Abstract
Background
This study aimed to identify NOTCH3 mutations and describe the genetic and clinical features and magnetic resonance imaging results in 11 unrelated patients with cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL) from Henan province in China.
Material/Methods
NOTCH3 was directly sequenced in 11 unrelated patients of Chinese descent. The clinical presentations and magnetic resonance imaging features were retrospectively analyzed in the 11 index patients with a definite diagnosis.
Results
Seven different mutations were identified in 11 unrelated patients, including 4 novel mutations (p.P167S, p.P652S, p.C709R, and p.R1100H) in China and 3 reported mutations (p.C117R, p.R578C, and p.R607C). Four novel mutations (p.P167S, p.P652S, p.C709R, and p.R1100H) were predicted to be probably pathogenic using an online pathogenicity prediction program through comprehensive analysis. Clinical presentations in symptomatic patients included stroke, cognitive decline, psychiatric disturbances, and migraine. Multiple lacunars infarcts and leukoaraiosis were detected on MRI in most symptomatic patients, while white-matter lesions were identified in the temporal pole or the external capsule in all affected patients.
Conclusions
The mutation spectrum of CADASIL patients from Henan province in China displayed some differences from that of those reported previously. DNA sequencing was used to diagnose all 11 patients as having CADASIL, and we found 4 novel mutations. The present results further contribute to the enrichment of NOTCH3 mutation databases.
MeSH Keywords: CADASIL; Magnetic Resonance Imaging; Mutation; Receptors, Notch
Background
Cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL) is an inherited disease of the microvasculature caused by NOTCH3 mutations, characterized by migraine with aura, ischemic stroke, mood disturbances, apathy, and cognitive impairment and dementia [1]; however, it is not strongly associated with typical cardiovascular risk factors. Magnetic resonance imaging (MRI) analyses have revealed leukoaraiosis with marked abnormal signals in the white matter, predominantly in the anterior part of the temporal lobes and the external capsule [2]. The existence of granular osmiophilic material (GOM) in the vascular smooth muscle cells observed under an electron microscope of skin biopsy samples suggests a diagnosis of CADASIL, for which NOTCH3 genetic testing is the criterion standard [1,3]. GOM has a high specificity and sensitivity [4]; however, surgery is a highly invasive treatment method. Hence, NOTCH3 gene analysis has greater applications.
NOTCH3 comprises 33 exons encoding a large transmembrane receptor with 34 extracellularly localized epidermal growth factor-like (EGF) repeat domains. Mutations are highly typical and result in the number of cysteine residues in the EGF-like domains in the extracellular regions of transmembrane NOTCH3. Each EGF-like domain contains 3 paired cysteine residues. Recent studies have reported the presence of a cluster of mutations with conserved cysteine residues; however, pathogenesis has not been confirmed yet [5].
Most studies have reported mutation hotspots in exons 3 and 4, which encode the first 5 EGF-like repeats of NOTCH3. However, some studies on different geographical populations report a different distribution of hotpot mutations. Mutations p.R544C in exon 11 in Taiwanese patients [6], p.R607C in exon 11 and p.G528C in exon 10 in Italian patients [7], p.R1006C in exon 19 and p.R1231C in exon 22 in Italian patients [8], and p.R133C in exon 4 in Finnish patients [9] are reported as hotspot mutations. The present study aimed to investigate the mutation spectrum of NOTCH3 in 11 unrelated Chinese patients with CADASIL from Henan province in China, and retrospectively analyze their clinical manifestation and MRI imaging data.
Material and Methods
Subjects
Eleven unrelated index patients of Chinese Han descent from Henan province were assessed. All these patients were hospitalized to Henan Provincial People’s Hospital Department of Neurology for a suspected diagnosis of CADASIL. Ethics approval was given by the Medical Ethics Committee of Henan Provincial People’s Hospital. All patients underwent brain MRI, which revealed markedly high-intensity signals in the temporal pole and/or the external capsule upon T2-weighted imaging (T2WI) or fluid-attenuated inversion recovery (FLAIR) imaging, and had any properties as follows: the onset of lacunar lesions or transient ischemic attacks at a young age, aura migraine, and ischemic stroke or cognitive impairment in the family history. All patients provided informed consent for genetic analysis, and NOTCH3 mutations were screened.
Clinical assessment
All participants were examined and interviewed by at least 1 experienced neurologist and were diagnosed according to the criteria of CADASIL [10]. The presence of vascular risk factors, CADASIL symptoms, and medication use were recorded.
Cognitive function was assessed using the mini-mental state examination (MMSE), Montreal cognitive assessment (MOCA), and memory and executive screening (MES). Further analyses were conducted in accordance with the scores of MMSE, MoCA, and MES.
Imaging analysis
Multimodal MRI was conducted using a Siemens Magnetom Trio, Tim 3.0 T system (http://www.siemensmriequipment.com), using commercially available hardware and software. The following sequences were obtained: DWI (TR=3100 ms, TE=82 ms, b-value=1000 s/mm2, acquisition matrix=128×128, FOV=240 mm, section thickness=6.0 mm, section gap=1.2 mm, duration=48 s). Susceptibility-weighted imaging (SWI) (TR/TE/flip angle=27 ms/20 ms/15°, FOV=240 mm, acquisition matrix=384×32, section thickness=3.0 mm, duration=3.0 min 27 s). Three-dimensional (3D) time-of-flight (TOF) MR angiography (MRA; TR/TE/flip angle=20 ms/3.2 ms/18°, FOV=230 mm, acquisition matrix=256×256, section thickness=0.9 mm, duration=3.0 min 6.0 s) was performed.
