Mutation of the chromatin regulator HDA19 causes age-dependent patterning defects in reproductive meristems. This effect is enhanced by mutation of FD, revealing a novel developmental role for this flowering time gene.
Keywords: Arabidopsis thaliana, developmental aging, FD, floral organ identity genes, flowering time, HDA19, inflorescence meristem, reproductive development, shoot apical meristem, transcriptional repression
Abstract
The shoot apical meristem (SAM) undergoes developmental transitions that include a shift from vegetative to reproductive growth. This transition is triggered by flowering time genes, which up-regulate floral meristem (FM) identity genes that, in turn, control flower development by activating floral organ identity genes. This cascade of transcriptional activation is refined by repression mechanisms that temporally and spatially restrict gene expression to ensure proper development. Here, we demonstrate that HISTONE DEACETYLASE 19 (HDA19) maintains the identity of the reproductive SAM, or inflorescence meristem (IM), late in Arabidopsis thaliana development. At late stages of growth, hda19 IMs display a striking patterning defect characterized by ectopic expression of floral organ identity genes and the replacement of flowers with individual stamenoid organs. We further show that the flowering time gene FD has a specific function in this regulatory process, as fd hastens the emergence of these patterning defects in hda19 growth. Our work therefore identifies a new role for FD in reproductive patterning, as FD regulates IM function together with HDA19 in an age-dependent fashion. To effect these abnormalities, hda19 and fd may accentuate the weakening of transcriptional repression that occurs naturally with reproductive meristem proliferation.
Introduction
To optimize growth and reproductive output, plants perceive and respond to various environmental cues. This requires great developmental plasticity, which is facilitated by continuously active stem cell populations that initiate new organs throughout the life of the plant. These are the shoot apical meristem (SAM) and root apical meristem (RAM) that are responsible for driving above- and below-ground growth, respectively. The SAM undergoes various developmental transitions over time and initiates different types of lateral organs at each stage (Bäurle and Dean, 2006). Post-embryonic growth begins with the vegetative stage, where the SAM initiates leaves to support photosynthetic activity. In Arabidopsis thaliana, a shift from juvenile to adult growth occurs as the plant becomes competent to respond to floral inductive signals (Poethig, 2003). Floral induction triggers an abrupt transition to the reproductive stage, where the SAM becomes an inflorescence meristem (IM) and produces flowers. Each flower arises from another distinct stem cell population termed the floral meristem (FM). While the SAM displays indeterminate growth, the FM is a determinate stem cell population that is consumed by the production of terminally differentiated floral organs.
Flowering time is controlled by both endogenous signals and environmental stimuli (Srikanth and Schmid, 2011). Genetic analyses in Arabidopsis and other plants have identified multiple pathways associated with these cues, including the aging pathway, of which the miR156 and miR172 miRNA families are major components. With developmental age, a decrease in the levels of miR156 [which targets SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE (SPL) transcription factors] is co-ordinated with an increase in the levels of miR172 [which targets APETALA2 (AP2) family transcription factors] (reviewed by Huijser and Schmid, 2011). These complementary patterns of expression influence multiple phase transitions, including the transition to flowering (Huijser and Schmid, 2011).
Among the most important pathways associated with environmental stimuli is the day-length-responsive photoperiod pathway. Inductive long-day conditions positively influence the activity of the transcription factor CONSTANS (CO), which up-regulates expression of the floral pathway integrator FLOWERING LOCUS T (FT) in leaves (Putterill et al., 1995; Kardailsky et al., 1999; Kobayashi et al., 1999). FT protein, a major component of the mobile flowering signal ‘florigen’, moves through the vasculature to the plant apex (Corbesier et al., 2007; Jaeger and Wigge, 2007; Mathieu et al., 2007). Here, it interacts with the basic leucine zipper transcription factor FD, which is already expressed in the vegetative SAM and provides spatial specificity to FT function (Abe et al., 2005; Wigge et al., 2005). The FT–FD interaction appears to be bridged by 14-3-3 proteins and to rely on phosphorylation of FD (Abe et al., 2005; Taoka et al., 2011). This protein complex promotes the expression of FM identity genes such as APETALA1 (AP1) (Abe et al., 2005; Wigge et al., 2005) to initiate the production of flowers from the flanks of the IM.
Following floral initiation, AP1 and the FM identity gene LEAFY (LFY) activate genes involved in specifying floral organ fate (Weigel and Meyerowitz, 1993; Parcy et al., 1998). These floral organ identity genes belong to different classes that function in combination to pattern the four whorls of the flower, as described by the ABC model (Bowman et al., 1991; Coen and Meyerowitz, 1991). The A-class gene AP1, which acts as both an FM and floral organ identity gene, specifies sepal fate in the first whorl. AP1 also works in conjunction with the B-class genes APETALA3 (AP3) and PISTILLATA (PI) to confer petal identity in the second whorl. The C-class gene AGAMOUS (AG) co-operates with B-class genes to specify stamens in the third whorl, and alone promotes carpel identity in the innermost fourth whorl (Bowman et al., 1991; Coen and Meyerowitz, 1991). Subsequent additions to the ABC model include the identification of D-class genes involved in conferring ovule identity and E-class genes involved in specifying the fates of all floral organs (reviewed in Krizek and Fletcher, 2005).
The cascade of transcriptional activation described above is refined by various repression mechanisms that spatially restrict gene expression. For instance, FM identity is repressed in the IM by a protein complex involving FD and TERMINAL FLOWER 1 (TFL1), a close homolog of FT (Abe et al., 2005; Wigge et al., 2005; Hanano and Goto, 2011). The IMs of tfl1 mutants misexpress both AP1 and LFY, and lose meristem indeterminacy by differentiating into terminal flowers (Shannon and Meeks-Wagner, 1991; Gustafson-Brown et al., 1994; Bradley et al., 1997). In developing flowers, a key tenet of the ABC model is that A- and C-class genes display mutual antagonism by restricting each other’s functional domains. For example, AP1 associates with the transcriptional co-repressor LEUNIG (LUG) and its binding partner SEUSS (SEU) to regulate AG expression negatively in the two outer flower whorls (Liu and Meyerowitz, 1995; Franks et al., 2002; Sridhar et al., 2006). Similarly, AP2 forms a complex with the co-repressor TOPLESS (TPL) and the RPD3-like HISTONE DEACETYLASE 19 (HDA19) to repress AG in the flower, consistent with the characterization of AP2 as an A-class gene (Bowman et al., 1991; Drews et al., 1991; Krogan et al., 2012). Moreover, this AP2–TPL–HDA19 complex represses other floral organ identity genes, including B-class genes in the first whorl (Krogan et al., 2012). As predicted by the ABC model, disruption of these repression mechanisms leads to partial or complete homeotic conversion of floral organ identity.
The chromatin regulator HDA19 associates with TPL [and/or other members of the TPL/TOPLESS RELATED (TPR) co-repressor family] to modulate additional aspects of stem cell function in Arabidopsis. For example, within the determinate FM, HDA19 and TPL participate in the termination of stem cell activity through repression of the transcription factor gene WUSCHEL (WUS) (Bollier et al., 2018). In the RAM, the WUS-related WUSCHEL HOMEOBOX 5 protein recruits HDA19 and TPL/TPR proteins to repress differentiation-promoting genes in root columella stem cells (Pi et al., 2015). Finally, HDA19 and TPL facilitate the correct establishment of the SAM by co-operatively repressing basal fate in the apical region of the developing embryo (Long et al., 2006). Apart from these stem cell-related functions, HDA19 also controls other diverse aspects of plant development, including root cell patterning and elongation (Chen et al., 2015; Chen et al., 2016) and the repression of embryonic traits and hormone signaling in the seedling (Tanaka et al., 2008; Zhou et al., 2013; Ryu et al., 2014; Gao et al., 2015).
In the present work, we describe a new role for HDA19 in regulating meristem activity during reproductive growth. Specifically, we show that HDA19 preserves the identity of the IM in an age-dependent manner. In older hda19 inflorescence apices, floral organ identity genes become broadly misexpressed in the IM, and the specification of FM identity is severely disrupted. We further demonstrate that mutation of the flowering time gene FD enhances the timing of these reproductive defects in hda19. This indicates that following its participation in floral induction, FD is redeployed in the IM to execute novel patterning roles.
Materials and methods
Plant material
Plants were grown on soil in a growth chamber under a 16 h light/8 h dark cycle. The Landsberg erecta ecotype of A. thaliana (L.) Heynh served as wild type. Genetic analyses used the previously described mutants hda19-1, tpl-2, tpr1, tpr3, tpr4 (Long et al., 2006), tpr2 (Krogan et al., 2012), fd-1, ft-2, co-4, fca-1 (Koornneef et al., 1991), tfl1-2 (Alvarez et al., 1992), pi-1, ag-1 (Bowman et al., 1989), and ap3-3 (Jack et al., 1992). The fd-6 allele was identified by a genetic enhancer screen of hda19-1 using ethylmethane sulfonate as a mutagen. All analyzed reproductive tissue was exclusively from primary inflorescence stems.
Histology
Inflorescence tissue was fixed in 4% formaldehyde, embedded in paraffin, and sectioned to a thickness of 8 µm. Sections were then deparaffinized, rehydrated through a reverse ethanol series (100% to 30%), incubated in water, and stained in 0.1% toluidine blue solution. Tissue was then destained in water, mounted, and imaged.
