Abstract
Objectives
The necrotrophic fungal pathogen Pyrenophora tritici-repentis (Ptr) is the causal agent of tan spot a major disease of wheat. We have generated a new genome resource for an Australian Ptr race 1 isolate V1 to support comparative ‘omics analyses. In particular, the V1 PacBio Biosciences long-read sequence assembly was generated to confirm the stability of large-scale genome rearrangements of the Australian race 1 isolate M4 when compared to the North American race 1 isolate Pt-1C-BFP.
Results
Over 1.3 million reads were sequenced by PacBio Sequel small-molecule real-time sequencing (SRMT) cell to yield 11.4 Gb for the genome assembly of V1 (285X coverage), with median and maximum read lengths of 8959 bp and 72,292 bp respectively. The V1 genome was assembled into 33 contiguous sequences with a of total length 40.4 Mb and GC content of 50.44%. A total of 14,050 protein coding genes were predicted and annotated for V1. Of these 11,519 genes were orthologous to both Pt-1C-BFP and M4. Whole genome alignment of the Australian long-read assemblies (V1 to M4) confirmed previously identified large-scale genome rearrangements between M4 and Pt-1C-BFP and presented small scale variations, which included a sequence break within a race-specific region for ToxA, a well-known necrotrophic effector gene.
Keywords: Pyrenophora tritici-repentis, Genome assembly, PacBio sequel, Comparative analysis, ToxA, Tan spot, Yellow spot
Introduction
The necrotrophic fungal pathogen Pyrenophora tritici-repentis (Ptr) is the causal agent of tan (or yellow) spot disease of wheat (Triticum aestivum), which has a significant economic impact on the grain industry worldwide [1]. Ptr, a necrotrophic fungal pathogen is an ascomycete within the order Pleosporales, which also contains other important crop pathogens [2, 3]. The production of necrotrophic host-specific effectors contributes to the pathogenicity of this fungus, and three Ptr effectors have been described to date. ToxA and ToxB, are both well-characterised small effector proteins that produce necrosis and chlorosis symptoms respectively [4, 5], while ToxC, which also causes chlorosis, remains to be identified and may be the product of a secondary metabolite gene cluster [6].
The ToxA gene, believed to have been horizontally transferred to Ptr from Parastagonospora nodorum [7], can occupy different loci positions in different isolates and races of Ptr [8]. It has been proposed that this type of translocation could be the result of gene proximity to a chromosomal break point [9]. Recently, we confirmed large scale chromosomal rearrangements/fusions between two race 1 isolates sourced from North America (Pt-1C-BFP) [10] and Australia (M4) [11]. We therefore undertook PacBio Biosciences (PacBio) Sequel system for long-read sequencing of a second Australian Ptr isolate (V1), collected from a different geographic state, for whole genome comparison necessary to confirm these larger rearrangements and examine any further genomic variations. Furthermore, a high quality PacBio genome assembly enables the characterization of predicted secondary metabolite gene clusters and effectors often contained in highly complex genomic regions.
Main text
Methods
Isolate collection and sequencing
The pathogenic isolate V1 was isolated from tan spot infected leaves collected from Horsham, Victoria, Australia in 2015. V1 was cultured in vitro from a single spore [12].
V1 genomic DNA was extracted from 3-day old mycelia grown in vitro in Fries 3 liquid medium, using a BioSprint 15 DNA Plant Kit (Qiagen, Hilden, Germany) and automated workstation according to the manufacturer’s instruction [11]. DNA was further treated with 50 μg/ml of RNase enzyme (Qiagen, Hilden, Germany) for 1 h followed by phenol/chloroform extraction, followed by precipitation with sodium acetate and ethanol, and finally resuspension in TE buffer [11].