All patients underwent MRI, including conventional T1WI, T2WI, diffusion-weighted imaging, and fluid-attenuated inversion recovery and SWI sequences.
NOTCH3 mutation screening and sequencing analysis
The 23 exons (exons 2–24) encoding the 34 EGF-like repeats were amplified via polymerase chain reaction (PCR) of genomic DNA with 23 sets of oligonucleotide primers specific for NOTCH3 (NG_009819.1, http://www.ncbi.nlm.nih.gov/nuccore/223972661). The sequences of these primers are shown in Supplementary Table 1. PCR was performed in a total reaction volume of 50 μl comprising 25 μl polymerase (Bioindustry, TaKaRa, BIO INC, Japan), 16 μl sterile water, 5 μl DNA (100 ng/μl), and optimized primer mix. PCR conditions were as follows: initial denaturation for 5 min at 94°C, followed by 35 cycles at 94°C for 30 s (denaturation), 65°C for 30 s (annealing) and 72°C for 1 min (extension), followed by final extension for 7 min at 72°C. PCR products were assessed via electrophoresis on a 2% low-melting-point agarose gel, followed by phenol-chloroform extraction and ethanol precipitation.
Purified PCR products were directly sequenced using an ABI3100 automated sequencer (Applied Biosystems, Foster City, CA, USA). Sequencing reads and lists were compared using the Chromas software and NCBI blast. Human NOTCH3 cDNA and protein sequences were obtained from GenBank (accession NG_009819.1). Genetic analysis of NOTCH3 mutations was performed in all 11 patients. We analyzed co-segregation if any variant was found, and direct DNA sequencing was used to examine the presence of all identified, novel, disease-associated variants in 300 controls. Pathogenicity of the identified missense variants was predicted using the following bioinformatics tools: PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/), sorting intolerant from tolerant (SIFT) (http://sift.jcvi.org/) and MutationTaster (http://www.mutationtaster.org/). Evolutionary conservation of the mutated amino acids was evaluated using NCBI gene HomoloGene (http://www.ncbi.nlm.nih.gov/pubmed) and MutationTaster (http://www.mutationtaster.org/). The structures of native and mutant NOTCH3 were generated by the SWISS-MODEL prediction program (https://www.swissmodel.expasy.org/).
Results
NOTCH3 mutations analysis
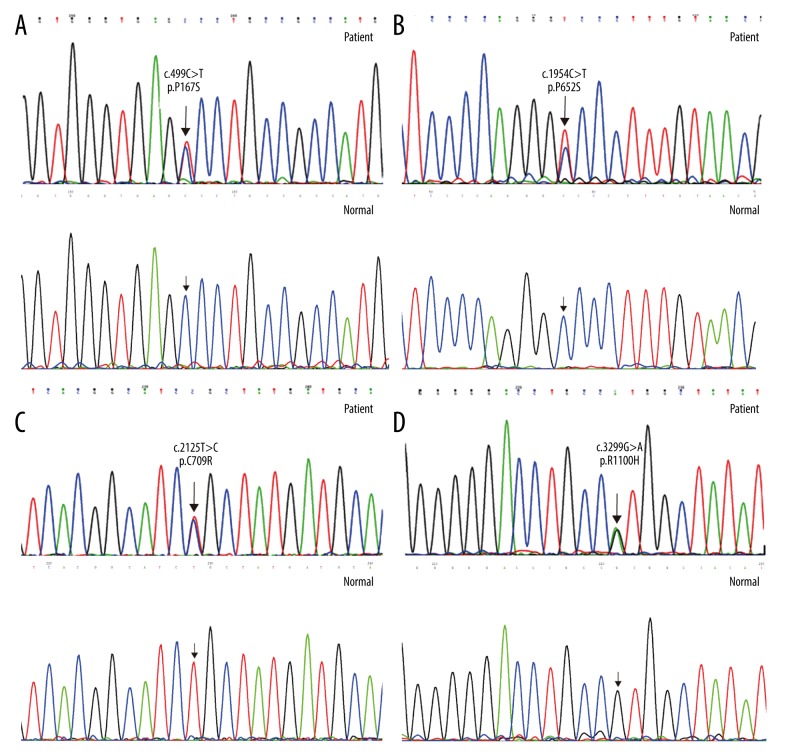
NOTCH3 mutations were found in 11 (52.3%) out of 21 patients with clinically suspected CADASIL. Seven different mutations located in exons 3, 4, 11, 13, and 20, respectively, were detected in 11 index patients (Table 1). Three mutations (p.C117R, p.R578C, and p.R607C) were reported in the literature, while 3 novel mutations (p.P652S, p.C709R, and p.R1100H) were rarely reported in the literature. In addition, although p.P167S mutation was previously reported in Japan and Korea, it was detected for the first time in the Chinese population. Mutations in patients 2, 9, and 11 did not alter the number of cysteine residues; however, they occurred at codons proximal to others where changes in cysteine residues reportedly occur. Mutation in patient 10 altered the number of cysteine residues. In patient 2, c.499C>T resulted in the replace of a proline residue for a serine residue in codon 167. The cysteine-sparing mutation p.P167S was detected in only 2 patients in Japan and Korea, but it is the first time it has been reported in a Chinese population. In patient 9, c.1954 C>T resulted in the replace of a proline residue for a serine residue in codon 652. In patient 11, c.3299G>A resulted in the replace of an arginine residue for a histidine residue in codon 1100. These 2 cysteine-sparing mutations (p.P652S and p.R1100H) have not yet been recorded in the Human Gene Mutation Database (HGMD) and therefore were described as novel mutations. In patient 10, c.2125 T>C resulted in the replacement of an arginine residue for a histidine residue in codon 709. p.C709R is a novel mutation altering cysteine residues and was not recorded in HGMD. The Sanger Sequencing chromatograms for these 4 novel mutations and their wild types are shown in Figure 1.