RNA in situ hybridization
RNA in situ hybridizations using digoxigenin-labeled riboprobes were performed as previously reported (Krogan et al., 2012). Probe sequences for detecting expression of HDA19 (Long et al., 2006), AP3 (Jack et al., 1992), PI (Goto and Meyerowitz, 1994), AG (Yanofsky et al., 1990), FD (Searle et al., 2006), and AP1 (Gustafson-Brown et al., 1994) were previously described. Two AGL6 antisense probes were generated using primer pairs AGL6-A-fw (5'-GAAAGCACAATCGAACGGTATAATCG-3') and AGL6-A-rv (5'-AAGAACCCAACCTTGGACGAAATTAG-3') or AGL6-B-fw (5'-CTAGGAGACATAAACAAACAACTCAAG-3') and AGL6-B-rv (5'-GTTTTAGATCAAGTAGGAGTAAGAGG-3'). Both probes produced comparable expression patterns. The control AGL6 sense probe was complementary to antisense probe AGL6-A.
Chromatin immunoprecipitation
ChIP experiments on reproductive tissue of HDA19p::HDA19-GFP (green fluorescent protein) and TPLp::TPL-GFP transgenic lines (Long et al., 2006) were performed as previously described (Krogan et al., 2012). Analyzed tissue consisted of the IM and young, unopened floral buds. Enrichment was calculated as the ratio of the signal from ChIP samples to that from input samples. Fold enrichment was calculated as the ratio of HDA19p::HDA19-GFP or TPLp::TPL-GFP enrichment to non-transgenic control sample enrichment and was normalized against ACTIN2 data. Primer sequences and positions are given in Supplementary Table S1 at JXB online.
Yeast two-hybrid assays and western blotting
Yeast two-hybrid assays were performed as previously described (Krogan et al., 2012). Anti-GAL4 DBD (sc-577) and anti-GAL4 AD (sc-1663) antibodies (Santa Cruz Biotechnologies, Dallas, TX, USA) were used in western blotting to verify yeast protein expression.
Quantitative real-time reverse transcription–PCR (RT–PCR)
An Invitrogen SuperScript first-strand synthesis system (Thermo Fisher Scientific, Waltham, MA, USA) was used for reverse transcription of total RNA samples. Real-time PCR on cDNA samples was performed with a Mx3005P QPCR system (Agilent Technologies, Santa Clara, CA, USA) using PerfeCTa SYBR Green SuperMix (Quanta Biosciences Inc., Beverly, MA, USA). Data analysis was carried out with MxPro QPCR software (Agilent Technologies). Primers for HDA19 amplification were 5'-CCTCCTAAAACATAAGACTCGGAGC-3' and 5'-TAAATACATATCCGTGCTCAATCCTC-3', while FD primers were previously described (Searle et al., 2006). Relative expression levels were normalized against ACTIN7 (Krogan et al., 2016).
Transgenic plant lines
To test for complementation, the Agrobacterium-mediated floral dip method (Clough and Bent, 1998) was used to introduce a genomic fragment of FD into hda19 fd-6. The FD fragment was amplified by PCR (primers 5'-GTCTAAGACGATCTAGTTATCCAAGGC-3' and 5'-AATGGTCAGAGTGAAGGTATCAGC-3'), cloned into pCR-Blunt II-TOPO (Thermo Fisher Scientific), digested, introduced into the HindIII–XhoI sites of plasmid pBJ36 (Eshed et al., 2001), and subcloned into the NotI site of binary vector pART27 (Gleave, 1992).
Microscopy
Olympus SZX16 dissecting and BX61 compound microscopes (Olympus, Center Valley, PA, USA) were used to capture images of live plant tissues and sectioned tissues, respectively. The scanning electron micrograph was acquired using a Zeiss EVO LS 15 analytical environmental scanning electron microscope (Carl Zeiss Inc., Thornwood, NY, USA).
Results
HDA19 is required for the patterning of the Arabidopsis reproductive apex
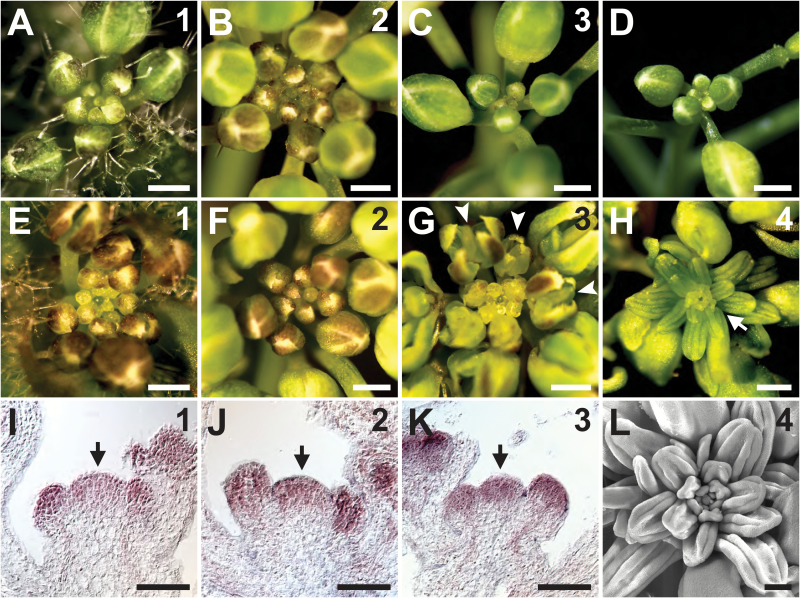
Because HDA19 is an important regulator of floral patterning (Krogan et al., 2012), we examined the hda19 mutant for defects in other aspects of reproductive development, including IM function. In the wild type, the IM initiates flowers in a spiral phyllotaxy throughout the reproductive stage (Fig. 1A–C). We arbitrarily separated this stage into successive temporal phases based on the number of FMs initiated by the primary IM (Supplementary Fig. S1). IM phase 1 is characterized by the visible emergence of the first floral bud cluster (Fig. 1A), while IM phases 2 and 3 (Fig. 1B,C) are marked by the appearance of ~10 and 20 mature flowers/siliques on the primary inflorescence stem, respectively (Supplementary Fig. S1). Shortly after phase 3, the IM and its most recently initiated flowers enter a state of quiescence and cease growth (Fig. 1D). In hda19 mutants, the appearance of reproductive tissues produced at IM phases 1 and 2 generally resembles that of the wild type (Fig. 1E, F; Supplementary Fig. S2A–C). However, the appearance of hda19 reproductive apices at IM phase 3 begins to differ. These mutant apices appear disorganized because of precocious bud opening and aberrant floral patterning (Fig. 1G; Supplementary Fig. S2D), which results from misexpression of floral organ identity genes (Krogan et al., 2012). Furthermore, unlike the wild type, hda19 inflorescences do not quiesce shortly after IM phase 3. Instead, hda19 IMs maintain indeterminate growth and continue to initiate flowers with progressively more severe patterning defects (Supplementary Fig. S2E, F).
Fig. 1.
HDA19 modulates Arabidopsis inflorescence development in an age-dependent manner. (A–H) Apical views of wild-type (A–D) and hda19 (E–H) reproductive apices. Numbers in the upper right indicate the inflorescence meristem (IM) phase. (D) A quiescent reproductive apex at the end of its life cycle. (G) Abnormally patterned and precociously opened flowers (arrowheads) are initiated in phase 3 hda19 IMs. (H) Individual stamenoid organs (arrow) are initiated in place of flowers in phase 4 hda19 IMs. (I–K) RNA in situ hybridizations of HDA19 in wild-type IMs (arrows) at phases 1, 2, and 3 (denoted in the upper right). (L) Scanning electron micrograph of the reproductive apex of a phase 4 hda19 IM. Note the anther-like morphology of abnormal lateral organs, indicative of stamen identity. Scale bars: (A–H) 0.5 mm; (I–K) 50 µm; (L) 200 µm.
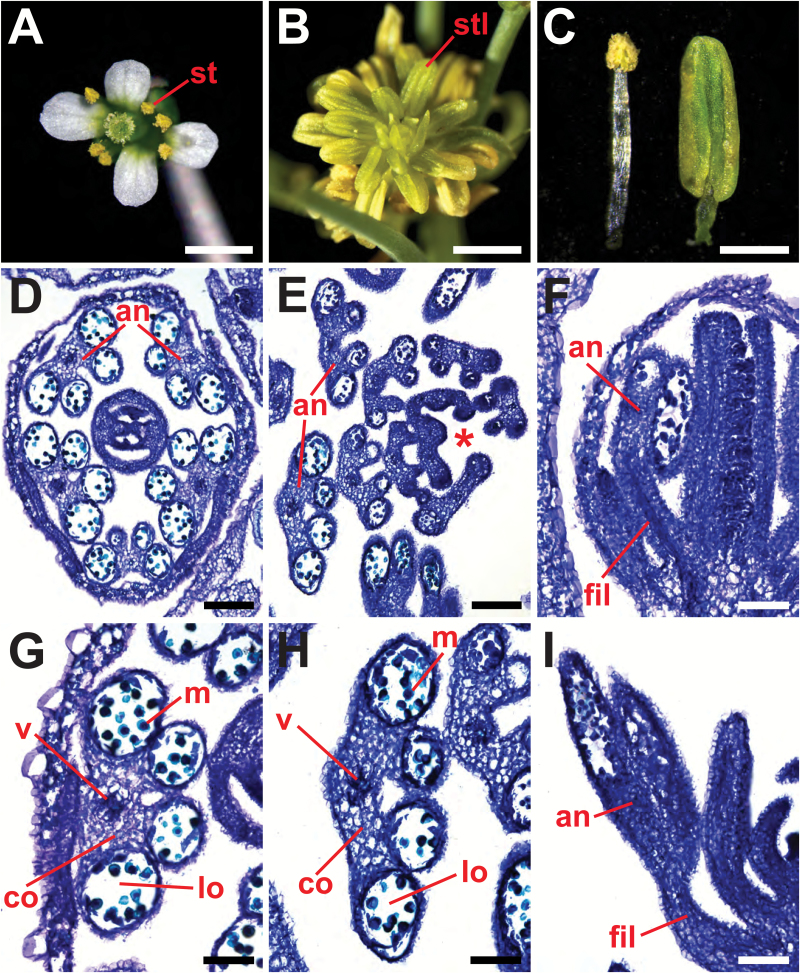
After producing ~30 mature flowers/siliques, hda19 IMs enter a fourth phase characterized by a distinct phenotype that we have termed the stamenoid inflorescence apex (SIA) (Fig. 1H, L; Supplementary Fig. S1). The SIA phenotype consists of a dysfunctional IM that initiates an indeterminate, spiral arrangement of individual stamen-like organs in the place of FMs (Figs 1H, L, 2B). Each of these stamenoid organs consists of a filament attached to a vastly enlarged anther-like region that fails to undergo dehiscence (Fig. 2A–C, F, I). Despite these morphological abnormalities, the internal composition of the SIA lateral organs closely resembles that of wild-type stamens, as both exhibit anthers with locules, connective and vascular tissue (Fig. 2D–I). This confirms that the lateral organs that replace FMs during the SIA phenotype are stamen-like in nature. Collectively, these results indicate that HDA19 is required to maintain the normal activity of the IM during later stages of reproductive growth.