The V1 genome was sequenced using PacBio Biosciences (PacBio) Sequel small-molecule real-time sequencing (SMRT) Cell Technology at 283X to yield 11.4 Gb (1,319,569 long reads) by Novogene Co., Ltd, Hong Kong. The V1 genome was also sequenced via Illumina HiSeq 150 bp PE by Novogene Co., Ltd (Hong Kong) to yield 3.2 Gb at 80X coverage. Illumina read Phred quality score distribution was greater than 30, and reads containing adaptors were removed.
Plant materials and pathogenicity assays
To assess pathogenicity and race classification differential wheat genotypes, which differ in their effector sensitivities were used for inoculation [5, 13]. The wheat lines used were Glenlea and BG261 (both ToxA-sensitive), 6B662 (ToxB-sensitive) and 6B365 (ToxC-sensitive). Two week-old wheat (Triticum aestivum L.) seedlings were spore-inoculated by spraying the whole plants evenly with approximately 2000 conidia/ml and grown under controlled growth conditions [14]. The second leaves were harvested 7-days post-inoculation, visually inspected for symptoms and photographed. Infection experiments were repeated twice with four replicate plants per wheat line to demonstrate the pathogenicity and race classification.
Genome assembly of V1 and comparative analysis
PacBio sequence data was error-corrected and assembled using linux-amd64 Canu 1.8 software [15] guided by a genome size of 40 Mb. Illumina PE reads were quality trimmed for random hexamer primers on the 5′ read end using Trimmomatic v0.22 [16]. The high quality trimmed Illumina reads were aligned to the Canu genome assembly using BWA 0.7.14-r1138 [17] and filtered for concordant PE read alignments using samtools 0.1.19-96b5f2294a [18]. The Canu genome assembly was additionally corrected with the high quality Illumina alignments using Pilon 1.2 [19] to generate a final polished V1 sequence assembly with SNP and INDEL corrections.
V1 was then aligned to Pt-1C-BFP [10] and M4 [11] scaffolded chromosomes using NUCmer v3.1 (-maxmatch -coords). The sequence alignment for Pt-1C-BFP (NCBI Accession DS231618.1), M4 (chr6) and V1 (C11 reverse complemented and C12 reverse complemented) was conducted using Progressive Mauve 2015-02-25 [20], and EasyFig Version 2.2.3 [21], BLASTN, filter 500 bp length.
Gene prediction and functional annotation
M4 Illumina RNA-seq data [11] was aligned to V1 using TopHat v2.0.12 [22] (-N 2 -i 10 -I 5000 -p 16 –no-discord- ant –no-mixed –report-secondary-alignments –micro- exon-search –library-type fr-firststrand) for supporting ab initio gene predictions by CodingQuarry v1.2 [23] in pathogen mode (PM). Ab initio gene predictions were also made with GeneMark-ES v4.33 [24].
Pt-1C-BFP and M4 reference proteins [10] [11] were aligned to V1 using Exonerate v2.2.0 [25] (–showvulgar no –showalignment no –minintron 10 –maxintron 3000) in mode protein2genome. The ab initio gene predictions and exonerate alignments were then combined using EvidenceModeller v1.1.1 [26] with a minimum intron length of 10 bp and weightings of CodingQuarry:1, GeneMark.hmm:1, protein exonerate:2.
Gene annotations were assigned by BLASTX v2.2.26 searches across NCBI RefSeq and NR (taxon = Ascomycota) (October 2018) databases and RPS-BLAST v2.2.26 of Pfam, Smart and CDD domain databases (October 2018). Final gene annotations were summarised by AutoFACT v3.4 [27].
Pt-1C-BFP, M4 and V1 proteins were clustered to identify orthologous genes using OrthoFinder version 1.1.4 [28]. Predicted gene proteins for isolates Pt-1C-BFP, M4 and V1 were also assessed using BUSCO version3 [29] against Fungi OrthoDB version 9.