Table 1.
Molecular characteristics and clinical data of the 11 index patients with cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy mutations.
| Index patients (proband) | Sex | Age | Family history | Sign/symptoms | CADASIL mutation Exon/amino acid change | Stroke risk factors | MMSE(30)/MoCA(30) |
|---|---|---|---|---|---|---|---|
| 1 | M | 51 | − | Stroke, pseudobulbar palsy | 3/p.C117R | Smoking | 29/27 |
| 2 | F | 72 | − | Stroke, migraine, cognitive decline, gait disturbance, pseudobulbar palsy | 4/p.P167S | No | 22/15 |
| 3 | F | 66 | − | Migraine, emotional disorders | 11/p.R578C | No | 26/23 |
| 4 | F | 39 | + | Stroke | 11/p.R607C | Diabetes | - |
| 5 | M | 53 | − | Stroke, hemorrhage | 11/p.R607C | Hypertension | 29/26 |
| 6 | F | 52 | + | Stroke, migraine | 11/p.R607C | No | 30/28 |
| 7 | M | 72 | − | Stroke, pseudobulbar palsy, cognitive decline, gait disturbance | 11/p.R607C | Diabetes, hypertension smoking | 17/9 |
| 8 | M | 43 | − | Stroke, pseudobulbar palsy | 11/p.R607C | No | 19/13 |
| 9 | F | 81 | − | Stroke, cognitive decline | 13/p.P652S | No | 10/2 |
| 10 | M | 47 | − | Stroke, cognitive decline, pseudobulbar palsy, emotional disorders | 13/p.C709R | Smoking | 25/14 |
| 11 | F | 52 | − | Memory impairment, migraine, urgent urination | 20/p.R1100H | Hypertension | 30/28 |
Figure 1.
The DNA sequencing chromatograph of the NOTCH3 gene. (A) c.499C>T (p. P167S) mutation (patient) and wild type (normal); (B) c.1594C>T (p. P652S) mutation (patient) and wild type (normal); (C) c.2125C>T (p. C709R) mutation (patient) and wild type (normal); (D) c.3299G>A (p. R1100H) mutation (patient) and wild type (normal).
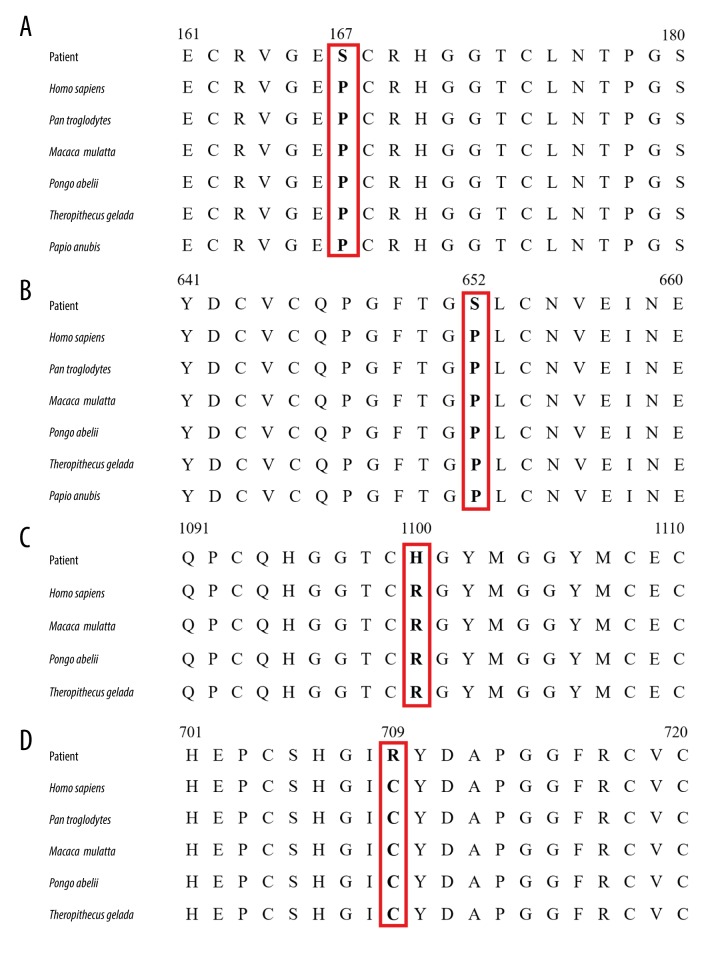
Since these 4 novel mutations were not found in the dbSNP database, Genome Aggregation Database (gnomAD) database, Exome Aggregation Consortium (ExAC) database (ExAC-ALL and ExAC-EAS), and 1000 Genomes Project, we also screened these mutations among 300 unrelated healthy control subjects, and none of them harbored these mutations. Using SIFT, PolyPhen_2, and Mutation Taster online program, p.C709R mutation was predicted to be deleterious, probably damaging and disease-causing, respectively; p.P167S mutation was predicted to be deleterious, benign and polymorphism, respectively. p.P652S mutation was predicted to be neutral, possibly damaging and disease-causing, respectively; and p.R1100H mutation was predicted to be neutral, benign and disease-causing, respectively. Through comprehensive analysis, 4 mutations were predicted to be probably pathogenic using the online prediction program. The evolutionary conservation analysis revealed that the 4 novel mutations resulted in a highly conserved amino acid change (Figure 2).
Figure 2.