Fig. 2.
Stamenoid inflorescence apices of hda19 produce lateral organs resembling male reproductive structures. (A) Mature stamens (st) of a wild-type flower. (B) Stamen-like organs (stl) of hda19 stamenoid inflorescence apex (SIA) tissue. (C) Wild-type stamen (left) and hda19 SIA lateral organ (right). (D–I) Toluidine blue-stained tissue sections showing internal morphologies of lateral organs. (D) Transverse section of a wild-type flower showing the arrangement of stage 9 anthers (an) (according to Sanders et al., 1999). (G) Transverse section of a wild-type stage 9 anther depicting locules (lo), microspores (m), connective (co), and vascular tissue (v). (E) Transverse section of distal SIA tissue of hda19. An asterisk denotes the position of the inflorescence meristem. (H) Transverse section of a stamen-like SIA lateral organ, with internal structures resembling those of (G). (F, I) Longitudinal sections of a wild-type flower (F) and SIA lateral organs (I). The anther and filament (fil) of a wild-type stamen (F) and SIA lateral organ (I) are denoted. Scale bars: (A, B) 1 mm; (C) 0.5 mm; (D, E) 100 µm; (F–I) 50 µm.
HDA19 is expressed in the IM and represses floral organ identity genes in an age-dependent manner
Since hda19 reproductive defects worsen with developmental age, we assessed HDA19 expression levels throughout IM phases in the wild type. RNA in situ hybridizations showed high HDA19 expression in the IM and young FMs throughout reproductive growth (Fig. 1I–K). Quantitative RT–PCR on wild-type reproductive apices also demonstrated relatively consistent HDA19 expression throughout IM phases 1–3 (Supplementary Fig. S3A). This expression profile is in agreement with HDA19 playing a role in IM function, including in later stages of reproductive growth. Furthermore, the age-dependent worsening of hda19 defects does not reflect broad, phase-specific fluctuations of HDA19 expression levels in wild-type apices.
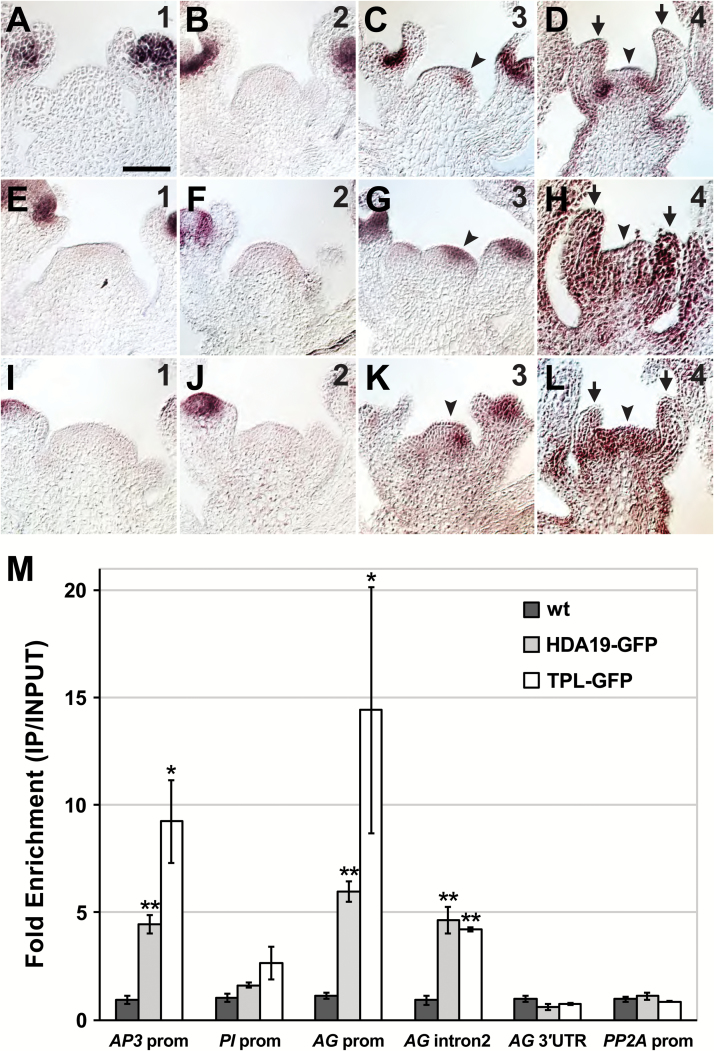
The late-arising reproductive defects in hda19 suggest that floral gene misregulation may increase with developmental age. Based on the morphology of SIA organs, we analyzed the expression patterns of genes that specify stamen identity, namely the B-class genes AP3 and PI and the C-class gene AG. In the wild type, B- and C- class gene expression is excluded from the IM and newly initiated FMs, and first becomes detectable in flowers when sepal primordia arise (Drews et al., 1991; Jack et al., 1992; Goto and Meyerowitz, 1994). The exclusion of expression from the IM is maintained throughout all phases (Supplementary Fig. S4). In hda19, however, ectopic expression of AP3, PI, and AG becomes apparent in phase 3 IMs, with AG displaying the most widespread misexpression at this IM phase (Fig. 3A–C, E–G, I–K). This initial ectopic expression is often asymmetrically distributed within the IM, possibly reflecting positions of incipient lateral primordia. By phase 4, hda19 IMs exhibit gross misregulation of B- and C-class gene expression, which is apparent throughout the IM and stamenoid lateral organs, as well as in underlying stem regions (Fig. 3D, H, L). Therefore, the age-dependent progression of hda19 reproductive defects, which culminates with the emergence of SIA, correlates with the extent of floral organ gene misexpression.
Fig. 3.
Floral organ identity genes are misexpressed in the hda19 inflorescence meristem and are direct targets of HDA19 and TOPLESS. (A–L) RNA in situ hybridizations of AP3 (A–D), PI (E–H), and AG (I–L) in hda19 inflorescence apices. Numbers in the upper right indicate the inflorescence meristem (IM) phase. Arrowheads denote ectopic expression of floral organ identity genes in IMs. Stamenoid organs (arrows) of phase 4 IMs show strong expression of floral organ identity genes. Scale bar: 50 µm. (M) Anti-GFP ChIP showing specific binding of HDA19 and the co-repressor TOPLESS (TPL) to the promoter of AP3 and to the promoter and second intron of AG. A control ChIP was performed on non-transgenic wild-type (wt) tissue. Data were normalized relative to input and ACT2 abundance. Data are represented as the mean ±SE of at least two biological replicates. Student’s t-test was used to determine the significance of target enrichment relative to wt IP (*P<0.05; **P<0.01).
HDA19 forms a complex with the co-repressor TPL and the transcription factor AP2 to bind the AP3 promoter and second intron of AG directly during flower development (Krogan et al., 2012). We aimed to determine whether HDA19 binds floral organ identity genes independently of AP2, which is not reliably expressed in the IM (Wollmann et al., 2010). Since genome-wide assessment of AP2 binding in reproductive apices failed to show association with the promoters of PI and AG (Yant et al., 2010), we tested whether HDA19 could bind these regions. As HDA19 does not interact with DNA directly, such binding would be consistent with recruitment of HDA19 to repress genes in the IM by transcription factor(s) other than AP2. We performed ChIP on HDA19 using reproductive apical tissue and failed to detect binding to the PI promoter (Fig. 3M), indicating that HDA19-mediated regulation of PI in the IM is likely to be indirect. Conversely, HDA19 showed specific binding to the AG promoter (Fig. 3M), suggesting that HDA19 directly binds to multiple sites at this locus, consistent with broad AG misexpression in hda19 SIA tissues. A similar binding profile was determined for the co-repressor TPL (Fig. 3M), further verifying a close association between HDA19 and TPL in the repression of floral organ identity genes. We also confirmed HDA19 and TPL binding to the AP3 promoter and AG second intron (Fig. 3M). These latter interactions are only partially disrupted in an ap2 mutant background (Krogan et al., 2012), indicating that HDA19 and TPL may complex with other transcription factors at these specific sites. Overall, these results suggest that SIA defects arise in hda19 reproductive tissues because HDA19 is broadly required for the direct repression of AG (and possibly AP3) and the indirect repression of PI.
To test whether the SIA phenotype could be suppressed by removing B- or C-class gene function, we crossed hda19 with the floral homeotic mutants ap3-3, pi-1, and ag-1. These three single mutants exhibit prolonged reproductive growth compared with the wild type but, despite reaching IM phase 4, do not display SIA defects (Supplementary Fig. S5E, F, I, J, M, N). Each B- and C-class mutant suppressed the severity of floral organ homeotic conversions, thereby reducing the extent of precocious bud opening and the overall disorganization of hda19 reproductive apices at IM phase 4 (Supplementary Fig. S5C, G, K, O). Notably, hda19 ag-1 double mutant floral buds were rescued to the greatest extent, showing no signs of precocious opening (Supplementary Fig. S5O). This observation is consistent with widespread misregulation of AG appearing at an earlier phase of hda19 reproductive growth relative to the misexpression of other floral organ genes (Fig. 3C, G, K). Despite this suppression, each hda19 double mutant combination displayed a SIA-like phenotype at IM phase 4, initiating individual lateral organs in place of FMs (Supplementary Fig. S5H, L, P). Therefore, despite their vast misregulation in hda19 reproductive tissues, mutation of individual B- or C-class genes was not sufficient to suppress SIA emergence.