Results
Plant infection assays confirmed V1 as a race 1 isolate (producing ToxA and ToxC), by the presence of necrosis and chlorosis symptoms on the differential wheat lines Glenlea and 6B365 respectively, and absence of tan spot symptoms on Auburn and 6B662 (Fig. 1a). Furthermore, the induced chlorosis symptoms on the tan spot susceptible Australian commercial wheat cultivar Yitpi, also confirmed V1 pathogenicity (Fig. 1b).
Fig. 1.
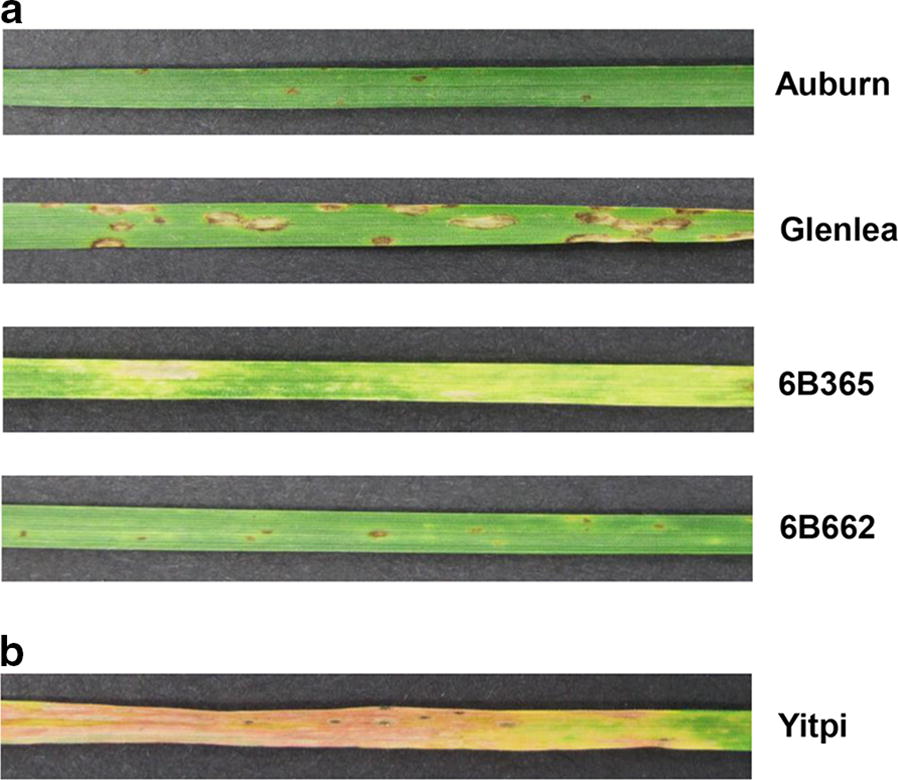
Pathogenicity assays of V1. a The presence of symptoms on the differential wheat lines Glenlea (ToxA sensitive) and 6B365 (ToxC sensitive), and the absence of tan spot symptoms on Auburn (insensitive) and 6B662 (ToxB sensitive) confirms a race 1 (ToxA and ToxC) classification for V1. b Susceptible disease reaction displayed on the Australian commercial wheat variety, Yitpi
The genomic sequence of V1 was assembled into 33 contiguous sequences with a of total length of 40,408,077 bp and a N50 of 3,421,861 bp. The mean and longest contig sizes were 1,224,487 bp and 9,664,470 bp respectively. V1 contig length statistics showed an improvement in comparison to M4 and Pt-1C-BFP (Table 1). A total of 14,050 protein coding genes were annotated for V1, which included the major effector ToxA (PtrV1_13859) positioned on contig12: 1,348,464–1,349,050. A total of 10,398 genes were orthologous to Pt-1C-BFP and M4. The V1 annotated genome has been deposited with National Center for Biotechnology Information (NCBI) GenBank under the accession SAXQ00000000.
Table 1.