Evolutionary conservation analysis of 4 NOTCH3 mutations (p.P167S, p.P652S, p.R1100H, p.C709R; red boxes) (A–D). Protein sequences of Homo sapiens (NP_000426.2), Pan troglodytes (XP_009433184.1), Macaca mulatta (XP_014978652.1), Pongo abelii (XP_002828866.1), Theropithecus gelada (XP_025221819.1) and Papio anubis (XP_021786500.1) were retrieved from GenBank.
Clinical and MRI features
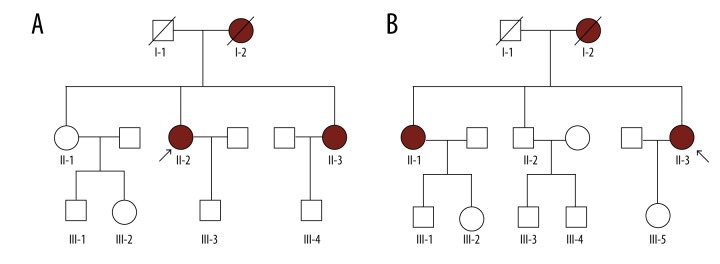
MRI and clinical features of these 11 index patients are presented in Tables 1 and 2. All patients were identified to harbor NOTCH3 mutations. None of the 11 index patients had a family except patients 4 and 6, whose family pedigrees are shown in Figure 3.
Table 2.
The primary imaging findings of the 11 index patients with cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy mutations.
| Index patients (proband) | Sex | Age | Stroke | White matter hyperintensity (WMH) | Lacunar lesions | Hemorrhage | Cerebral microbleeds (CMBs) | Dilated perivascular spaces (dPVS) | Brain atrophy |
|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 51 | Corpus callosum, centrum semiovale | Temporal pole, external capsule, periventricular area | Frontal lobe, basal ganglia | – | – | Basal ganglia | – |
| 2 | F | 72 | Centrum semiovale, corona radiata, posterior watershed | Temporal pole, external capsule, periventricular area | Frontal lobe, parietal lobe, basal ganglia | – | – | Basal ganglia, subcortex | – |
| 3 | F | 66 | Periventricular area | Temporal pole, external capsule, centrum semiovale | Frontal lobe, parietal lobe, basal ganglia | – | – | Basal ganglia, subcortex | – |
| 4 | F | 39 | Centrum semiovale, corona radiata, thalamus | Temporal pole, centrum semiovale, periventricular area | Occipital lobe | – | – | Basal ganglia | – |
| 5 | M | 53 | Centrum semiovale, corona radiata | Temporal pole, external capsule, centrum semiovale | Frontal lobe, basal ganglia | Brainstem, cerebellum, thalamus, basal ganglia, external capsule | – | Basal ganglia | – |
| 6 | F | 52 | Periventricular area, thalamus, frontal lobe | Temporal pole, centrum semiovale | Frontal lobe, basal ganglia | Brainstem, thalamus | – | Basal ganglia | – |
| 7 | M | 72 | Centrum semiovale, corona radiata, thalamus, basal ganglia, pons | Temporal pole, periventricular area, centrum semiovale | Frontal lobe, parietal lobe, basal ganglia | – | frontal lobe | Basal ganglia, subcortex | + |
| 8 | M | 43 | Basal ganglia, brainstem, callosum, frontotemporal lobe, hemicerebrum | Temporal pole, basal ganglia, external capsule | Occipital lobe | Basal ganglia | – | Basal ganglia | – |
| 9 | F | 81 | Thalamus, basal ganglia | Temporal pole, external capsule, periventricular area, centrum semiovale | Frontal lobe, parietal lobe, basal ganglia | – | – | Basal ganglia, subcortex | + |
| 10 | M | 47 | Pons, basal ganglia, thalamus, corona radiata, centrum semiovale | Temporal pole, centrum semiovale, corpus callosum, occipital lobe | Occipital lobe | – | – | Basal ganglia | + |
| 11 | F | 52 | Basal ganglia, thalamus, brainstem, cerebellum | Temporal pole, external capsule | Frontal lobe, occipital lobe, basal ganglia | – | – | Basal ganglia | – |
Figure 3.
Family pedigree based on a novel mutation in NOTCH3. (A) Family 4 I-2D: female, 35 years old, dead; II-2P (index patient 4, exon11 mutation/p.R607C): female, 39 years old; III-3: female, 22 years old. (B) Family 6 I-2D: female, 67 years old, dead; II-1: female, 62 years old; II-3P (index patient 6, exon20 mutation/p.R1100H): female, 52 years old. P – proband; D – dead.
The 11 patients included 5 men and 6 women, aged 39 to 81 years [57.1±13.5 years]. As shown in Tables 1 and 2, 2 patients had a history of smoking, 2 patients had hypertension, 1 patient had diabetes, and one patient had diabetes, hypertension, and a history of smoking. All patients presented a loss of white matter, 11 patients had a history of cerebral infarction, 4 patients had symptoms of migraine, 4 patients presented cognitive decline, notably 2 patients in their 40s. One patient initially experienced cognitive decline during the first stroke at the age of 47 years, while another patient with a long history of infarction at the age of 43 years did not have a good recovery. Two patients experienced psychiatric disturbances in the form of depression. Seven patients had symptoms of fatigue ascribed to subcortical lesions.