FD is a second site enhancer of the hda19 SIA phenotype
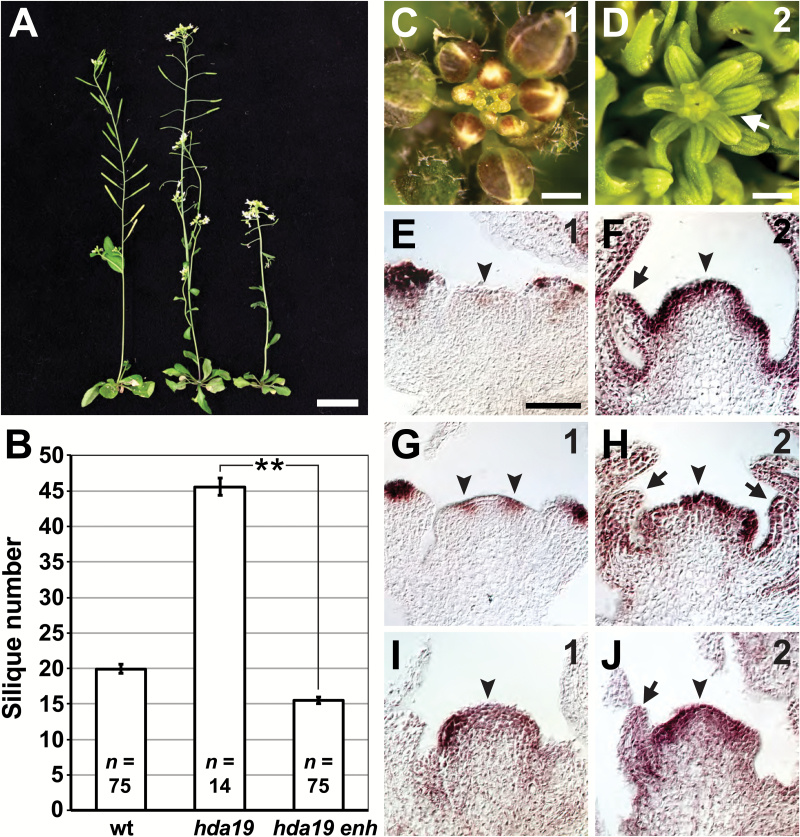
Our results indicate that HDA19 participates in an age-dependent regulatory mechanism that maintains IM function in Arabidopsis. We sought to elucidate other factors involved in this regulation through a genetic enhancer screen of hda19. We identified an enhancer mutation (enh) that hastened the timing of hda19 SIA development (Fig. 4A, B). In this double mutant, floral organ patterning defects were apparent already in the first initiated flowers and worsened with each subsequent flower (Supplementary Fig. S2G–I) before transitioning to the SIA phenotype at IM phase 2 (Fig. 4C, D). As a result, primary inflorescence stems of hda19 enh produced far fewer siliques than those of hda19 before exhibiting the SIA defect (Fig. 4B). RNA in situ hybridizations also showed earlier misexpression of floral organ identity genes in hda19 enh IMs. Specifically, ectopic expression became apparent at IM phase 1 and increased at phase 2, resembling hda19 IM phases 3 and 4, respectively (Fig. 4E–J, compare with Fig. 3C, D, G, H, K, L).
Fig. 4.
Enhancement of hda19 reproductive defects by a second-site mutation. (A) Wild-type (wt) (left), hda19 (middle), and an hda19 enhancer (hda19 enh) mutant (right) at 40 d after germination. (B) Numbers of siliques produced before growth termination (for wt) or before the appearance of the stamenoid inflorescence apex (SIA) phenotype (for hda19 and hda19 enh) are shown. Data are represented as the mean ±SE. A statistically significant difference is indicated (**P<0.001; two-tailed t-test). The sample size (n) of each genotype is provided. (C, D) hda19 enh reproductive apices. Inflorescence meristem (IM) phase denoted in the upper right. The SIA phenotype (arrow) is evident at IM phase 2. (E–J) RNA in situ hybridizations of AP3 (E, F), PI (G, H), and AG (I, J) in hda19 enh IMs at phases 1 and 2 (indicated in the upper right). Arrowheads denote ectopic expression of floral organ identity genes in IMs. Arrows indicate stamenoid organs produced by phase 2 IMs. Scale bars: (A) 2 cm; (C, D) 0.5 mm; (E–J) 50 µm.
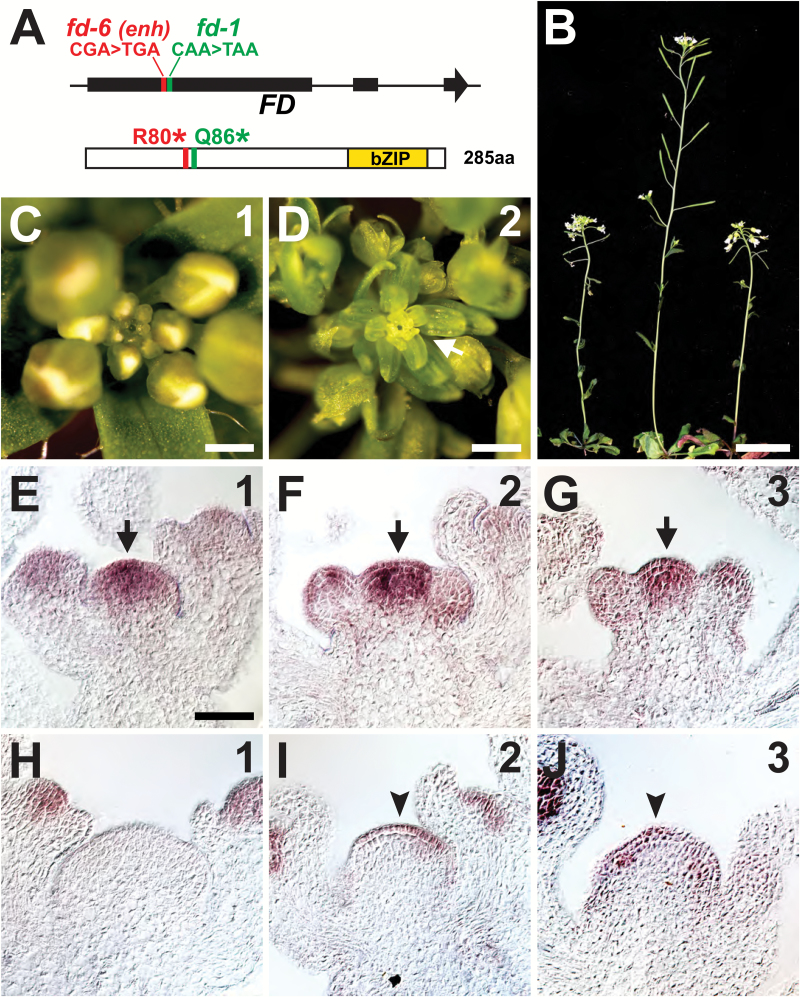
Using classical map-based cloning, we narrowed the genetic location of enh to a small interval on the lower arm of chromosome 4 (Supplementary Fig. S6A). Since hda19 enh flowered later than hda19, we concentrated on the flowering time gene FD within this mapping interval as a possible candidate for enh. Sequencing of FD in hda19 enh revealed a premature stop codon at a position just upstream of the lesion found in the well-characterized fd-1 allele (Fig. 5A) (Koornneef et al., 1991; Abe et al., 2005). To test for genetic complementation, we transformed hda19 enh with a wild-type copy of the FD gene (Supplementary Fig. S6B). Multiple independent transformants showed restoration of reproductive defects to an hda19 appearance, implicating fd as enh (Supplementary Fig. S6C). As a final confirmation, we crossed hda19 to fd-1 and found that this double mutant combination very closely resembled hda19 enh with respect to the timing of floral organ patterning defects (Supplementary Fig. S2J–L), SIA emergence (Fig. 5B–D), and floral gene misexpression in the IM (Supplementary Fig. S7). Therefore, we conclude that enh is a new allele of fd which we have called fd-6 based on standard naming conventions (Wigge et al., 2005).
Fig. 5.
fd is an enhancer of hda19 reproductive defects. (A) Schematic of the FD gene (top) showing the positions of fd-6 (identified as hda19 enhancer mutation enh) and fd-1 genetic lesions. FD exons are depicted as rectangles, with arrowhead showing gene orientation. Amino acid positions converted to stop codons in fd-6 (R80*) and fd-1 (Q86*) are shown on the FD protein (bottom), which contains a basic leucine zipper (bZIP) domain. (B) hda19 fd-6 (left), fd-1 (middle), and hda19 fd-1 (right) at 40 d after germination. (C, D) Apical views of reproductive apices of hda19 fd-1. IM phase number is given in the upper right. The stamenoid inflorescence apex (arrow) is evident at IM phase 2. (E–G) FD RNA in situ hybridizations on the wild type showing IM expression (arrows). IM phase is given in the upper right. (H–J) AG RNA in situ hybridizations on fd-1 IMs (phase number in the upper right). Arrowheads denote ectopic AG expression in IMs. Scale bars: (B) 2 cm; (C and D) 0.5 mm; (E–J) 50 µm.