Pyrenophora tritici-repentis race 1 isolate genome information and assembly statistics
| V1 | M4a | Pt-1C-BFPa | |
|---|---|---|---|
| Isolate information | |||
| Sequencing Platform | PacBio Sequel | PacBio RSII | Sanger, Illumina |
| Genome accession | SAXQ00000000 | NQIK00000000 | AAXI00000000 |
| Collection site | Victoria, Australia | Western Australia, Australia | South Dakota, USA |
| Collection year | 2015 | 2009 | 1994 |
| Contig assembly statistics | |||
| Total length (Mb) | 40.4 | 40.9 | 38.0 |
| Number | 33 | 51 | 48 |
| N50 (Mb) | 3.4 | 2.9 | 1.9 |
| Mean (Kb) | 1224 | 802 | 778 |
| Max (Mb) | 9.6 | 5.6 | 6.7 |
| GC % | 50.4 | 50.7 | 50.8 |
| Gene predictions | |||
| Number | 14,050 | 13,797 | 12,171 |
| %bBUSCO gene | 96.55 | 98.62 | 98.62 |
aPreviously published genome assemblies
bComplete and fragmented gene
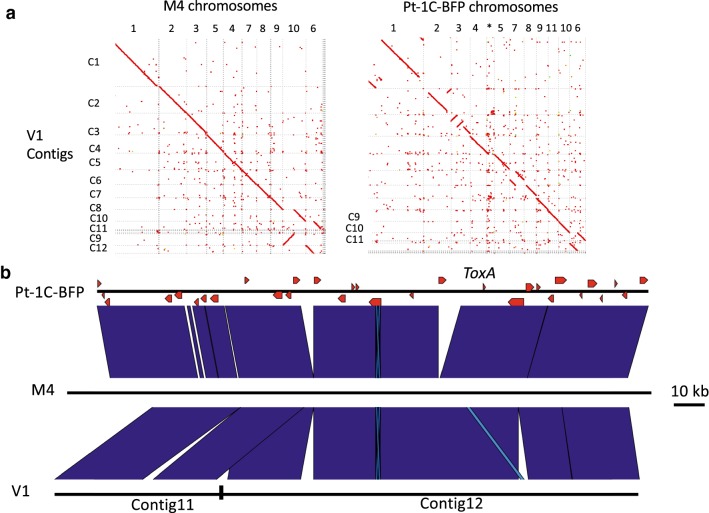
Whole genome alignment of V1 to M4 (PacBio RSII) and Pt-1C-BFP (Sanger) confirmed the same large-scale rearrangements in V1 that were found in M4 for chr3, chr7 and the distal regions of chr1 and 2 when compared to Pt-1C-BFP (Fig. 2a). However, V1 had sequence breaks at points not observed for M4 chr6 and observed for chr10 (which were confirmed previously by M4 optical mapping) [11]. The sequence alignment of M4 (chr6:1.57–1.75 Mb), Pt-1C-BFP (chr6 1.32–1.50) and V1 (contigs 11 and 12) highlight the sequence break point in V1 76,077 bp upstream of the ToxA effector gene (Fig. 2b), that was not found in M4.
Fig. 2.
a Dot plots showing the whole genome nucleotide alignments for V1 contigs (vertical axis) to M4 and Pt-1C-BFP scaffolded genomes (horizontal axes). *Pt-1C-BFP contigs that are not assigned to a Pt-1C-BFP chromosome. b Sequence alignment showing Pt-1C-BFP Chr6:1.32–1.50 Mb, M4 Chr6:1.57–1.75 Mb and V1 contigs11 and 12 (both reverse complemented). The V1 contig break point is indicated 76 kb upstream of the ToxA locus (red arrow labelled as ToxA)
Discussion
The PacBio sequence for V1 had longer assembly statistics when compared to the recent M4 PacBio RSII (six SMRT cells) assembly, possibly due to the higher depth of coverage obtained from the PacBio Sequel system for overlap assembly.
