan (L.) Millsp.) and wild Cajanus relatives. PLoS ONE. 2012;7(6):e39563. doi: 10.1371/journal.pone.0039563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie Z, et al. Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol. 2005;137:176–189. doi: 10.1104/pp.104.054312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ross CA, Liu Y, Shen QJ. The WRKY gene family in rice (Oryza sativa) J. Inter. Plant Biol. 2007;49:827–842. doi: 10.1111/j.1744-7909.2007.00504.x. [DOI] [Google Scholar]
- 21.Yamasaki K, et al. Solution structure of an Arabidopsis WRKY DNA binding domain. Plant Cell. 2005;17:944–956. doi: 10.1105/tpc.104.026435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duan MR, et al. DNA binding mechanism revealed by high resolution crystal structure of Arabidopsis thaliana WRKY1 protein. Nucleic Acids Res. 2007;35:1145–1154. doi: 10.1093/nar/gkm001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ke J, et al. Structural basis for recognition of diverse transcriptional repressors by the TOPLESS family of corepressors. Sci. Adv. 2015;1:e1500107. doi: 10.1126/sciadv.1500107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ikai Thermostability and aliphatic index of globular proteins. J. Biochem. 1980;88:1895–1898. [PubMed] [Google Scholar]
- 25.Zhou Q, et al. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 2008;6:486–503. doi: 10.1111/j.1467-7652.2008.00336.x. [DOI] [PubMed] [Google Scholar]
- 26.Zhang ZL, et al. A rice WRKY gene encodes a transcriptional repressor of the gibberellin signaling pathway in aleurone cells. Plant Physiol. 2004;134:1500–1513. doi: 10.1104/pp.103.034967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mangelsen E, et al. Phylogenetic and comparative gene expression analysis of barley (Hordeum vulgare) WRKY transcription factor family reveals putatively retained functions between monocots and dicots. BMC Genomics. 2008;9:194–211. doi: 10.1186/1471-2164-9-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Verk MC, Pappaioannou D, Neeleman L, Bol JF, Linthorst HJM. A novel WRKY transcription factor is required for induction of PR-1a gene expression by salicylic acid and bacterial elicitors. Plant Physiol. 2008;146:1983–1995. doi: 10.1104/pp.107.112789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang B, Jiang Y, Rahman MH, Deyholos MK, Nat NVK. Identification and expression analysis of WRKY transcription factor in canola (Brassica napus L.) in response to fungal pathogens and hormone treatments. BMC Plant Biol. 2009;9:68. doi: 10.1186/1471-2229-9-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giacomelli JI, Ribichich KF, Dezar CA, Chan RL. Expression analyses indicate the involvement of sunflower WRKY transcription factors in stress responses, and phylogenetic reconstructions reveal the existence of a novel clade in the Asteraceae. Plant Sci. 2010;178:398–410. doi: 10.1016/j.plantsci.2010.02.008. [DOI] [Google Scholar]
- 31.Guo C, et al. Evolution and expression analysis of the grape (Vitis vinifera L.) WRKY gene family. J. Exp. Bot. 2014;65:1513–1528. doi: 10.1093/jxb/eru007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Srivastava S, Patel JS, Singh HB, Sinha A, Sarma BK. Streptomyces rochei SM3 induces stress tolerance in chickpea against Sclerotinia sclerotiorum and NaCl. J. Phytopathol. 2015;163:583–592. doi: 10.1111/jph.12358. [DOI] [Google Scholar]
- 33.Ilangumaran G, Smith DL. Plant growth-promoting rhizobacteria in amelioration of salinity stress: a systems biology perspective. Front. Plant Sci. 2017;8:1768. doi: 10.3389/fpls.2017.01768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarma BK, Yadav SK, Singh S, Singh HB. Microbial consortium-mediated plant defense against phytopathogens: readdressing for enhancing efficacy. Soil Biol. Biochem. 2015;87:25–33. doi: 10.1016/j.soilbio.2015.04.001. [DOI] [Google Scholar]
- 35.Bai Y, Sunarti S, Kissoudis C, Visser RGF, van der Linden CG. The role of tomato WRKY genes in plant responses to combined abiotic and biotic stresses. Front. Plant Sci. 2018;9:801. doi: 10.3389/fpls.2018.00801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramamoorthy, R., Jiang, S. Y., Kumar, N., Venkatesh, P. N. & Ramachandran, S. A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant Cell Physiol. 49, 865–879. [DOI] [PubMed]
- 37.Wang F, et al. A novel cold inducible gene from Pak-choi (Brassica campestris ssp. chinensis), BcWRKY46, enhances the cold, salt and dehydration stress tolerance in transgenic tobacco. Mol. Biol. Rep. 2012;39:4553–4564. doi: 10.1007/s11033-011-1245-9. [DOI] [PubMed] [Google Scholar]
- 38.Yadav SK, Dave A, Sarkar A, Singh HB, Sarma BK. Co-inoculated biopriming with Trichoderma, Pseudomonas and Rhizobium improves crop growth in Cicer arietinum and Phaseolus vulgaris. Intern. J. Agri. Environ. Biotechnol. 2013;6:255–259. [Google Scholar]
- 39.Patel JS, et al. A Simple and efficient protocol for isolation of RNA from different tissues of chickpea and pea. Intern. J. Agri. Environ. Biotechnol. 2016;9:39–43. doi: 10.5958/2230-732X.2016.00006.1. [DOI] [Google Scholar]
- 40.Sambrook, J. & Russell, D. W. Molecular cloning: a laboratory manual. CSHL Press (2001).
- 41.Jin JP, et al. Plant TFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017;45(D1):D1040–D1045. doi: 10.1093/nar/gkw982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thornton B, Basu C. Real-Time PCR (qPCR) primer design using free online software. Biochem. Mol. Biol. Educ. 2011;39:145–154. doi: 10.1002/bmb.20461. [DOI] [PubMed] [Google Scholar]
- 44.Marone M, Mozzetti S, Ritis DD, Pierelli L, Scambia G. Semi-quantitative RT-PCR analysis to assess the expression levels of multiple transcripts from the same sample. Biol. Proceed. 2001;3:19–25. doi: 10.1251/bpo20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.