While it is known that FD is expressed in the SAM where it facilitates the transition from vegetative to reproductive growth (Abe et al., 2005; Wigge et al., 2005), we assessed whether its expression is maintained in the apex throughout reproductive development. RNA in situ hybridizations of wild-type IM phases 1–3 invariably showed strong FD expression in the IM and weaker expression in FMs (Fig. 5E–G). Relatively consistent FD expression in wild-type reproductive apices was also detected by quantitative RT–PCR throughout all IM phases (Supplementary Fig. S3B). These results are in agreement with a role for FD in IM function later in reproductive growth. Such a role is not obvious in the fd single mutant which lacks overt SIA-related defects (Supplementary Fig. S8A–C). Notably, although B-class genes are not appreciably misexpressed in fd (Supplementary Fig. S8D–I), ectopic expression of AG is apparent in phase 2 and 3 IMs of fd (Fig. 5I, J). Thus, the importance of FD in meristem function extends beyond the floral transition, not only because its mutation hastens SIA emergence and floral gene misregulation in hda19, but also because AG is ectopically expressed in the fd single mutant.
Enhancement of SIA is not simply a consequence of late flowering
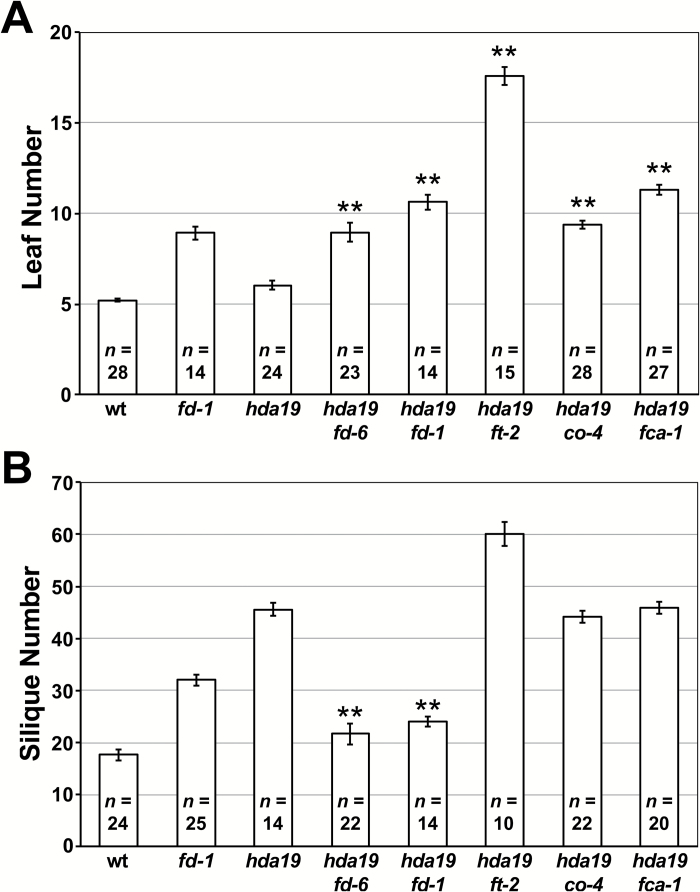
Since the SIA defect is age-dependent and because fd delays flowering, it is possible that fd-mediated enhancement of hda19 is indirectly caused by a prolonged vegetative stage. In this scenario, SIA emergence occurs after the initiation of fewer FMs in hda19 fd because the vegetative SAM has undergone extended growth and initiated more lateral organs (leaves) prior to its transition to an IM. If this is correct, crossing hda19 to other late-flowering mutants should similarly hasten SIA emergence following the transition to reproductive growth. To test this, we crossed hda19 to mutations of FT, CO, and FCA, the last of which is a component of the autonomous floral pathway that operates independently of environmental cues (Koornneef et al., 1991; Srikanth and Schmid, 2011). Like hda19 fd double mutants, each of these mutant combinations (hda19 ft-2, hda19 co-4, and hda19 fca-1) exhibited a delay in flowering time relative to hda19, as assessed by the numbers of vegetative leaves produced (Fig. 6A). Unlike hda19 fd, however, these three double mutant combinations did not hasten the timing of SIA emergence, producing numbers of siliques comparable with or greater than the hda19 single mutant (Fig. 6B). Therefore, the enhancement of hda19 by fd is not simply a product of delayed flowering, but rather results from separable functions specific to FD. This specificity is particularly apparent given that mutation of FT, whose protein product associates with FD to promote flowering (Abe et al., 2005; Wigge et al., 2005), failed to enhance the timing of SIA defects (Fig. 6B).
Fig. 6.
Reproductive defects of hda19 are not enhanced by other late-flowering mutants. (A) Numbers of vegetative leaves initiated before flowering and (B) total siliques produced by the primary inflorescence stem are given for the wild type (wt), fd-1, hda19, and combinations of hda19 with late-flowering mutants. Data are represented as the mean ±SE. For each double mutant, statistically significant increases in leaf number relative to hda19 (A) and decreases in silique number relative to hda19 (B) are indicated (**P<0.001; one tailed t-test). The sample size (n) of each genotype is provided. Plants were grown under long-day conditions.
The SIA defect is not due to ectopic AP1 or AGL6 expression
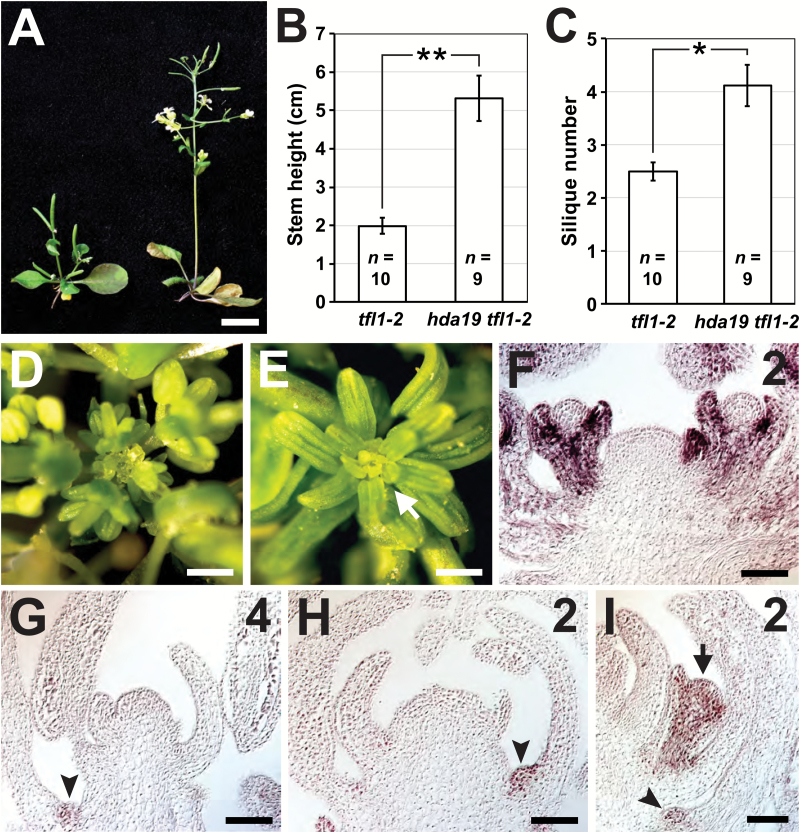
The FD–FT complex induces flowering by up-regulating FM identity genes such as AP1, which subsequently activates floral organ identity genes (Weigel and Meyerowitz, 1993; Abe et al., 2005; Wigge et al., 2005). Conversely, FD is believed to associate with TFL1 to repress genes in the IM (Hanano and Goto, 2011), as tfl1 IMs show ectopic AP1 expression (Gustafson-Brown et al., 1994). As a result, inflorescence stems of tfl1-2 mutants are very short and exhibit IMs that produce only a few FMs before being consumed into terminally differentiated flowers (Fig. 7A–C) (Alvarez et al., 1992). Similar to tfl1 defects, the SIA phenotype is associated with ectopic gene expression in the IM. We therefore hypothesized that a synergistic effect may be displayed by the hda19 tfl1 double mutant, particularly if the enhancement of hda19 by fd is a result of disrupted FD–TFL1 function. However, relative to tfl1-2, the hda19 tfl1-2 double mutant was more similar to the wild type, displaying taller inflorescence stems and producing more siliques (Fig. 7A–C). This suggests that FD has functions in IM maintenance that are separable from those of TFL1.
Fig. 7.
The stamenoid inflorescence apex phenotype is not due to derepression of AP1. (A) tfl1-2 (left) and hda19 tfl1-2 (right) at 40 d after germination (DAG). (B, C) Quantification of the height (B) and number of siliques (C) on the primary inflorescence stems of tfl1-2 and hda19 tfl1-2 at 45 DAG. Data are represented as the mean ±SE. Statistically significant differences are indicated (*P<0.005, **P<0.001; two-tailed t-test). The sample size (n) of each genotype is provided. (D, E) Apical views of ap1-1 (D) and hda19 ap1-1 (E) reproductive apices. Arrow denotes the stamenoid inflorescence apex. (F–I) AP1 RNA in situ hybridizations on the wild type (F), hda19 (G), and hda19 fd-6 (H, I). Numbers in the upper right indicate the inflorescence meristem phase. Arrowheads indicate expression in the axils of stamenoid organs, while an arrow denotes expression in an emerging axillary floral meristem. Scale bars: (A) 1 cm; (D, E) 0.5 mm; (F–I) 50 µm.