The sequence comparison of V1 to both M4 and Pt-1C-BFP confirmed the chromosomal rearrangements for M4 chr1, chr2, chr3 and chr7 for the Australia isolates, which appears stable despite isolates being collected from different geographic states. Smaller scale differences between V1 and M4 were however detected in chr4, chr6, chr7 and chr8. Also, V1 had a sequence break upstream of ToxA with a significant sequence variation between Pt-1C-BFP and M4 in both length and complexity that was not resolved by assembly. These larger and smaller scale rearrangements can impact gene clusters, especially when they are proximal to complex sub-telomeric regions and breakpoints. This resource will therefore be useful for future ‘omics experiments and comparative Ptr genomic analyses.
Limitations
Although all methods have been made as consistent as possible for comparative analyses, this analysis has used databases, software and PacBio sequencing versions currently available, which may be updated in the future. The comparison of two Australian long-read assemblies is only an indication of potential genome stability in Australia.
Acknowledgements
We thank the Australian Government National Collaborative Research Infrastructure Strategy (NCRIS) for providing access to Pawsey Supercomputing under a National Computational Merit Allocation Scheme (NCMAS), Nectar Research/Pawsey Nimbus Cloud resources. We also thank Mr John Robertson for the provision of tan spot leaf samples from Horsham, Victoria, and Kalai Marathamuthu (Curtin University) for assistance with the pathogenicity assays.
Abbreviations
- Ptr
Pyrenophora tritici-repentis
- PacBio
PacBio Biosciences
- SRMT
small-molecule real-time sequencing
- NCBI
National Centre for Biotechnology Information
Authors’ contributions
PM conducted the bioinformatics analysis and wrote the draft manuscript. PTS and CM conducted the molecular analysis. CM led the project conceptualization. All authors contributed to reviewing and editing this manuscript. All authors read and approved the final manuscript. All authors agree to the publication policies of BMC Research Notes.
Funding
The analysis, data interpretation and writing of the manuscript were generously funded through the Grains and Research Development Corporation (project code CUR0023).
Availability of data and materials
All data generated or analysed during this study are included in this article. The assembled and annotated genome for isolate V1 was submitted to NCBI GenBank repository under accession SAXQ00000000.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Paula Moolhuijzen, Email: paula.moolhuijzen@curtin.edu.au.
Pao Theen See, Email: paotheen.see@curtin.edu.au.
Caroline S. Moffat, Email: caroline.moffat@curtin.edu.au
References
- 1.Moffat CS, Santana MF. Diseases affecting wheat: tan spot. In: Oliver R, editor. Integrated disease management of wheat and barley. Cambridge: Burleigh dodds; 2018. [Google Scholar]
- 2.Oliver R, Tan K, Moffat C. Necrotrophic pathogens of wheat. Encycl Food Grains. 2016;4:273–278. doi: 10.1016/B978-0-12-394437-5.00240-0. [DOI] [Google Scholar]
- 3.Ellwood SR, Syme RA, Moffat CS, Oliver RP. Evolution of three Pyrenophora cereal pathogens: recent divergence, speciation and evolution of non-coding DNA. Fungal Genet Biol. 2012;49(10):825–829. doi: 10.1016/j.fgb.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Strelkov SE, Lamari L, Ballance GM. Characterization of a host-specific protein toxin (Ptr ToxB) from Pyrenophora tritici-repentis. Mol Plant Microb Interact. 1999;12(8):728–732. doi: 10.1094/MPMI.1999.12.8.728. [DOI] [Google Scholar]
- 5.Ciuffetti LM, Tuori RP, Gaventa JM. A single gene encodes a selective toxin causal to the development of tan spot of wheat. Plant Cell. 1997;9(2):135–144. doi: 10.1105/tpc.9.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Effertz RJ, Meinhardt SW, Anderson JA, Jordahl JG, Francl LJ. Identification of a chlorosis-inducing toxin from Pyrenophora tritici-repentis and the chromosomal location of an insensitivity locus in wheat. Phytopathology. 2002;92(5):527–533. doi: 10.1094/PHYTO.2002.92.5.527. [DOI] [PubMed] [Google Scholar]