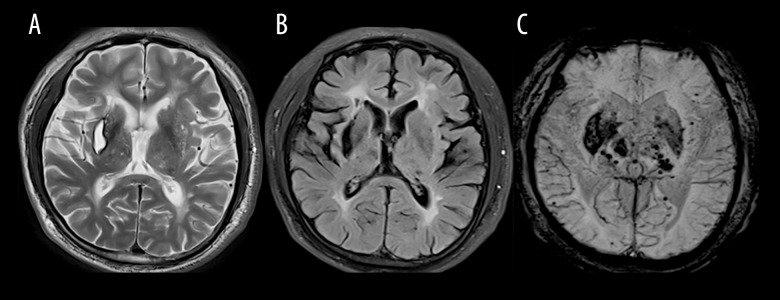
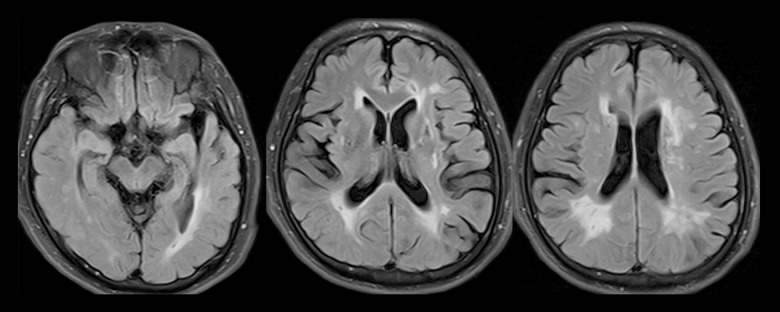
Brain MR images obtained for all 11 index patients revealed largely symmetric, diffuse white-matter abnormalities and lesions suggestive of small, subcortical infarcts. Affected areas of infarction were as follows: 5 cases in the centrum semiovale, 6 cases in the thalamus, 6 cases in the basal ganglia, 2 cases in the brainstem, 2 cases in the corpus callosum, 3 cases in the periventricular, and 1 case in the cerebral watershed (Table 2). WMH mainly occurred in the temporal pole (11 cases), external capsule (7 cases), periventricular area (5 cases), centrum semiovale (7 cases), corpus callosum (1 case), occipital lobe (1 case), and basal ganglia (1 case) (Table 2). The areas of lacunar lesions were as follows: 8 cases in the basal ganglia, 7 cases in the frontal lobe, 4 cases in the parietal lobe, and 4 cases in the occipital lobe (Table 2). The areas of WMH were as follows: 8 cases in the basal ganglia. Dilated perivascular spaces (dPVS) were located in the basal ganglia (11 cases) and partly in the subcortex (4 cases) (Table 2). The index patient 5 with hypertension experienced cerebral hemorrhage in the external capsule, which is a common location for hypertensive cerebral hemorrhage (Figure 4). The index patient 10 presented more atrophy of the cerebral hemisphere on the right side than on the left side on MRI (Figure 5). All these patients had typical radiologic markers for the temporal pole or capsular external hyperintensity on MRI.
Figure 4.
(A) T2-weighted magnetic resonance (MR) image showing an old hemorrhage in the right external capsule in index patient 5; (B) FLAIR sequence showing severe leukoencephalopathy in the anterior and posterior horn of lateral ventricle and an old hemorrhage in the right external capsule; (C) Susceptibility-weighted imaging (SWI) showing an old hemorrhage in the right external capsule and previous hemorrhage in the thalamus-capsular area and numerous microbleeds (MBs) in the basal ganglia.
Figure 5.
Fluid-attenuated inversion recovery magnetic resonance image showing asymmetric atrophy (Right side >Left side) and severe leukoencephalopathy in the anterior and posterior horns of the lateral ventricle in index patient 10 (p.C709R).
Discussion
In the present study, we identified 7 NOTCH3 mutations in 11 patients. To date, 3 large-scale studies have been conducted on CADASIL patients of Chinese descent [6,11,12]. There were 10 NOTCH3 p.R544C mutations in 21 unrelated CADASIL Taiwanese families, mainly presenting in exon 11 [6]. Wang et al. also reported that in 33 mainland Chinese CADASIL families, the mutation frequency was the highest in exon 4 (55%) and exon 3 (30%) [11]. Yin et al. investigated 35 unrelated Eastern Chinese families with CADASIL, and their results agreed with those of Wang et al. [11,12]. However, the present results are significantly different from previous results. e found that mutation frequency was the highest in exon 11 and the most common mutation was p.R607C. Five patients with the mutation p.R607C experienced unilateral limb weakness, which did not affect their activities of daily living; those patients were neither from the same village nor town, and did not have blood lineage and consanguineous marriage, and thus it is unclear why they carried the same mutation. Other primary features included pseudobulbar palsy, cognitive decline, gait disturbance, and emotional disorders in different individuals. p.R607C is most probably the ancestral mutation in Henan province in China. This study supports the assumption that NOTCH3 mutations in CADASIL resulted from one and possibly more ancestors in China [11]. Further cases have been reported from around China and further investigations are required to confirm the ancestral mutations in mainland China.
p.C709R is a novel mutation that alters cysteine residues by destabilizing disulfide linkages in cysteine residues, consequently altering the structure of NOTCH3. According to the online prediction program, the p.C709R mutation is probably pathogenic. The predicted structure of the 3-dimensional (3-D) protein for the mutation was different from that of the wild type. The index patient with mutation C709R was 47 years old when he first experienced a stroke. The clinical-onset manifestation was pseudobulbar palsy without gait disturbance. Later, he experienced a wide decline in memory, visuospatial ability, calculation, and executive function. Furthermore, he experienced psychiatric disturbances in the form of depression. We investigated his history and found that his father had the same clinical manifestation and symptoms before death. Brain MRI revealed asymmetric cerebral atrophy with atypical features of CADASIL. The abnormal signal of white matter in the occipital lobe was less severely affected [13].