In SIA tissues, IMs display FM-like traits, including expression of floral organ genes and initiation of individual floral organs. In addition, the B-class genes that are ectopically expressed in SIA tissues are transcriptionally activated by the FM identity gene AP1 (Ng and Yanofsky, 2001). These observations suggest that AP1 misregulation could contribute to the formation of these abnormal IMs. To investigate this possibility, we first crossed hda19 with ap1-1 and found that removal of AP1 function did not suppress SIA defects (Fig. 7D, E). Secondly, we assessed AP1 expression which, in the wild type, is absent from the IM but present throughout young FMs and in the outer whorls of developing flowers (Fig. 7F) (Mandel et al., 1992). In hda19 and hda19 fd SIA tissues, AP1 expression remained absent from the IM but was also excluded from newly initiated lateral organs (Fig. 7G–I). AP1 expression was only detected in the axils of stamenoid organs, which on rare occasions correlated with the emergence of FMs (Fig. 7I). Collectively, these results indicate that the SIA defect is not the result of ectopic AP1 expression and, therefore, the observed misexpression of floral organ identity genes is independent of AP1. Moreover, SIA lateral organs lack FM identity, evidenced not only by the absence of AP1 expression, but also by occasional axillary FMs, which are common in mutants with defective FM specification (Irish and Sussex, 1990; Weigel et al., 1992; Bowman et al., 1993). This suggests that SIA tissues are the product of two overlapping defects: broad misregulation of B- and C-class floral organ identity genes and incomplete FM specification. Together, these produce an IM that initiates spirally arranged stamenoid lateral organs.
SIA-like defects have also been reported to result from the expression of a translational fusion between AGAMOUS-LIKE6 (AGL6) and the VP16 activation domain (Koo et al., 2010). We therefore sought to determine if AGL6, described as a regulator of lateral organ development and flowering (Koo et al., 2010), is misexpressed in SIA tissues, possibly contributing to patterning abnormalities. To this end, we performed AGL6 RNA in situ hybridizations and observed expression at the base of floral lateral organs and in ovules of wild-type flowers (Supplementary Fig. S9B–D), consistent with previous reports (Schauer et al., 2009; Koo et al., 2010). Notably, AGL6 expression was not detected in the wild-type IM (Supplementary Fig. S9A). A similar pattern of expression was seen in SIA tissues of hda19, hda19 fd-6, and hda19 fd-1, which also lacked detectable AGL6 expression in the IM (Supplementary Fig. S9E–P). This demonstrates that despite a previous connection between modified AGL6 activity and SIA-like defects, SIA abnormalities in hda19 mutant backgrounds are not associated with misexpression of AGL6.
The co-repressor TPL is involved in age-dependent maintenance of IM identity
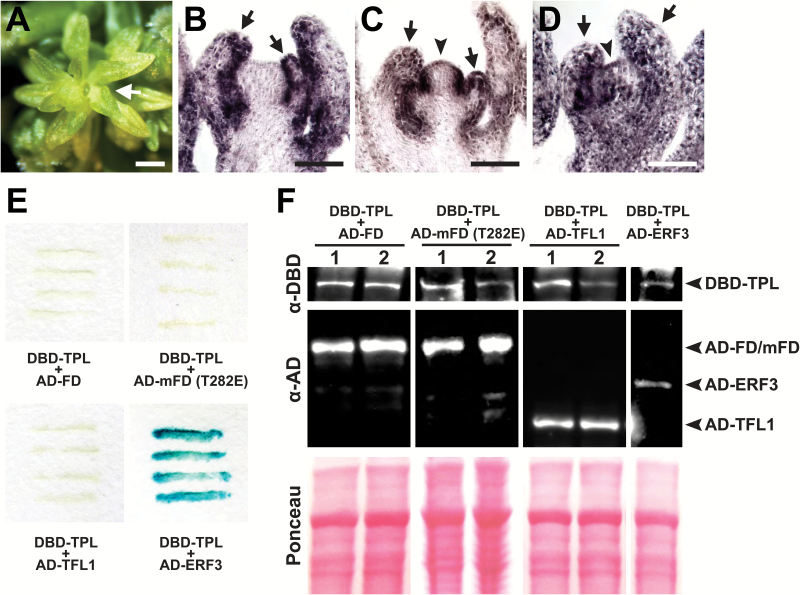
Since HDA19 forms a complex with the co-repressor TPL to restrict the expression of floral organ identity genes spatially in developing flowers (Krogan et al., 2012), they may similarly co-operate to regulate genes in the IM. TPL is a member of a multigene family that includes TPR1–TPR4, and mutation of all family members results in severe disruptions in floral patterning (Krogan et al., 2012). Examination of later reproductive stages of tpl tpr1 tpr2 tpr3 tpr4 revealed SIA defects (Fig. 8A–D), implicating this co-repressor family in age-dependent IM regulation. These observations are consistent with our ChIP analyses which show that TPL and HDA19 bind to the same non-coding regulatory regions of floral organ identity genes (Fig. 3M). Our genetic analyses suggest that FD may recruit TPL, and in turn HDA19, to repress floral organ identity genes. We therefore performed a yeast two-hybrid assay to test for possible binding between FD and TPL. Despite efficient protein expression, we could not detect an appreciable interaction (Fig. 8E, F). We also tested whether TPL could bind a mutant FD (mFD) harboring a phospho-mimic substitution (T282E) of a threonine residue important for FD–FT complex formation and FD activity in planta (Abe et al., 2005). This residue appears to be targeted by calcium-dependent protein kinases, and its phosphorylation is believed to be critical for protein–protein interactions (Abe et al., 2005; Kawamoto et al., 2015). However, the phospho-mimic mFD variant also failed to interact with TPL (Fig. 8E, F). Since the FD-interacting partner TFL1 appears to function as a transcriptional repressor (Hanano and Goto, 2011), it is possible that TFL1 facilitates FD–TPL association. However, binding between TPL and TFL1 was not apparent by yeast two-hybrid assay (Fig. 8E, F). Although the molecular mechanism remains unknown, our results nevertheless support that the TPL/TPR co-repressor family is also important for maintaining IM function during reproductive growth.
Fig. 8.
Role of TPL in the maintenance of IM identity. (A) tpl-2 tpr1 tpr2 tpr3 tpr4 quintuple mutant displaying the stamenoid inflorescence apex (SIA) phenotype (arrow). (B–D) RNA in situ hybridizations of AP3 (B), PI (C), and AG (D) on tpl-2 tpr1 tpr2 tpr3 tpr4 SIA tissue. Floral organ identity genes show misexpression in the IM (arrowheads) and strong expression in stamenoid organs (arrows). (E) Yeast two-hybrid assays testing interaction between TPL [fused to the GAL4 DNA-binding domain (DB)] and FD, mFD (T282E), TFL1, and ERF3 [each fused to the GAL4 activation domain (AD)]. ERF3, a known interactor of TPL (Causier et al., 2012), serves as a positive control as indicated by the darkening of the yeast streaks. (F) Western blot showing appreciable expression of all bait and prey constructs tested. Two independent yeast transformants are shown for TPL+FD, TPL+mFD (T282E), and TPL+TFL1 combinations. Ponceau staining shows equal protein loading. Scale bars: (A) 0.5 mm; (B–D) 50 µm.
Discussion
Arabidopsis post-embryonic growth relies on the continuous activity of the SAM to transition from vegetative to reproductive stages. We have identified a regulatory process that maintains the identity of the reproductive SAM in an age-dependent manner. Disruption of this process leads to a dysfunctional meristem that ectopically expresses B- and C-class floral organ identity genes and initiates stamenoid organs in place of FMs. We have named this striking patterning defect the stamenoid inflorescence apex, or SIA. AP1 expression is absent from SIA lateral organs but is detectable in their axils where secondary meristems occasionally emerge. Thus, SIA exhibits two defects: extensive misregulation of floral organ identity genes throughout the reproductive apex and disruption of FM fate.
During normal development, multiple transcriptional regulators prevent the emergence of SIA. This includes the histone deacetylase HDA19, plausibly in conjunction with TPL/TPR co-repressors. While it has been reported that an AP2–TPL–HDA19 complex spatially restricts floral gene expression in developing flowers (Krogan et al., 2012), our current work indicates that TPL and HDA19 perform similar roles in the IM through unknown biochemical interactions. The replacement of FMs with individual floral organs in older hda19 inflorescences suggests that HDA19 also participates directly or indirectly in the control of FM identity. This work adds to a growing list of meristem functions for HDA19 that include the embryonic SAM (Long et al., 2006), FM (Krogan et al., 2012; Bollier et al., 2018), and RAM (Pi et al., 2015). Notably, such widespread involvement of HDA19 in meristem regulation mirrors the importance of HDAC-mediated repression in the maintenance of stem cells in animals (Liang et al., 2008; Jamaladdin et al., 2014).
Our determination that FD represses floral organ identity genes in the reproductive apex has uncovered a novel developmental role for this flowering time gene. Along with hastening B- and C-class gene misexpression in hda19 (Fig. 4E–J; Supplementary Fig. S7), mutation of FD results in ectopic AG expression in the IM (Fig. 5I, J). The timing of hda19 FM identity defects was also enhanced by fd, possibly reflecting a continued role for FD in the activation of AP1 in later stages of reproductive development. However, the primary defect in AP1 expression displayed by fd mutants is simply a delay in its activation during the transition to flowering (Abe et al., 2005; Wigge et al., 2005). This does not explain why the enhancement of hda19 by fd is most apparent in stages of reproductive development much later than the floral transition. Furthermore, FD activates AP1 in conjunction with FT, yet ft did not enhance the timing of SIA emergence in hda19 (Fig. 6B). Combining other late-flowering mutations with hda19 also did not exacerbate SIA defects (Fig. 6B), further indicating that the enhancement of hda19 by fd is specific to FD function, and not simply the result of delayed flowering.