- 7.Friesen TL, Stukenbrock EH, Liu Z, Meinhardt S, Ling H, Faris JD, et al. Emergence of a new disease as a result of interspecific virulence gene transfer. Nat Genet. 2006;38(8):953–956. doi: 10.1038/ng1839. [DOI] [PubMed] [Google Scholar]
- 8.Aboukhaddour R, Cloutier S, Ballance GM, Lamari L. Genome characterization of Pyrenophora tritici-repentis isolates reveals high plasticity and independent chromosomal location of ToxA and ToxB. Mol Plant Pathol. 2009;10(2):201–212. doi: 10.1111/j.1364-3703.2008.00520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bertazzoni S, Williams AH, Jones DA, Syme RA, Tan KC, Hane JK. Accessories make the outfit: accessory chromosomes and other dispensable DNA regions in plant-pathogenic fungi. Mol Plant Microb Interact. 2018;31(8):779–788. doi: 10.1094/MPMI-06-17-0135-FI. [DOI] [PubMed] [Google Scholar]
- 10.Manning VA, Pandelova I, Dhillon B, Wilhelm LJ, Goodwin SB, Berlin AM, et al. Comparative genomics of a plant-pathogenic fungus, Pyrenophora tritici-repentis, reveals transduplication and the impact of repeat elements on pathogenicity and population divergence. G3 (Bethesda) 2013;3(1):41–63. doi: 10.1534/g3.112.004044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moolhuijzen P, See PT, Hane JK, Shi G, Liu Z, Oliver RP, et al. Comparative genomics of the wheat fungal pathogen Pyrenophora tritici-repentis reveals chromosomal variations and genome plasticity. BMC Genomics. 2018;19(1):279. doi: 10.1186/s12864-018-4680-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moffat CS, See PT, Oliver RP. Leaf yellowing of the wheat cultivar Mace in the absence of yellow spot disease. Australas Plant Pathol. 2015;44(2):161–166. doi: 10.1007/s13313-014-0335-2. [DOI] [Google Scholar]
- 13.Lamari L, Strelkov SE, Yahyaoui A, Orabi J, Smith RB. The identification of two new races of Pyrenophora tritici-repentis from the host center of diversity confirms a one-to-one relationship in tan spot of wheat. Phytopathology. 2003;93(4):391–396. doi: 10.1094/PHYTO.2003.93.4.391. [DOI] [PubMed] [Google Scholar]
- 14.Moffat CS, See PT, Oliver RP. Generation of a ToxA knockout strain of the wheat tan spot pathogen Pyrenophora tritici-repentis. Mol Plant Pathol. 2014;15(9):918–926. doi: 10.1111/mpp.12154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27(5):722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE. 2014;9(11):e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14(7):1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27(7):1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Testa AC, Hane JK, Ellwood SR, Oliver RP. CodingQuarry: highly accurate hidden Markov model gene prediction in fungal genomes using RNA-seq transcripts. BMC Genomics. 2015;16:170. doi: 10.1186/s12864-015-1344-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borodovsky M, Lomsadze A. Eukaryotic gene prediction using GeneMark.hmm-E and GeneMark-ES. Curr Protoc Bioinformatics. 2011;6:1–10. doi: 10.2174/157489311795222356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slater GS, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haas BJ, Salzberg SL, Zhu W, Pertea M, Allen JE, Orvis J, et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 2008;9(1):R7. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koski LB, Gray MW, Lang BF, Burger G. AutoFACT: an automatic functional annotation and classification tool. BMC Bioinformatics. 2005;6:151. doi: 10.1186/1471-2105-6-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emms DM, Kelly S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015;16:157. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seppey M, Manni M, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness. Methods Mol Biol. 2019;1962:227–245. doi: 10.1007/978-1-4939-9173-0_14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this article. The assembled and annotated genome for isolate V1 was submitted to NCBI GenBank repository under accession SAXQ00000000.