p.P167S, p.P652S, and p.R1100H are 3 novel mutations with unaffected cysteine residues in China. p.P652S and p.R1100H have not yet been reported. Although the p.P167S mutation has been reported in Japan and Korea [14,15], this is its first report in the Chinese population. There are many arguments about whether the novel p.P167S mutation predicts the patient’s state of illness. Online pathogenicity prediction of the functional effect of p.P167S showed different results (deleterious, benign, and polymorphism, respectively). Mutations not altering cysteine residues are associated with CADASIL, especially those altering an arginine residue in the extracellular domain [16]. All these patients presented clinical presentations and MRI features typical of CADASIL. These patients showed typical CADASIL clinical manifestations and positive family history, and were confirmed to show genealogy co-segregation. The predicted structures of the 3-dimensional protein for 3 mutations were different from that of the wild type. A comprehensive comparison of the results of SIFT, PolyPhen_2, and MutationTaster online program in combination with clinical features revealed that the 3 cysteine-sparing mutations (p.P167S, p.P652S, and p.R1100H) are probably pathogenic. Because of the limitation of online programs, we still needed to collect more clinical data and perform basic animal experiments to prove whether these mutations are pathogenic. Regrettably, these 3 patients refused to undergo skin biopsy, so we could not obtain definite pathologic results.
There were 3 reported mutations (p.C117R, p.R578C, and p.R607C) with mutation of cysteine residues, 3 novels, cysteine-sparing residue mutations (p.P167S, p.P652S, and p.R1100H), and a novel mutation p.C709R in the Chinese population. However, the clinical manifestations of CADASIL display great heterogeneity [3]. Affected individuals experience recurrent ischemic episodes with accumulating motor, sensory, and cognitive deficits [17,18]. However, none of these features are specific. More than one-third were reported to be migraine with aura in Caucasian CADASIL patients [19,20]. Migraine is often present without aura. In the present study, 36.3% (4/11) of patients had migraine without aura, and the percentage of migraine was higher than that reported previously in Asians. Cognitive decline is the second most common characteristic of CADASIL. Most patients eventually experience dementia, except for some patients, but the relatives of these patients do not experience such outcomes. Dementia in CADASIL patients is frequently accompanied by gait disturbance, urinary incontinence, and pseudobulbar palsy. In the present study, 5 patients had dementia characterized by memory impairment or cognitive decline, 2 patients had gait disturbance and pseudobulbar palsy, 1 had pseudobulbar palsy, and 1 had mild memory impairment with urinary incontinence. Two patients with p.C117R and p.C709R mutations displayed similar pathophysiology for pseudobulbar palsy without gait disturbance and they shared the same risk factors for stroke, including smoking. However, they had 2 different clinical outcomes. One patient with mutation p.C117R had no sequelae and no complaint of cognitive decline. We could not obtain evidence, despite infarcts in the callosum based on brain MRI. The patient with p.R1100H mutation experienced memory loss and had a positive family history of stroke and dementia. She displayed favorable outcomes upon neuropsychological assessment via MMSE, MoCA, and MES. We could not establish a specific association between clinical features and mutation sites owing to the retrospective nature of the study.
Brain MRI is a sensitive method for CADASIL diagnosis. MRI reveals leukoaraiosis with marked abnormal signals in the white matter, especially in the temporal poles and external capsule. In our study, all these patients had the typical radiologic marker for temporal pole hyperintensity (100%). Most MR images of these patients displayed focal infarcts. The infarcts were located in the centrum semiovale, thalamus, and basal ganglia. Four patients displayed microbleeds in the brainstem, cerebellum, thalamus, basal ganglia, external capsule, and frontal lobe in SWI, but not in the cerebral cortex, wherein microbleeds were not observed in the anterior temporal lobe or the external capsule [16].
Our study has the following limitations. We enrolled only 11 patients who had no blood relationship, of which patients 4 and 6 had a family history, and 2 family pedigrees were drawn in this study. The number of patients was relatively small, and was not enough to study mutation distribution; therefore, studies with large sample sizes are needed in the future. In addition, contradistinction could not be made regarding the characteristics of the family members. Further, asymptomatic mutation carriers were not enrolled in the study. Finally, we could not identify associations between the mutation sites and clinical characteristics owing to the small cohort size.
Conclusions
In conclusion, we identified 4 novel mutations and found that the spectrum of NOTCH3 mutations in the Henan province in China is quite distinct from that of other studies. The primary clinical features of CADASIL patients in the present study were similar to those reported previously in mainland China.
Supplementary Data
Supplementary Table 1.
Primers for amplification of NOTCH3 exons.