The developmental age of mutant IMs clearly influences the extent of gene misregulation and timing of SIA emergence, which occurs only after the primary IM has initiated numerous functional FMs. In hda19, SIA abnormalities become evident after the production of >30 FMs, a number not achieved by wild-type plants under our experimental conditions. This extended reproductive phase indicates that hda19 delays the co-ordinated arrest of IM activity, known as global proliferative arrest (GPA), which is a function of plant age and reproductive output (Hensel et al., 1994; Balanzá et al., 2018). Notably, a recent report shows that AP2 and AP2-like factors act downstream of the transcription factor FRUITFULL to control the timing of GPA in an age-dependent fashion (Balanzá et al., 2018). Therefore, AP2, which can associate with HDA19 and TPL (Krogan et al., 2012), may also influence the timing of SIA defects, but would probably do so indirectly as it does not show consistent expression in the IM (Wollmann et al., 2010).
It could be argued that the SIA defect is a product of abnormally prolonged IM proliferation in hda19, which results in an aberrant meristematic state not displayed by wild-type IMs. Interestingly, reproductive growth can be artificially prolonged in wild-type plants by surgically removing siliques or secondary inflorescences, thereby delaying GPA (Hensel et al., 1994). Such treatments result in the eventual conversion of the IM into terminally differentiated floral structures (Hensel et al., 1994). This is consistent with the notion that extended meristematic activity can lead to gene misregulation and the disruption of IM function. However, these IM abnormalities occurred after the production of >60 flowers, an extension of reproductive growth vastly beyond that displayed by hda19. Furthermore, other Arabidopsis backgrounds that display a prolonged reproductive stage do not exhibit the SIA condition, including ap3-3, pi-1, and ag-1 mutants, each of which does not completely suppress SIA in hda19 (Supplementary Fig. S5F, H, J, L, N, P). The hda19 fd double mutant also indicates that SIA does not strictly rely on abnormally prolonged IM activity, as this background exhibits SIA after initiating FM numbers comparable with the wild type (Fig. 4B).
The hda19 and hda19 fd backgrounds are reminiscent of other reproductive mutants whose patterning defects and misexpression of floral genes (particularly AG) increase with age. These include the transcriptional regulators lug, seu, ap2, bellringer (blr), and rabbit ears (rbe), all of which display floral patterning defects that worsen in an acropetal fashion along the inflorescence stem (Bowman et al., 1989; Liu and Meyerowitz, 1995; Franks et al., 2002; Bao et al., 2004; Krizek et al., 2006). The nature of SIA emergence is particularly similar to the terminal carpelloid flower defect displayed by older blr inflorescence stems that results from derepression of AG (Bao et al., 2004). In both cases, ectopic floral organ gene expression presages a striking shift in IM patterning near the end of reproductive development. Interestingly, both conditions also show defects in FM identity, as abnormal blr flowers are often subtended by carpelloid bracts (Bao et al., 2004). This suggests that misregulation of AG in reproductive apices can counteract the acquisition of FM fate, and is consistent with previous observations of ectopic AG expression resulting in the formation of bracts with carpel-like traits (Cartolano et al., 2009). This relationship may be due to reduced expression of the FM identity gene AP1 (as seen in SIA tissues) resulting from the ability of AG to antagonize AP1 (Gustafson-Brown et al., 1994).
The mechanisms underlying age-dependent enhancement of floral gene misexpression have yet to be fully characterized, despite the prevalence of this effect in various mutants. Since floral defects worsen as IM cellular proliferation advances, it is possible that epigenetic repression naturally weakens at the reproductive apex as rounds of cell division accumulate. This progression could be enhanced by mutants which, in some cases, surpass a threshold of gene misregulation that triggers abnormalities such as SIA. Such a relationship could explain why SIA only emerges in older IMs of hda19 and hda19 fd, even though both HDA19 and FD are expressed at the apex throughout reproductive growth (Figs 1I–K, 5E–G; Supplementary Fig. S3). The common association of AG misexpression with age-related reproductive defects is also consistent with epigenetic deregulation, as AG transcription is repressed by numerous chromatin regulators, including Polycomb group proteins (reviewed in Kaufmann et al., 2010). Indeed, even on wild-type inflorescences, the last-formed flowers can show aberrant expansion of carpel fate (Bowman et al., 1989), indicative of a natural age-dependent loss of AG repression. In this scenario, mutants with compromised histone deacetylase (hda19) and co-repressor (tpl/tpr) functions would exhibit escalated IM defects, as greater disruption of repressive chromatin modifications would exacerbate natural age-related floral gene derepression. Our ChIP results that show direct binding of HDA19 and TPL to floral organ identity genes such as AG (Fig. 3M) is consistent with such a molecular mechanism.
The enhancement of hda19 by fd suggests that FD also contributes to a general weakening of transcriptional repression of floral organ identity genes. It is noteworthy that this enhancement does not involve the emergence of patterning abnormalities not already seen in hda19. Instead, fd causes a temporal shift in the appearance of gene misexpression and SIA, as defects characteristic of hda19 fd IM phases 1 and 2 resemble those of hda19 IM phases 3 and 4, respectively. If SIA emergence results from reaching a threshold level of gene derepression, it is likely that loss of FD exacerbates the gene misregulation of hda19 to reach this threshold at an earlier developmental age. The mechanism by which FD represses B- and C-class floral organ identity genes is not yet clear. One attractive possibility is that FD participates in a TPL–HDA19 transcriptional repressor complex in the IM, potentially mediated by its association with TFL1. However, our genetic and protein interaction analyses do not support such a relationship, raising the possibility that a TFL1-independent mode of FD-conferred repression operates in this context. Clarifying the nature of FD-mediated floral gene repression will probably be complicated by the multifaceted nature of FD protein interactions, which may require FD phosphorylation and the involvement of 14-3-3 proteins for stable association (Abe et al., 2005; Taoka et al., 2011; Kawamoto et al., 2015). Additionally, a group of SPL proteins interacts with FD to facilitate the integration of developmental age and photoperiodic flowering (Jung et al., 2016), broadening the network of factors that FD may associate with to fulfill its developmental roles.
Another future challenge will be to determine how well conserved the roles of FD in repressing floral organ identity genes and maintaining IM identity are among angiosperms. The function of FD in regulating flowering is highly conserved among plants (Muszynski et al., 2006; Tsuji et al., 2013; Randoux et al., 2014), suggesting that the processes we have described may be similarly conserved. Finally, the identification of HDA19 and FD as regulators of an age-dependent patterning process in the IM offers inroads into understanding the connections between the duration of stem cell proliferation and the overall maintenance of meristem function.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Temporal progression of Arabidopsis inflorescence meristem (IM) phases.
Fig. S2. Floral defects worsen with developmental age in hda19 mutants.
Fig. S3. Temporal expression of HDA19 and FD in reproductive apices.
Fig. S4. Floral organ identity genes are not expressed in wild-type inflorescence meristems.
Fig. S5. Floral homeotic mutants only partially suppress hda19 reproductive defects.
Fig. S6. The hda19 enhancer enh is an allele of the flowering time gene fd.
Fig. S7. Floral organ identity genes are misexpressed in hda19 fd-1 IMs.
Fig. S8. B-class floral organ identity genes are not misexpressed in fd-1 inflorescence meristems.
Fig. S9. AGL6 is not misexpressed in reproductive tissues of hda19 mutant backgrounds.
Table S1. List of primers used in ChIP experiments.
Supplementary Material
Acknowledgements
We thank J. Long for valuable advice and support, B. Chow for helpful comments on the manuscript, and M. Joens and J. Fitzpatrick for technical assistance with the scanning electron microscopy. This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R15GM114733 to NTK. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- Abe M, Kobayashi Y, Yamamoto S, et al. 2005. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056. [DOI] [PubMed] [Google Scholar]
- Alvarez J, Guli CL, Yu XH, Smyth DR. 1992. terminal flower: a gene affecting inflorescence development in Arabidopsis thaliana. The Plant Journal 2, 103–116. [Google Scholar]
- Balanzà V, Martínez-Fernández I, Sato S, Yanofsky MF, Kaufmann K, Angenent GC, Bemer M, Ferrándiz C. 2018. Genetic control of meristem arrest and life span in Arabidopsis by a FRUITFULL–APETALA2 pathway. Nature Communications 9, 565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X, Franks RG, Levin JZ, Liu Z. 2004. Repression of AGAMOUS by BELLRINGER in floral and inflorescence meristems. The Plant Cell 16, 1478–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäurle I, Dean C. 2006. The timing of developmental transitions in plants. Cell 125, 655–664. [DOI] [PubMed] [Google Scholar]
- Bollier N, Sicard A, Leblond J, et al. 2018. At-MINI ZINC FINGER2 and Sl-INHIBITOR OF MERISTEM ACTIVITY, a conserved missing link in the regulation of floral meristem termination in Arabidopsis and tomato. The Plant Cell 30, 83–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. 1989. Genes directing flower development in Arabidopsis. The Plant Cell 1, 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. 1991. Genetic interactions among floral homeotic genes of Arabidopsis. Development 112, 1–20. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR. 1993. Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119, 721–743. [Google Scholar]
- Bradley D, Ratcliffe O, Vincent C, Carpenter R, Coen E. 1997. Inflorescence commitment and architecture in Arabidopsis. Science 275, 80–83. [DOI] [PubMed] [Google Scholar]
- Cartolano M, Efremova N, Kuckenberg M, Raman S, Schwarz-Sommer Z. 2009. Enhanced AGAMOUS expression in the centre of the Arabidopsis flower causes ectopic expression over its outer expression boundaries. Planta 230, 857–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causier B, Ashworth M, Guo W, Davies B. 2012. The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiology 158, 423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Wu K, Schmidt W. 2015. The histone deacetylase HDA19 controls root cell elongation and modulates a subset of phosphate starvation responses in Arabidopsis. Scientific Reports 5, 15708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WQ, Li DX, Zhao F, Xu ZH, Bai SN. 2016. One additional histone deacetylase and 2 histone acetyltransferases are involved in cellular patterning of Arabidopsis root epidermis. Plant Signaling and Behavior 11, e1131373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Coen ES, Meyerowitz EM. 1991. The war of the whorls: genetic interactions controlling flower development. Nature 353, 31–37. [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, et al. 2007. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030–1033. [DOI] [PubMed] [Google Scholar]
- Drews GN, Bowman JL, Meyerowitz EM. 1991. Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell 65, 991–1002. [DOI] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Perea JV, Bowman JL. 2001. Establishment of polarity in lateral organs of plants. Current Biology 11, 1251–1260. [DOI] [PubMed] [Google Scholar]
- Franks RG, Wang C, Levin JZ, Liu Z. 2002. SEUSS, a member of a novel family of plant regulatory proteins, represses floral homeotic gene expression with LEUNIG. Development 129, 253–263. [DOI] [PubMed] [Google Scholar]
- Gao MJ, Li X, Huang J, et al. 2015. SCARECROW-LIKE15 interacts with HISTONE DEACETYLASE19 and is essential for repressing the seed maturation programme. Nature Communications 6, 7243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave AP. 1992. A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Molecular Biology 20, 1203–1207. [DOI] [PubMed] [Google Scholar]
- Goto K, Meyerowitz EM. 1994. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes and Development 8, 1548–1560. [DOI] [PubMed] [Google Scholar]
- Gustafson-Brown C, Savidge B, Yanofsky MF. 1994. Regulation of the arabidopsis floral homeotic gene APETALA1. Cell 76, 131–143. [DOI] [PubMed] [Google Scholar]
- Hanano S, Goto K. 2011. Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. The Plant Cell 23, 3172–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel LL, Nelson MA, Richmond TA, Bleecker AB. 1994. The fate of inflorescence meristems is controlled by developing fruits in Arabidopsis. Plant Physiology 106, 863–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijser P, Schmid M. 2011. The control of developmental phase transitions in plants. Development 138, 4117–4129. [DOI] [PubMed] [Google Scholar]
- Irish VF, Sussex IM. 1990. Function of the apetala-1 gene during Arabidopsis floral development. The Plant Cell 2, 741–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack T, Brockman LL, Meyerowitz EM. 1992. The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68, 683–697. [DOI] [PubMed] [Google Scholar]
- Jaeger KE, Wigge PA. 2007. FT protein acts as a long-range signal in Arabidopsis. Current Biology 17, 1050–1054. [DOI] [PubMed] [Google Scholar]
- Jamaladdin S, Kelly RDW, O’Regan L, et al. 2014. Histone deacetylase (HDAC) 1 and 2 are essential for accurate cell division and the pluripotency of embryonic stem cells. Proceedings of the National Academy of Sciences, USA 111, 9840–9845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Lee HJ, Ryu JY, Park CM. 2016. SPL3/4/5 integrate developmental aging and photoperiodic signals into the FT–FD module in Arabidopsis flowering. Molecular Plant 9, 1647–1659. [DOI] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. 1999. Activation tagging of the floral inducer FT. Science 286, 1962–1965. [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Pajoro A, Angenent GC. 2010. Regulation of transcription in plants: mechanisms controlling developmental switches. Nature Reviews. Genetics 11, 830–842. [DOI] [PubMed] [Google Scholar]
- Kawamoto N, Sasabe M, Endo M, Machida Y, Araki T. 2015. Calcium-dependent protein kinases responsible for the phosphorylation of a bZIP transcription factor FD crucial for the florigen complex formation. Scientific Reports 5, 8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. 1999. A pair of related genes with antagonistic roles in mediating flowering signals. Science 286, 1960–1962. [DOI] [PubMed] [Google Scholar]
- Koo SC, Bracko O, Park MS, et al. 2010. Control of lateral organ development and flowering time by the Arabidopsis thaliana MADS-box gene AGAMOUS-LIKE6. The Plant Journal 62, 807–816. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, van der Veen JH. 1991. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Molecular and General Genetics 229, 57–66. [DOI] [PubMed] [Google Scholar]
- Krizek BA, Fletcher JC. 2005. Molecular mechanisms of flower development: an armchair guide. Nature Reviews. Genetics 6, 688–698. [DOI] [PubMed] [Google Scholar]
- Krizek BA, Lewis MW, Fletcher JC. 2006. RABBIT EARS is a second-whorl repressor of AGAMOUS that maintains spatial boundaries in Arabidopsis flowers. The Plant Journal 45, 369–383. [DOI] [PubMed] [Google Scholar]
- Krogan NT, Hogan K, Long JA. 2012. APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development 139, 4180–4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NT, Marcos D, Weiner AI, Berleth T. 2016. The auxin response factor MONOPTEROS controls meristem function and organogenesis in both the shoot and root through the direct regulation of PIN genes. New Phytologist 212, 42–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Wan M, Zhang Y, et al. 2008. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nature Cell Biology 10, 731–739. [DOI] [PubMed] [Google Scholar]
- Liu Z, Meyerowitz EM. 1995. LEUNIG regulates AGAMOUS expression in Arabidopsis flowers. Development 121, 975–991. [DOI] [PubMed] [Google Scholar]
- Long JA, Ohno C, Smith ZR, Meyerowitz EM. 2006. TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312, 1520–1523. [DOI] [PubMed] [Google Scholar]
- Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF. 1992. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360, 273–277. [DOI] [PubMed] [Google Scholar]
- Mathieu J, Warthmann N, Küttner F, Schmid M. 2007. Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Current Biology 17, 1055–1060. [DOI] [PubMed] [Google Scholar]
- Muszynski MG, Dam T, Li B, Shirbroun DM, Hou Z, Bruggemann E, Archibald R, Ananiev EV, Danilevskaya ON. 2006. delayed flowering1 encodes a basic leucine zipper protein that mediates floral inductive signals at the shoot apex in maize. Plant Physiology 142, 1523–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M, Yanofsky MF. 2001. Activation of the Arabidopsis B class homeotic genes by APETALA1. The Plant Cell 13, 739–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy F, Nilsson O, Busch MA, Lee I, Weigel D. 1998. A genetic framework for floral patterning. Nature 395, 561–566. [DOI] [PubMed] [Google Scholar]
- Pi L, Aichinger E, van der Graaff E, Llavata-Peris CI, Weijers D, Hennig L, Groot E, Laux T. 2015. Organizer-derived WOX5 signal maintains root columella stem cells through chromatin-mediated repression of CDF4 expression. Developmental Cell 33, 576–588. [DOI] [PubMed] [Google Scholar]
- Poethig RS. 2003. Phase change and the regulation of developmental timing in plants. Science 301, 334–336. [DOI] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G. 1995. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80, 847–857. [DOI] [PubMed] [Google Scholar]
- Randoux M, Davière JM, Jeauffre J, et al. 2014. RoKSN, a floral repressor, forms protein complexes with RoFD and RoFT to regulate vegetative and reproductive development in rose. New Phytologist 202, 161–173. [DOI] [PubMed] [Google Scholar]
- Ryu H, Cho H, Bae W, Hwang I. 2014. Control of early seedling development by BES1/TPL/HDA19-mediated epigenetic regulation of ABI3. Nature Communications 5, 4138. [DOI] [PubMed] [Google Scholar]
- Sanders PM, Bui AQ, Weterings K, McIntire KN, Hsu YC, Lee PY, Truong MT, Beals TP, Goldberg RB. 1999. Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sexual Plant Reproduction 11, 297–322. [Google Scholar]
- Schauer SE, Schlüter PM, Baskar R, Gheyselinck J, Bolaños A, Curtis MD, Grossniklaus U. 2009. Intronic regulatory elements determine the divergent expression patterns of AGAMOUS-LIKE6 subfamily members in Arabidopsis. The Plant Journal 59, 987–1000. [DOI] [PubMed] [Google Scholar]
- Searle I, He Y, Turck F, Vincent C, Fornara F, Kröber S, Amasino RA, Coupland G. 2006. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes and Development 20, 898–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon S, Meeks-Wagner DR. 1991. A mutation in the Arabidopsis TFL1 gene affects inflorescence meristem development. The Plant Cell 3, 877–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar VV, Surendrarao A, Liu Z. 2006. APETALA1 and SEPALLATA3 interact with SEUSS to mediate transcription repression during flower development. Development 133, 3159–3166. [DOI] [PubMed] [Google Scholar]
- Srikanth A, Schmid M. 2011. Regulation of flowering time: all roads lead to Rome. Cellular and Molecular Life Sciences 68, 2013–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Kikuchi A, Kamada H. 2008. The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiology 146, 149–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoka K, Ohki I, Tsuji H, et al. 2011. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476, 332–335. [DOI] [PubMed] [Google Scholar]
- Tsuji H, Nakamura H, Taoka K, Shimamoto K. 2013. Functional diversification of FD transcription factors in rice, components of florigen activation complexes. Plant and Cell Physiology 54, 385–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM. 1992. LEAFY controls floral meristem identity in Arabidopsis. Cell 69, 843–859. [DOI] [PubMed] [Google Scholar]
- Weigel D, Meyerowitz EM. 1993. Activation of floral homeotic genes in Arabidopsis. Science 261, 1723–1726. [DOI] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D. 2005. Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309, 1056–1059. [DOI] [PubMed] [Google Scholar]
- Wollmann H, Mica E, Todesco M, Long JA, Weigel D. 2010. On reconciling the interactions between APETALA2, miR172 and AGAMOUS with the ABC model of flower development. Development 137, 3633–3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldmann KA, Meyerowitz EM. 1990. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346, 35–39. [DOI] [PubMed] [Google Scholar]
- Yant L, Mathieu J, Dinh TT, Ott F, Lanz C, Wollmann H, Chen X, Schmid M. 2010. Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. The Plant Cell 22, 2156–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Tan B, Luo M, et al. 2013. HISTONE DEACETYLASE19 interacts with HSL1 and participates in the repression of seed maturation genes in Arabidopsis seedlings. The Plant Cell 25, 134–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.