| Exons | Primer sequence (from 5′ to 3′) | Target (bp) | Annealing temperature (°C) |
|---|---|---|---|
| Exon 2 | AGACCTCCCTGTAACACTCAGCACC | 616 | 55 |
| AGAGCCCCTGCTCAGTTTCCAAT | |||
| Exon 3 | GCAAATGTGTGTTTGCTGCT | 482 | 55 |
| CACTCATCCACGTCGCTTC | |||
| Exon 4 | TCCTAAACTCACCCTGTCCTG | 559 | 60 |
| TTATAGGTGTTGACGCCATCC | |||
| Exon 5 | GGCGACCTCACTTACGACTG | 454 | 56 |
| GTCCAGCCATTGACACACAC | |||
| Exon 6 | CTGGACTGCTGCATCTGTGT | 283 | 55 |
| TGCAGAGAAAACGGCCACTC | |||
| Exon 7 | TACTCAAGGGGTGTGGGC | 319 | 55 |
| GATGGAGTGCGATCGGTGT | |||
| Exon 8 | TGTGGACGAGTGCTCTATCG | 400 | 57 |
| CCCACTTACACCCCATTCTG | |||
| Exon 9 | GAGTGTAGAGGTGGGGACGA | 407 | 55 |
| TGGGTGGAAAAACCCATTTA | |||
| Exon 10 | CAGGGTGGGAACCTGTAAAA | 481 | 56 |
| ACTTTGGCTCCACACGTAGC | |||
| Exon 11 | GAGCTGAACCAGGATTGGTC | 453 | 60 |
| TTCACTTCGCAGTTCACACC | |||
| Exon 12 | AAATGCCTAGACCTGGTGGA | 385 | 55 |
| CGTTGGACAAGAGTCTGCAA | |||
| Exon 13 | GGTGTGCTAAGTGGGGTCAC | 485 | 57 |
| CAGTGGAAACCCATTCCATC | |||
| Exon 14 | AGGGAGGTTCCTGTGTGGAT | 499 | 56 |
| AGCTCCTGGAGGGAAATGAT | |||
| Exon 15 | CTCTCTCCTCCCGCTATGTT | 578 | 58 |
| GCCGTCTCCCTGTCTCAGTA | |||
| Exon 16 | TGACAGCACGGCTTATTTTG | 476 | 55 |
| CCAGCTCATTCCCTAACTCG | |||
| Exon 17 | CTGTCCTTGTCCTGTTCCAA | 659 | 60 |
| CAGTCATCAGAGTCCGCAGT | |||
| Exon 18 | CTTGCCGAGATAAGGGTCAG | 450 | 55 |
| AGGGGAAGCACTCAGAGTCA | |||
| Exon 19 | GGCACAGTTTCTCCCAGACTA | 455 | 51 |
| CCTTCAACGCTCACAGCAG | |||
| Exon 20 | GACTCATTCCACCAAGGATGTT | 430 | 51 |
| CACCCACAGATACACCAAGAGT | |||
| Exon 21–23 | ACCTACTGGTAGTGCGCTGAACATC | 937 | 55 |
| TGTCAGCATTTTCCAGAAACTCCCT | |||
| Exon 24 | GGAGCGGTTGGGTGGGACA | 982 | 55 |
| GGCAGACAAACGGGGCAGAG |
Footnotes
Source of support: This study was supported by the National Natural Science Foundation of China (No. 81671068 and No. 81873727) and by the Health Commission of Henan Province (No. 201701020)
Ethics statement
All procedures were approved by the Ethics Committee of Henan Provincial People’s Hospital. The title of the research: Gene sequencing of NOTCH3 in CADASIL patients; The date of approval: 20150108; the authorization number: 17180214.
Conflict of interest
None.
References
- 1.Chabriat H, Joutel A, Dichgans M, et al. Cadasil. Lancet Neurol. 2009;8:643–53. doi: 10.1016/S1474-4422(09)70127-9. [DOI] [PubMed] [Google Scholar]
- 2.O’Sullivan M, Jarosz JM, Martin RJ, et al. MRI hyperintensities of the temporal lobe and external capsule in patients with CADASIL. Neurology. 2001;56:628–34. doi: 10.1212/wnl.56.5.628. [DOI] [PubMed] [Google Scholar]
- 3.Coupland K, Lendahl U, Karlstrom H. Role of NOTCH3 mutations in the cerebral small vessel disease cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy. Stroke. 2018;49:2793–800. doi: 10.1161/STROKEAHA.118.021560. [DOI] [PubMed] [Google Scholar]
- 4.Tikka S, Mykkanen K, Ruchoux MM, et al. Congruence between NOTCH3 mutations and GOM in 131 CADASIL patients. Brain. 2009;132:933–39. doi: 10.1093/brain/awn364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ampuero I, Alegre-Abarrategui J, Rodal I, et al. On the diagnosis of CADASIL. J Alzheimers Dis. 2009;17:787–94. doi: 10.3233/JAD-2009-1112. [DOI] [PubMed] [Google Scholar]
- 6.Lee YC, Liu CS, Chang MH, et al. Population-specific spectrum of NOTCH3 mutations, MRI features and founder effect of CADASIL in Chinese. J Neurol. 2009;256:249–55. doi: 10.1007/s00415-009-0091-3. [DOI] [PubMed] [Google Scholar]
- 7.Dotti MT, Federico A, Mazzei R, et al. The spectrum of Notch3 mutations in 28 Italian CADASIL families. J Neurol Neurosurg Psychiatry. 2005;76:736–38. doi: 10.1136/jnnp.2004.048207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bianchi S, Zicari E, Carluccio A, et al. CADASIL in central Italy: A retrospective clinical and genetic study in 229 patients. J Neurol. 2015;262:134–41. doi: 10.1007/s00415-014-7533-2. [DOI] [PubMed] [Google Scholar]
- 9.Mykkanen K, Savontaus ML, Juvonen V, et al. Detection of the founder effect in Finnish CADASIL families. Eur J Hum Genet. 2004;12:813–19. doi: 10.1038/sj.ejhg.5201221. [DOI] [PubMed] [Google Scholar]
- 10.Davous P. CADASIL: A review with proposed diagnostic criteria. Eur J Neurol. 1998;5:219–33. doi: 10.1046/j.1468-1331.1998.530219.x. [DOI] [PubMed] [Google Scholar]
- 11.Wang Z, Yuan Y, Zhang W, et al. NOTCH3 mutations and clinical features in 33 mainland Chinese families with CADASIL. J Neurol Neurosurg Psychiatry. 2011;82:534–39. doi: 10.1136/jnnp.2010.209247. [DOI] [PubMed] [Google Scholar]
- 12.Yin X, Wu D, Wan J, et al. Cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy: Phenotypic and mutational spectrum in patients from mainland China. Int J Neurosci. 2015;125:585–92. doi: 10.3109/00207454.2014.951929. [DOI] [PubMed] [Google Scholar]
- 13.Stojanov D, Vojinovic S, Aracki-Trenkic A, et al. Imaging characteristics of cerebral autosomal dominant arteriopathy with subcortical infarcts and leucoencephalopathy (CADASIL) Bosn J Basic Med Sci. 2015;15:1–8. doi: 10.17305/bjbms.2015.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi BW, Park S, Kim HJ. Possible role of a missense mutation of p.P167S on NOTCH3 gene associated with cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy. Dement Neurocogn Disord. 2016;15:52–54. doi: 10.12779/dnd.2016.15.2.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomimoto H, Ohtani R, Wakita H, et al. Small artery dementia in Japan: Radiological differences between CADASIL, leukoaraiosis and Binswanger’s disease. Dement Geriatr Cogn Disord. 2006;21:162–69. doi: 10.1159/000090677. [DOI] [PubMed] [Google Scholar]
- 16.Kim Y, Choi EJ, Choi CG, et al. Characteristics of CADASIL in Korea: A novel cysteine-sparing Notch3 mutation. Neurology. 2006;66:1511–16. doi: 10.1212/01.wnl.0000216259.99811.50. [DOI] [PubMed] [Google Scholar]
- 17.Dichgans M, Mayer M, Uttner I, et al. The phenotypic spectrum of CADASIL: Clinical findings in 102 cases. Ann Neurol. 1998;44:731–39. doi: 10.1002/ana.410440506. [DOI] [PubMed] [Google Scholar]
- 18.Chabriat H, Vahedi K, Iba-Zizen MT, et al. Clinical spectrum of CADASIL: A study of 7 families. Cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy. Lancet. 1995;346:934–39. doi: 10.1016/s0140-6736(95)91557-5. [DOI] [PubMed] [Google Scholar]
- 19.Kalimo H, Ruchoux MM, Viitanen M, Kalaria RN. CADASIL: A common form of hereditary arteriopathy causing brain infarcts and dementia. Brain Pathol. 2002;12:371–84. doi: 10.1111/j.1750-3639.2002.tb00451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Desmond DW, Moroney JT, Lynch T, et al. The natural history of CADASIL: A pooled analysis of previously published cases. Stroke. 1999;30:1230–33. doi: 10.1161/01.str.30.6.1230. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1.
Primers for amplification of NOTCH3 exons.
| Exons | Primer sequence (from 5′ to 3′) | Target (bp) | Annealing temperature (°C) |
|---|---|---|---|
| Exon 2 | AGACCTCCCTGTAACACTCAGCACC | 616 | 55 |
| AGAGCCCCTGCTCAGTTTCCAAT | |||
| Exon 3 | GCAAATGTGTGTTTGCTGCT | 482 | 55 |
| CACTCATCCACGTCGCTTC | |||
| Exon 4 | TCCTAAACTCACCCTGTCCTG | 559 | 60 |
| TTATAGGTGTTGACGCCATCC | |||
| Exon 5 | GGCGACCTCACTTACGACTG | 454 | 56 |
| GTCCAGCCATTGACACACAC | |||
| Exon 6 | CTGGACTGCTGCATCTGTGT | 283 | 55 |
| TGCAGAGAAAACGGCCACTC | |||
| Exon 7 | TACTCAAGGGGTGTGGGC | 319 | 55 |
| GATGGAGTGCGATCGGTGT | |||
| Exon 8 | TGTGGACGAGTGCTCTATCG | 400 | 57 |
| CCCACTTACACCCCATTCTG | |||
| Exon 9 | GAGTGTAGAGGTGGGGACGA | 407 | 55 |
| TGGGTGGAAAAACCCATTTA | |||
| Exon 10 | CAGGGTGGGAACCTGTAAAA | 481 | 56 |
| ACTTTGGCTCCACACGTAGC | |||
| Exon 11 | GAGCTGAACCAGGATTGGTC | 453 | 60 |
| TTCACTTCGCAGTTCACACC | |||
| Exon 12 | AAATGCCTAGACCTGGTGGA | 385 | 55 |
| CGTTGGACAAGAGTCTGCAA | |||
| Exon 13 | GGTGTGCTAAGTGGGGTCAC | 485 | 57 |
| CAGTGGAAACCCATTCCATC | |||
| Exon 14 | AGGGAGGTTCCTGTGTGGAT | 499 | 56 |
| AGCTCCTGGAGGGAAATGAT | |||
| Exon 15 | CTCTCTCCTCCCGCTATGTT | 578 | 58 |
| GCCGTCTCCCTGTCTCAGTA | |||
| Exon 16 | TGACAGCACGGCTTATTTTG | 476 | 55 |
| CCAGCTCATTCCCTAACTCG | |||
| Exon 17 | CTGTCCTTGTCCTGTTCCAA | 659 | 60 |
| CAGTCATCAGAGTCCGCAGT | |||
| Exon 18 | CTTGCCGAGATAAGGGTCAG | 450 | 55 |
| AGGGGAAGCACTCAGAGTCA | |||
| Exon 19 | GGCACAGTTTCTCCCAGACTA | 455 | 51 |
| CCTTCAACGCTCACAGCAG | |||
| Exon 20 | GACTCATTCCACCAAGGATGTT | 430 | 51 |
| CACCCACAGATACACCAAGAGT | |||
| Exon 21–23 | ACCTACTGGTAGTGCGCTGAACATC | 937 | 55 |
| TGTCAGCATTTTCCAGAAACTCCCT | |||
| Exon 24 | GGAGCGGTTGGGTGGGACA | 982 | 55 |
| GGCAGACAAACGGGGCAGAG |